Abstract
What good are transposable elements (TEs)? Although their activity can be harmful to host genomes and can cause disease, they nevertheless represent an important source of genetic variation that has helped shape genomes. In this review, we examine the impact of TEs, collectively referred to as the mobilome, on the transcriptome. We explore how TEs—particularly retrotransposons—contribute to transcript diversity and consider their potential significance as a source of small RNAs that regulate host gene transcription. We also discuss a critical role for the mobilome in engineering transcriptional networks, permitting coordinated gene expression, and facilitating the evolution of novel physiological processes.
Introduction
The 1983 Nobel Prize for Physiology or Medicine was awarded to Barbara McClintock for her seminal discovery of transposable elements (TEs). McClintock's studies of colour patterns in maize kernels led her to conclude that “controlling elements” that could jump around the genome regulate gene expression (reviewed in [1]). Although the response of the scientific community to her work was initially cautious, the discovery of similar elements in flies, bacteria, and yeast underlined its significance. Today, TEs are recognised as important components of genomes that have helped shape their evolution.
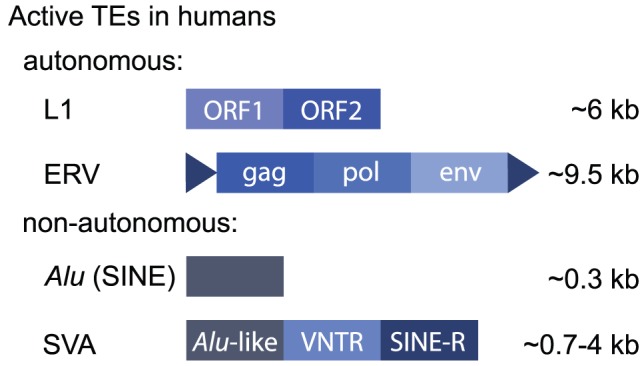
Approximately half of the human genome is derived from TEs [2], although recent work suggests this may be closer to two-thirds [3]. Most human TEs are retrotransposons, and some are still active today (Box 1 and Figure 1). Consequently, TEs represent a significant source of genetic variation [4]–[7].
Box 1. The Human Mobilome
Human TEs may be classified as retrotransposons, which replicate using an mRNA intermediate to “copy and paste,” or DNA transposons, which transpose using a DNA intermediate. Retrotransposons constitute ∼42% of the human genome [2], and some elements are still active today, meaning they are still capable of retrotransposing. DNA transposons represent ∼3% of the human genome but are no longer transposition-competent. Retrotransposons can be further classified based on their structure. LTR elements are characterised by long terminal repeats and include endogenous retroviruses (ERVs) that encode gag, pol, and env genes. These evolved as a consequence of retroviral infection of germ cells, so that they are inherited through generations (reviewed in [77]). Other LTRs are “solo” LTRs, meaning they exist alone. These result from a recombination event that deletes the intervening retroviral genes [78]. Non-LTR retrotransposons include long and short interspersed repeat elements (LINEs and SINEs) and SVA elements that are a composite of sequences derived from other repeats (SINE, VNTR [variable number tandem repeat], and Alu). There are only three highly active elements in the human genome: (1) a subset of L1 LINEs, (2) Alu elements that are a family of primate-specific SINEs, and (3) SVA elements (Figure 1). Alu elements are the most active, with approximately one de novo germline insertion per 20 births [79]. De novo insertions of L1 and SVA elements occur at the rate of approximately 1 in 108 births and 1 in 916 births, respectively [4], [80]. Some ERVs may still be active in humans [81], although the vast majority are nonfunctional, in contrast to their relatively high levels of activity in mice. Only L1 elements encode the enzymes required for retrotransposition, and these preferentially (but not exclusively) recognise L1 mRNA molecules. Alu and SVA elements co-opt the L1 machinery to retrotranspose. The mechanism of retrotransposition has been expertly reviewed elsewhere recently [82], [83].
Figure 1. Active human retrotransposons.
How might TEs influence gene expression? It is easy to imagine how an insertion into a gene might disrupt an open reading frame (ORF), preventing the synthesis of a protein (Figure 2). Indeed, examples of human diseases caused in this manner have been reported [8], [9]. However, the impact of an insertion may not be so dramatic or deleterious. TEs can influence host genes by providing novel promoters, splice sites, or polyadenylation signals (Figure 2). An important consequence is the generation of transcript diversity. There are many more different mRNA molecules in the human transcriptome than the 20,000 protein-coding genes in the genome, and this transcript diversity is thought to be key for promoting phenotypic diversity in higher eukaryotes [10], [11]. Additionally, genome-scale studies have revealed the importance of TEs in dispersing transcription factor binding sites, linking genes in transcriptional networks (e.g., [12], [13]), and facilitating the evolution of novel traits.
