Abstract
Early T-cell precursor acute lymphoblastic leukemia (ETP-ALL) has been identified as high-risk subgroup of acute T-lymphoblastic leukemia (T-ALL) with a high rate of FLT3-mutations in adults. To unravel the underlying pathomechanisms and the clinical course we assessed molecular alterations and clinical characteristics in a large cohort of ETP-ALL (n = 68) in comparison to non-ETP T-ALL adult patients. Interestingly, we found a high rate of FLT3-mutations in ETP-ALL samples (n = 24, 35%). Furthermore, FLT3 mutated ETP-ALL was characterized by a specific immunophenotype (CD2+/CD5-/CD13+/CD33-), a distinct gene expression pattern (aberrant expression of IGFBP7, WT1, GATA3) and mutational status (absence of NOTCH1 mutations and a low frequency, 21%, of clonal TCR rearrangements). The observed low GATA3 expression and high WT1 expression in combination with lack of NOTCH1 mutations and a low rate of TCR rearrangements point to a leukemic transformation at the pluripotent prothymocyte stage in FLT3 mutated ETP-ALL. The clinical outcome in ETP-ALL patients was poor, but encouraging in those patients with allogeneic stem cell transplantation (3-year OS: 74%). To further explore the efficacy of targeted therapies, we demonstrate that T-ALL cell lines transfected with FLT3 expression constructs were particularly sensitive to tyrosine kinase inhibitors. In conclusion, FLT3 mutated ETP-ALL defines a molecular distinct stem cell like leukemic subtype. These data warrant clinical studies with the implementation of FLT3 inhibitors in addition to early allogeneic stem cell transplantation for this high risk subgroup.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive leukemia accounting for 10–15% of childhood and 25% of adult ALL cases. Based on molecular studies, T-ALL can be divided into at least four molecular-cytogenetic subgroups, i.e. the TAL/LMO, the TLX/HOX11, the TLX3/HOX11L2 and the HOXA subgroups [1]–[3]. Apart from these genetic subgroups, a fifth subgroup of T-ALL cases with developmental arrest at a very early stage of T-cell development was defined by a characteristic early T-cell precursor (ETP) signature in pediatric T-ALL [4]. This T-ALL subtype, termed as ETP-ALL, is described by an immature surface immunophenotype: absence of CD1a and CD8 expression, weak CD5 expression and expression of one or more myeloid-associated and/or stem cell-associated markers. In addition, an increased genomic instability, a high frequency of remission failures and hematologic relapse characterize this highly unfavorable T-ALL subgroup in pediatric patients [4].
Oncogenic alterations that lead to a differentiation arrest at specific stages of T-cell development are well known for specific subgroups of T-ALL. Of these, the overexpression of the orphan homeobox proteins TLX1 and TLX3 lead to a maturation block due to ETS1-mediated TLX recruitment to the Eα core [5]. Most recently, for the group of ETP-ALL a mutational spectrum similar to acute myeloid leukemia (AML) was observed, however no single genetic alterations could be tracked down [6]. For the majority of T-ALL, activation of NOTCH1 signalling is a driving force in the pathogenesis [7]. Activating NOTCH1 mutations have been found in more than 60% of T-lineage leukemias and result in a ligand-independent release of the intracellular NOTCH1 domain, which subsequently translocates to the nucleus, where it acts as transcriptional co-activator [8]–[11]. Various groups have shown that activated NOTCH1 signalling causes activation of downstream targets including HES1, DTX1, PTCRA, and MYC and clinical studies have explored gamma secretase inhibitors (GSI) as targeted therapeutic strategy in T-ALL [12]–[14].
In contrast to the high frequency of NOTCH1 mutations, activating FLT3 mutations (FLT3mut) occur only in a very low frequency of T-ALL cases (1–3%), but were evaluated in only limited patient series [15]–[17]. In contrast, mutations of the FLT3 gene, including internal tandem duplications (ITD) and tyrosine kinase domain (TKD) mutations, are one of the most frequent somatic alterations in AML. About one third of AML patients harbor these alterations, which are associated with a poor prognosis in both, adult and pediatric AML [18], [19]. These findings have promoted the use of tyrosine kinase inhibitors (TKI) in patients with FLT3 mutated AML [20], [21].
