Abstract
Hybridisation among taxa with different ploidy levels is often associated with hybrid sterility. Clonal reproduction can stabilise these hybrids, but pervasive clonality may have a profound impact on the distribution of genetic diversity in natural populations. Here we investigate a widespread triploid taxon resulting from hybridisation between diploid Mimulus guttatus and tetraploid Mimulus luteus, two species that were introduced into the United Kingdom (UK) in the nineteenth century. This hybrid, Mimulus x robertsii, is largely sterile but capable of prolific vegetative propagation and has been recorded in the wild since 1872. We surveyed 40 Mimulus populations from localities across the UK to examine the current incidence of hybrids, and selected seventeen populations for genetic analysis using codominant markers. Cluster analyses revealed two main groups of genetically distinct individuals, corresponding to either diploid (M. guttatus) or polyploid (M. luteus and M. x robertsii) samples. Triploid hybrids were found in around 50% of sampled sites, sometimes coexisting with one of the parental species (M. guttatus). The other parent, M. luteus, was restricted to a single locality. Individual populations of M. x robertsii were genetically variable, containing multiple, highly heterozygous clones, with the majority of genetic variation distributed among- rather than within populations. Our findings demonstrate that this largely sterile, clonal taxon can preserve non-negligible amounts of genetic variation. The presence of genetically variable hybrid populations may provide the material for the continued success of asexual taxa in diverse environments.
Keywords: clonal reproduction, genetic markers, Mimulus guttatus , Mimulus luteus , Mimulus x robertsii , polyploidy
Introduction
By combining differentiated genomes, hybridisation generates novel genetic combinations that can result in phenotypic diversity, potentially producing ecological and evolutionary change (Grant, 1971; Arnold, 1997; Mallet, 2007). In cases where previously geographically isolated taxa come into secondary contact via human-assisted dispersal (Cox, 2004), hybridisation may also be important for the evolution of invasiveness (Ellstrand and Schierenbeck, 2000) and the origin of new hybrid taxa (Abbott, 1992). Therefore, the study of non-native hybrids is of relevance for understanding ecological and evolutionary processes as well as current biological invasions.
Hybridisation among taxa of differing ploidy levels is often limited by strong postzygotic barriers including early-acting hybrid inviability (Ramsey and Schemske, 1998; Köhler et al., 2010). In cases where interploidy hybrids are viable, these often suffer from severe reductions in sexual fertility that limit their ability to become established in the wild (reviewed in Petit et al., 1999). However, it has long been accepted that asexual (clonal) reproduction may allow sterile hybrids to form persistent populations and stabilise hybrid lineages (Grant, 1971; Mallet, 2007; Marques et al., 2011).
Strong sterility in clonal, hybrid taxa is expected to have an impact on the pattern of genetic variation present within and between populations (Bengtsson, 2003). Given the limited ability to create new genetic combinations in the absence of sex, the standing levels of genetic variation in clonal hybrids are likely to be relevant for understanding how sterile taxa persist and spread after their origination. Contrary to previous assumptions that asexual lineages are devoid of genetic variation, studies of asexual taxa have generally uncovered a substantial number of genetically diverse genotypes (Ellstrand and Roose, 1987; Rogstad et al., 2002; Meirmans et al., 2003; Van der Hulst et al., 2003; Vallejo-Marin et al., 2010). Levels of genetic variability in sterile hybrids are determined by the complex interaction of multiple processes including the amount of genetic diversity initially present in the parents, the frequency of hybrid formation events, the intensity of competitive exclusion among clones, the reduced effective population size in clonal populations and somatic mutation (Ellstrand and Roose, 1987; Balloux et al., 2003; Symonds et al., 2010). As a consequence, the structure of genetic diversity in asexual populations can be expected to vary widely depending on the relative importance of these factors.
Human-assisted dispersal and interploidy hybridisation of Mimulus (Phrymaceae) in the last 200 years, provides an ideal opportunity to study the early consequences of hybridisation and clonality for genetic diversity. In the United Kingdom (UK), several Mimulus taxa were introduced in the nineteenth century as garden plants. Among them, Mimulus guttatus DC (2n=2 × =28) was brought from North America in 1812, closely followed by the introduction of South American taxa belonging to the Mimulus luteus complex a few decades later (2n=4 × =60–62; hereafter M. luteus sensu lato) (Roberts, 1964; McArthur, 1974; Stace, 2010). Both species belong to the section Simiolus and can form vegetatively vigorous, but largely sterile, triploid hybrids (2n=3 × =44–46) (Roberts, 1964; Parker, 1975; Stace, 1975). The hybrid between M. guttatus and M. luteus L. (=Mimulus x robertsii Silverside) (Silverside, 1990) quickly escaped cultivation and became naturalised and widespread, being found in riparian habitats across the UK (Preston et al., 2002). Currently, M. x robertsii appears to constitute a relatively large component of the Mimulus taxa present in the UK (Preston et al., 2002; Stace, 2010). The genetic diversity of M. x robertsii populations is unknown and it remains to be established to what extent sexual sterility in this hybrid taxon has resulted in genetically invariant populations.
Here, we use microsatellite and intron-based markers to compare the genetic diversity between introduced populations of M. guttatus and a single introduced population of M. luteus s.l., against their triploid hybrid derivative, M. x robertsii in the UK. This study represents the first attempt to genetically compare introduced Mimulus populations of contrasting ploidy levels, and provides insight into the availability of standing genetic variation in sexually sterile hybrid lineages.
Materials and methods
Population sampling
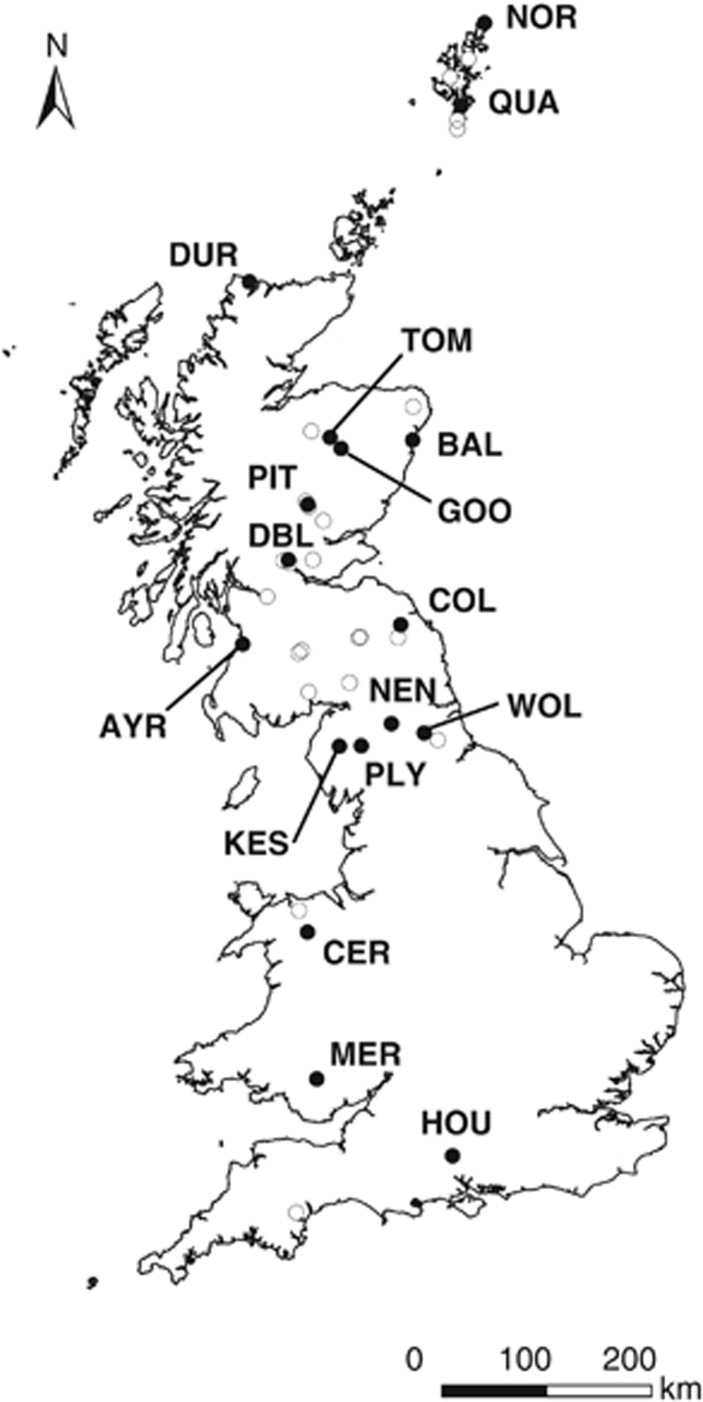
To determine the rate of occurrence of hybrids in extant Mimulus populations in the UK, we conducted a field survey across a ∼1200 km latitudinal transect from the Lizard Peninsula in Cornwall to the Isle of Unst in the Shetland Islands. Before embarking on the survey, a route was planned based on previous records of Mimulus spp. (Preston et al., 2002) and information from local contacts, with the aim of providing the best possible coverage of the latitudinal range of Mimulus within the UK. During the survey, we also explored the banks of freshwater bodies including streams, rivers and lakes along the survey route in order to increase the number of sampled populations and in an attempt to provide better coverage in areas where previous records were scarce. Between 21 May 2010 and 11 June 2011, we located 40 sites in which Mimulus spp. were growing (Figure 1). These sites, hereafter populations, were identified without taking into account species identity.
