Abstract
Large-scale genome rearrangements have been observed in cells adapting to various selective conditions during laboratory evolution experiments. However, it remains unclear whether these types of mutations can be stably maintained in populations and how they impact the evolutionary trajectories. Here we show that chromosomal rearrangements contribute to extremely high copper tolerance in a set of natural yeast strains isolated from Evolution Canyon (EC), Israel. The chromosomal rearrangements in EC strains result in segmental duplications in chromosomes 7 and 8, which increase the copy number of genes involved in copper regulation, including the crucial transcriptional activator CUP2 and the metallothionein CUP1. The copy number of CUP2 is correlated with the level of copper tolerance, indicating that increasing dosages of a single transcriptional activator by chromosomal rearrangements has a profound effect on a regulatory pathway. By gene expression analysis and functional assays, we identified three previously unknown downstream targets of CUP2: PHO84, SCM4, and CIN2, all of which contributed to copper tolerance in EC strains. Finally, we conducted an evolution experiment to examine how cells maintained these changes in a fluctuating environment. Interestingly, the rearranged chromosomes were reverted back to the wild-type configuration at a high frequency and the recovered chromosome became fixed in less selective conditions. Our results suggest that transposon-mediated chromosomal rearrangements can be highly dynamic and can serve as a reversible mechanism during early stages of adaptive evolution.
Author Summary
Large-scale chromosomal rearrangements are often associated with dramatic phenotypic changes such as cancer cell formation. It has been speculated that large-scale chromosomal rearrangements may play a crucial role at the early stages of adaptation, since they can quickly change the expression level of multiple genes or even a whole pathway by changing the gene copy number. Nonetheless, it remains unclear whether such mutations can be stably maintained in populations, especially in a fluctuating environment. Here we characterize an adaptive copper-tolerant phenotype in a wild yeast population. We discovered that the adaptive phenotype was contributed to by two large-scale chromosomal rearrangements, which increased the copy number of key components of copper regulation, including a crucial transcriptional activator, Cup2. We further identified three previously unknown downstream targets of Cup2 that also contributed to copper tolerance. Finally, we conducted an evolution experiment to test the stability of the rearranged chromosomes under conditions of relaxed selection. We found that the rearranged chromosomes returned back to the original configuration at a high frequency, and the wild type-like chromosome became fixed in all the evolved cultures. Our results suggest that chromosomal rearrangements can provide a reversible mechanism for cells when adapting to a fluctuating environment.
Introduction
Organisms have different ways to respond to environmental stresses and evolve corresponding adaptive functions [1]. At the genic level, adaptation can be achieved by subtle, small-scale nucleotide changes (base insertions, deletions or substitutions) that alter gene expression, protein structure or protein interactions. Alternatively, at the genomic level, large-scale genome rearrangements (chromosome duplications, translocations and aneuploidy) create copy number variations that may change gene dosage so as to shape adaptive evolution. Although a similar adaptive phenotype can be achieved by both mechanisms [2], it is still unclear whether one type of mutations is specifically preferred under certain conditions, especially in natural populations.
Unicellular organisms can quickly adapt to different environmental challenges in diverse niches. Comparing different populations of the same microbes that have adapted to distinct environments allows us to identify the underlying mechanisms of adaptive evolution [3], [4]. Studies using the budding yeast Saccharomyces cerevisiae have revealed that both small- and large-scale adaptive changes have occurred in natural and laboratory yeast populations [5]. For example, in a natural yeast strain, a few point mutations in the transcriptional factors, IME1, RME1 and RSF1, were found to improve sporulation efficiency [6]. Another study in yeast isolated from sherry wines showed that this yeast strain carries two types of mutations in the gene encoding a cell surface glycoprotein. The mutations include a 111-bp deletion in the promoter region that increases its expression level and duplications of a tandem repeat in the coding region that enhance the protein's hydrophobicity [7].
Large-scale changes that involve chromosome duplication, translocation or aneuploidy have been observed in yeast populations during short-term evolution experiments [5], [8]–[13]. Under glucose-limited conditions, evolved strains carry an amplified region that encodes a high-affinity hexose transporter [14]. Under sulfate-limited conditions, amplifications of a high-affinity sulfate transporter locus (SUL1) were observed [15]. Beyond changes observed in experimental populations, a chromosome translocation resulting in overexpression of SSU1, a gene encoding a sulfite efflux pump, was identified in a sulfate-tolerant wine strain [16]. In other yeast species, such as the clinical isolates of pathogenic Candida spp., large-scale chromosomal rearrangements also play an important role in drug resistance. For example, aneuploidy and isochromosome formation increase the copy number and expression of critical genes for fluconazole resistance in Candida albicans [17], [18]. Segmental duplications and new chromosome formation were found to be correlated with fluconazole tolerance in Candida glabrata [19]. These studies indicate that large-scale changes allow yeast to quickly adapt to different environments. Despite this wealth of experimental data, it is less clear how cells maintain these mutations over a long evolutionary timescale since large-scale rearrangements are often accompanied by extra costs. In sexual populations, large-scale rearrangements can also result in gamete lethality when they are heterozygous unless they localize near the telomeres and do not carry regions with essential genes [20].
The mutations that cause large-scale chromosomal rearrangements occur at a high frequency in yeast populations. In mutation-accumulation lines of haploid budding yeast, the estimated spontaneous mutation rate of large-scale changes was 4.8-fold higher than that of small-scale changes (0.019 and 0.004 per genome per cell division, respectively) [21]. In another similar experiment in diploid yeast cells, it was shown that most structural variations occurred in the subtelomeric regions [22]. Like other types of mutations, most large-scale changes are probably deleterious and will quickly vanish from the population [23]. However, even in large evolving populations isochromosome formation and segmental duplication can be detected after as few as 5 or 100 generations, respectively [24], indicating that large-scale mutations supply the population with genetic variation that could facilitate adaptation to novel environments.
It has been suggested that Ty transposons may play an important role in the formation of large-scale chromosomal changes in yeast [25]. Although the yeast genome is relatively compact compared to other eukaryotic genomes, about 1–4% of the yeast genome is comprised of Ty sequences [26]. In addition, Ty sequences are often found in clusters [25]. Inverted arrays of transposon sequences can cause replication fork stalling that leads to chromosome breakage, especially when the replication machinery or checkpoints are compromised [27]–[29]. Those Ty-rich regions may constitute a preferred double-strand break site similar to the fragile sites observed in mammalian chromosomes [28], [30]. Previous studies in budding yeast suggested that many observed chromosomal rearrangements might result from ectopic recombination between Ty sequences [14], [30]–[33]. It is likely that Ty sequences often serve as initiation sites for generating chromosomal rearrangements.
Our knowledge about natural adaptation of budding yeast is often complicated due to human interference in the natural history of yeast. Yeast strains collected from Evolution Canyon (EC) provide an excellent model for studying how yeast populations adapt to natural environments. EC is an east-west-oriented canyon at Lower Nahal Oren, Israel, that originated 3–5 million years ago and is believed to have experienced minimal human disturbance [34]. Its microclimates provide ideal conditions for diverse local adaptations of many organisms [35]–[37]. In previous work, we employed a panel of phenotypic assays to characterize 14 diploid yeast strains collected from different locations within EC. We observed that a specific group of EC yeast strains (EC-C1) could tolerate a high concentration of cadmium. The cadmium-resistant phenotype was shown to be caused by an ancient allele of PCA1 (PCA1-C1), which encodes a metal efflux pump [38].
Here, we show that the same group of EC yeast strains was also highly resistant to another metal, copper. However, the copper-tolerant phenotype is not correlated with the PCA1-C1 mutant allele. Instead, the copper-tolerant phenotype mainly results from chromosomal rearrangements that increase the copy numbers of CUP1 and CUP2, two major genes involved in copper regulation [39], [40]. By analyzing the whole-genome expression pattern of cells carrying different copy numbers of CUP2, we found three previously unidentified genes, PHO84, SCM4 and CIN2, whose expression was regulated by Cup2 dosage and contributed to copper tolerance. Finally, we observed that the chromosomal rearrangements in EC-C1 cells were highly reversible. When cells were growing in medium with 1 mM of copper sulfate, a wild type-like chromosome reappeared and was fixed in the population within 300 generations. These results suggest that large-scale chromosomal rearrangements provide not only a fast arising but also readily reversible source of variation during early stages of adaptive evolution.
Results
One subset of EC diploid strains is highly tolerant to copper
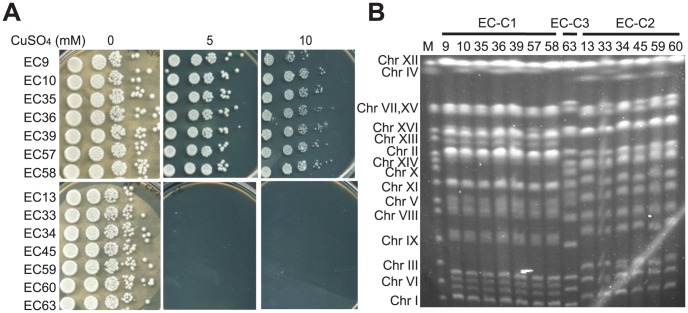
Yeast strains collected from Evolution Canyon have been shown to adapt to various environmental stresses, such as oxidative stress, UV radiation, and high concentrations of cadmium [37], [38], [41]. In addition, most of the EC strains are heterothallic [42]. To further examine if EC strains have evolved other adaptive phenotypes, we tested the growth of EC diploid strains on several metal-containing plates. Interestingly, those cadmium-resistant strains (EC-C1 strains, including EC9, 10, 35, 36, 39, 40, 57 and 58) could also tolerate high concentrations of copper sulfate (Figure 1A). However, when we crossed the copper-tolerant haploids with a copper-sensitive strain and analyzed the meiotic products, we found that the copper-tolerant phenotype did not co-segregate with the PCA1-C1 mutation responsible for the cadmium resistance (data not shown). In our previous study, we also showed that the PCA1-C1 allele did not increase the copper tolerance when it was put into a copper-sensitive strain [38]. Together, these results suggest that other genes are responsible for the tolerance to copper in the EC-C1 strains.