Recently, we have characterized ETP-ALL as a subgroup of early T-ALL in adults [22]. To unravel the underlying pathomechanisms of ETP-ALL and to extend our insights on FLT3mut ETP-ALL, we performed a comprehensive molecular and clinical study on a large cohort of adult ETP-ALL patients. We were able to demonstrate that FLT3mut ETP-ALL could be classified by its specific immunophenotype and distinctive stem cell like characteristics. Moreover, T-lymphoblastic cells transfected with FLT3-ITD constructs were particular sensitive to tyrosine kinase inhibition making this a new and potentially useful therapeutic option.
Materials and Methods
Patients and treatment
We screened 1241 peripheral blood and bone marrow samples of T-ALL patients that were sent to the central diagnostic reference laboratory of the German Acute Lymphoblastic Leukemia Multicenter Study Group (GMALL). Most cases were characterized with monoclonal antibodies to precursor cells (CD10, CD34, CD117, TdT and HLA-DR) and with a selection of lymphoid-associated antigens including surface and cytoplasmic (c) antigens (cCD79a, CD22, cIgM, CD19, CD20, CD24, CD3, TCR, CD2, CD5, CD4, CD8, CD7, CD1a) and myeloid-associated antigens including myeloperoxidase (MPO), CD13, CD33, CD65s, CD15, CD14, CD64. An antigen was considered positive, if they were expressed in ≥20% of leukemic cells (10% for cytoplasmic antigens). Classification of ETP-ALL was based on the immunophenotypic diagnostic criteria as originally described [4]: CD5 <75%; CD1a and CD8 <5%; CD117, CD34, HLA-DR, CD13, CD33, and CD65s >25%. CD11b was not determined (Suppl. Table S1). Of all immunophenotypically identified ETP-ALL patients (n = 142), sufficient material for further investigations was available in 68 cases. Sixteen of these 68 patients were already included in a previous work [22]. For 52 of these 68 patients clinical follow-up data were available. The median follow-up was 9.4 months (range: 0–124.6 months). Most patients were treated according to protocols of the GMALL study group (43/46, 93% by medical report, Table 1). In addition, 94 T-ALL patients from the GMALL trial 07/2003 were used as reference group, of which nine patients showed an ETP-ALL immunophenotype and were included in the cohort of 68 ETP-ALL patients [23], [24]. Of the remaining 85 non-ETP T-ALL patients, 17 had an immunophenotype of early T-ALL, 15 of mature T-ALL, and 53 of thymic T-ALL. All patients gave written informed consent to participate in the study according to the Declaration of Helsinki. The studies were approved by the ethics board of the Johann Wolfgang Goethe-Universität Frankfurt/Main, Germany.
Table 1. Gene expression levels in ETP-ALL compared to non-ETP T-ALL.
| Expression | ETP-ALL (N = 68) | non-ETP T-ALL (N = 85) | P-value | |
| BAALC | median (range) | 0.69 (0.0–27.1) | 0.08 (0.0–160.3) | <.001 |
| IGFBP7 | median (range) | 1.24 (0.01–4.2) | 0.49 (0.0–16.2) | .009 |
| WT1 | median (range) | 0.53 (0.0–4.2) | <0.01 (0.0–1.6) | <.001 |
| ERG | median (range) | 1.16 (0.0–18.6) | 10.69 (0.5–136.7) | <.001 |
| MN1 | median (range) | 4.59 (0.0–33.1) | 0.66 (0.01–2.7) | <.001 |
| BCL11B | median (range) | 0.09 (0.0–1.4) | 0.44 (0.0–9.9) | <.001 |
| GATA3 | median (range) | 2.11 (0.0–27.3) | 3.91 (0.3–32.4) | .005 |
| MEF2C | median (range) | 0.50 (0.0–5.1) | 0.20 (0.0–1.7) | .001 |
P values were calculated by Mann-Whitney-U-test.
Nucleic acid preparation and molecular characterization
Pretreatment bone marrow and peripheral blood samples from patients were used for DNA and total RNA extraction using TRIzol (Life Technologies, Grand Island, NY, USA) according to the manufacturer's protocol with minor modifications. Complementary DNA (cDNA) was synthesized using 500 ng of total RNA and avian myeloblastosis virus reverse transcriptase (RT-AMV; Roche, Mannheim, Germany) in the presence of RNase inhibitor (RNasin; Roche, Mannheim, Germany).