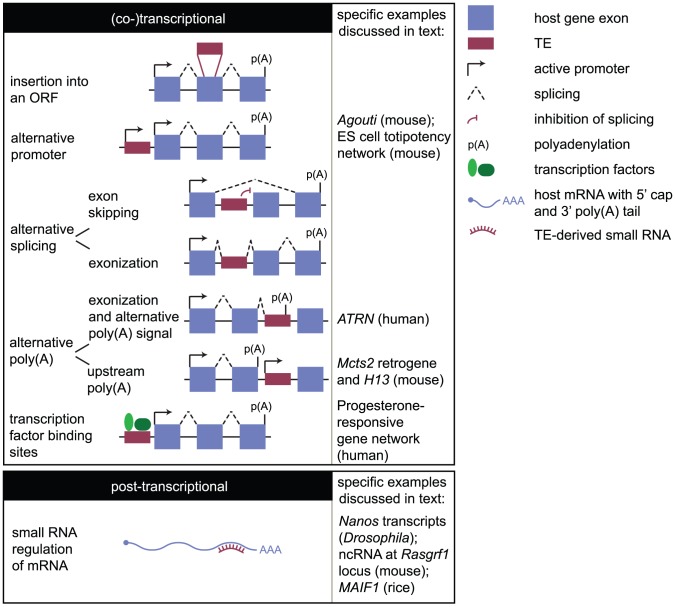
Figure 2. How the mobilome can impact the transcriptome.
Impacts on the transcriptome may be considered transcriptional (or co-transcriptional) and posttranscriptional. The former mechanisms include insertion of a TE into an ORF; provision of an alternative promoter that may be tissue- or stage-specific in its activity; promotion of alternative splicing either through prevention of the splicing machinery from recognising a splice acceptor site in an endogenous exon (exon skipping) or through incorporation of the TE into the mature transcript (exonization); promotion of alternative polyadenylation (poly(A)) either by providing an alternative polyadenylation signal or by promoter activity interfering with host gene transcription and causing upstream polyadenylation; and by introducing transcription factor binding sites that may confer tissue- or stage-specific expression, or link a gene into a transcriptional network. Posttranscriptional regulation involves TE-derived small RNAs binding to host transcripts. In the case of Drosophila Nanos transcripts, small RNAs destabilise the transcript by recruiting the deadenylation machinery. In the case of murine Rasgrf1, the binding of small RNAs to an ncRNA associated with one allele results in the recruitment of the de novo methylation machinery to that allele, causing allele-specific Rasgrf1 expression. The events occurring downstream of small RNA binding are therefore diverse and locus-specific.
In this review, we consider how TEs, collectively referred to as the mobilome, have impacted the transcriptome. This includes elements active today as well as those no longer transposition-competent. Although not nearly an exhaustive account, we draw on specific examples from a range of organisms to illustrate the variety of mechanisms through which this can occur. We aim to highlight the importance of the mobilome in shaping both the diversity and regulation of the transcriptome.
Generating Transcriptome Diversity
One surprising finding from sequencing the human genome was that humans have a similar number of genes to the model nematode Caenorhabditis elegans: about 20,000 and 19,000, respectively. This was unexpected because of the apparent complexity of humans relative to nematodes; indeed, earlier estimates for the number of human genes ranged from 60,000–150,000. Several factors may account for this discrepancy [14], but one important consideration is alternative mRNA processing. This includes alternative splicing and polyadenylation, enabling multiple mRNA species to be generated from a single gene. These mRNA isoforms can encode proteins with different functions or may be differentially regulated. More than 95% of human multi-exonic genes are alternatively spliced [15], while this is around 25% in C. elegans [16].
TEs, particularly L1 and Alu elements, can introduce novel splice sites [17], [18]. Indeed, Alu elements inserted into a gene can provide both splice acceptor and donor sites, creating new exons [19], [20]. Moreover, most Alu-derived exons are alternatively spliced [21], contributing to transcript diversity. They are enriched in the 5′ untranslated regions of human genes, where they regulate mRNA translation [22]. Furthermore, many alternative splicing events of Alu-derived exons are tissue-specific, suggesting TEs contribute to the transcriptome differences that define cell types [23]–[25]. Polyadenylation stabilises mRNA transcripts and influences nuclear export and translation efficiency. The majority of human genes utilise alternative polyadenylation sites [26], and the signals for some of these are embedded in TEs [27], suggesting TEs can influence the 3′ end processing of host gene transcripts.