Figure 1.
Locations of populations sampled across the UK. Closed circles indicate populations for which genetic analysis was carried out.
In each population, we identified Mimulus taxa in the field, using the taxonomic keys provided by Stace (2010) and Silverside (1998) for British Mimulus. These keys use floral characters (for example, colour patterns of the corolla lobes, extent to which the corolla throat is open or closed, pollen fertility), leaf morphology (teeth shape on leaf margins) and fertility (for example, the production of abundant pollen) to distinguish British Mimulus. In each population, at least 20 randomly chosen individuals were identified using these morphological keys. In addition, for a subset of 1–10 sampled individuals from five populations thought to represent all sampled taxa (COL, DBL, MUK, NEN and PLY), we confirmed taxonomic identity in the lab using pollen size, flow cytometry and chromosome counts (Vallejo-Marin, 2012; M Vallejo-Marín and J Bailey, unpublished).
We selected a subset of 17 populations for genetic analysis with the aim of encompassing the latitudinal range covered during the field survey (Figure 1). These populations were separated by a minimum of 15 km and included parental and/or hybrid Mimulus taxa. Four populations contained more than one ploidy level, resulting in 21 population–ploidy sample sets. We sampled between 15 and 30 randomly chosen individuals in each population, with the exception of one site (PIT) for which only 12 individuals were available. We collected leaf tissue from individuals at least 2 m apart to maximise the probability of sampling different genotypes, and kept the tissue in plastic bags with silica gel until the DNA was extracted.
Genetic markers
To genotype British populations of Mimulus we used 14 codominant markers, which included seven microsatellite loci previously used to genotype North American populations (Kelly and Willis, 1998; Lowry et al., 2008), and seven markers revealing length polymorphisms in the introns of single-copy nuclear genes in M. guttatus (cf. Fishman and Willis, 2005; Sweigart et al., 2006; Lowry et al., 2008; Supplementary Table S2). The intron-based markers (M. guttatus sequence-tagged sites or MgSTSs) were chosen from a database of 461 candidate loci that have been genetically and/or physically mapped to the M. guttatus genome (Y-W Lee, unpublished results). To identify suitable MgSTS markers, we conducted a selection strategy of candidate loci as described in Vallejo-Marín et al. (2011). Briefly, in order to allow effective multiplexing, we categorised loci in one of six expected PCR product size classes based on previous amplification in North American material (Y-W Lee, unpublished results), and ranked them according to their allele number. We then selected 24 primer pairs representing all six size classes, and tested their amplification in simplex in a panel of 16 M. guttatus and 1 M. luteus s.l. individuals from the UK. Seven MgSTS loci were found to amplify, to be variable in the test samples, and to be theoretically compatible with multiplexing as assessed with Multiplex Manager (Holleley and Geerts, 2009).
DNA extraction and PCR amplification
To extract DNA from field-collected material we used a modified CTAB protocol (Doyle and Doyle, 1990), and quantified DNA yield using a Nanodrop 2000 (Thermo Scientific, Wilmington, DE, USA). The 14 loci were amplified in two multiplex reactions (Supplementary Table S2), using 1 × Qiagen Type-it Microsatellite PCR Kit (Qiagen, Crawley, West Sussex, UK), 2 μℳ of each of the fluorescent forward primers labelled with one of 6-FAM (Eurofins MWG Operon, Ebersberg, Germany), VIC, PET or NED (Applied Biosystems, Foster City, CA, USA) dyes and 2 μℳ of each reverse primer, and approximately 5–50 ng of template DNA. PCR cycles consisted of a denaturing step of 5 min at 95 °C, followed by 30 cycles of 95 °C for 30 s, 55 °C for 180 s and 72 °C for 30 s and a final elongation step of 30 min at 60 °C. We checked PCR products in a 3% agarose 1 × Tris-Borate-EDTA electrophoresis gel, and sent them to DNA Sequencing and Services (Dundee, UK) for fragment analysis using an ABI 3730xl capillary sequencer with a GeneScan 500 LIZ internal size standard (Applied Biosystems).
Genetic analysis
We analysed fluorescence profiles using STRand 2.4.59 (Toonen and Hughes, 2001), which allows calling and exporting of more than two peaks per locus, as can occur in triploid and tetraploid individuals. Raw peak sizes were analysed in a modified version of MsatAllele (Alberto, 2009) in R version 2.15.0 (R Development Core Team, 2012) to determine suitable allele bin range.
As the assumptions of traditional population genetic techniques cannot be easily met in mixed ploidy hybrid data sets, we used a presence–absence approach where the observed multilocus phenotypes are transformed to an allele matrix in which each allele is scored as either present or absent in each individual, similar to an amplified fragment length polymorphism (AFLP) profile. Further justification for use of this type of analysis for codominant markers in polyploid species can be found in Kloda et al. (2008), Vanderpoorten et al. (2011), and Sampson and Byrne (2012).
Population genetic structure
We applied three complementary methods to analyse the patterns of population genetic structure in British Mimulus: (1) a discriminant analysis of principal components (DAPCs; Jombart et al., 2010); (2) a principal coordinate analyses of genetic similarity indices (Kloda et al., 2008); and (3) a hierarchical analysis of molecular variance (AMOVA; Excoffier et al., 1992). We describe each of these approaches below.
We used a DAPCs to identify and describe clusters of genetically similar individuals (Jombart et al., 2010). DAPC is a multivariate method that uses sequential k-means and model selection to infer genetic clusters. DAPC is a recently proposed alternative to Bayesian clustering methods such as STRUCTURE (Pritchard et al., 2000) and BAPS (Corander and Marttinen, 2006), to analyse genetic data from large or complex genetic data sets. Similarly to STRUCTURE, each individual is assigned a probability of belonging to each of k-clusters (membership probability), which can be useful for detecting admixed individuals (Pritchard et al., 2000; Jombart et al., 2010). In addition, the relationships between clusters (for example, differentiation between groups) can be characterised through the synthetic variables produced in the discriminant analysis (DA) (Jombart et al., 2010). However, unlike STRUCTURE, DAPC does not require a population genetic model to identify genetic clusters (Jombart et al., 2010). Therefore DAPC is well suited for the analysis of the current data set, in which the combination of different ploidy levels, ambiguity in allele-copy number in polyploid heterozygotes, uncertainty of the inheritance patterns of markers in triploids and tetraploids (disomic vs polysomic), and potential departures from random mating (all studied Mimulus can reproduce through both selfing and clonal propagation), complicate the application of traditional population genetic models.
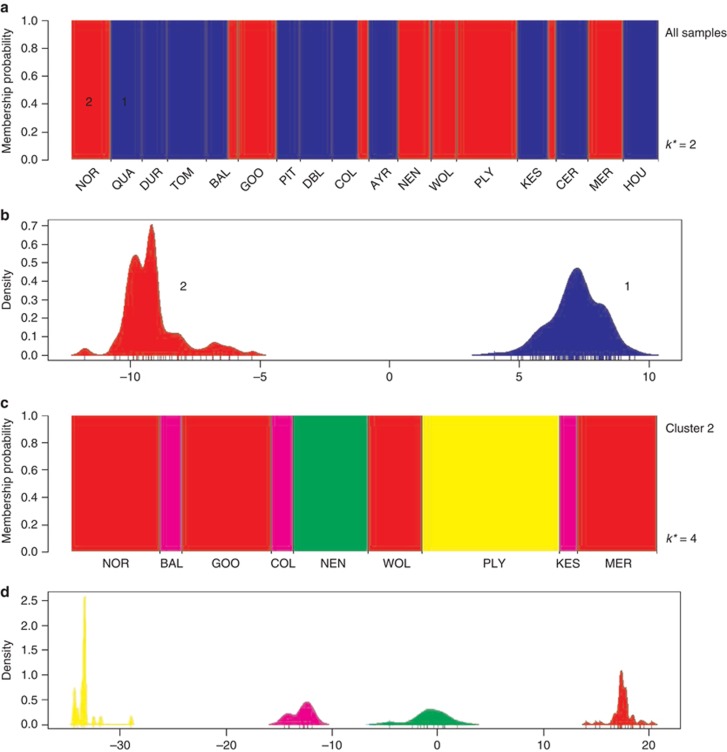
The DAPC analysis was conducted using the package adegenet ver. 1.3–4 (Jombart and Ahmed, 2011) in the statistical program R ver. 2.15.1 (R Development Core Team, 2012) using the presence–absence (binary) genetic data. To find the optimal number of clusters in our data, we used k-means clustering of the principal components and calculated the statistical fit of the data for a given k, using the function find.clusters in adegenet. The optimal number of clusters in the data was determined using the diffNgroup option, which identifies sharp changes in the fit of models (measured using Bayesian Information Criterion) with different numbers of clusters. We used 106 iterations of the model to search for convergence and obtained the likelihood associated with each value of k between 1 and 21. The clustering analysis identified two genetically divergent groups, representing the diploid M. guttatus and the polyploid M. robertsii and M. luteus s.l. respectively.
