Standing between us and global food shortage are high yielding varieties of wheat, oilseed rape, potato, maize and sugarcane. These come from diverse plant families, but share the fact that they have undergone recent hybridisation and whole genome duplication (that is, they are allopolyploids). How the possession of two complete sub-genomes from different parental species might contribute to their high yields is not fully understood, but it certainly does contribute to complexity in bioinformatic analyses of the genomes and transcriptomes of these crops (for example, Harper et al., 2012).
Few laboratories have studied the complexities of allopolyploid crops as intently as the group of Jonathan Wendel at Iowa State University. Taking cotton as their study system over the past decade, the Wendel lab have applied a portfolio of newly emerging technologies to characterise patterns of gene expression in wild and cultivated species of diploid and allopolyploid cotton. Starting with single-stranded conformation polymorphism analysis (Adams et al., 2003), they moved on to two generations of Nimblegen microarrays (Udall et al., 2006, 2007; Flagel et al., 2008; Hovav et al., 2008; Rapp et al., 2009), Sequenom MassARRAYs (Chaudhary et al., 2009), proteomics (Hu et al., 2011), and as reported by Yoo et al. (2012), high coverage Illumina sequencing.
Such a portfolio of methods is necessary to understand gene expression in allopolyploids, because we need to take into account not just the overall level of expression from a given locus, but also the relative contribution made at each locus by copies of genes derived from each diploid parental species. These gene copies are known as homoeologs. Studying the relative expression of homoeologs in polyploids is methodologically similar to studying allelic expression in diploids, but it is biologically dissimilar because the homoeologs are set in the context of two different sub-genomes, which seldom recombine; they are also themselves present as two allelic copies that may or may not differ.
During the past 6 years, the Wendel lab has used two types of microarray to study gene expression in cotton allopolyploids. The first type of microarray has long oligonucleotides that measure the overall gene expression level at thousands of loci, but cannot distinguish between homoeologs at these loci because both homeologs bind to the same oligonucleotide (Udall et al., 2007; Hovav et al., 2008). The second type has short oligonucleotides that are designed in pairs differing by one base at the central nucleotide—these allow the measurement of expression levels of each homoeolog at a locus independently, because each homoeolog binds to a unique oligonucleotide (Udall et al., 2006; Flagel et al., 2008; Hovav et al., 2008).
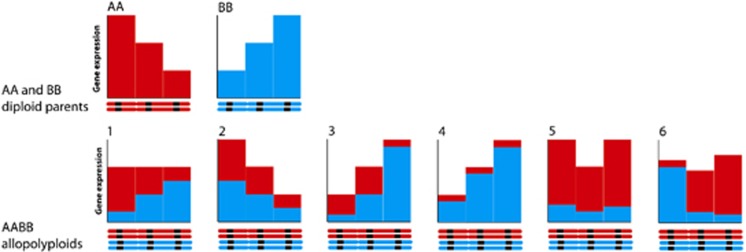
The homoeolog-specific microarrays showed that homoeolog expression bias is frequently found in allopolyploids, often favouring one sub-genome across the majority of loci (Flagel et al., 2008; Flagel and Wendel, 2010). The microarrays measuring total expression of each locus showed a new phenomenon: the expression level of each locus in allopolyploid genomes was often similar to the level found at that locus in one and not the other parental diploid species (Rapp et al., 2009). Both of these findings represented the expression dominance of one genome over the other, but they are different phenomena: one is in terms of which homoeologous gene copy is most expressed and the other is in terms of the overall expression of all gene copies at a locus (see Figure 1). There has been some confusion in the literature since this discovery, as both phenomena have been referred to as ‘genome dominance' (for example, Rapp et al., 2009; Schnable et al., 2011). To clarify this, the Yoo et al. (2012) paper in this issue suggests that ‘expression-level dominance' should be used to refer to patterns of overall gene expression (termed ‘genome dominance' in Rapp et al., 2009), and ‘homoeolog expression bias' should be used to refer to patterns in the ratio of homoeologous expression (referred to as ‘genome dominance' in Schnable et al., 2011).