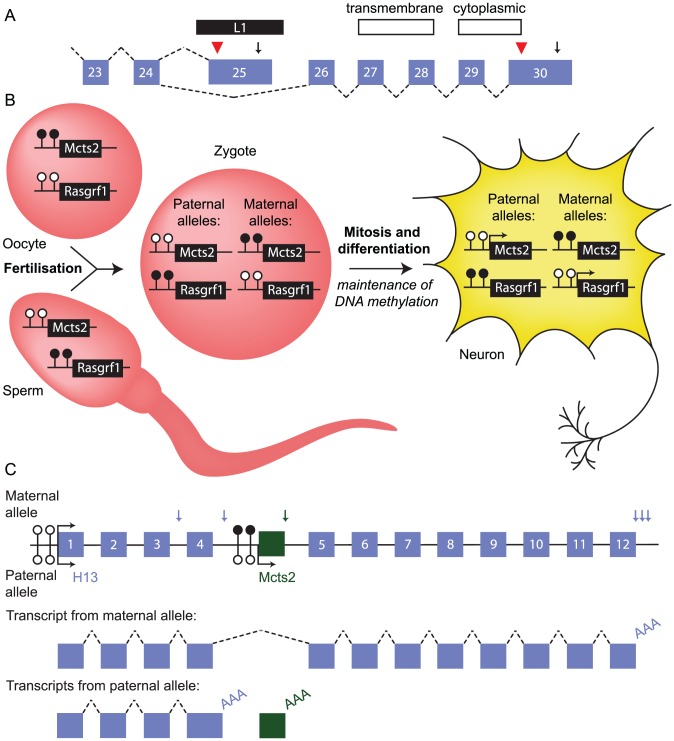
The human ATRN gene provides a good example of how TE-induced alternative mRNA processing can enable functional diversification of one gene. A subset of ATRN transcripts are cleaved and polyadenylated within an L1 element that has retrotransposed into an intron (Figure 3A) [28]. Other transcripts splice around the L1 element and incorporate an additional five exons. Transcripts polyadenylated within the L1 element encode a soluble form of Attractin that is released by activated T lymphocytes as part of the basic inflammatory response [29]. The alternative transcripts encode a protein with transmembrane and cytoplasmic domains that is membrane-bound. This isoform is similar to murine Atrn, which functions as a receptor involved in pigmentation and energy metabolism [28], [30], [31]. This is a clear example of how a single retrotransposition event can increase transcript diversity with direct consequences on cellular function.
Figure 3. Retrotransposition can influence mRNA processing.
(A) Schematic of the 3′ end of the human ATRN gene. An L1 element (black bar) inserted between exons 24 and 26 (numbered boxes) provides a terminal exon, translation termination site (red arrowhead), and polyadenylation signal (arrow) for a subset of transcripts. Alternative splicing produces an mRNA isoform that is polyadenylated in exon 30; only this isoform encodes transmembrane and cytoplasmic domains. Dashed lines, splicing event. (B) Inheritance of DNA methylation at the imprinted Mcts2 and Rasgrf1 genes in mouse. The promoter of Mcts2 is methylated (filled lollipops) in the oocyte and unmethylated (empty lollipops) in sperm. This is opposite to the Rasgrf1 promoter. After fertilisation, these differences persist, marking the origin of the parental alleles even in terminally differentiated cell types, where the unmethylated promoters are transcriptionally active (arrows). (C) Relationship between the retrogene Mcts2 and the gene H13. (Top) Locus structure. Mcts2 (green box) is situated between exons 4 and 5 of H13. Allele-specific differences in methylation at the Mcts2 promoter result in expression of Mcts2 from the paternal allele only. The H13 promoter is unmethylated and active on both alleles. H13 transcripts use alternative polyadenylation sites (vertical blue arrows). Vertical green arrow, single Mcts2 polyadenylation site. (Middle) Representative transcript produced from transcription of the maternal allele. H13 transcripts splice around Mcts2 and use one of three downstream polyadenylation signals (one transcript is shown for clarity). (Bottom) Representative transcripts produced from transcription of the paternal allele. Mcts2 is transcribed and the mRNA is polyadenylated (AAA). H13 transcripts use one of two upstream polyadenylation signals (one transcript is shown for clarity). Transcription of the retrogene Mcts2 is associated with upstream polyadenylation of H13 transcripts.
mRNA processing can also be impacted by TEs indirectly. Although the L1-encoded retrotransposition enzymes preferentially recognise L1 mRNA molecules, host protein-coding mRNAs can also be retrotransposed by the L1 machinery, creating a copy of the original gene [32]. In most cases the copy is nonfunctional, but it can potentially evolve into a retrogene with a novel function or expression pattern. About 120 retroposed sequences have evolved into bona fide genes in the human genome [33]. Retrogenes embedded in introns of other genes can influence transcription of that gene, causing upstream transcript polyadenylation. This is a mechanism through which TEs can indirectly influence mRNA processing, further promoting transcript diversity.