We conducted a further DAPC using only the polyploid samples identified in the first DAPC (that is, cluster 2). This hierarchical approach allowed us to describe the pattern of genetic variation seen within polyploids after accounting for the overall genetic divergence between diploid and polyploid samples. In this second analysis, we evaluated k-values between 1 and 9 (that is, number of populations with individuals from cluster 2). The likelihood of different k-values was compared using Bayesian Information Criteria as recommended by Jombart et al., (2010).
To complement the DAPC results, we conducted a principal coordinate analysis of a matrix of pairwise genetic distances between individuals, as has been done for other studies of codominant data in polyploids (Kloda et al., 2008; Sampson and Byrne, 2012). This analysis does not require a priori assumptions of population membership, but simply represents the genetic (dis-)similarity among individuals. We calculated pairwise dissimilarity coefficients among individuals using the R-library ade4 (Dray and Dufour, 2007). The metric of dissimilarity used here (‘Dice genetic distance') is based on Dice's (1945) similarity index S. The results of the principal coordinate analysis were also used to produce visual summaries of the relationships among each of the 21 population–ploidy combinations. To do this, we estimated mean genetic distances per population and used this mean distance to produce a neighbour-joining tree using the R-library ape (Paradis, 2006). The mean genetic distances were then used to conduct a Mantel test of the correlation between genetic and geographic distance (rm), calculated separately for diploid and polyploid samples.
Finally, we performed a hierarchical AMOVA on the binary data in Genalex 6.4 (Peakall and Smouse, 2006) using the predefined population–ploidy combinations described above. AMOVAs were done by nesting populations within ploidy, as well as by analysing each ploidy-level separately, except for M. luteus s.l. for which no separate analysis was done because of the fact that only one population was collected. The AMOVA was used to calculate Φ-statistics, which are equivalent to F fixation indices (for example, FST) for dominant data (Peakall and Smouse, 2006). In these analyses, one of the population–ploidy combinations contained just a single individual, and was thus excluded (WOL: diploid). The significance of Φ-statistics was calculated based on 1000 permutations.
Genetic diversity within groups
To estimate genetic diversity within population–ploidy combinations, we calculated the following statistics (Sampson and Byrne, 2012): the number of unique alleles summed across all loci (A); the number of alleles per locus per population (A′); the number of alleles per locus per individual (H′); and the proportion of observed heterozygotes per locus (Ho). Alleles from duplicated loci in triploid and tetraploid individuals—that is, homeologous loci that amplify with the same primer pair—were combined to represent a single locus.
To quantify clonal diversity, we assigned individuals of each population–ploidy combination to clonal lineages using a distance matrix of individual pairwise genetic distance (Meirmans and Van Tienderen, 2004). Starting with the binary data, we calculated pairwise dissimilarity coefficients (Dice's genetic distance) among individuals using ade4 (Dray and Dufour, 2007). Pairs of individuals with non-zero genetic distance were considered to belong to different multilocus genotypes (clones). Multilocus genotype assignment was done with the R-library polysat (Clark and Jasieniuk, 2011). For each population–ploidy combination, we calculated genotypic richness (R=G−1/N−1; where G is the number of multilocus genotypes and N is the number of genotyped individuals; Dorken and Eckert, 2001), and the complement of Simpson's diversity index (D; equation 11 in Arnaud-Haond et al. 2007).
Results
Species composition of sampled populations
The 40 Mimulus populations surveyed here were distributed across Great Britain from the Isle of Unst in Shetland (N 60.81°, W 0.80°) to Chudleigh in Devon (N 50.60°, W 3.62°), covering an elevation range from sea level to 405 m (Figure 1, Supplementary Table S1). All of these populations contained M. guttatus or M. guttatus-hybrids, whereas only one population also included individuals of tetraploid M. luteus s.l. (identified as M. luteus var. luteus × M. luteus var. variegatus=M. x smithii Paxton; 2n=60–62). In two northern populations (Muckle Roe, Shetland and Durness, Sutherland), we also found putative hybrids of M. guttatus and M. cupreus (=M. x burnetii), which can be easily distinguished from M. x robertsii by their petaloid calyx (Stace, 2010). However, here we focus on M. x robertsii hybrids as the analysis of samples collected from M. x burnetti individuals is beyond the scope of this investigation.
Approximately 52% of the Mimulus populations surveyed (21/40) contained at least some individuals that were identified as hybrids between M. guttatus and M. luteus (=M. x robertsii) based on morphological characteristics (Stace, 2010; Supplementary Table S1). Of the 17 populations chosen for genetic analysis, field identification suggested that 35% (6) contained only M. guttatus, 12% (2) contained only M. x robertsii, 47% (8) included both M. guttatus and M. x robertsii, and one population (6%) consisted of M. guttatus and M. luteus s.l. but no M. x robertsii hybrids.
Genetic markers
All 14 primer pairs successfully amplified PCR products across all taxa. Two of the 14 primer pairs assayed were excluded from further analyses because of the widespread presence of PCR artefacts during multiplexing (AAT356) or failure to amplify any alleles in a large number of populations (MgSTS657). Analysis is thus based on 12 loci.
We first describe the structure of genetic variation without a priori assumptions about taxonomic identity or ploidy level, and then use the knowledge generated by these analyses to present the results of the levels of genetic variation in the different groups identified.
Population genetic structure
The k-means clustering analysis of 300 genotyped individuals revealed a major separation of the genetic data set into two groups (Figure 2a). Populations represented within cluster 1 corresponded closely with those populations for which the presence of M. guttatus had been identified in the field based on morphological features. An examination of the genotypes of cluster 2 individuals showed that they always contained three or more alleles at one or more loci, clearly revealing the polyploid nature of individuals in this group. Only 2 out of the 168 individuals belonging to cluster 1 contained >2 alleles at any locus. In addition, populations represented within cluster 2 generally corresponded to those within which M x. robertsii had been identified based on morphological features. Exceptions were: (1) three populations that were characterised in the field survey as containing a mixture of M. guttatus and M. x robertsii (GOO, MER and NOR) but for which individuals sampled for genetic analysis were found to consist entirely of cluster 2 polyploids (M. x robertsii); and (2) one population (QUA) also characterised in the field survey as a mixture but for which only cluster 1 (diploid) genotypes were detected in the genetic analysis. Cluster 2 also included five individuals previously identified as M. luteus s.l. based on both morphological and ploidy-level analysis (COL population; Vallejo-Marin, 2012).
Figure 2.
Results of the DAPCs showing the genetic clustering of 300 Mimulus spp. individuals from 17 populations across Great Britain. (a) Individual probability of membership to either of two genetic clusters (k*=2) identified in the DAPC analysis. Cluster 1 (blue) is associated with diploid individuals (M. guttatus), whereas cluster 2 (red) represents polyploid individuals (M. x robertsii and M. luteus s.l.). The x axis shows the population of origin from north to south, with population codes as given in Table 4. (b) Discriminant analysis using the first 15 principal components shows a clear ability to distinguish the two genetic clusters. The x axis shows the values of the first discriminant function for each individual (vertical bars), and the y axis indicates the smoothed density of observations. (c, d) Analysis of cluster 2 samples only. (c) Individual probabilities of membership to either of four genetic clusters (k*=4) identified in the DAPC analysis. Notice that the tetraploid individuals (M. luteus s.l.) from population COL are not uniquely separated from triploid (M. x robertsii) individuals in other populations (BAL and KES). (d) Discriminant analysis of cluster 2 samples showing the values of the first discriminant function. Colours correspond to the four genetic clusters in (c).
The distinction between clusters 1 and 2 was evident in the DAPC analysis as the first discriminant function uniquely separated the membership of individuals to either of these two clusters (Figure 2b). From the total of 300 individuals from 17 populations analysed, 56% of genotypes belonged to cluster 1 (diploid; M. guttatus), and 44% to cluster 2 (polyploid: 42% M. x robertsii, and 2% M. luteus s.l.). Among the 17 populations analysed, 47% (8) were found to contain only cluster 1 (diploid) genotypes, 29% (5) only cluster 2 (polyploid) and 24% (4) contained a mixture of genotypes from both clusters (Figure 2).
The DAPC analysis conducted within the main cluster 2, detected four genetic clusters that were clearly differentiated in the first discriminant function (Figure 2d). Two clusters were composed exclusively of individuals from single populations (NEN and PLY), whereas the other two clusters contained individuals from three or more populations (Figure 2c). Individuals of M. luteus s.l. (COL population) showed the closest genetic affinity with triploid hybrids from populations BAL and KES (Figure 2c).