Figure 1.
Patterns of gene expression in allopolyploids and their diploid parents. In the upper row, example levels of gene expression for three homologous genes in two diploids are shown. The lower row shows six example patterns of gene expression in an allotetraploid formed from the two diploids, with the contribution of the two homoeologs shown in red and blue: (1) additive patterns of gene expression, (2) expression-level dominance by the A sub-genome at all loci, with equal expression of homeologs at each locus, (3) expression-level dominance by the B sub-genome at all loci, with additive expression of homeologs at each locus, (4) expression-level dominance by the B sub-genome at all loci, with homoeologous expression bias in favour of the B sub-genome, (5) expression-level dominance by different sub-genomes at different loci, and homoeologous expression bias in favour of the A sub-genome at all loci, (6) no expression-level dominance, and homeologous expression bias differing among loci.
Until now, the two levels of expression dominance have been hard to link up, as different assays were needed to measure each level. It was difficult to tell if biases in homoeolog expression might be governing biases in overall gene expression. To investigate this, Yoo et al. (this issue) report a study of gene expression in leaves of allopolyploid cotton using high-throughput sequencing of gene transcripts, using the Illumina Genome Analyzer (Illumina Inc, San Diego, CA, USA). High depth of 80 bp reads produced by this method allows the overall expression of each locus to be measured relative to expression of all other loci in the genome, and also allows expression of each locus to be partitioned into transcripts from the different homoeologs. Thus, the same data set is used to quantify both expression-level dominance and homoeolog expression bias at the same time, allowing us to understand how they interact.
For the first time, the authors discover a link between expression-level dominance and homoeolog expression bias in cotton. They find that expression-level dominance by one parental sub-genome at a particular locus is commonly due to an alteration in the expression level of the homoeolog from the other parental genome, relative to its expression level in its parental species. For example if a gene is highly expressed in parent A, little expressed in parent B and highly expressed in an allopolyploid formed between them (that is, expression level dominance by parent A), this is frequently due to raised expression of the homoeolog from parent B (that is, homoeolog expression bias towards homoeolog B relative to the additive pattern of expression). In this example, it would appear that trans elements from the parent A sub-genome are activating expression in the parent B sub-genome.
Expression-level dominance can involve not only increases of homoeologous gene expression in the other sub-genome, but also decreases: if a gene is little expressed in parent A, and highly expressed in parent B, expression-level dominance by parent A will cause the gene to be little expressed in an allopolyploid, due to lowered expression of the homoeolog from parent B. In the results of Yoo et al. (this issue) trans-repression of gene expression in allopolyploids is almost as common as trans-activation. This provides an interesting contrast to a recent study of tissue-specific homoeolog expression bias in young Tragopogon miscellus allopolyploids, where there seemed to be frequent trans-activation of gene expression in early generations of allopolyploidy, but little trans-repression (Buggs et al., 2011).
To keep these fascinating interactions in perspective, it is important to note that the most commonly found gene expression states in allopolyploid loci are those with no change from parental expression states. A little more than half of genes in Yoo et al.'s study show changes in expression, and of these, just under half show expression-level dominance. Interestingly, domesticated cotton allopolyploids, used for crop production, show more transcriptome changes relative to diploids than do wild allopolyploids. Thus, it appears that patterns of duplicated gene expression are evolutionarily labile in allopolyploids and can be selected for.
The painstaking work of the Wendel lab over several years has produced huge insights into the evolution of gene expression in complex allopolyploid crop genomes. Many of these insights have been corroborated in other systems: expression-level dominance has been found in Coffea arabica (Bardil et al., 2011) and Spartina anglica (Chelaifa et al., 2010) and homeologous expression bias has been found in most allopolyploids where it has been studied (for example, Wang et al., 2006; Buggs et al., 2011; Schnable et al., 2011). Whilst many questions remain to be answered about the contribution of allopolyploidy to plant productivity (Soltis et al., 2010), it seems likely that homeologous expression bias and genome level dominance have an important role. Nailing down exactly how these transcriptomic patterns map to improved crop yields is a promising area for further research.