In some cases, the impact of the retrogene on host mRNA processing is specific to only one of the two parental alleles [34], [35]. For example, the retrogene Mcts2 is embedded in an intron of H13. Mcts2 is subject to genomic imprinting. It is expressed exclusively from the allele inherited through the paternal line because its promoter is silenced on the maternally inherited allele by DNA methylation (Figure 3B). Silencing of Mcts2 on the maternally inherited allele permits transcription of H13 to continue through the retrogene, which is spliced out of mature transcripts, and downstream polyadenylation sites are used (Figure 3C). Mcts2 transcription on the paternally inherited allele is associated with H13 transcripts using upstream polyadenylation sites. This is not a consequence of introducing alternative polyadenylation signals, but may involve the elongation complexes that are transcribing H13 “crashing” into those at the transcribing retrogene, a process termed “transcriptional interference” [36]. This interference may promote H13 transcript cleavage and polyadenylation. Intronic ERVs may influence transcription in a similar manner at many loci, impacting on the levels of protein produced from the endogenous gene [37], [38].
TEs further promote transcript diversity by providing alternative promoters for host genes. Perhaps one of the most elegant examples of this can be found in viable yellow agouti (Avy) mice, in which an ERV intracisternal A particle (IAP) upstream of the Agouti gene provides an alternative promoter than can drive ectopic Agouti expression, producing yellow fur [39]. Although the overt phenotype means this is a well-studied specific example, high-throughput approaches have demonstrated widespread use of TEs as alternative gene promoters in normal tissues, and these contribute to tissue-specific expression profiles [24], [40]. Inappropriate activation of promoters embedded in TEs, perhaps resulting from the relaxation of repressive epigenetic marks, can drive ectopic gene expression, and this mechanism has been implicated in human diseases, including cancer [24], [41].
These examples illustrate the impacts of retrotransposition events that occurred in the germline and are stably inherited. Recently there has been much debate about the extent and significance of somatic retrotransposition. L1 expression increases as neural stem cells commit to a neuronal lineage, and this was reported to be associated with elevated L1 retrotransposition [42]. The proposed outcome of this is increased transcriptome heterogeneity among neurons, contributing to interindividual variation [43]. However, the true extent of this mechanism in vivo is debated, with recent estimates ranging from ∼80 to fewer than 0.6 somatic L1 insertions per neuron in the human brain [44], [45], depending on the experimental approach used. Additional work will be required to confirm the true significance of this mechanism for promoting somatic transcriptome diversity. However, the generation of transcript diversity by alternative mRNA processing and the utilisation of alternative promoters in the genome inherited through the germline is likely to be a key factor in the evolution of higher eukaryotes. The mobilome has played a significant role in this process, both directly, by introducing regulatory sequences, and indirectly, by interfering with host gene transcription.
The Mobilome as a Source of Small Regulatory RNAs
Mcts2 is one example of ∼150 genes that are subject to genomic imprinting in the mouse. Although the mechanisms for regulating imprinting vary between loci, silencing of one allele by DNA methylation is a common theme. DNA methylation is likely to have evolved initially as a host defence mechanism against TE expression. Indeed, male mouse germ cells lacking the de novo methyltransferase Dnmt3L exhibit elevated expression of L1 elements and IAPs, resulting in meiotic catastrophe [46]. Thus, the need to minimise the impacts of retrotransposons may have driven the evolution of a novel mode of gene regulation—genomic imprinting—that is critical for mammalian development ([47] and reviewed in [48], [49]).
In addition to DNA methylation, small regulatory RNAs, including PIWI-interacting RNAs (piRNAs) and small interfering RNAs (siRNAs), may also have evolved as a host defence mechanism, repressing the translation of TE transcripts or promoting their decay. Like DNA methylation, this mechanism has been adopted by the host to regulate endogenous genes. Indeed, TEs are major players in the production of small RNAs that regulate host gene transcripts. For example, TE-encoded piRNAs are required to establish a gradient of maternal Nanos mRNA transcripts in the early Drosophila embryo [50]. This is achieved by the binding of piRNAs to a specific sequence in the 3′ untranslated region of Nanos transcripts, which promotes removal of the polyadenylate tail and transcript degradation. This process is essential for establishing correct anterior-posterior patterning in the embryo.