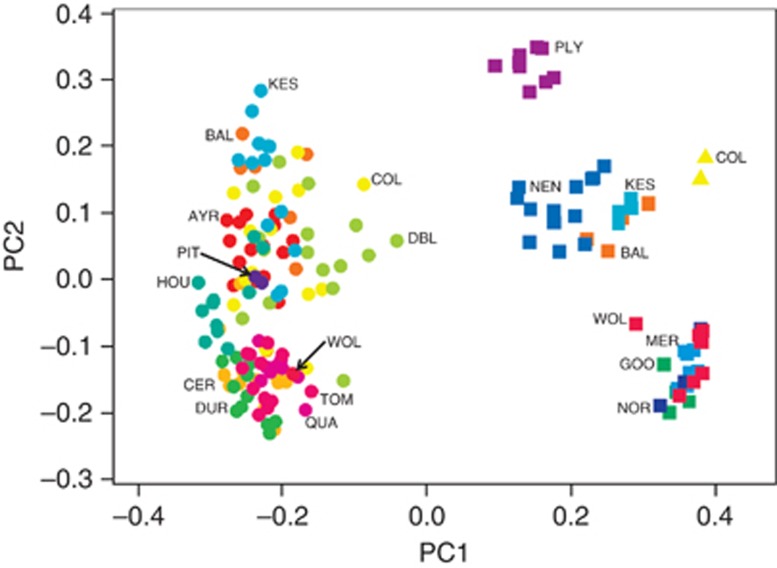
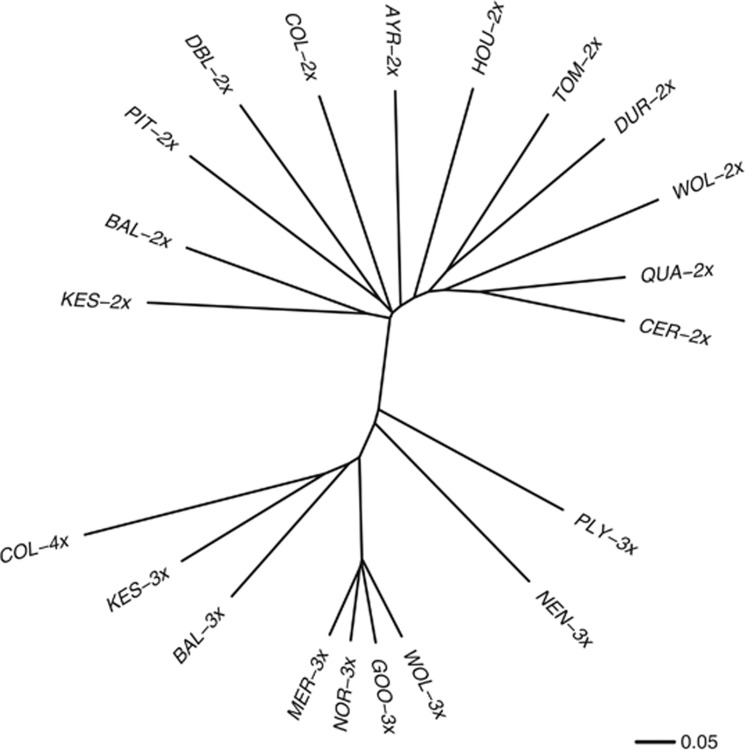
The principal coordinate analysis of the genetic dissimilarity matrix of binary genotypes supported the findings obtained through the DAPC approach. The two clusters identified using DAPC were clearly separated in this coordinate space (Figure 3). In addition, the results of the principal coordinate analysis were consistent with the indication of further genetic structure within polyploid genotypes. Triploid genotypes (squares, Figure 3) fell into three distinctive groups along the first two principal component axes. One group consisted of individuals from one population (PLY), whereas the other two groups contained individuals from three (BAL, KES and NEN) or four populations (GOO, MER, NOR and WOL; Figure 3). In addition, individuals of M. luteus s.l., represented by two distinct genotypes (each composed by either two or three individuals; triangles, Figure 3), were distinguishable from other polyploid individuals. The neighbour-joining tree summarising the pairwise mean genetic distance among population–ploidy combinations, shows the separation between diploid and triploid samples (Figure 4). There was no distinction of the M. luteus s.l. genotypes (COL, 2n=4 × ) from other polyploids using this mean distance metric (Figure 4).
Figure 3.
Relationships among individuals sampled as inferred by principal coordinate analysis of binary data using Dice's genetic distance (see Materials and methods). The goodness of fit of the first two principal components was 43%. Symbols: circles=diploid (M. guttatus), squares=triploid (M. x robertsii), triangles=tetraploid (M. x smithii=M. luteus var. luteus × M. luteus var. variegatus). Geographic populations are represented with different colours and identified with their respective population codes. For mixed populations, codes are shown for both diploid and polyploid samples.
Figure 4.
Neighbour-joining tree summarising the relationship among studied populations based on Dice genetic distance. The ploidy level is indicated for each population (2 × , 3 × , 4 × ).
In order to assess genetic structuring within and among ploidy levels, we used the results of the DAPC clustering analysis to assign individual genotypes to one of three taxa: M. guttatus (diploid genotypes, Cluster 1), M. x robertsii (triploid genotypes, cluster 2) and M. luteus s.l. (tetraploid genotypes, cluster 2). An AMOVA on the binary genetic data showed very strong differentiation between ploidy levels with 46% of the variance arising from differences among the three taxa (index of fixation between taxa: ΦTaxon,Total=0.457, P<0.01), as well as significant population differentiation (33% of variance, ΦPopulation,Taxon=0.601, P<0.01; Supplementary Table S3). A comparison of AMOVAs performed separately on M. x robertsii (cluster 2, triploid) and M. guttatus (cluster 1, diploid) samples, showed stronger population differentiation in the hybrids (M. x robertsii: ΦST=0.766 vs M. x guttatus: ΦST=0.468; both indices P<0.01; Supplementary Table S3).
A Mantel test of the correlation between mean genetic and geographic distance (rm) indicated a weak but significant negative correlation for both the triploid (M. x robertsii; rm=−0.489, P<0.05), and diploid genotypes (M. guttatus; rm=−0.352, P<0.05). Therefore, we did not detect a classic pattern of isolation by distance.
Genetic diversity within groups
To analyse the genetic variation within taxa and population ploidy combinations, we used the results of the DAPC clustering analysis to assign individual genotypes to either of three taxa as above. All loci were polymorphic across populations of M. guttatus and M. x robertsii and 9 out of 12 loci were polymorphic in the five sampled individuals of M. luteus s.l. (Table 1). In total, 77 different alleles were amplified at 12 loci in the 300 individuals studied. Within taxa, 56 alleles were found in M. guttatus, 44 in M. robertsii and 23 in M. luteus s.l. (Table 2). Of the 77 unique alleles jointly present in M. x robertsii and M. guttatus, 30% (23/77) were found in both taxa. Of the 44 unique alleles found across M. x robertsii and M. luteus s.l., 52% (23/44) were common to both. Finally, of the 71 unique alleles found across M. guttatus and M. luteus s.l., 11% (8/71) were shared among the two taxa. The number of private alleles (present in no other taxa) for M. guttatus was 33, 6 for M. x robertsii, and none for M. luteus s.l. Every allele in M. luteus s.l. was found in M. x robertsii, including eight alleles that were shared among the three taxa.
Table 1. Number of alleles and heterozygosity (H o ) in three Mimulus taxa at six microsatellite loci (AAT), and six intron-based length-polymorphism markers (MgSTS).
| Locus |
M. guttatus
|
M. x robertsii
|
M. luteus s.l.
|
All taxa | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of alleles | Alleles per individual (mean±s.e.) | Ho | Total no. of alleles | Alleles per individual (mean±s.e.) | Ho | Total no. of alleles | Alleles per individual (mean±s.e.) | Ho | Total no. of alleles | |
| AAT217 | 4 | 1.46 (±0.04) | 0.46 | 5 | 2.98 (±0.01) | 1.00 | 2 | 2.00 (±0.00) | 1.00 | 6 |
| AAT225 | 3 | 1.32 (±0.04) | 0.32 | 2 | 1.42 (±0.04) | 0.42 | 1 | 1.00 (±0.00) | 0.00 | 4 |
| AAT230 | 8 | 1.37 (±0.04) | 0.37 | 3 | 1.76 (±0.04) | 0.76 | 2 | 2.00 (±0.00) | 1.00 | 10 |
| AAT240 | 3 | 1.07 (±0.02) | 0.07 | 2 | 1.83 (±0.03) | 0.83 | 2 | 2.00 (±0.00) | 1.00 | 4 |
| AAT267 | 4 | 1.02 (±0.02) | 0.01 | 3 | 1.70 (±0.08) | 0.44 | 2 | 2.00 (±0.00) | 1.00 | 6 |
| AAT278 | 2 | 1.33 (±0.04) | 0.33 | 4 | 2.59 (±0.04) | 1.00 | 3 | 3.00 (±0.00) | 1.00 | 4 |
| MgSTS234 | 6 | 1.47 (±0.04) | 0.45 | 2 | 1.25 (±0.04) | 0.25 | 1 | 1.00 (±0.00) | 0.00 | 6 |
| MgSTS321 | 5 | 1.34 (±0.04) | 0.34 | 4 | 2.17 (±0.03) | 1.00 | 2 | 2.00 (±0.00) | 1.00 | 7 |
| MgSTS430 | 6 | 1.30 (±0.04) | 0.29 | 7 | 1.33 (±0.04) | 0.33 | 1 | 1.00 (±0.00) | 0.00 | 9 |
| MgSTS681 | 4 | 1.35 (±0.04) | 0.33 | 2 | 1.80 (±0.04) | 0.80 | 2 | 2.00 (±0.00) | 1.00 | 5 |
| MgSTS685 | 5 | 1.48 (±0.04) | 0.48 | 3 | 2.00 (±0.00) | 1.00 | 2 | 2.00 (±0.00) | 1.00 | 7 |
| MgSTS84 | 6 | 1.45 (±0.04) | 0.44 | 7 | 3.51 (±0.05) | 0.99 | 3 | 2.00 (±0.00) | 1.00 | 9 |
| Mean | 4.67 (±0.46) | 1.33 (±0.04) | 0.32 (±0.04) | 3.67 (±0.53) | 2.03 (±0.20) | 0.73 (±0.09) | 1.92 (±0.19) | 1.83 (±0.17) | 0.75 (±0.13) | 6.42 (±0.60) |
| Total | 56 | 44 | 23 | 77 | ||||||
Abbreviation: MgSTS, M. guttatus sequence-tagged site.