Figure 1. A subset of EC diploid strains is highly tolerant to copper.
(A) Strains EC9, 10, 35, 36, 39, 57 and 58 can tolerate a high level of copper (10 mM CuSO4). Diploid EC strains were grown in YPD media overnight, serially diluted and plated on YPD plates containing different concentrations of CuSO4. Plates were incubated at 28°C until obvious colonies were formed. (B) Pulsed-field gel electrophoresis analysis reveals that the EC strains comprised three major karyotypes: EC cluster 1 (EC-C1) containing seven strains (EC9, 10, 35, 36, 39, 57 and 58), EC cluster 2 (EC-C2) containing six strains (EC13, 33, 34, 45, 59 and 60), and EC cluster 3 (EC-C3) containing only one strain (EC63). All copper-tolerant strains belong to EC-C1. M, yeast chromosomal DNA from a standard laboratory strain.
In our previous study, we observed that the diploid S. cerevisiae strains isolated from Evolution Canyon comprised three major karyotypes (with some minor deviations), including EC cluster 1 (EC-C1), EC cluster 2 (EC-C2) and EC cluster 3 (EC-C3) (Figure 1B) [38]. This karyotype clustering pattern is consistent with the results from the phylogenetic analyses [42], [43]. Because all copper-tolerant strains belong to EC-C1, it suggests that the metal-tolerant phenotypes had already evolved before the EC-C1 populations split. Therefore, we chose EC9 from EC-C1 as representative of this clade for subsequent genetic analyses.
The copper-tolerant strains have gross chromosomal rearrangements
Laboratory evolution experiments have shown that chromosomal rearrangements can result in adaptive changes to gene copy number [14], [15], [44]. To further examine each individual chromosome, chromosomes separated by pulsed-field gel electrophoresis (PFGE) were subjected to Southern blotting using chromosome-specific DNA probes. The result showed that EC-C1 strains have high chromosome heterozygosity. They carry at least four heterozygous chromosome pairs (chromosomes 5, 6, 8 and 14), as revealed by length differences between homologous chromosomes. In addition, we observed several large chromosomal rearrangements in EC-C1 strains that had resulted in an elongated chromosome 10, an elongated chromosome 8 of almost twice its original size, and a novel chromosome that was hybridized by probes from both chromosomes 7 and 8 (Figure S1).
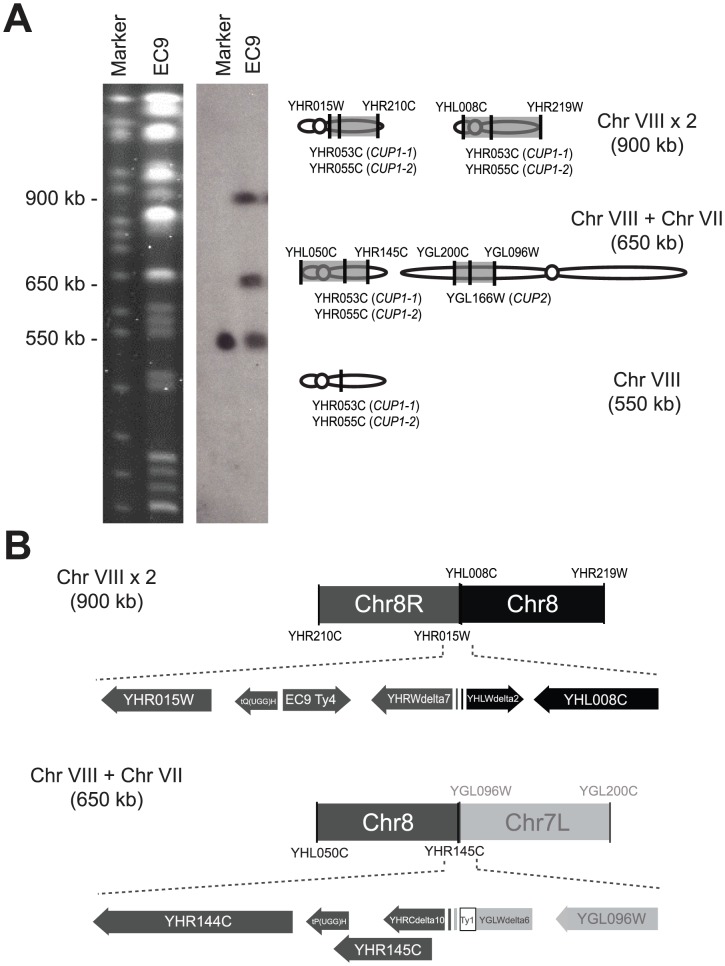
The fact that the latter two chromosomal rearrangement events that we observed both involved chromosome 8 prompted closer examination. The rearranged chromosomes were purified from PFGE gels and subjected to array-based comparative genomic hybridization (aCGH) using S. cerevisiae oligonucleotide microarrays. These experiments revealed that the aberrant 900-kb chromosome 8 is a fusion product of two chromosome 8 fragments (between YHR015W to YHR210C and YHL008C to YHR219W) and that the novel 650-kb chromosome is a fusion product of a small chromosome 7 fragment (between YGL096W and YGL200C), a large chromosome 8 fragment (between YHL050C and YHR145C) and the telomere of chromosome 8 (between YHR210C to YHR217C) (Figure 2). We also conducted aCGH using genomic DNA isolated from EC-C1 diploid cells (EC9) and haploid cells that carry both rearranged chromosomes (EC9-7 in Figure 3B). The results confirmed that the copy numbers were indeed increased in the duplicated regions.
Figure 2. EC-C1 strains contain large-scale chromosomal rearrangements.
(A) A diagram showing the structures of the rearranged chromosomes 8 in EC-C1 strains. EC-C1 strains contain three chromosomes that can be hybridized by a chromosome 8 probe (left panel and Figure S1). These chromosomes were isolated and subjected to array CGH analysis. The results showed that the 900-kb chromosome was a fusion product of two chromosome 8 fragments (between YHR015W to YHR216W and YHL008C to YHR219W) while the 650-kb chromosome contained a chromosome 7 fragment (between YGL096W and YGL200C) and a chromosome 8 fragment (between YHL050C and YHR144C). The 550-kb fragment is a wild type copy of chromosome 8. The shaded areas are the fragments contained in the rearranged chromosomes. The positions of the CUP1 and CUP2 genes are also indicated. (B) The junction sites of two rearranged chromosomes. The 900-kb chromosome was formed by fusing the regions near YHR015W and YHL008C. The 650-kb chromosome was formed by fusing the regions near YHR145C and YGL096W. EC9-Ty4, a Ty4 sequence found only in EC-C1 strains. Ty1, a Ty1 sequence found only in the junction site. The detailed DNA sequences can be found in Figure S2 and in GenBank under the accession numbers JX101633 and JX101634. The scale of this illustration is not proportional to the base pair size of genes.
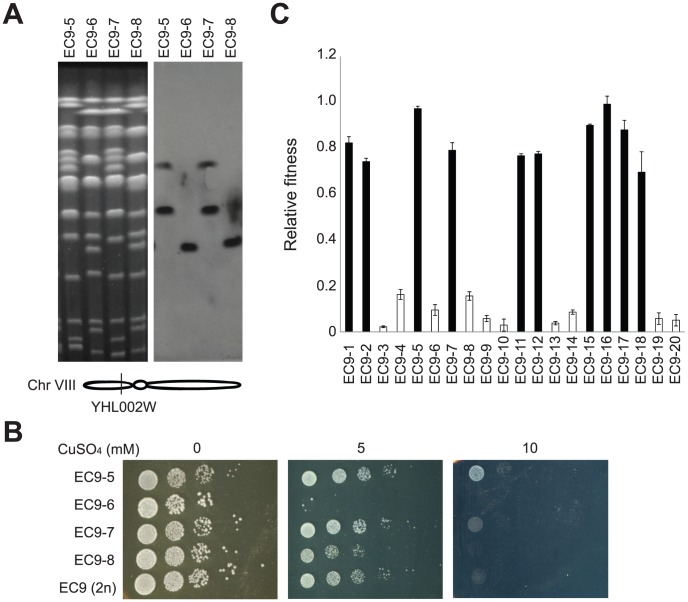
Figure 3. Haploid segregants of EC9 carrying rearranged chromosomes are more tolerant to copper.
(A) EC9-5, EC9-6, EC9-7 and EC9-8 are haploid segregants from a single EC9 tetrad. Two of them (EC9-5 and -7) carry two rearranged copies of chromosome 8, and the other two carry a wild type copy of chromosome 8. A gene (YHL002W) located on the left arm of chromosome 8 was used as a probe for Southern blotting. M, yeast chromosomal DNA from a standard laboratory strain. (B) Segregants carrying the rearranged chromosomes are more copper tolerant. (C) Rearranged chromosomes are responsible for about 60% of the observed copper-tolerant phenotype. Twenty EC9 haploid segregants were grown in medium with or without 1.5 mM CuSO4 and their growth rates were measured. Relative fitness represents the ratio of cell growth rate in medium with 1.5 mM copper to that in medium without 1.5 mM copper.