Mammals also utilise TE-encoded small RNAs to regulate host gene expression. In mice, the imprinted gene Rasgrf1, like Mcts2, is under the control of differential DNA methylation on the two alleles. At this locus, DNA methylation is established in the paternal germline (Figure 3B), opposite to that for Mcts2, and this requires TE-encoded piRNAs [51]. During spermatogenesis, these piRNAs bind to a noncoding (nc)RNA transcribed from the locus; specifically, they recognise an LTR-type retrotransposon RMER4B embedded within the ncRNA. Targeting of this ncRNA results in recruitment of the de novo methylation machinery, through an unknown mechanism. Disruption of the piRNA pathway or expression of the ncRNA perturbs methylation and imprinting of Rasgrf1, and mice defective for Rasgrf1 imprinting exhibit impaired postnatal growth [52].
These examples from Drosophila and mice demonstrate the importance of TEs as a source of small RNAs for regulating host transcripts. As such, the host depends upon TEs to provide these regulatory molecules, illustrating their intimate relationship. This relationship is not exclusive to animals, with plants utilising the same system to fine-tune gene expression. For example, in rice, siRNAs originating from the miniature inverted-repeat TE (MITE) Stowaway1 regulate tolerance to abiotic stress [53]. These siRNAs may function by targeting transcripts of the growth regulator MAIF1 and stalling growth, a common physiological response in plants to abiotic stresses.
A defensive response to TEs is critical to guard against uncontrolled transposition. Hosts have evolved several mechanisms for tackling this, including transcriptional repression by DNA methylation and posttranscriptional repression by small RNAs. The evolution of these systems has dramatically impacted the transcriptome because they have been adopted for more general gene regulation.
TEs as Engineers of Transcriptional Networks
TEs can influence alternative mRNA processing or generate small regulatory RNAs, but these effects on the transcriptome are locus-specific. How is transcription regulated on a global scale, say in response to an environmental cue? In the yeast Saccharomyces cerevisiae, genes involved in common metabolic pathways are physically clustered in the genome [54], [55], permitting co-ordinated expression [56] and tight gene regulation in response to a stimulus. However, not all genes that must be co-ordinately expressed are physically clustered. In higher eukaryotes, the binding of transcription factors can co-ordinately activate the expression of genes dispersed throughout the genome. Genes linked in this manner can be considered part of a single transcriptional network. The mobilome has been vital for linking genes in this manner. Regulatory elements required for TE expression can be co-opted by endogenous genes, or the TE may harbour transcription factor binding sites (TFBSs; Figure 2) [12], [13], [57], [58]. TE-derived TFBSs evolve rapidly relative to non-repeat-derived sites [59], suggesting they are important drivers in conferring species-specific gene expression profiles.
A good example of the importance of TEs in linking genes in a network can be found in embryonic stem (ES) cells. ES cells are pluripotent but can enter a transient phase of totipotency from which they can generate both embryonic and extra-embryonic lineages [60]. This switch depends on the activation of a network of transcripts that initiate within ERV LTRs and is controlled by epigenetic modifications. In the pluripotent state, ERVs are transcriptionally repressed, in part by histone 3 lysine 9 methylation [61]. This is established by the histone methyltransferase SETDB1 that is recruited to ERVs by KAP1 [62], [63]. ES cells deficient for Kap1 switch more readily to the totipotent state, consistent with the idea that relaxation of ERV repression drives network activation [60]. These studies highlight a critical role for ERVs in contributing to host cell fate decisions by activating a transcriptional network. This is mediated by epigenetic marks that are established and removed by endogenous cellular machinery.
Earlier, we discussed how a TE could contribute to the evolution of novel functions at a single gene, such as human ATRN. However, by re-wiring networks, the mobilome can facilitate the evolution of complex physiological processes involving gene expression on a global scale. The evolution of pregnancy, the trait that defines mammals, is an intriguing example of this. The hormone progesterone triggers the differentiation of endometrial stromal cells to form the decidua, the maternal component of the placenta [64]. This relies on the activation of a network of transcripts linked by MER20 elements that provide binding sites for progesterone-responsive signalling molecules [65].
An important progesterone-responsive gene is prolactin. In addition to being linked in this network by MER20, the promoter for human prolactin is derived from an independent TE, MER39 [66]. This TE is primate-specific, yet other mammals activate prolactin expression during pregnancy. Emera et al. [67] demonstrated that the endometrial stromal cell-specific promoters of human, mouse, and elephant prolactin are all distinct and are all derived from different TEs (MER77 for mouse, L1 for elephant), suggesting TEs can contribute to convergent evolution. Similarly, the syncytin genes, essential for formation of the syncytiotrophoblast layer that mediates fetal-maternal exchange, are derived from ERV env genes and have been independently acquired in the human, mouse, and rabbit genomes [68].