Number of individuals analysed per taxon (number of populations): M. guttatus: 168 (12), M. x robertsii: 127 (8); M. luteus s.l. (M. luteus var. luteus × M. luteus var. variegatus=M. x smithii Paxton): 5 (1). Note: For each locus, only individuals amplifying at least one allele were used in the calculation of means and heterozygosity.
Table 2. Genetic diversity in M. guttatus, M. luteus s.l. (M. luteus var. luteus × M. luteus var. variegatus=M. x smithii), and M. x robertsii in the UK.
| Population | A | A′ | H′ | HO | Mean pairwise dissimilarity |
|---|---|---|---|---|---|
| M. guttatus (2 × ) | |||||
| AYR | 24 | 2.08 | 1.28 | 0.24 | 0.46 |
| BAL | 18 | 1.50 | 1.19 | 0.18 | 0.35 |
| CER | 23 | 1.92 | 1.70 | 0.64 | 0.35 |
| COL | 37 | 3.17 | 1.32 | 0.29 | 0.62 |
| DBL | 36 | 3.08 | 1.32 | 0.30 | 0.61 |
| DUR | 19 | 1.58 | 1.57 | 0.45 | 0.32 |
| HOU | 20 | 1.67 | 1.25 | 0.24 | 0.38 |
| KES | 23 | 1.92 | 1.09 | 0.09 | 0.47 |
| PIT | 15 | 1.25 | 1.01 | 0.01 | 0.07 |
| QUA | 27 | 2.33 | 1.72 | 0.63 | 0.30 |
| TOM | 26 | 2.25 | 1.40 | 0.39 | 0.43 |
| WOL | 17 | 1.42 | 1.42 | 0.42 | — |
| Population mean | 23.75±2.10 | 2.01±0.18 | 1.36±0.07 | 0.32*±0.05 | 0.39*±0.05 |
| Within taxon total | 56 | ||||
| M. x robertsii (3 × ) | |||||
| BAL | 24 | 2.17 | 1.90 | 0.62 | 0.24 |
| GOO | 25 | 2.25 | 2.06 | 0.65 | 0.16 |
| KES | 26 | 2.33 | 2.23 | 0.77 | 0.22 |
| MER | 27 | 2.42 | 2.09 | 0.74 | 0.20 |
| NEN | 28 | 2.42 | 2.01 | 0.77 | 0.37 |
| NOR | 26 | 2.33 | 2.14 | 0.74 | 0.19 |
| PLY | 25 | 2.33 | 2.07 | 0.77 | 0.19 |
| WOL | 26 | 2.33 | 2.06 | 0.72 | 0.25 |
| Population mean | 25.87±0.47 | 2.32±0.03 | 2.08±0.03 | 0.72*±0.02 | 0.23*±0.02 |
| Within taxon total | 44 | ||||
| M. luteus s.l. (4 × ) | |||||
| COL | 23 | 1.92 | 1.92 | 0.75 | 0.13 |
| Overall population Mean | 26.00±1.37 | 2.17±0.11 | 1.62±0.08 | 0.52±0.05 | 0.31±0.03 |
| Across taxa total | 77 | ||||
Abbreviations: A, number of unique (different) alleles across all loci; A′, number of alleles per locus in the population (mean across loci); H′, number of alleles per locus in an individual (mean over loci); HO, proportion of observed heterozygotes (mean across loci).
* Denotes statistically differences among taxa as assessed with a t-test; P<0.01.
Mean pairwise genetic dissimilarity calculated from a binary matrix.
In the three populations where both M. guttatus and M. x robertsii co-occur (BAL, KES and WOL) the two taxa shared in common 33% (11/33), 24% (10/41) and 26% (9/35), respectively, of the total alleles present within the population. For the COL population where both M. guttatus and M. luteus s.l. co-occur, these taxa shared in common 15% (8/53) of the total alleles present within the population. These values were similar to the overall mean proportion of shared alleles between taxa (30% between M. guttatus and M. x robertsii; and 11% between M. guttatus and M. luteus s.l.).
Despite having fewer alleles in total, M x robertsii amplified a greater number of alleles per individual per locus than M. guttatus (M. x robertsii=2.02±0.21 vs M. guttatus=1.33±0.07), whereas the mean number of alleles amplified for the tetraploid M. luteus s.l. was 1.92±0.20 alleles (Table 1). At a population level, the mean numbers of alleles per locus (A′) was more variable among M. guttatus than among M. x robertsii populations, although their overall means are similar (Table 2). As might be expected because of their hybrid origin, M. x robertsii populations had significantly higher heterozygosity than M. guttatus populations (Ho=0.72±0.02 vs 0.32±0.05; t=−5.77, P<0.001). Mean heterozygosity of the single population of M. luteus s.l. was similar to that of M. x robertsii (Ho=0.75). Interestingly, all but one individual in five populations containing M. x robertsii (GOO, KES, MER, NOR and WOL), amplified four alleles at one of the markers (MgSTS84; Table 1). As these populations clearly lack tetraploid M. luteus s.l., this observation suggests a duplication event at this locus.
The analysis of pairwise genetic distance calculated from the binary data in each population indicated a lower mean dissimilarity among individuals within M. x robertsii populations compared with M. guttatus (mean of population-level mean dissimilarities: 0.23±0.02 vs 0.39±0.05; t=3.21, P<0.01; Table 2). However, one population of M. guttatus in Perthshire (PIT) had the lowest mean pairwise dissimilarity (0.07) of all populations, indicating high levels of similarity of genotypes in this particular case. The mean pairwise differentiation for the single M. luteus s.l. population sampled was 0.13. The mean number of unique multilocus genotypes per population was 10.25±1.43 in M. guttatus individuals, vs 7.88±1.19 in M. x robertsii hybrids (Table 3). As we used a non-zero genetic similarity criterion to identify unique multilocus genotypes, this represents an upper bound of the number of distinct genets. In M. guttatus populations genotypic richness (R), a measure of the proportion of distinguishable genotypes in the sample, ranged from R=0.09–1.00 with a population mean to R=0.70±0.08. The mean genotypic richness was lower in M. robertsii (R=0.58±0.09; range=0.27–1.00) and in M. luteus s.l. (R=0.25). We found no difference in the complement of Simpson's diversity indices (D) between M. guttatus and M. x robertsii (Table 3). Together our results indicate that all populations studied here are polyclonal, but that M. guttatus populations show the highest genotypic richness.
Table 3. Genotypic diversity in M. guttatus, M. luteus s.l. (M. luteus var. luteus × M. luteus var. variegatus) and M. x robertsii.
| Population | N | G | R | D |
|---|---|---|---|---|
| M. guttatus (2 × ) | ||||
| AYR | 15 | 15 | 1.00 | 1.00 |
| BAL | 11 | 6 | 0.50 | 0.73 |
| CER | 16 | 13 | 0.80 | 0.95 |
| COL | 14 | 14 | 1.00 | 1.00 |
| DBL | 16 | 16 | 1.00 | 1.00 |
| DUR | 13 | 9 | 0.67 | 0.96 |
| HOU | 18 | 13 | 0.71 | 0.95 |
| KES | 16 | 13 | 0.80 | 0.95 |
| PIT | 12 | 2 | 0.09 | 0.17 |
| QUA | 16 | 9 | 0.53 | 0.88 |
| TOM | 20 | 12 | 0.58 | 0.89 |
| WOL | 1 | 1 | — | — |
| Total | 168 | 123 | — | — |
| Population mean | 14.00±1.38 | 10.25±1.43 | 0.70±0.08 | 0.86±0.07 |
| M. x robertsii (3 × ) | ||||
| BAL | 5 | 4 | 0.75 | 0.90 |
| GOO | 20 | 8 | 0.37 | 0.69 |
| KES | 4 | 4 | 1.00 | 1.00 |
| MER | 18 | 12 | 0.65 | 0.92 |
| NEN | 17 | 13 | 0.75 | 0.96 |
| NOR | 20 | 6 | 0.26 | 0.81 |
| PLY | 31 | 9 | 0.27 | 0.74 |
| WOL | 12 | 7 | 0.55 | 0.88 |
| Total | 127 | 63 | — | — |
| Population mean | 15.88±3.11 | 7.88±1.19 | 0.58±0.09 | 0.86±0.04 |
| M. luteus s.l. (4 × ) | ||||
| COL | 5 | 2 | 0.25 | 0.60 |
| Overall total | 300 | 188 | — | — |
| Overall mean | 14.29±1.47 | 8.95±1.01 | 0.63±0.06 | 0.85±0.04 |
Abbreviations: D, complement of Simpson's diversity index; G, number of unique (different) multilocus genotypes (genets) using a zero threshold of genetic distance to identify different clones; N, number of individuals analysed; R, genotypic richness: (G−1)/(N−1) (Dorken and Eckert 2001).