Breakpoints of the rearranged chromosomes contain many transposon sequences
To understand how novel chromosomes were formed, we fine-mapped the junction sites of the rearranged chromosomes. We designed primers near each possible breakpoint according to the aCGH data (i.e., regions close to YHR015W, YHR210C, YHL008C, YHR219W, YHL050C, YHR145C, YGL200C and YGL096W) and used these primers to find out the junction site of two chromosomal fragments (see Materials and Methods). As shown in Figure 2C and Figure S2, the aberrant 900-kb chromosome 8 was formed by fusing the regions near YHR015W and YHL008C and the novel 650-kb chromosome was formed by fusing the regions near YHR145C and YGL096W. Interestingly, we found that the breakpoints were all flanked by Ty sequences (next to YHR015W, YHL008C, YHR145C and YGL096W), indicating that transposable elements might be the mediator of these chromosomal rearrangements. Moreover, three out of the four flanking regions (YGL096W, YHR015W and YHL008C) contain multiple Ty long terminal repeats (LTRs) including at least one inverted LTR pair. It is possible that the double-strand break hotspots formed in these Ty arrays allow chromosomes to be rearranged at a high frequency. We also sequenced the breakpoints of the other EC-C1 strains. The result confirmed that the same chromosomal rearrangements exist in all EC-C1 strains (Table S1).
The copper-tolerant phenotype is correlated with chromosomal rearrangements
To assess the contribution of the rearranged chromosomes to copper tolerance, we dissected tetrads (meiotic products) of EC9 and measured the copper resistance of individual haploid segregants (spores). The spore viability of EC9 is about 60% due to the chromosomal rearrangement of EC-C1 strains. In eight four-viable-spore tetrads yielding 32 haploid segregants, all sixteen segregants containing both rearranged chromosomes showed higher copper tolerance than the other sixteen segregants containing only wild type copies of chromosomes (Figure 3 and Figure S3). However, we noticed that within these two groups of segregants, there were different levels of copper tolerance between individual clones, indicating that the copper-tolerant phenotype in EC-C1 strains was polygenic and genes on other chromosomes might also be involved in copper tolerance with minor effects. We measured the relative fitness of twenty EC9 haploid segregants in copper-containing medium (Figure 3C). By comparing the fitness between stains carrying rearranged chromosomes and the wild type chromosome, we estimated that rearranged chromosomes are responsible for about 60% of the observed copper-tolerant phenotype.
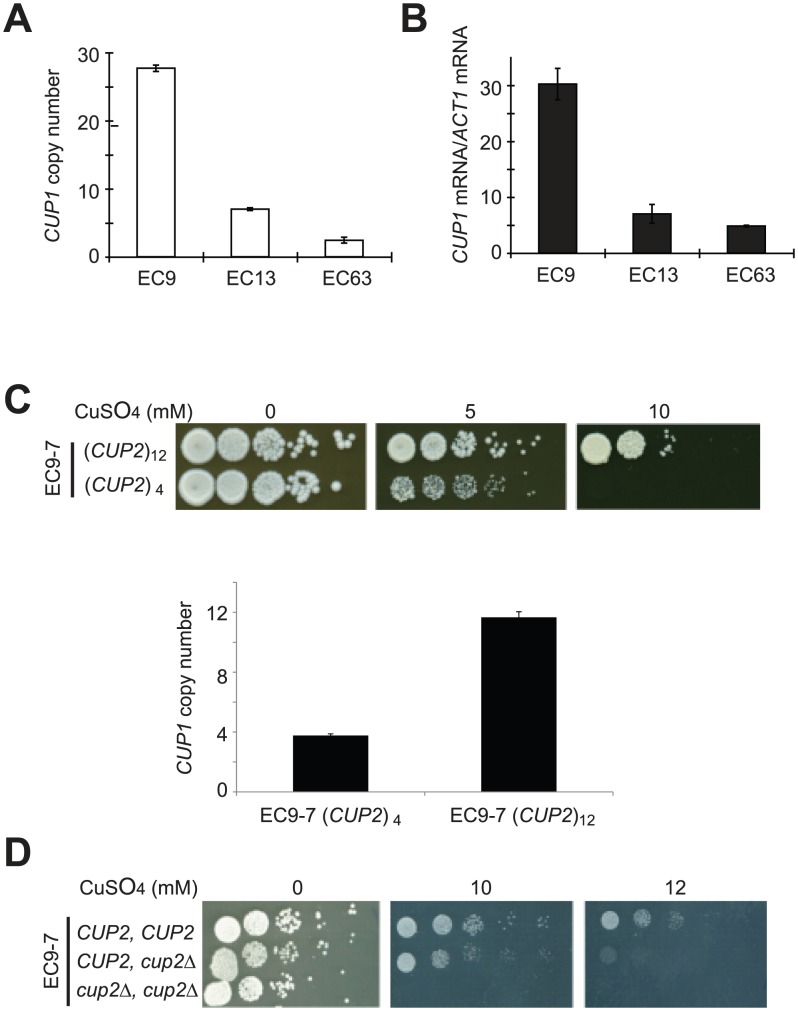
When the rearranged chromosomes were inspected, we observed that genes involved in response to copper ions (CUP1, CUP2 and COX23) were significantly enriched. CUP1 is a gene encoding a metallothionein and its expression level has been shown to play an important role in copper tolerance [39]. We measured the CUP1 gene copy number and expression level using quantitative PCR. The results showed that the CUP1 copy number and mRNA level in EC9 (an EC-C1 strain) were about 5–6-fold higher than expression in EC34 and EC63 (EC-C2 and EC-C3 strains) after cells were treated with CuSO4 (Figure 4A and 4B). To confirm that the increased copies of CUP1 are important for copper tolerance in EC-C1 strains, we deleted eight copies of CUP1 in an EC9 haploid segregant (EC9-7 in Figure 3B) and measured their copper sensitivity. The results showed that cells with fewer copies of CUP1 were indeed less copper-tolerant (Figure 4C).
Figure 4. Increased copy numbers of CUP1 and CUP2 were correlated with enhanced copper tolerance in EC-C1 strains.
(A) The copy number of CUP1 in EC9 (an EC-C1 strain) is higher than that in EC13 (an EC-C2 strain) and EC63 (an EC-C3 strain). Total genomic DNA was isolated from EC9, EC13 and EC63, and subjected to quantitative PCR using CUP1-specific and ACT1-specific primers. The CUP1 DNA levels were normalized to the ACT1 DNA levels. (B) EC9 has a higher CUP1 expression level after copper treatment. EC9, EC13 and EC63 log-phase cells were treated with 1 mM CuSO4 for 2 h. Total RNA was isolated from the Cu-treated cells, reverse-transcribed and subjected to quantitative PCR using CUP1-specific and ACT1-specific primers. The CUP1 mRNA levels were normalized to the ACT1 mRNA levels. (C) Amplification of CUP1 has a major contribution to copper tolerance in EC-C1 cells. Haploid cells carrying different copy numbers of CUP1 were grown in YPD media overnight, serially diluted and plated on YPD plates containing different concentrations of CuSO4. Total genomic DNA was isolated from EC9-7 and EC9-7 with CUP1 deletion, and subjected to quantitative PCR as previously described. (D) Copper tolerance is correlated with the copy number of CUP2. Haploid cells (EC9-7) carrying two, one or no copies of CUP2 were grown in YPD media overnight, serially diluted and plated on YPD plates containing different concentrations of CuSO4.
Cup2 is a copper-binding transcriptional factor that activates CUP1 expression [40], [45]. The chromosome rearrangements increase the CUP2 copy number to three in EC-C1 cells. To determine whether the increased copy number of CUP2 contributes to copper tolerance in EC-C1 strains, we used EC9-7 to construct yeast strains carrying zero, one or two copies of CUP2 and tested their copper sensitivity. The sensitivity of cells to copper was negatively correlated with the copy number of CUP2 (Figure 4D), indicating that an increase in the copy number of CUP2 is also important for copper tolerance. Our results indicate that amplification of both CUP1 and CUP2 genes are required for cells to achieve high copper tolerance.
PHO84, SCM4, and CIN2 are involved in copper tolerance in EC-C1 strains
The dosage effect of CUP2 prompted us to investigate the downstream targets of the Cup2 transcription factor. In previous studies, Cup2 has been shown to regulate three metal-responsive genes, including two metallothionein genes, CUP1 and CRS5, and the copper-zinc superoxide dismutase gene SOD1 [40], [45]–[47]. To identify more candidate genes under the regulation of Cup2, we performed a whole-genome expression pattern analysis of the EC9 haploid segregant carrying different copy numbers of CUP2 (EC9-7, EC9-7 cup2Δ and EC9-7 cup2Δ cup2Δ in Figure 4D). Cells were treated with 1 mM CuSO4 for 1 h, which resulted in no obvious effects on the growth of cup2 null mutant cells. Total RNA from treated samples was collected and analyzed using microarrays. In cup2 double deletion cells we found 39 genes with reduced (1.5-fold or more) expression compared with wild type cells (Table S2). Among these genes, 18 showed a positive correlation between their expression levels and the copy number of CUP2.