The author declares no conflict of interest.
References
- Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Nat Acad Sci USA. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardil A, de Almeida JD, Combes MC, Lashermes P, Bertrand B. Genomic expression dominance in the natural allopolyploid Coffea arabica is massively affected by growth temperature. New Phytol. 2011;192:760–774. doi: 10.1111/j.1469-8137.2011.03833.x. [DOI] [PubMed] [Google Scholar]
- Buggs RJA, Zhang L, Miles N, Tate JA, Gao L, Wei W, et al. Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr Biol. 2011;21:551–556. doi: 10.1016/j.cub.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Chaudhary B, Flagel L, Stupar RM, Udall JA, Verma N, Springer NM, et al. Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium) Genetics. 2009;182:503–517. doi: 10.1534/genetics.109.102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelaifa H, Monnier A, Ainouche M. Transcriptomic changes following recent natural hybridization and allopolyploidy in the salt marsh species Spartina x townsendii and Spartina anglica (Poaceae) New Phytol. 2010;186:161–174. doi: 10.1111/j.1469-8137.2010.03179.x. [DOI] [PubMed] [Google Scholar]
- Flagel LE, Udall J, Nettleton D, Wendel J. Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 2008;6:16. doi: 10.1186/1741-7007-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 2010;186:184–193. doi: 10.1111/j.1469-8137.2009.03107.x. [DOI] [PubMed] [Google Scholar]
- Harper AL, Trick M, Higgins J, Fraser F, Clissold L, Wells R, et al. Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat Biotech. 2012;30:798–802. doi: 10.1038/nbt.2302. [DOI] [PubMed] [Google Scholar]
- Hovav R, Udall JA, Chaudhary B, Rapp R, Flagel L, Wendel JF. Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc Natl Acad Sci USA. 2008;105:6191–6195. doi: 10.1073/pnas.0711569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Houston NL, Pathak D, Schmidt L, Thelen JJ, Wendel JF. Genomically biased accumulation of seed storage proteins in allopolyploid cotton. Genetics. 2011;189:1103–1115. doi: 10.1534/genetics.111.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp RA, Udall JA, Wendel JF. Genomic expression dominance in allopolyploids. BMC Biol. 2009;7:18. doi: 10.1186/1741-7007-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA. 2011;108:4069–4074. doi: 10.1073/pnas.1101368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Buggs RJA, Doyle JJ, Soltis PS. What we still don't know about polyploidy. Taxon. 2010;59:1387–1403. [Google Scholar]
- Udall JA, Flagel LE, Cheung F, Woodward AW, Hovav R, Rapp RA, et al. Spotted cotton oligonucleotide microarrays for gene expression analysis. BMC Genomics. 2007;8:81. doi: 10.1186/1471-2164-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall JA, Swanson JM, Nettleton D, Percifield RJ, Wendel JF. A novel approach for characterizing expression levels of genes duplicated by polyploidy. Genetics. 2006;173:1823–1827. doi: 10.1534/genetics.106.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee H-S, Wei NE, Jiang H, Watson B, et al. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 2006;172:507–517. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo M-J, Szadkowski E, Wendel JF. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity. 2013;110:171–180. doi: 10.1038/hdy.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Editor's suggested reading
- Ng DW-K, Zhang C, Miller M, Briggs SP, Chen ZJ. Proteomic divergence in Arabidopsis autopolyploids and allopolyploids and their progenitors. Heredity. 2012;108:419–430. doi: 10.1038/hdy.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan AC, Barker D, Hiscock SJ, Abbott RJ. Molecular genetic and quantitative trait divergence associated with recent homoploid hybrid speciation: a study of Senecio squalidus (Asteraceae) Heredity. 2012;108:87–95. doi: 10.1038/hdy.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyford AD, Ennos RA. Next-generation hybridization and introgression. Heredity. 2012;108:179–189. doi: 10.1038/hdy.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]