Discussion
Human activities have resulted in a rapid increase in the rate at which species are transported beyond their native ranges (Mooney and Cleland, 2001). The resulting change in the distribution of the global biota brings about a higher potential for hybridisation between previously isolated taxa (Abbott et al., 2003). The application of genetic tools to the study of these neo-hybrids continue to provide an opportunity to study the extent to which novel taxa are able to become naturalised and widespread despite sometimes strong fertility barriers (Ellstrand and Schierenbeck, 2000; Hollingsworth and Bailey, 2000; Huotari et al., 2011). Our genetic study of Mimulus hybrids, which are mostly sterile yet capable of vegetative propagation, demonstrates that even taxa in which sexual reproduction is impaired can maintain polyclonal populations with non-negligible levels of allelic variation, and become relatively widespread. Given the limited opportunities for recurrent formation of M. robertsii in the wild and their sexual infertility (for example, Roberts, 1964), determining the functional significance of standing levels of genetic variation will be fundamental to our understanding of how these populations are able to persist and colonise new environments.
Utility of genetic markers for further study
The markers used in this study have proven to be variable across taxa, providing a set of genetic tools suitable for the study of introduced Mimulus populations. As a first approximation, we have treated these markers as dominant molecular traits, as has been done in other similar studies (for example, Kloda et al., 2008; Vanderpoorten et al., 2011; Sampson and Byrne, 2012). This resulted in simpler mixed-ploidy analyses and interpretation at the cost of a potential loss of information that could be obtained by exploiting the codominant nature of microsatellites and STS's. In order to fully exploit this set of genetic tools, it would be necessary to investigate inheritance pattern in artificial crosses. This could help to resolve the ability of different primers to amplify in both M. guttatus and M. luteus genomes, and establish the type of inheritance (that is, polysomic, disomic or intermediate) of each locus (Obbard et al., 2006; Stift et al., 2008), which would allow incorporating explicit genetic models of marker inheritance to population genetic analyses.
Despite its current limitations, the analysis carried out here provides a first approximation of relative levels of genetic diversity in Mimulus populations of mixed ploidy. In addition, we have shown that genotyping individuals can be useful to establish the identity of field-collected individuals. The DAPC results show that the combined use of multiple genetic markers analysed here are a powerful tool to distinguish genetic clusters corresponding to different Mimulus taxa in the UK. The two main clusters identified here show a clear association with either diploid or polyploid genotypes, and can thus be used to distinguish M. guttatus from polyploid taxa in the UK, including M. x robertsii and M. luteus s.l. The fact that M. luteus s.l. could not be clearly distinguished from M. x robertsii based on genetic data alone could simply be a reflection of the very small number of genotypes of M. luteus s.l. found in the current survey. Comparisons of individual genotypes of M. luteus s.l. clearly show a lack of most alleles that are common in M. guttatus and M. x robertsii, and support the idea that the lack of resolution is probably associated with low statistical power because of limited sampling. Future studies should attempt to increase sampling across the range of M. luteus using other British populations and perhaps native South American material. Increasing the sample size in this study was unfortunately not possible, as we could not detect any other M. luteus s.l., across the 40 populations sampled, and no wild-collected material from South America was available. However, the genetic analyses performed here unambiguously distinguished M. gutattus from polyploid taxa even in mixed ploidy populations. The morphology of M. guttatus and M. x robertsii is highly variable, and taxa overlap for some characters used in taxonomic identification (Silverside, 1990; Stace, 2010). For example, pollen sterility associated with M. x robertsii (Roberts, 1964) can also occur in M. guttatus as a result of the expression of recessive sterility mutations in inbred populations (Willis, 1999). Thus taxonomic identification in mixed populations can present a significant challenge. The molecular tools used here provide another resource for future characterisation and identification of introduced populations of Mimulus and its hybrids.
Occurrence and distribution of Mimulus in the UK
The field identification of Mimulus taxa based on phenotypic characters was generally consistent with individual identification based on genetic data. The discrepancies found between the phenotypic characterisation of field populations and the genetic identification of individuals could be explained by the presence of one of the taxa at low-frequency within populations, meaning that the rarer taxon may have gone unsampled during collection of material for genotyping. For example, three of the populations that were characterised in the field survey as containing a mixture of M. guttatus and M. x robertsii based on colour polymorphism of the corolla and pollen production (GOO, MER and NOR; Figure 2) were found to be made entirely of Cluster 2 polyploids (M. x robertsii). In contrast, M. x robertsii is likely rarer in the phenotypically polymorphic population QUA where only cluster 1 (diploid) genotypes were detected. Alternatively, characters thought to be diagnostic of M. guttatus (fertility, absence of petal blotches) may also occur in hybrids. Given the large phenotypic variability in Mimulus hybrids, and the fact that not all individuals may be flowering at the time of sampling, we suggest caution with field identification based on morphology alone.
Both M. guttatus and M. x robertsii were found to be widespread in our survey, whereas M. luteus s.l. was only found in a single population. The paucity of M. luteus populations in Great Britain is somewhat surprising given that historical records suggest that this taxon should be relatively widespread (Preston et al., 2002). The dearth of populations of M. luteus in the current survey could be explained in different ways, including potential over-recording of M. luteus in previous botanical surveys, perhaps in error for M. x robertsii, which sometimes present large petal blotches that superficially resemble M. luteus (MVM personal observation). A more intriguing possibility is that M. luteus may be becoming less common, for example, by being outcompeted by other Mimulus taxa. Analysis of historical herbarium records and of the competitive abilities of the different taxa may help distinguishing between these possibilities.
Consistent with historical records of the distribution of M. x robertsii (Preston et al., 2002), this study carried out in 2010–2011 showed that hybrids are widespread and constitute a large proportion (∼50%) of extant Mimulus populations in the UK. These hybrids grow in broadly similar environments to other British Mimulus (Truscott et al., 2006), and in fact hybrids coexist in several populations with one of the parental taxa (M. guttatus). The continuous records of naturalised hybrids in the UK since at least 1872 (Preston et al., 2002), and its current widespread distribution highlights the ecological success of this largely sterile (Roberts, 1964) triploid plant.
It has been suggested that M. guttatus × M. luteus s.l. hybrids are predominantly plants of garden origin but may occasionally form in the wild (Stace, 2010). Although M. guttatus is widespread, the very low frequency of M. luteus s.l. in extant populations indicates that the potential for recurrent formation of M. x robertsii is currently fairly limited. In addition, even when both parental taxa co-occur, hybrids do not seem to be exceedingly common, as we did not detect hybrids, either through morphology or genetic comparisons, in the single population containing both parental taxa found in this study (COL). Thus, spread of hybrids from extant populations and subsequent introductions from cultivated individuals is likely to be the most important source of new populations for M. x robertsii.
The exact parentage of the M. x robertsii individuals studied here remains unknown. One of the parents, M. luteus s.l. includes several tetraploid and interfertile varieties (Cooley and Willis 2009; Stace, 2010), which could in principle be involved in the formation of these triploid hybrids. The triploid hybrids analysed showed a large degree of variability between populations in the extent and pattern of colour blotches in the corolla. Some populations (for example, NOR, GOO, WOL, PLY and MER; Table 4) showed a single colour blotch in the lower petal lobe, reminiscent of M. luteus var. rivularis (Silverside, 1990; Stace, 2010), whereas others (for example, NEN and BAL; Table 4) presented colour blotches in multiple lobes, which resembles the pattern seen in M. luteus var. variegatus and in crosses between M. luteus varieties (for example, M. x smithii; Stace, 2010). The identification of multiple genetic clusters in the DAPC analysis (Figures 2c and d) is consistent with origins of the triploid hybrids from diverse genetic stock. It is likely that multiple varieties are involved in the hybridisation of M. luteus s.l. with M. guttatus. in the UK (Stace, 2010), but determining their identity remains to be established.
Table 4. Location and morphology of Mimulus populations used for genetic analysis.