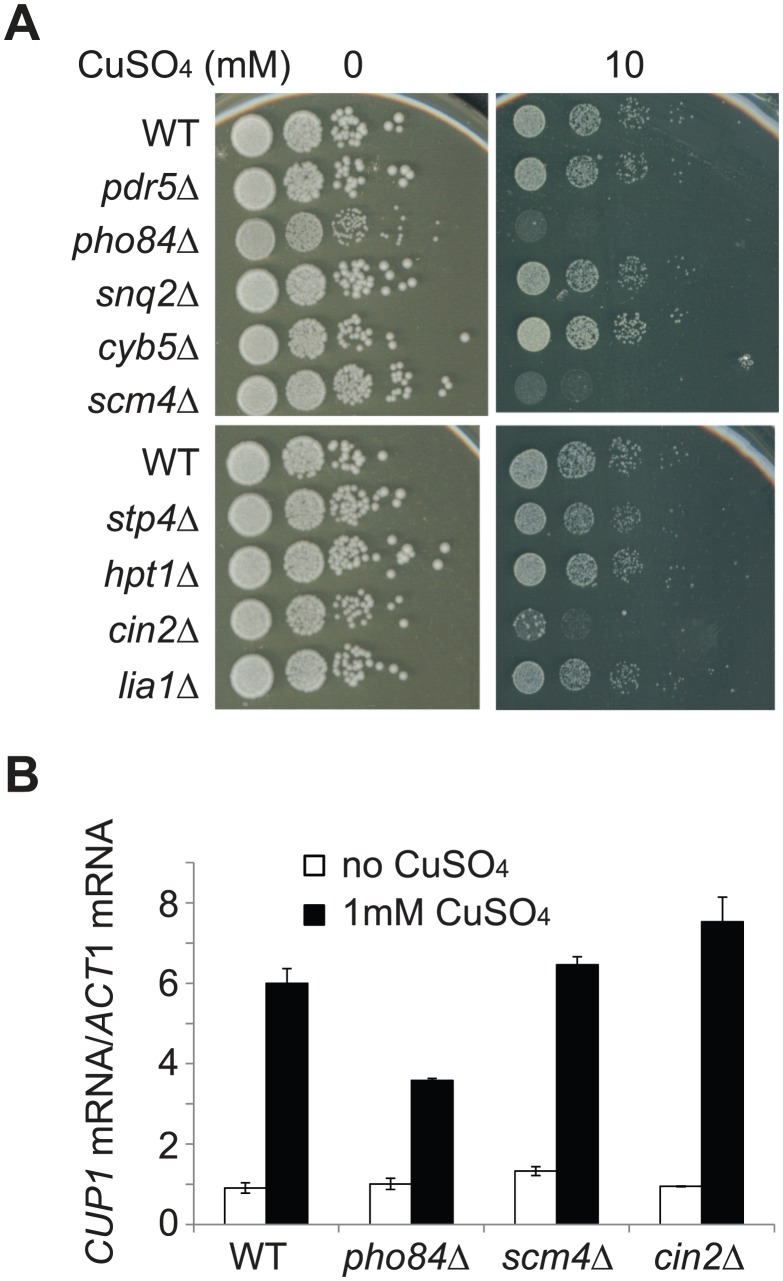
To directly test the effect of these candidate genes on copper tolerance, we examined eight non-essential genes in the aforementioned group (PDR5, SNQ2, CYB5, SCM4, STP4, HPT1, CIN2 and LIA1) and PHO84, a gene whose expression showed more than a 3-fold difference between wild type and cup2 double deletion cells but did not correlate with the CUP2 copy number. We found that when individual genes were deleted, three mutant strains (pho84Δ, scm4Δ and cin2Δ) showed reduced tolerance to copper sulfate (Figure 5A). To rule out the possibility that the mutant cells were sensitive to sulfates instead of copper, the deletion strains were also tested on plates containing copper chloride (Figure S4). Our result indicates that pho84Δ, scm4Δ and cin2Δ mutant cells were indeed sensitive to copper despite no reported linkage between these genes and copper tolerance. We further tested the effect of increased expression of these three genes by introducing a CEN plasmid carrying PHO84, SCM4 or CIN2 into EC9-8 haploid cells and examining their copper tolerance. However, we were unable to detect obvious growth differences using spot assays (data not shown). We also tried to measure cell fitness using a more sensitive competitive fitness assay, but it was unsuccessful as the EC cells tended to clump together in copper-containing medium. Our results suggest that PHO84, SCM4 and CIN2 are involved in copper tolerance, but it is less clear how much the increased expression of these three genes contributes to the elevated copper tolerance in EC-C1 cells.
Figure 5. PHO84, SCM4, and CIN2 are involved in copper tolerance.
(A) Eight non-essential genes (PDR5, SNQ2, CYB5, SCM4, STP4, HPT1, CIN2, and LIA1), whose expression levels correlated with their CUP2 copy number, and PHO84, whose expression showed more than a 3-fold difference between wild type and cup2 double deletion cells but did not correlate with CUP2 copy number, were tested for their contributions to copper tolerance. Individual genes were deleted in the EC9-8 haploid background (WT). Cells were grown in YPD media overnight, serially diluted and plated on YPD plates containing different concentrations of CuSO4. (B) CUP1 expression is reduced in pho84Δ cells. Total RNA was isolated from cells with or without copper treatments, reverse-transcribed and subjected to quantitative PCR using CUP1-specific and ACT1-specific primers. The CUP1 mRNA levels were normalized to the ACT1 mRNA levels.
To investigate the molecular mechanisms about how PHO84, SCM4 and CIN2 affect copper tolerance, we tested whether expression of the CUP1 gene was affected by mutations in these three genes. Total RNA was isolated from cells treated with 1 mM CuSO4 and the expression level of CUP1 was measured using quantitative PCR. In pho84Δ cells, CUP1 expression was significantly reduced, suggesting that Pho84 may influence copper tolerance through a Cup1-dependent mechanism (Figure 5B).
Rearranged and wild-type chromosomes share the same sequence
In yeast, it has been shown that large-scale chromosomal rearrangements occur frequently and beneficial ones can become fixed rapidly in the population [14], [21], [24]. It is therefore of interest whether the observed rearranged chromosomes have existed in the EC strains for a long time or have been formed recently. We sequenced a 6628-bp fragment (corresponding to positions 188,179 to 194,679 bp on chromosome 7) from the rearranged chromosome 7 and a 6602-bp fragment (corresponding to positions 496,154 to 502,755 on chromosome 8) from the aberrant chromosome 8 and compared them with wild-type chromosomes (see Materials and Methods for details). Both fragments had the same sequence as that on wild-type chromosomes. From the genome sequence divergence between two closely related yeast species, S. cerevisiae and S. paradoxus, we obtained an estimation that it may require about 0.25 million years to accumulate 1% of sequence divergence in intergenic regions [48]. If we applied this estimation to the case of the rearranged chromosomes in EC-C1 strains, it suggests that the rearranged chromosomes occurred in the last 3800 years assuming that there was no recombination between the rearranged and wild type chromosomes.
Rearranged chromosomes revert to wild-type-like chromosomes in laboratory evolution experiments
Large-scale chromosomal rearrangements can spread through a population if they are beneficial. However, the rearranged chromosomes can be quite unstable when growth conditions change and selective pressure is relieved [24], [49]. We set up a laboratory evolution experiment to investigate the stability of rearranged chromosomes. Five individual colonies derived from an EC9 haploid segregant carrying rearranged chromosomes (EC9-7 in Figure 3) were used to set up 10 independent evolving lines, 5 with relaxed selection (YPD containing 1 mM CuSO4) and 5 with strong selection (YPD containing 5 mM CuSO4). We regarded the medium containing 1 mM CuSO4 as a more relaxed growth condition because EC9 haploid segregants without rearranged chromosomes (such as EC9-6 in Figure 3B) could grow efficiently under such conditions. These cells were grown and diluted daily in fresh medium. After 400 generations, we observed that evolved cells with relaxed selection all exhibited improved growth on YPD or YPD with 1 mM CuSO4, but decreased tolerance to high concentrations of copper (Figure 6A).
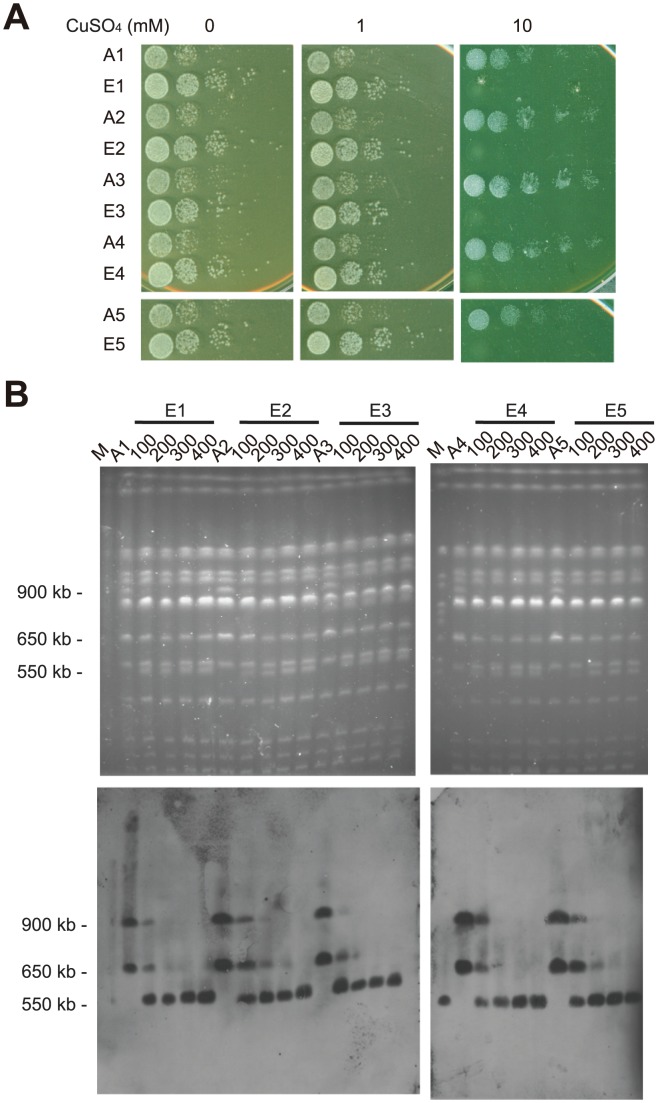
Figure 6. Rearranged chromosomes revert back to wild-type-like chromosomes in laboratory evolution experiments.