Other aspects of the physiological changes required for pregnancy may have evolved by TEs re-wiring networks, such as the tolerance of the maternal immune system to a fetus expressing paternal antigens [69]. Together, these examples illustrate the requirement for TEs in pregnancy: engineering a transcriptional network, providing cell type-specific promoters, and contributing to gene function. Thus, the impact of TEs can extend well beyond single-locus effects, making vital contributions to the evolution of complex physiological processes. This role is not confined to animals; TEs in plants have had similar impacts [70].
Conclusions and Future Perspectives
The impacts of TEs on the host transcriptome are diverse. At the single locus level, a transposition event may result in an alternatively processed transcript that can evolve a new function. At the genome level, TEs may disperse regulatory elements that rewire transcriptional networks. The significance of TEs is becoming increasingly apparent with more widespread application of next-generation sequencing technologies. At first, repetitive elements were a nuisance in the analysis of genome-wide datasets; now new experimental protocols and computational pipelines are being utilised to ask questions specifically about TE distribution [71], [72]. The 1000 Genomes Project will provide a valuable tool for interrogating the extent and functional importance of insertional polymorphisms in humans, and indeed has already yielded some intriguing findings; for example, the insertion rates of TEs differ between populations [73].
Many important questions remain unanswered. For example, what is the extent and biological significance of somatic retrotransposition? What is the contribution of this mechanism to the transcriptome differences between neurons, and how does this influence behaviour? Additionally, inappropriate activation of TEs has been associated with somatic cancers [71]. It is too early to say if this mechanism is truly causative, but the current data are provocative. Another exciting area with important outstanding questions is the influence of epigenetic silencing of TEs on the host. For example, could methylated TEs act as “messengers,” transmitting epigenetic information between generations? During primordial germ cell development, most DNA methylation is erased and reset, but a small fraction of the genome is resistant to erasure. This includes IAPs, suggesting these are candidates for mediating transgenerational epigenetic inheritance [74], [75]. Indeed, such a role has been demonstrated for at least two specific IAPs [39], [76], but whether this represents a more global mechanism is undetermined.
Are the evolutionary benefits conferred by TEs purely accidental? Most point mutations arising in the germline have a deleterious or neutral effect on the host, but some do introduce innovative changes that are beneficial. Likewise, TE insertions may be deleterious but can also provide opportunities for increasing transcript diversity or rewiring the transcriptome. Such advantageous insertion events can be selected for and fixed in a population. This “fine-tuning” of the transcriptome could explain why organisms have evolved mechanisms to regulate TE activity without completely silencing all types.
Our understanding of the diverse impacts of the mobilome on the transcriptome has come a long way since the finding that TEs could cause insertional mutations leading to disease. TEs have been fundamental players in evolution and are intimately associated with the regulation of host gene transcription.
Acknowledgments
The authors apologise for the omission of important research due to space limitations. The authors thank Mike Malim, Thomas Schlitt, Ruth McCole, and Adam Prickett for very helpful comments and discussion on the manuscript.
Funding Statement
MC is supported by a King's College London/The London Law Trust Medal Fellowship. RJO is supported by the Wellcome Trust (085448/Z/08/Z) and the Medical Research Council (G1001689). The funders had no role in the preparation of the article.
References
- 1. Fedoroff N (2001) How jumping genes were discovered. Nat Struct Biol 8: 300–301. [DOI] [PubMed] [Google Scholar]
- 2. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- 3. de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD (2011) Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384 doi:10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang CR, Schneider AM, Lu Y, Niranjan T, Shen P, et al. (2010) Mobile interspersed repeats are major structural variants in the human genome. Cell 141: 1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, et al. (2010) LINE-1 retrotransposition activity in human genomes. Cell 141: 1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ewing AD, Kazazian HH Jr (2010) High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res 20: 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, et al. (2010) Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141: 1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kazazian HH Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, et al. (1988) Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332: 164–166. [DOI] [PubMed] [Google Scholar]
- 9. Chen JM, Stenson PD, Cooper DN, Ferec C (2005) A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet 117: 411–427. [DOI] [PubMed] [Google Scholar]
- 10. Graveley BR (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17: 100–107. [DOI] [PubMed] [Google Scholar]
- 11. Nilsen TW, Graveley BR (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bourque G, Leong B, Vega VB, Chen X, Lee YL, et al. (2008) Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, et al. (2012) Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148: 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hodgkin J (2001) What does a worm want with 20,000 genes? Genome Biol 2: COMMENT2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415. [DOI] [PubMed] [Google Scholar]