| Population | Code | Latitude | Longitude | Elevation (m) | Reddish blotch on lower petal lobe | Reddish marks on other petal lobes | Corolla throat | Presence of hybrids based on morphology | Taxa present based on genetic data |
|---|---|---|---|---|---|---|---|---|---|
| Norwick beach, Isle of Unst, Shetland | NOR | 60.8091 | −0.8048 | 3 | Polymorphic | Absent | Variable | Yes | M. x robertsii |
| Quarff, Shetland | QUA | 60.1046 | −1.2268 | 36 | Polymorphic | Absent | Variable | Yes | M. guttatus |
| Durness, Sutherland | DUR | 58.5684 | −4.7471 | 43 | Polymorphic | Absent | Open | Yes | M. guttatus |
| Tomintoul, Banffshire | TOM | 57.2550 | −3.3678 | 318 | Absent | Absent | Open | No | M. guttatus |
| Balmedie, Aberdeenshire | BAL | 57.2375 | −2.0639 | 19 | Polymorphic | Polymorphic | Open | Yes | M. guttatus/M. x robertsii |
| Goodbrand and Ross Woollen Shop, Colnabaichin, Aberdeenshire | GOO | 57.1620 | −3.1864 | 357 | Polymorphic | Absent | Variable | Yes | M. x robertsii |
| Pitlochry, Perthshire | PIT | 56.6755 | −3.6953 | 72 | Absent | Absent | Closed | No | M. guttatus |
| Dunblane, Perthshire | DBL | 56.1965 | −3.9686 | 62 | Absent | Absent | Closed | No | M. guttatus |
| Coldstream, Berwickshire | COL | 55.6550 | −2.2401 | 9 | Polymorphic | Polymorphic | Variable | No | M. guttatus/M. luteus s.l. |
| Ayr, River Ayr, Ayrshire | AYR | 55.4612 | −4.6253 | 14 | Absent | Absent | Closed | No | M. guttatus |
| Nenthall, Cumbria | NEN | 54.8061 | −2.3765 | 355 | Fixed | Fixed | Open | Yes | M. x robertsii |
| Wolsingham, Durham | WOL | 54.7269 | −1.8879 | 138 | Polymorphic | Absent | Closed | Yes | M. guttatus/M. x robertsii |
| Pooley Bridge, Cumbria | PLY | 54.6117 | −2.8213 | 154 | Fixed | Absent | Variable | Yes | M. x robertsii |
| Keswick, Cumbria | KES | 54.6052 | −3.1431 | 85 | Polymorphic | Absent | NA | Yes | M. guttatus/M. x robertsii |
| Cerrigydrudion, Conwy | CER | 53.0060 | −3.5493 | 227 | Absent | Absent | Closed | No | M. guttatus |
| Merthyr Tydfil, Glamorgan | MER | 51.7475 | −3.3826 | 175 | Polymorphic | Absent | Variable | Yes | M. x robertsii |
| Houghton Lodge, Hampshire | HOU | 51.0970 | −1.5084 | 33 | Polymorphic | Absent | Closed | No | M. guttatus |
Abbreviation: NA, not available.
A full list of the 40 populations surveyed is presented in the Supplementary Table S1.
Genetic diversity in introduced Mimulus
To the extent that parental taxa have different sets of alleles, hybrids are expected to display high levels of heterozygosity. In an extreme case, where parental taxa have no alleles in common at any loci, observed heterozygosity should be equal to one, that is, fixed heterozygosity (Brochmann et al., 2004). We found that across loci, the heterozygosity of M. x robertsii individuals was Ho=0.72±0.02. Values of Ho<1 could arise because of shared alleles between parental taxa. In fact, the number of alleles shared by M. guttatus and M. luteus was approximately 10%. Shared alleles could reflect ancestral polymorphisms or, in rapidly mutating markers, homoplasy (Estoup et al., 2002). Another possibility is that the primer pairs used do not amplify all homeologous copies, perhaps due to mutations in the primer regions that have accumulated because the divergence of the parental taxa. Such mutations may prevent amplification of some or all alleles at homeologous loci (null alleles), reducing the apparent heterozygosity and number of bands observed in triploid hybrids.
The relatively high number of multilocus genotypes detected in M. x robertsii (R=0.58) is perhaps surprising given the high sterility of this triploid (Roberts, 1964), and the likelihood that hybrid production in the wild may not be currently common given the paucity of populations where both parental taxa co-occur. Genotyping error and somatic mutation may explain detection of small amounts of genetic variation among clones (Meirmans and Van Tienderen, 2004), but clearly genotypic variation within all hybrid populations cannot be explained away in this manner. Within-populations genotypic diversity should reflect, in part, variation that has persisted because the original colonisation of the local population. It is also possible that genotypic diversity may be maintained by recurrent hybrid introductions through garden escapes. However, within populations, individuals clustered together in the principal coordinate analyses (Figure 3), suggesting that individuals within a population probably originated from a single colonisation event. The rate at which the genotypes present in the initial population are lost should be influenced by both genetic drift and competitive exclusion among clones. In the UK, Mimulus occurs as a perennial herb (Truscott et al., 2006; Stace, 2010), which combined with vegetative propagation could result in long-lived lineages composed of multiple ramets, potentially slowing down loss through stochastic events.
Theoretical expectations suggest that even low levels of sexual reproduction in highly sterile populations can result in patterns of allelic variation similar to sexual populations (Bengtsson, 2003). Although so far unconfirmed, it is possible that M. x robertsii may occasionally be able to produce viable gametes and engage in sexual reproduction (Ramsey and Schemske, 2002). Pollen viability in Mimulus x robertsii (measured as stained pollen) has been reported in some cases (Parker, 1975). However, naturalised populations of M. x robertsii are usually sterile (Stace, 1975, 2010), and artificial pollination using field-collected material fails to produce viable seed (Roberts, 1964). Further work is required to explore the possibility of sexual reproduction in the wild.
Ecological and evolutionary fate of Mimulus hybrids
The introduction of Mimulus into the UK has resulted in hybridisation between closely related, but previously geographically isolated species (Roberts, 1964; Stace, 2010). The resulting triploids have proven to be very successful in escaping their garden origin and forming persistent and widespread naturalised populations. The success of these hybrids despite the high sterility usually associated with odd-ploidy individuals (Ramsey and Schemske, 2002) is at first glance surprising. However, there are well-known examples of ecologically successful plant species that have been able to spread following human-assisted introductions despite being functionally asexual (for example, Hollingsworth and Bailey, 2000). In hybrid Mimulus, the ability to proliferate in new environments could be associated with the genetic changes resulting from genome merging, as well as multiple and recurrent origins that may create an influx of genetic variation (Symonds et al., 2010).
Sterile hybrids could have limited evolutionary significance because of their lack of sexual reproduction and recombination. However, it is well known that sterile hybrids can recover fertility, for example, through polyploidisation, sometimes soon after the breakdown of geographic barriers (for example, Abbott et al., 2009; Symonds et al., 2010). The recent discovery in Scotland of M. peregrinus, a hexaploid and fertile hybrid of M. guttatus × M. luteus s.l. (Vallejo-Marin, 2012), suggests that M. x robertsii may not be necessarily an evolutionary dead-end.
Conclusions
We have shown that the triploid hybrid M. x robertsii compose a large fraction of extant Mimulus populations in the UK (∼50%). Populations of this largely sterile taxon show high levels of heterozygosity as expected from their hybrid origin, and display variable levels of clonal diversity. It would be of interest to determine the extent to which this genetic variation is paralleled by performance differences between genotypes. Understanding the functional significance of genetic variation in this and other clonal taxa will help us predict the ability of largely sterile taxa (and their fertile polyploid derivatives), to persist and spread in changing environments.
Data archiving
Genotype data have been submitted to Dryad: doi:10.5061/dryad.mj5gb.
Acknowledgments
We would like to thank Young-Wha Lee and the Mimulus community for providing sequence data and advice for the MgSTS markers and John Willis, Arielle Cooley, Carrie Wu and the rest of the Willis lab for sharing their Mimulus expertise. We also thank Mark McNair, Alan Forrest, Nigel Wilby and the BSBI for advice on British Mimulus, and John Bailey for his continued support with chromosome counts since the initial stages of the project. Sampling would not have been possible without the help of numerous volunteers and botanists throughout the UK, in particular Antoine Keruzoré, Bob Dawson, Elizabeth Lye, Roy Sexton, Ilaria Marengo and Chris Chandler. For assistance with molecular work and plant maintenance we thank Daniel Souto, Michelle Lee, Jessica Scriven, James Weir and Pauline Monteith. We are grateful for the detailed comments provided by the Associate Editor and two anonymous reviewers on previous versions of the manuscript. This project was funded in part by a Carnegie Trust travel grant to MVM and by the Department of Biological and Environmental Sciences at the University of Stirling.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abbott RJ. Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol Evol. 1992;7:401–405. doi: 10.1016/0169-5347(92)90020-C. [DOI] [PubMed] [Google Scholar]
- Abbott RJ, Brennan AC, James JK, Forbes DG, Hegarty MJ, Hiscock SJ. Recent hybrid origin and invasion of the British Isles by a self-incompatible species, Oxford ragwort (Senecio squalidus L., Asteraceae) Biol Invasions. 2009;11:1145–1158. [Google Scholar]
- Abbott RJ, James JK, Milne RI, Gillies ACM. Plant introductions, hybridization and gene flow. Phil Trans Royal Soc London Series B-Biological Sciences. 2003;358:1123–1132. doi: 10.1098/rstb.2003.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberto F. MsatAllele_1.0: an R package to visualize the binning of microsatellite alleles. J Hered. 2009;100:394–397. doi: 10.1093/jhered/esn110. [DOI] [PubMed] [Google Scholar]