(A) Experimentally evolved cells exhibited improved growth on YPD or YPD with 1 mM CuSO4 but decreased their tolerance to high concentrations of copper after 400 generations of growth in medium containing 1 mM CuSO4. Ancestral cells (A1–A5) and evolved cells (E1–E5) were grown in YPD media overnight, serially diluted and plated on YPD plates containing different concentrations of CuSO4. (B) Pulsed-field gel electrophoresis analysis reveals that a wild type-like chromosome 8 (approximately 550 kb) replaced the rearranged chromosomes (in the size range of 900 kb and 650 kb) in the evolved cell populations. Evolved cultures from generations 100, 200, 300 and 400 were examined. A gene (YHL002W) located on the left arm of chromosome 8 was used as a probe for Southern blotting. M, yeast chromosomal DNA from a standard laboratory strain.
We examined the karyotype of evolved cells collected from generations 100, 200, 300 and 400. In all five evolved cultures with relaxed selection, the rearranged chromosomes (in the size range of 900 kb and 650 kb) were replaced by a novel wild type-like chromosome 8 (approximately 550 kb) during the course of evolution (Figure 6B). Since each evolved culture was initiated from an independent single colony, it is likely that the novel chromosome 8 repeatedly evolved at least five times in our evolution experiment. In addition, this novel chromosome 8 could be detected in the populations collected from generation 100, suggesting that it already existed in the population at a very early stage of our experiment (Figure 6B). In contrast, four out of five evolved cultures under strong selection pressure retained the rearranged chromosomes in the majority of populations even after 400 generations (Figure S5A). We also set up evolution experiments using EC9 diploid cells. Similar results were observed except that it took a longer time to fix the wild type-like chromosome 8 in diploid populations (Figure S5B). One possible explanation for the difference between haploid and diploid populations is that the cost of carrying extra chromosomal fragments is relatively lower in diploid cells [50]. Together, these results suggest that these chromosomal rearrangements are highly dynamic and reversible.
To further investigate its structure, the novel 550-kb chromosome 8 of the evolved cells was purified from PFGE gels and subjected to array-based comparative genomic hybridization (aCGH). The result showed that the novel chromosome 8 had almost the same gene content as the wild type chromosome 8 except for some telomeric genes; YHR217C, YHR218W and YHR219W in the right telomere and YHL044W, YHL045W, YHL048W, YHL049C and YHL050C in the left telomere were undetected in our assay. On the other hand, we detected signals of other telomeric regions, including YAR062W, YAR064W, YAR066W, YAR068W, YAR069C, YAR070C and YAR073W from the telomere of chromosome 1 and YFL065C, YFL066C and YFL067W from the telomere of chromosome 6. Telomeric regions have been known to be very dynamic. It is possible that recombination between different telomeres occurred during our evolution experiment.
Discussion
Adaptive phenotypes can be generated by chromosomal rearrangements in natural populations
It has been observed that yeast can adapt to various nutrient-limited conditions [15], [44], [51]. Recent surveys on yeast strains collected from different continents also revealed that S. cerevisiae populations exhibit a high degree of phenotypic variance, suggesting that they have adapted to diverse ecological niches [26], [52]. However, unlike experimentally evolved cells, adaptations in natural populations are more difficult to study. It has remained elusive whether the types of mutations commonly observed in laboratory adaptation are also involved in natural adaptation.
Copper is an essential cofactor for many enzymes such as the cytochrome c oxidase in the respiratory chain. Nonetheless, an excess of copper is deleterious to cells [53]. The toxicity of copper may come from the generation of reactive oxygen species, the competition with other metals for their native binding sites, the alteration of protein conformations or interference with biochemical reactions [53], [54]. Cells have evolved multiple mechanisms to regulate copper homeostasis including different metal transporters, sequestration factors, and detoxification enzymes [54], [55].
The EC-C1 strains carry two rearranged chromosomes that significantly enrich the copy number of genes involved in copper regulation. Evolved phenotypes often arise from duplicated chromosomal fragments that contain the critical genes for adaptation in experimental yeast populations [14], [15], [44]. We speculate that the rearranged chromosomes in EC-C1 strains might result from selection for higher copper tolerance for the following reasons. First, among all the diploid yeast strains collected from Evolution Canyon, the EC-C1 strains constitute a major group (8/21 or 38%). In addition, the EC-C1 strains show very low levels of polymorphism in their microsatellite loci compared with other EC groups [43], suggesting that EC-C1 strains carry some adaptive phenotypes allowing them to quickly spread in Evolution Canyon. Second, when the copper content in the soil samples collected from different sites of Evolution Canyon was measured using inductively coupled plasma-atomic emission spectroscopy (see Materials and Methods), we found that the copper levels of most EC sites are above 30 ppm (with an average of 38 ppm) and in one area it even reaches 95 ppm, which are higher than the average copper content (20 ppm) in soil [56]. Previous studies have suggested that increased copper levels in vineyard soil caused the wine yeast strains to evolve higher copper tolerance [57], [58]. A similar adaptive process might also occur in the EC-C1 strains. Third, a previous study has shown that increased expression of CUP1 also enhanced cadmium resistance of cells. When we examined cadmium sensitivity of twenty EC9 haploid segregants, the cadmium-resistant phenotype was not co-segregated with rearranged chromosomes. This suggests that increased copies of CUP1 and CUP2 were not a result of selection for the pleiotropic effect on cadmium resistance (Figure S6). In our lab, we have examined the fitness of all EC strains under more than 30 different growth conditions (including different temperatures, nutrient starvation, chemicals and metal ions). Only in two conditions, medium containing either high levels of copper or cadmium, EC-C1 strains showed higher growth rates ([38] and our unpublished results). Nonetheless, we cannot completely rule out the possibility that chromosomal rearrangements in EC-C1 strains were caused by adaptive effects resulting from other amplified genes or other unknown pleiotropy of Cup1 and Cup2. In the future, it will be interesting to compare the whole-genome gene expression patterns between haploid cells carrying rearranged or wild type chromosomes under different conditions. If differentially regulated genes are enriched in biological pathways other than copper tolerance, it may provide us a clue to further test other possible causes of chromosomal rearrangements in EC-C1 strains.
Increased CUP1 copy number has been observed in copper-resistant strains isolated from laboratory evolution experiments, industry or natural habitats [59]–[61]. Nonetheless, EC-C1 diploid cells carry more than 20 copies of CUP1 that are much higher than the CUP1 amplification reported in previous cases (ranging from 2 to 15 copies). In addition, we showed that increased CUP1 copy number alone was not enough to achieve the high copper tolerance observed in EC-C1 cells. Amplification of the copper-binding transcriptional factor CUP2 was also critical, suggesting that a more complex adaptive strategy has occurred in EC-C1 strains. Increasing the dosage of a transcriptional factor may influence the expression of its downstream target genes to different levels depending on its feedback regulation or other compensatory mechanisms. Although the CUP1 amplification clearly plays a major role in copper tolerance of EC-C1 cells, other downstream targets of Cup2 probably also contribute to the observed phenotype.
By combining the whole-genome gene expression analysis of cells carrying different copy numbers of CUP2 and the functional assay, we identified and confirmed three previously unidentified genes, PHO84, SCM4 and CIN2, that were involved in copper tolerance. In two of them (SCM4 and CIN2) we also observed a conserved Cup2-binding motif sequence in their promoters. CIN2 encodes a GTPase-activating protein involved in tubulin folding [62], and cin2Δ mutant cells are also sensitive to another metal, arsenic, suggesting that Cin2 is involved in metal regulation [63]. SCM4 was previously identified as a suppressor of a cell cycle mutant of CDC4 [64]. Nonetheless, the Scm4 protein contains four transmembrane domains and localizes to the mitochondria, an organelle involved in many metal metabolic pathways. It will be interesting to determine whether Scm4 affects copper tolerance through its function in mitochondria. PHO84 encodes a high-affinity inorganic phosphate transporter that also functions in manganese homeostasis [65], [66]. The enhanced tolerance of pho84Δ mutants to several metal ions (including manganese, zinc, cobalt and copper) has been attributed to defects in the uptake of metal ions [66]. However, we found that deletion of PHO84 in EC-C1 strains decreases tolerance to high concentrations of copper in a Cup1-dependent manner. This suggests that genetic background may have a strong influence on the regulatory network of metal metabolism.
Dynamic large-scale chromosomal rearrangements in a fluctuating environment
Large-scale chromosomal rearrangements can quickly change the expression level of multiple genes or even a whole pathway by changing the gene copy number. In addition, the spontaneous rate of chromosomal rearrangements is higher than the spontaneous rate of point mutations [21]. This class of mutations is most likely to be found at the early stage of adaptation since they allow a brute force change in phenotype by changing multiple genes in one step. Many such examples have been reported in short-term experimental evolution in S. cerevisiae and C. albicans [8]–[11], [15], [24], [67]. But in addition to increasing the copy numbers of beneficial genes, rearrangement also increases copy numbers of other genes in the same chromosomal segments that may not be beneficial. It has been shown that when compared with euploid cells, aneuploid cells have higher fitness under certain conditions but have reduced fitness in general [50], [68]. Thus, chromosomal rearrangement is unlikely to be an optimal form of mutation, but may allow a population to survive until either better mutations appear or until a population's environment becomes more permissive. In the EC-C1 strains, we examined two loci on the rearranged chromosomes 7 and 8 (∼7 kb/each) and found that they had identical sequences as wild type chromosomes, supporting the idea that the rearranged chromosomes were recently generated rather than an ancient relic. A lingering question is then how cells adjust to the cost of these crude adaptive changes on longer evolutionary timescales, especially in a fluctuating environment.