- 16. Ramani AK, Calarco JA, Pan Q, Mavandadi S, Wang Y, et al. (2011) Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res 21: 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li WH, Gu Z, Wang H, Nekrutenko A (2001) Evolutionary analyses of the human genome. Nature 409: 847–849. [DOI] [PubMed] [Google Scholar]
- 18. Nekrutenko A, Li WH (2001) Transposable elements are found in a large number of human protein-coding genes. Trends Genet 17: 619–621. [DOI] [PubMed] [Google Scholar]
- 19. Lev-Maor G, Sorek R, Shomron N, Ast G (2003) The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300: 1288–1291. [DOI] [PubMed] [Google Scholar]
- 20. Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, et al. (2007) Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol 8: R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sorek R, Ast G, Graur D (2002) Alu-containing exons are alternatively spliced. Genome Res 12: 1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, et al. (2011) Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci U S A 108: 2837–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin L, Shen S, Tye A, Cai JJ, Jiang P, et al. (2008) Diverse splicing patterns of exonized Alu elements in human tissues. PLoS Genet 4: e1000225 doi:10.1371/journal.pgen.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, et al. (2009) The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571. [DOI] [PubMed] [Google Scholar]
- 25. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, et al. (2012) Landscape of transcription in human cells. Nature 489: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, et al. (2012) A quantitative atlas of polyadenylation in five mammals. Genome Res 22: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee JY, Ji Z, Tian B (2008) Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res 36: 5581–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang W, Gunn TM, McLaughlin DF, Barsh GS, Schlossman SF, et al. (2000) Secreted and membrane attractin result from alternative splicing of the human ATRN gene. Proc Natl Acad Sci U S A 97: 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Duke-Cohan JS, Gu J, McLaughlin DF, Xu Y, Freeman GJ, et al. (1998) Attractin (DPPT-L), a member of the CUB family of cell adhesion and guidance proteins, is secreted by activated human T lymphocytes and modulates immune cell interactions. Proc Natl Acad Sci U S A 95: 11336–11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nagle DL, McGrail SH, Vitale J, Woolf EA, Dussault BJ Jr, et al. (1999) The mahogany protein is a receptor involved in suppression of obesity. Nature 398: 148–152. [DOI] [PubMed] [Google Scholar]
- 31. Gunn TM, Miller KA, He L, Hyman RW, Davis RW, et al. (1999) The mouse mahogany locus encodes a transmembrane form of human attractin. Nature 398: 152–156. [DOI] [PubMed] [Google Scholar]
- 32. Esnault C, Maestre J, Heidmann T (2000) Human LINE retrotransposons generate processed pseudogenes. Nat Genet 24: 363–367. [DOI] [PubMed] [Google Scholar]
- 33. Vinckenbosch N, Dupanloup I, Kaessmann H (2006) Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A 103: 3220–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, et al. (2008) Regulation of alternative polyadenylation by genomic imprinting. Genes Dev 22: 1141–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cowley M, Wood AJ, Bohm S, Schulz R, Oakey RJ (2012) Epigenetic control of alternative mRNA processing at the imprinted Herc3/Nap1l5 locus. Nucleic Acids Res 40: 8917–8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shearwin KE, Callen BP, Egan JB (2005) Transcriptional interference–a crash course. Trends Genet 21: 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li J, Akagi K, Hu Y, Trivett AL, Hlynialuk CJ, et al. (2012) Mouse endogenous retroviruses can trigger premature transcriptional termination at a distance. Genome Res 22: 870–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E (2004) Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res 32: 5800–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morgan HD, Sutherland HG, Martin DI, Whitelaw E (1999) Epigenetic inheritance at the agouti locus in the mouse. Nat Genet 23: 314–318. [DOI] [PubMed] [Google Scholar]
- 40. Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, et al. (2004) Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606. [DOI] [PubMed] [Google Scholar]
- 41. Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, et al. (2010) Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet 6: e1000917 doi:10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, et al. (2005) Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435: 903–910. [DOI] [PubMed] [Google Scholar]
- 43. Thomas CA, Paquola AC, Muotri AR (2012) LINE-1 Retrotransposition in the Nervous System. Annu Rev Cell Dev Biol 28: 555–573. [DOI] [PubMed] [Google Scholar]
- 44. Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, et al. (2009) L1 retrotransposition in human neural progenitor cells. Nature 460: 1127–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, et al. (2012) Single-neuron sequencing analysis of l1 retrotransposition and somatic mutation in the human brain. Cell 151: 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bourc'his D, Bestor TH (2004) Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99. [DOI] [PubMed] [Google Scholar]
- 47. Suzuki S, Ono R, Narita T, Pask AJ, Shaw G, et al. (2007) Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet 3: e55 doi:10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barlow DP (1993) Methylation and imprinting: from host defense to gene regulation? Science 260: 309–310. [DOI] [PubMed] [Google Scholar]