- Arnaud-Haond S, Duarte CM, Alberto F, Serrao EA. Standardizing methods to address clonality in population studies. Molecular Ecology. 2007;16:5115–5139. doi: 10.1111/j.1365-294X.2007.03535.x. [DOI] [PubMed] [Google Scholar]
- Arnold ML. Natural Hybridization and Evolution. Oxford University Press: Oxford; 1997. [Google Scholar]
- Balloux F, Lehmann L, de Meeus T. The population genetics of clonal and partially clonal diploids. Genetics. 2003;164:1635–1644. doi: 10.1093/genetics/164.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson BO. Genetic variation in organisms with sexual and asexual reproduction. J Evol Biol. 2003;16:189–199. doi: 10.1046/j.1420-9101.2003.00523.x. [DOI] [PubMed] [Google Scholar]
- Brochmann C, Brysting A, Alsos I, Borgen L, Grundt H, Scheen A, et al. Polyploidy in arctic plants. Biol J Linn Soc. 2004;82:521–536. [Google Scholar]
- Clark LV, Jasieniuk M. POLYSAT: an R package for polyploid microsatellite analysis. Mol Ecol Resour. 2011;11:562–566. doi: 10.1111/j.1755-0998.2011.02985.x. [DOI] [PubMed] [Google Scholar]
- Cooley AM, Willis JH. Genetic divergence causes parallel evolution in Chilean Mimulus. New Phytologist. 2009;183:729–739. doi: 10.1111/j.1469-8137.2009.02858.x. [DOI] [PubMed] [Google Scholar]
- Corander J, Marttinen P. Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol. 2006;15:2833–2843. doi: 10.1111/j.1365-294X.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- Cox GW. Alien Species and Evolution. Island Press: Washington; 2004. [Google Scholar]
- Dorken ME, Eckert CG. Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticilatus (Lythraceae) J Ecol. 2001;89:339–350. [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Dray S, Dufour AB. The ade4 package: implementing the duality diagram for ecologists. J Stat Software. 2007;22:1–20. [Google Scholar]
- Ellstrand NC, Roose ML. Patterns of genotypic diversity in clonal plant species. Am J Bot. 1987;74:123–131. [Google Scholar]
- Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants. Proc Natl Acad Sci USA. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estoup A, Jarne P, Cornuet JM. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol Ecol. 2002;11:1591–1604. doi: 10.1046/j.1365-294x.2002.01576.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial-DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Willis JH. A novel meiotic drive locus almost completely distorts segregation in Mimulus (monkeyflower) hybrids. Genetics. 2005;169:347–353. doi: 10.1534/genetics.104.032789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V. Plant Speciation. Columbia University Press: New York; 1971. [Google Scholar]
- Holleley CE, Geerts PG. Multiplex manager 1.0: a cross-platform computer program that plans and optimizes multiplex PCR. BioTechniques. 2009;46:511–517. doi: 10.2144/000113156. [DOI] [PubMed] [Google Scholar]
- Hollingsworth ML, Bailey JP. Evidence for massive clonal growth in the invasive weed Fallopia japonica (Japanese Knotweed) Bot J Linn Soc. 2000;133:463–472. [Google Scholar]
- Huotari T, Korpelainen H, Leskinen E, Kostamo K. Population genetics of the invasive water weed Elodea canadensis in Finnish waterways. Plant Syst Evol. 2011;294:27–37. [Google Scholar]
- Jombart T, Ahmed I. Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AJ, Willis JH. Polymorphic microsatellite loci in Mimulus guttatus and related species. Mol Ecol. 1998;7:769–774. [Google Scholar]
- Kloda JM, Dean PDG, Maddren C, MacDonald DW, Mayes S. Using principle component analysis to compare genetic diversity across polyploidy levels within plant complexes: an example from British Restharrows (Ononis spinosa and Ononis repens) Heredity. 2008;100:253–260. doi: 10.1038/sj.hdy.6801044. [DOI] [PubMed] [Google Scholar]
- Köhler C, Mittelsten Scheid O, Erilova A. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 2010;26:142–148. doi: 10.1016/j.tig.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Rockwood RC, Willis JH. Ecological reproductive isolation of coast and inland races of Mimulus guttatus. Evolution. 2008;62:2196–2214. doi: 10.1111/j.1558-5646.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- Marques I, Nieto Feliner G, Martins-Loucao MA, Fuertes Aguilar J. Fitness in Narcissus hybrids: low fertility is overcome by early hybrid vigour, absence of exogenous selection and high bulb propagation. J Ecol. 2011;99:1508–1519. [Google Scholar]
- McArthur ED. The cytotaxonomy of naturalized British. Mimulus. Watsonia. 1974;10:155–158. [Google Scholar]
- Meirmans PG, Van Tienderen PH. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes. 2004;4:792–794. [Google Scholar]
- Meirmans PG, Vlot EC, Den Nijs JCM, Menken SBJ. Spatial ecological and genetic structure of a mixed population of sexual diploid and apomictic triploid dandelions. J Evol Biol. 2003;16:343–352. doi: 10.1046/j.1420-9101.2003.00515.x. [DOI] [PubMed] [Google Scholar]
- Mooney H, Cleland E. The evolutionary impact of invasive species. Proc Natl Acad Sci USA. 2001;98:5446–5451. doi: 10.1073/pnas.091093398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Harris SA, Pannell JR. Simple allelic-phenotype diversity and differentiation statistics for allopolyploids. Heredity. 2006;97:296–303. doi: 10.1038/sj.hdy.6800862. [DOI] [PubMed] [Google Scholar]
- Paradis E. Analysis of Phylogenetics and Evolution with R. Springer: New York; 2006. [Google Scholar]
- Parker PF. Mimulus in Great Britain: a cytotaxonomic note. New Phytol. 1975;74:155–160. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Bretagnolle F, Felber F. Evolutionary consequences of diploid-polyploid hybrid zones in wild species. Trends Ecol Evol. 1999;14:306–311. doi: 10.1016/s0169-5347(99)01608-0. [DOI] [PubMed] [Google Scholar]
- Preston CD, Pearman DA, Dines TD. New Atlas of the British and Irish Flora. Oxford University Press: Oxford; 2002. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R. A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna; 2012. [Google Scholar]
- Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst. 1998;29:467–501. [Google Scholar]
- Ramsey J, Schemske DW. Neopolyploidy in flowering plants. Annu Rev Ecol Syst. 2002;33:589–639. [Google Scholar]
- Roberts RH. Mimulus hybrids in Britain. Watsonia. 1964;6:70–75. [Google Scholar]
- Rogstad SH, Keane B, Beresh J. Genetic variation across VNTR loci in central North American Taraxacum surveyed at different spatial scales. Plant Ecol. 2002;161:111–121. [Google Scholar]
- Sampson JF, Byrne M. Genetic diversity and multiple origins of polyploid Atriplex nummularia Lindl. (Chenopodiaceae) Biol J Linn Soc. 2012;105:218–230. [Google Scholar]
- Silverside AJ. A new binomial in Mimulus. Watsonia. 1990;18:210–212. [Google Scholar]
- Silverside AJ.1998Mimulus Section simiolusIn Rich T, Jermy A, (eds)Plant Crib Botanical Society of the British Isles: London; 259–261. [Google Scholar]
- Stace CA. New Flora of the British Isles. Cambridge University Press: Cambridge; 2010. [Google Scholar]
- Stace CA. Hybridization and the Flora of the British Isles. Academic Press: London; 1975. [Google Scholar]
- Stift M, Berenos C, Kuperus P, van Tienderen PH. Segregation models for disomic, tetrasomic and intermediate inheritance in tetraploids: a general procedure applied to Rorippa (Yellow cress) microsatellite data. Genetics. 2008;179:2113–2123. doi: 10.1534/genetics.107.085027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart AL, Fishman L, Willis JH. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics. 2006;172:2465–2479. doi: 10.1534/genetics.105.053686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds VV, Soltis PS, Soltis DE. Dynamics of polyploid formation in Tragopogon (Asteraceae): recurrent formation, gene flow, and population structure. Evolution. 2010;64:1984–2003. doi: 10.1111/j.1558-5646.2010.00978.x. [DOI] [PubMed] [Google Scholar]
- Toonen RJ, Hughes S. Increased throughput for fragment analysis on an ABI PRISM (R) automated sequencer using a membrane comb and STRand software. BioTechniques. 2001;31:1320–1324. [PubMed] [Google Scholar]
- Truscott AM, Soulsby C, Palmer SCF, Newell L, Hulme PE. The dispersal characteristics of the invasive plant Mimulus guttatus and the ecological significance of increased occurrence of high-flow events. J Ecol. 2006;94:1080–1091. [Google Scholar]
- Vallejo-Marin M. Mimulus peregrinus (Phrymaceae): a new British allopolyploid species. PhytoKeys. 2012;14:1–14. doi: 10.3897/phytokeys.14.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo-Marin M, Dorken ME, Barrett SCH. The ecological and evolutionary consequences of clonality for plant mating. Annu Rev Ecol Evol Syst. 2010;41:193–213. [Google Scholar]
- Vallejo-Marín M, Solis-Montero L, Bacles CFE, Lepais O. Thirteen microsatellites developed by SSR-enriched pyrosequencing for Solanum rostratum (Solanaceae) and related species. Am J Bot. 2011;98:e296–e299. doi: 10.3732/ajb.1100159. [DOI] [PubMed] [Google Scholar]
- Van der Hulst RGM, Mes THM, Falque M, Stam P, Den Nijs JCM, Bachmann K. Genetic structure of a population sample of apomictic dandelions. Heredity. 2003;90:326–335. doi: 10.1038/sj.hdy.6800248. [DOI] [PubMed] [Google Scholar]
- Vanderpoorten A, Hardy OJ, Lambinon J, Raspe O. Two reproductively isolated cytotypes and a swarm of highly inbred, disconnected populations: a glimpse into Salicornia's evolutionary history and challenging taxonomy. J Evol Biol. 2011;24:630–644. doi: 10.1111/j.1420-9101.2010.02198.x. [DOI] [PubMed] [Google Scholar]
- Willis JH. The contribution of male-sterility mutations to inbreeding depression in Mimulus guttatus. Heredity. 1999;83:337–346. doi: 10.1038/sj.hdy.6885790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.