When EC-C1 cells carrying the rearranged chromosomes were propagated in medium containing 1 mM copper sulfate, a wild type-like chromosome 8 quickly became fixed in all five individual populations in as early as 200 generations. The repetitive appearance of this novel chromosome 8 suggests that some large-scale chromosomal rearrangements are highly dynamic and reversible. This result is in agreement with a previous study showing that large-scale inter- and intra-chromosomal duplications were intrinsically unstable when no selective advantages were provided by those duplications [49]. Previous studies in budding yeast suggested that clustered Ty sequences might serve as double-strand break hotspots to initiate ectopic recombination in the yeast genome [28], [30]. This type of recombination allows cells to quickly adapt to stressful environments by duplicating chromosomal fragments that contain critical genes. Furthermore, when the stress is relieved or better mutations have evolved, the duplicated chromosomal fragments can revert back to the original configuration at a high frequency by another round of ectopic or homologous recombination. Such genome flexibility enables organisms to generate switch-like adaptive phenotypes. This would be especially valuable for sexual populations since large-scale chromosomal rearrangements often cause gamete lethality when they are heterozygous. This idea is indirectly supported by the observation that although large-scale chromosomal rearrangements occur frequently in laboratory evolution experiments or natural isolates, closely related yeast species such as S. cerevisiae and S. paradoxus still maintain colinear genomes [69]. Together with the facts that transposon expression is known to be activated under environmental stress and elevated transcription levels increase the rate of mitotic recombination [70]–[72], the abovementioned Ty-mediated chromosomal rearrangements supply the population with an effective mechanism to quickly respond to environmental changes.
Materials and Methods
Strains and genetic procedures
All EC diploid strains are Saccharomyces cerevisiae collected from an east-west facing canyon (Evolution Canyon) at Lower Nahal Oren, Israel [43]. In brief, EC33, 34, 35 and 36 were isolated from the south-facing slope (SFS), EC9, 10, 39 and 45 from the valley bottom (VB), and EC13, 57, 58, 59, 60 and 63 from the north-facing slope (NFS). Substitutive and integrative transformations were carried out by the lithium acetate procedure [73]. Media, microbial and genetic techniques were performed as described [74].
Karyotyping of EC strains and Southern blot
A total of 1∼2×108 yeast cells were used for plug preparation. Cells were washed with 1 ml EDTA/Tris (50 mM EDTA, 10 mM Tris, pH 7.5) and transferred into EDTA/Tris with 0.13 mg/ml zymolyase (Seikagaku America Inc., St. Petersburg, FL). The cell mixtures were incubated for 30 s at 42°C and then embedded in low melting point agarose (Sigma-Aldrich, St. Louis, MO). The agarose plugs were placed at 37°C overnight for zymolyase digestion. After digestion, the agarose plugs were placed in LET solution (0.5 M EDTA, 10 mM Tris, pH 7.5) containing 2 mg/ml protease K and 1% N-lauroylsarcosine at 50°C overnight. This step was repeated three times. The plugs were transferred to EDTA/Tris solution and dialyzed four times for 1 h at 37°C. Yeast chromosomes were separated on 0.7% agarose gels by pulsed field gel electrophoresis (PFGE) using a Rotaphor Type V apparatus (Biometra, Göttingen, Germany). Electrophoresis was performed for 48 h at 13°C in 0.5× TBE buffer at a fixed voltage of 120 V and an angle of 115° with pulse time intervals of 30 s.
After PFGE, the chromosomal DNA was depurinated and denatured by incubating the agarose gel in 0.25 N HCl and then in alkaline solution (0.5 M NaOH, 1.5% NaCl). The DNA was transferred to a charged nylon membrane, Immobilon-NY+ (Millipore, Billerica, MA). DNA probes for each chromosome were obtained by PCR using the primers listed in Table S1. The Digoxigenin-labeled DNA probes were prepared using the DNA labeling and detection kit (Roche, Indianapolis, IN).
Array-based comparative genomic hybridization
Oligonucleotide arrays were produced at the Microarray Core, Institute of Molecular Biology, Academia Sinica, using an Omnigrid 100 arrayer (Digilab, Holliston, MA) and the Yeast Genome Array-Ready Oligo Set (Version 1.1, Operon, Huntsville, AL). The printing protocol can be found at the Institute of Molecular Biology Microarray Facility web site (http://www.imb.sinica.edu.tw/mdarray/methods.html).
Yeast genomic DNA was extracted using the Qiagen Genomic-Tip 100/G kit (Qiagen, Valencia, CA). For individual-chromosome aCGH, DNA was excised from PFGE gel after EtBr staining and purified using the Geneaid Gel DNA Fragment Extraction kit (Geneaid, Taiwan). The purified chromosomal DNA was further amplified using GenomePlex Whole Genome Amplification Kit (Sigma-Aldrich, St. Louis, MO). Probe preparation and hybridization were performed as described [75]. The array data were analyzed using GeneSpring GX 7.3.1 (Agilent, Santa Clara, CA).
Quantitative PCR
After copper treatment (1 mM CuSO4) for 2 h at 28°C, total RNA was isolated by Qiagen RNeasy Midi Kit (Qiagen). First-strand cDNA was synthesized for 2 h at 37°C using the High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems, Foster City, CA). A 20-fold dilution of the reaction products was then subjected to real-time quantitative PCR using gene-specific primers, SYBR Green PCR master mix and an ABI-7000 sequence detection system (Applied Biosystems). Data were analyzed using the built-in analysis program.
Fitness assays
Fitness of individual strains was obtained by propagating replicate cultures in complete synthetic medium (CSM) with or without 1.5 mM CuSO4 in 96-well plates inside a temperature controlled, shaking plate reader Infinite F200 (Tecan, Mannedorf, Switzerland). Growth rates were calculated as the maximum slope that could be derived from any continuous 2-hour period during the 20-hr assay. Four replicate cultures were used per strain.
To determine competitive relative fitness, we measured the fitness of the experimental strains by competing them against a reference strain expressing PGK1::GFP in YPD media at 28°C. The testing cells and reference cells were inoculated in the YPD medium individually and acclimated for 24 h. The cells were subsequently diluted in fresh media and incubated for another 4 h. The reference and testing cells were then mixed (1∶1 ratio), diluted into fresh medium at a final cell concentration of 5×103 cells/ml, and allowed to compete for 17 h, which represents about 11 generations of growth. The ratio of the two competitors was quantified at the initial and final time points using a fluorescence activated cell sorter (FACSCalibur, Becton Dickinson, Franklin Lakes, NJ). Four independent replicates for each fitness measurement were performed.
Rearranged chromosome sequencing
We sequenced both wild type and rearranged chromosomes. To prevent cross-contamination between wild type and rearranged chromosomes, wild type chromosomes were purified from EC9-8 haploid cells that do not carry any rearranged chromosome, and rearranged chromosomes were purified from EC9-7 haploid cells that do not carry wild type chromosome 8 (Figure 3). Individual chromosomal DNA was purified from PFGE gels. Two regions on chromosome 7 (from 188,179 to 194,679 bp) and chromosome 8 (from 496,154 to 502,755 bp) were amplified by PCR using a set of primers. The PCR products were purified and then sequenced. The accession numbers for the sequences are JN835223 and JN835224.
To identify the junction sites of the rearranged chromosomes, we designed primers near each possible breakpoint (YHR015W, YHR210C, YHL008C, YHR219W, YHL050C, YHR145C, YGL200C and YGL096W) according to the aCGH data. For each rearranged chromosome, four different combinations of primer pairs were used to PCR the junction site. Only one pair of the primers could successfully amplify the junction site. The PCR products were purified and then sequenced. The accession numbers for the sequence are JX101633 and JX101634.
Whole-genome expression analysis
For the whole-genome gene expression analysis, log-phase cells were grown in YPD with 1 mM CuSO4 (which does not affect the growth of cup2 null mutant cells) for 1 h. Total RNA from the treated samples and the corresponding untreated control samples (in YPD) was isolated using the Qiagen RNeasy Midi Kit (Qiagen). Probe preparation and hybridization were performed as described [75]. The array data were analyzed using GeneSpring GX 7.3.1 (Agilent). We excluded the data with hybridization intensities lower than 500 as they were close to the background values (∼200). The intensities of each array were normalized using a LOWESS function [76].
Experimental evolution
EC9-7 haploid cells were streaked out on a YPD plate to form single colonies. Five individual colonies (A1–A5) were then used to initiate the evolution experiment. Cells were cultured in 3 ml YPD with 1 mM CuSO4 (E1–E5) or YPD with 5 mM CuSO4 (E6–E10) through a daily 1000-fold dilution (about 10 generations). Once every five transfers population samples from each line were stored in 20% glycerol at −80°C for later analysis. For diploid evolution experiments, EC9 diploid cells were diluted and plated on an YPD plate to grow to single colonies. Five individual colonies (AD1–AD5) were then used to initiate the evolution experiment (ED1–ED5) with a protocol similar to that of haploid evolution experiments.
Measurement of soil copper concentrations
Soil samples were collected at 7 locations of Evolution Canyon corresponding to the collection sites of the EC yeast strains (three at the SFS, one at the VB, and three at the NFS). Copper contents in soil were measured by inductively coupled plasma-atomic emission spectroscopy (ICP-AES) using at least 200 g of individual samples.
Data access
The array CGH data are available from the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE22431, GSE38034 and GSE33652. The expression data are available from GEO under accession number GSE31661.