- 49. Haig D (2012) Retroviruses and the placenta. Curr Biol 22: R609–613. [DOI] [PubMed] [Google Scholar]
- 50. Rouget C, Papin C, Boureux A, Meunier AC, Franco B, et al. (2010) Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467: 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, et al. (2011) Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332: 848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Drake NM, Park YJ, Shirali AS, Cleland TA, Soloway PD (2009) Imprint switch mutations at Rasgrf1 support conflict hypothesis of imprinting and define a growth control mechanism upstream of IGF1. Mamm Genome 20: 654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yan Y, Zhang Y, Yang K, Sun Z, Fu Y, et al. (2011) Small RNAs from MITE-derived stem-loop precursors regulate abscisic acid signaling and abiotic stress responses in rice. Plant J 65: 820–828. [DOI] [PubMed] [Google Scholar]
- 54. Wong S, Wolfe KH (2005) Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat Genet 37: 777–782. [DOI] [PubMed] [Google Scholar]
- 55. Lee JM, Sonnhammer EL (2003) Genomic gene clustering analysis of pathways in eukaryotes. Genome Res 13: 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cohen BA, Mitra RD, Hughes JD, Church GM (2000) A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet 26: 183–186. [DOI] [PubMed] [Google Scholar]
- 57. Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, et al. (2010) Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634. [DOI] [PubMed] [Google Scholar]
- 58. Jordan IK, Rogozin IB, Glazko GV, Koonin EV (2003) Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet 19: 68–72. [DOI] [PubMed] [Google Scholar]
- 59. Polavarapu N, Marino-Ramirez L, Landsman D, McDonald JF, Jordan IK (2008) Evolutionary rates and patterns for human transcription factor binding sites derived from repetitive DNA. BMC Genomics 9: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, et al. (2012) Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, et al. (2011) Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev 25: 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, et al. (2010) KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240. [DOI] [PubMed] [Google Scholar]
- 63. Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd (2002) SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gellersen B, Brosens IA, Brosens JJ (2007) Decidualization of the human endometrium: mechanisms, functions, and clinical perspectives. Semin Reprod Med 25: 445–453. [DOI] [PubMed] [Google Scholar]
- 65. Lynch VJ, Leclerc RD, May G, Wagner GP (2011) Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet 43: 1154–1159. [DOI] [PubMed] [Google Scholar]
- 66. Gerlo S, Davis JR, Mager DL, Kooijman R (2006) Prolactin in man: a tale of two promoters. Bioessays 28: 1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Emera D, Casola C, Lynch VJ, Wildman DE, Agnew D, et al. (2012) Convergent evolution of endometrial prolactin expression in primates, mice, and elephants through the independent recruitment of transposable elements. Mol Biol Evol 29: 239–247. [DOI] [PubMed] [Google Scholar]
- 68. Dupressoir A, Vernochet C, Harper F, Guegan J, Dessen P, et al. (2011) A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci U S A 108: E1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, Rudensky AY (2012) Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell 150: 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, et al. (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134. [DOI] [PubMed] [Google Scholar]
- 71. Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, et al. (2012) Landscape of somatic retrotransposition in human cancers. Science 337: 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fiston-Lavier AS, Carrigan M, Petrov DA, Gonzalez J (2011) T-lex: a program for fast and accurate assessment of transposable element presence using next-generation sequencing data. Nucleic Acids Res 39: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stewart C, Kural D, Stromberg MP, Walker JA, Konkel MK, et al. (2011) A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet 7: e1002236 doi:10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Popp C, Dean W, Feng S, Cokus SJ, Andrews S, et al. (2010) Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463: 1101–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lane N, Dean W, Erhardt S, Hajkova P, Surani A, et al. (2003) Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35: 88–93. [DOI] [PubMed] [Google Scholar]
- 76. Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, et al. (2003) Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A 100: 2538–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stoye JP (2012) Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nature reviews Microbiology 10: 395–406. [DOI] [PubMed] [Google Scholar]
- 78. Copeland NG, Hutchison KW, Jenkins NA (1983) Excision of the DBA ecotropic provirus in dilute coat-color revertants of mice occurs by homologous recombination involving the viral LTRs. Cell 33: 379–387. [DOI] [PubMed] [Google Scholar]
- 79. Cordaux R, Hedges DJ, Herke SW, Batzer MA (2006) Estimating the retrotransposition rate of human Alu elements. Gene 373: 134–137. [DOI] [PubMed] [Google Scholar]
- 80. Xing J, Zhang Y, Han K, Salem AH, Sen SK, et al. (2009) Mobile elements create structural variation: analysis of a complete human genome. Genome Res 19: 1516–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Moyes D, Griffiths DJ, Venables PJ (2007) Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet 23: 326–333. [DOI] [PubMed] [Google Scholar]
- 82. Burns KH, Boeke JD (2012) Human transposon tectonics. Cell 149: 740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Finnegan DJ (2012) Retrotransposons. Curr Biol 22: R432–437. [DOI] [PubMed] [Google Scholar]