Supporting Information
EC-C1 strains contain three rearranged chromosomes. (A) Three different chromosomes were hybridized in EC-C1 strains when a gene (YHR003W) located on the right arm of chromosome 8 was used as a probe for Southern blotting. A similar pattern was observed when two other genes (YHL002W and YHR102W) were used as probes. (B)(C) Two different chromosomes were hybridized in EC-C1 strains when a gene (YHL027W) located on the left arm of chromosome 8 or a gene (YGL175W) located on the left arm of chromosome 7 were used as a probe for Southern blotting. The 650-kb chromosome was hybridized by both probes. (D) EC-C1 strains contain an enlarged chromosome 10. A gene (YJL001W) located on the left arm of chromosome 10 was used as a probe for Southern blotting. A similar pattern was observed when another gene (YJR001W) was used as a probe.
(EPS)
Aligned DNA sequences of the junction sites. (A) The junction site sequence of the rearranged 900-kb chromosome. (B) The junction site sequence of the rearranged 650-kb chromosome. Ty1, a Ty1 sequence found only in the junction site. DNA sequences of the flanking regions can be found in GenBank under the accession numbers JX101633 and JX101634.
(EPS)
Haploid segregants of EC9 carrying rearranged chromosomes are more tolerant to copper. (A) Segregants carrying the rearranged chromosomes are more copper-tolerant. Haploid segregants from four-viable-spore tetrads of EC9 strains were grown in YPD media overnight, serially diluted and plated on YPD plates containing different concentrations of CuSO4. Plates were incubated at 28°C until obvious colonies were formed. (B) Pulsed-field gel electrophoresis analysis of haploid segregants from four four-viable-spore EC9 tetrads. In four haploid segregants from a single tetrad, two of them carry two rearranged copies of chromosome 8, and the other two carry a wild type copy of chromosome 8. A gene (YHL002W) located on the left arm of chromosome 8 was used as a probe for Southern blotting. M, yeast chromosomal DNA from a standard laboratory strain.
(EPS)
pho84, scm4, and cin2 mutant cells are sensitive to copper but not sulfates. Results in Figure 5 showed that pho84, scm4 and cin2 mutant cells are sensitive to CuSO4. To rule out the possibility that these mutants are sensitive to sulfates instead of copper, the same set of mutant cells were tested on YPD plates containing different concentrations of CuCl2.
(EPS)
Rearranged chromosomes are more likely to revert back to wild type-like chromosomes only in cultures with relaxed selection. (A) Most of the haploid cells evolving under strong selection (E6–E10) maintain the rearranged chromosomes even after 400 generations. (B) In diploid populations evolving under relaxed selection (ED1–ED5), a wild type-like chromosome 8 (approximately 550 kb) replaced the rearranged chromosomes (in the size range of 900 kb and 650 kb). Evolved cultures from generations 100, 200, 300 and 400 were examined. A gene (YHL002W) located on the left arm of chromosome 8 was used as a probe for Southern blotting. M, yeast chromosomal DNA from a standard laboratory strain.
(EPS)
Rearranged chromosomes do not correlate with the cadmium-resistant phenotype. Twenty EC9 haploid segregants were grown in YPD media overnight, serially diluted and plated on YPD plates containing different concentrations of CdCl2. Plates were incubated at 28°C until obvious colonies were formed.
(EPS)
Single nucleotide polymorphisms in the junction sites of EC-C1 strains.
(PDF)
Gene expression in cells under copper treatment.
(PDF)
Acknowledgments
We thank Justin Fay, Duncan Greig, Frank Rosenzweig, and members of the Leu lab for helpful discussions and comments on the manuscript. We also thank AndreAna Peña for manuscript editing and the Microarray and Bioinformatics Cores of IMB for technique help. We thank Eviatar Nevo and Gianni Liti for providing yeast strains.
Funding Statement
This work was supported by Academia Sinica of Taiwan (grant no. 100-CDA-L04) and the National Science Council of Taiwan (grant no. NSC100-2321-B-001-021). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nadeau NJ, Jiggins CD (2010) A golden age for evolutionary genetics? Genomic studies of adaptation in natural populations. Trends Genet 26: 484–492. [DOI] [PubMed] [Google Scholar]
- 2. Cowen LE (2008) The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol 6: 187–198. [DOI] [PubMed] [Google Scholar]
- 3. Kim HS, Fay JC (2007) Genetic variation in the cysteine biosynthesis pathway causes sensitivity to pharmacological compounds. Proc Natl Acad Sci U S A 104: 19387–19391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Will JL, Kim HS, Clarke J, Painter JC, Fay JC, et al. (2010) Incipient balancing selection through adaptive loss of aquaporins in natural Saccharomyces cerevisiae populations. PLoS Genet 6: e1000893 doi:10.1371/journal.pgen.1000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeyl C (2004) Capturing the adaptive mutation in yeast. Res Microbiol 155: 217–223. [DOI] [PubMed] [Google Scholar]
- 6. Gerke J, Lorenz K, Cohen B (2009) Genetic interactions between transcription factors cause natural variation in yeast. Science 323: 498–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fidalgo M, Barrales RR, Ibeas JI, Jimenez J (2006) Adaptive evolution by mutations in the FLO11 gene. Proc Natl Acad Sci U S A 103: 11228–11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adams J, Puskas-Rozsa S, Simlar J, Wilke CM (1992) Adaptation and major chromosomal changes in populations of Saccharomyces cerevisiae. Curr Genet 22: 13–19. [DOI] [PubMed] [Google Scholar]
- 9. Perepnikhatka V, Fischer FJ, Niimi M, Baker RA, Cannon RD, et al. (1999) Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J Bacteriol 181: 4041–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koszul R, Caburet S, Dujon B, Fischer G (2004) Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. Embo J 23: 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, et al. (2008) Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135: 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, et al. (2010) Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proc Natl Acad Sci U S A 107: 18551–18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dhar R, Sagesser R, Weikert C, Yuan J, Wagner A (2011) Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J Evol Biol 24: 1135–1153. [DOI] [PubMed] [Google Scholar]
- 14. Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, et al. (2002) Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 99: 16144–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, et al. (2008) The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet 4: e1000303 doi:10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perez-Ortin JE, Querol A, Puig S, Barrio E (2002) Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res 12: 1533–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Selmecki A, Forche A, Berman J (2006) Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313: 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J (2008) An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol 68: 624–641. [DOI] [PubMed] [Google Scholar]
- 19. Polakova S, Blume C, Zarate JA, Mentel M, Jorck-Ramberg D, et al. (2009) Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci U S A 106: 2688–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Argueso JL, Carazzolle MF, Mieczkowski PA, Duarte FM, Netto OV, et al. (2009) Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res 19: 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lynch M, Sung W, Morris K, Coffey N, Landry CR, et al. (2008) A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci U S A 105: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishant KT, Wei W, Mancera E, Argueso JL, Schlattl A, et al. (2010) The baker's yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLoS Genet 6: e1001109 doi:10.1371/journal.pgen.1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eyre-Walker A, Woolfit M, Phelps T (2006) The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics 173: 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J (2009) Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet 5: e1000705 doi:10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garfinkel DJ (2005) Genome evolution mediated by Ty elements in Saccharomyces. Cytogenet Genome Res 110: 63–69. [DOI] [PubMed] [Google Scholar]
- 26. Liti G, Carter DM, Moses AM, Warringer J, Parts L, et al. (2009) Population genomics of domestic and wild yeasts. Nature 458: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cha RS, Kleckner N (2002) ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606. [DOI] [PubMed] [Google Scholar]
- 28. Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD (2005) Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587–598. [DOI] [PubMed] [Google Scholar]
- 29. Lobachev KS, Gordenin DA, Resnick MA (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193. [DOI] [PubMed] [Google Scholar]
- 30. Mieczkowski PA, Lemoine FJ, Petes TD (2006) Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst) 5: 1010–1020. [DOI] [PubMed] [Google Scholar]
- 31. Rachidi N, Barre P, Blondin B (1999) Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol Gen Genet 261: 841–850. [DOI] [PubMed] [Google Scholar]
- 32. Casaregola S, Nguyen HV, Lepingle A, Brignon P, Gendre F, et al. (1998) A family of laboratory strains of Saccharomyces cerevisiae carry rearrangements involving chromosomes I and III. Yeast 14: 551–564. [DOI] [PubMed] [Google Scholar]
- 33. Umezu K, Hiraoka M, Mori M, Maki H (2002) Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160: 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nevo E (1995) Asian, African and European Biota Meet at Evolution-Canyon Israel - Local Tests of Global Biodiversity and Genetic Diversity Patterns. Proceedings of the Royal Society of London Series B-Biological Sciences 262: 149–155. [Google Scholar]
- 35. Nevo E (1997) Evolution in action across phylogeny caused by microclimatic stresses at “Evolution Canyon”. Theor Popul Biol 52: 231–243. [DOI] [PubMed] [Google Scholar]
- 36. Nevo E (2001) Evolution of genome-phenome diversity under environmental stress. Proc Natl Acad Sci U S A 98: 6233–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lidzbarsky GA, Shkolnik T, Nevo E (2009) Adaptive response to DNA-damaging agents in natural Saccharomyces cerevisiae populations from “Evolution Canyon”, Mt. Carmel, Israel. PLoS ONE 4: e5914 doi:10.1371/journal.pone.0005914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang SL, Leu JY (2011) A tradeoff drives the evolution of reduced metal resistance in natural populations of yeast. PLoS Genet 7: e1002034 doi:10.1371/journal.pgen.1002034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Welch JW, Fogel S, Cathala G, Karin M (1983) Industrial yeasts display tandem gene iteration at the CUP1 region. Mol Cell Biol 3: 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Welch J, Fogel S, Buchman C, Karin M (1989) The CUP2 gene product regulates the expression of the CUP1 gene, coding for yeast metallothionein. Embo J 8: 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miyazaki S, Nevo E, Bohnert HJ (2005) Adaptive oxidative stress in yeast Saccharomyces cerevisiae: interslope genetic divergence in ‘Evolution Canyon’. Biol J Linn Soc 84: 103–117. [Google Scholar]
- 42. Katz Ezov T, Chang SL, Frenkel Z, Segre AV, Bahalul M, et al. (2010) Heterothallism in Saccharomyces cerevisiae isolates from nature: effect of HO locus on the mode of reproduction. Mol Ecol 19: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ezov TK, Boger-Nadjar E, Frenkel Z, Katsperovski I, Kemeny S, et al. (2006) Molecular-genetic biodiversity in a natural population of the yeast Saccharomyces cerevisiae from “Evolution Canyon”: microsatellite polymorphism, ploidy and controversial sexual status. Genetics 174: 1455–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kao KC, Sherlock G (2008) Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet 40: 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buchman C, Skroch P, Welch J, Fogel S, Karin M (1989) The CUP2 gene product, regulator of yeast metallothionein expression, is a copper-activated DNA-binding protein. Mol Cell Biol 9: 4091–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jensen LT, Howard WR, Strain JJ, Winge DR, Culotta VC (1996) Enhanced effectiveness of copper ion buffering by CUP1 metallothionein compared with CRS5 metallothionein in Saccharomyces cerevisiae. J Biol Chem 271: 18514–18519. [DOI] [PubMed] [Google Scholar]
- 47. Gralla EB, Thiele DJ, Silar P, Valentine JS (1991) ACE1, a copper-dependent transcription factor, activates expression of the yeast copper, zinc superoxide dismutase gene. Proc Natl Acad Sci U S A 88: 8558–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254. [DOI] [PubMed] [Google Scholar]
- 49. Koszul R, Dujon B, Fischer G (2006) Stability of large segmental duplications in the yeast genome. Genetics 172: 2211–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, et al. (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. [DOI] [PubMed] [Google Scholar]
- 51. Ferea TL, Botstein D, Brown PO, Rosenzweig RF (1999) Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci U S A 96: 9721–9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kvitek DJ, Will JL, Gasch AP (2008) Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet 4: e1000223 doi:10.1371/journal.pgen.1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bruins MR, Kapil S, Oehme FW (2000) Microbial resistance to metals in the environment. Ecotoxicol Environ Saf 45: 198–207. [DOI] [PubMed] [Google Scholar]
- 54. De Freitas J, Wintz H, Kim JH, Poynton H, Fox T, et al. (2003) Yeast, a model organism for iron and copper metabolism studies. Biometals 16: 185–197. [DOI] [PubMed] [Google Scholar]
- 55. Haferburg G, Kothe E (2007) Microbes and metals: interactions in the environment. J Basic Microbiol 47: 453–467. [DOI] [PubMed] [Google Scholar]
- 56.Emsley J (2001) Nature's building blocks: An A-Z guide to the elemets. Oxford: Oxford University Press.
- 57. Besnard E, Chenu C, Robert M (2001) Influence of organic amendments on copper distribution among particle-size and density fractions in Champagne vineyard soils. Environ Pollut 112: 329–337. [DOI] [PubMed] [Google Scholar]
- 58. Mortimer RK (2000) Evolution and variation of the yeast (Saccharomyces) genome. Genome Res 10: 403–409. [DOI] [PubMed] [Google Scholar]
- 59. Adamo GM, Lotti M, Tamas MJ, Brocca S (2012) Amplification of the CUP1 gene is associated with evolution of copper tolerance in Saccharomyces cerevisiae. Microbiology 158: 2325–2335. [DOI] [PubMed] [Google Scholar]
- 60. Fogel S, Welch JW (1982) Tandem gene amplification mediates copper resistance in yeast. Proc Natl Acad Sci U S A 79: 5342–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Warringer J, Zorgo E, Cubillos FA, Zia A, Gjuvsland A, et al. (2011) Trait variation in yeast is defined by population history. PLoS Genet 7: e1002111 doi:10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hoyt MA, Stearns T, Botstein D (1990) Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol Cell Biol 10: 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pan X, Reissman S, Douglas NR, Huang Z, Yuan DS, et al. (2010) Trivalent arsenic inhibits the functions of chaperonin complex. Genetics 186: 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Smith SA, Kumar P, Johnston I, Rosamond J (1992) SCM4, a gene that suppresses mutant cdc4 function in budding yeast. Mol Gen Genet 235: 285–291. [DOI] [PubMed] [Google Scholar]
- 65. Bun-Ya M, Nishimura M, Harashima S, Oshima Y (1991) The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol 11: 3229–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jensen LT, Ajua-Alemanji M, Culotta VC (2003) The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J Biol Chem 278: 42036–42040. [DOI] [PubMed] [Google Scholar]
- 67. Kabir MA, Ahmad A, Greenberg JR, Wang YK, Rustchenko E (2005) Loss and gain of chromosome 5 controls growth of Candida albicans on sorbose due to dispersed redundant negative regulators. Proc Natl Acad Sci U S A 102: 12147–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, et al. (2010) Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fischer G, James SA, Roberts IN, Oliver SG, Louis EJ (2000) Chromosomal evolution in Saccharomyces. Nature 405: 451–454. [DOI] [PubMed] [Google Scholar]
- 70. Aguilera A (2002) The connection between transcription and genomic instability. Embo J 21: 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Morillon A, Springer M, Lesage P (2000) Activation of the Kss1 invasive-filamentous growth pathway induces Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol Cell Biol 20: 5766–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Servant G, Pennetier C, Lesage P (2008) Remodeling yeast gene transcription by activating the Ty1 long terminal repeat retrotransposon under severe adenine deficiency. Mol Cell Biol 28: 5543–5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guthrie C, Fink G (2004) Guide to yeast genetics and molecular and cell biology. San Diego: Elsevier Academic Press.
- 75. Lieu PT, Jozsi P, Gilles P, Peterson T (2005) Development of a DNA-labeling system for array-based comparative genomic hybridization. J Biomol Tech 16: 104–111. [PMC free article] [PubMed] [Google Scholar]
- 76. Robinson MD, Grigull J, Mohammad N, Hughes TR (2002) FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EC-C1 strains contain three rearranged chromosomes. (A) Three different chromosomes were hybridized in EC-C1 strains when a gene (YHR003W) located on the right arm of chromosome 8 was used as a probe for Southern blotting. A similar pattern was observed when two other genes (YHL002W and YHR102W) were used as probes. (B)(C) Two different chromosomes were hybridized in EC-C1 strains when a gene (YHL027W) located on the left arm of chromosome 8 or a gene (YGL175W) located on the left arm of chromosome 7 were used as a probe for Southern blotting. The 650-kb chromosome was hybridized by both probes. (D) EC-C1 strains contain an enlarged chromosome 10. A gene (YJL001W) located on the left arm of chromosome 10 was used as a probe for Southern blotting. A similar pattern was observed when another gene (YJR001W) was used as a probe.
(EPS)
Aligned DNA sequences of the junction sites. (A) The junction site sequence of the rearranged 900-kb chromosome. (B) The junction site sequence of the rearranged 650-kb chromosome. Ty1, a Ty1 sequence found only in the junction site. DNA sequences of the flanking regions can be found in GenBank under the accession numbers JX101633 and JX101634.
(EPS)
Haploid segregants of EC9 carrying rearranged chromosomes are more tolerant to copper. (A) Segregants carrying the rearranged chromosomes are more copper-tolerant. Haploid segregants from four-viable-spore tetrads of EC9 strains were grown in YPD media overnight, serially diluted and plated on YPD plates containing different concentrations of CuSO4. Plates were incubated at 28°C until obvious colonies were formed. (B) Pulsed-field gel electrophoresis analysis of haploid segregants from four four-viable-spore EC9 tetrads. In four haploid segregants from a single tetrad, two of them carry two rearranged copies of chromosome 8, and the other two carry a wild type copy of chromosome 8. A gene (YHL002W) located on the left arm of chromosome 8 was used as a probe for Southern blotting. M, yeast chromosomal DNA from a standard laboratory strain.
(EPS)
pho84, scm4, and cin2 mutant cells are sensitive to copper but not sulfates. Results in Figure 5 showed that pho84, scm4 and cin2 mutant cells are sensitive to CuSO4. To rule out the possibility that these mutants are sensitive to sulfates instead of copper, the same set of mutant cells were tested on YPD plates containing different concentrations of CuCl2.
(EPS)
Rearranged chromosomes are more likely to revert back to wild type-like chromosomes only in cultures with relaxed selection. (A) Most of the haploid cells evolving under strong selection (E6–E10) maintain the rearranged chromosomes even after 400 generations. (B) In diploid populations evolving under relaxed selection (ED1–ED5), a wild type-like chromosome 8 (approximately 550 kb) replaced the rearranged chromosomes (in the size range of 900 kb and 650 kb). Evolved cultures from generations 100, 200, 300 and 400 were examined. A gene (YHL002W) located on the left arm of chromosome 8 was used as a probe for Southern blotting. M, yeast chromosomal DNA from a standard laboratory strain.
(EPS)
Rearranged chromosomes do not correlate with the cadmium-resistant phenotype. Twenty EC9 haploid segregants were grown in YPD media overnight, serially diluted and plated on YPD plates containing different concentrations of CdCl2. Plates were incubated at 28°C until obvious colonies were formed.
(EPS)
Single nucleotide polymorphisms in the junction sites of EC-C1 strains.
(PDF)
Gene expression in cells under copper treatment.
(PDF)