The samples were investigated by comparative real-time PCR (RT-PCR) for expression of eight genes (BAALC, ERG, IGFBP7, WT1, MN1, GATA3, BCL11B, and MEF2C). The mRNA expression levels for WT1 [25], BAALC [24], IGFBP7 [26], and ERG [24] were determined by RT-PCR as previously described. MN1-primers were designed as reported [27]. Primer sequences for the expression analysis of GATA3, BCL11B and MEF2C are available on request.
The NOTCH1 mutation status was defined by direct sequencing of the N-terminal and the C-terminal region of the HD domain, the N-terminal and the C-terminal region of the PEST domain, and the TAD domain [28], [29].
WT1 mutations were analysed as recently reported [25]. FLT3mut (ITD/TKD) were analyzed using a commercially available FLT3 mutation assay (InVivoScribe Technologies, San Diego, USA). The TCR rearrangement status was assessed by the IdentiCloneTM TCRG Gene Clonality Assay (InVivoScribe Technologies, San Diego, USA).
Statistical analysis
Differences in the clinical characteristics were tested by the Pearson χ2 test. For overall survival (OS) in the different subgroups, Kaplan-Meier curves were created and compared by the Log-rank test. OS was calculated from the time-point of study entry to the time-point of death or last follow-up (censored).
The statistical difference of gene expression between two independent groups was tested by the nonparametric Mann-Whitney-U-test. Differences in the mutation rates were analyzed by the Pearson χ2 or the Fisher's exact test. For all tests a P-value<0.05 (two-sided) was considered to indicate a significant difference. All calculations were performed using the SPSS software version 19 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism® software version 5 (GraphPad Software Inc., LA Jolla, CA, USA).
Cell culture and chemicals
The human mature T-cell leukemia cell lines Jurkat, BE13 and MOLT-4 were obtained from the German Resource Center for Biological Material, DSMZ (Braunschweig, Germany) and previously characterized on a molecular level [30]. They were grown in RPMI media with 10% fetal bovine serum. All cell lines were cultured at 37°C in a 5% CO2 humidified chamber. TKI258 was a kind gift from Novartis (Basel, Switzerland). Tyrosine kinase inhibitors Sorafenib and PKC412 were purchased from Alexis/Enzo Life Sciences (BAY 43-9006; Loerrach, Germany) and LC Laboratories (Woburn, MA, USA) respectively. The chemotherapeutic agent cytarabine (AraC) was provided by Merck Chemicals (Darmstadt, Germany).
Plasmid constructs and transfection
For transduction of Jurkat, BE13 and MOLT-4, 2×106 cells were transfected with either FLT3-ITD, or FLT3-wt expression constructs and with the empty vector as a control, using the Nucleofector systems (Lonza Cologne AG, Cologne, Germany) according to the manufacturer's recommendations. The final concentration of the constructs and the empty vector control was 2 µg. FLT3-wt and FLT3-ITD expression constructs were previously described [31].
Cell proliferation assay
Cell proliferation was measured with the WST-1 reagent according to the manufacturer's instructions (Roche Diagnostics GmbH, Germany). Briefly, 48 hours (hrs) after transfection with FLT3 constructs and empty vector control, the cells were seeded in a 96-well plate with 2×105/well. Subsequently, the cells were cultured for 48 hrs with Sorafenib, PKC412, TKI258 and AraC as a chemotherapy agent or Dimethylsulfoxid (DMSO) as negative control. Absorbance was measured after 48 hrs by optical density absorption analyses at 450 nm using an ELISA multiplate reader.
The 50% growth inhibitory concentrations (IC50) of Sorafenib, PKC412 and TKI258 were determined by plotting the logarithm of the drug concentrations (Sorafenib: 0–500 µM, PKC412: 0–18 nM, TKI258: 0–500 nM) and the growth rate of the cells treated with FLT3-ITD or FLT3-wt constructs and empty vector, using the WST-1 assay.
Apoptosis assay
The cellular apoptosis was measured transfected with FLT3-ITD or FLT3-wt constructs and with the empty vector. Briefly, after 48 hrs treatment with Sorafenib, PKC412, TKI258, and AraC cells were labelled with Annexin V and 7-amino-actinomycin D (7-AAD), using Annexin V Apoptosis Detection Kit (BD Pharmingen, Heidelberg, Germany) and then analyzed by FACS Calibur (Becton-Dickinson) to determine the percentage of apoptotic cells.
Results
Characteristics and clinical outcome of adult ETP-ALL patients
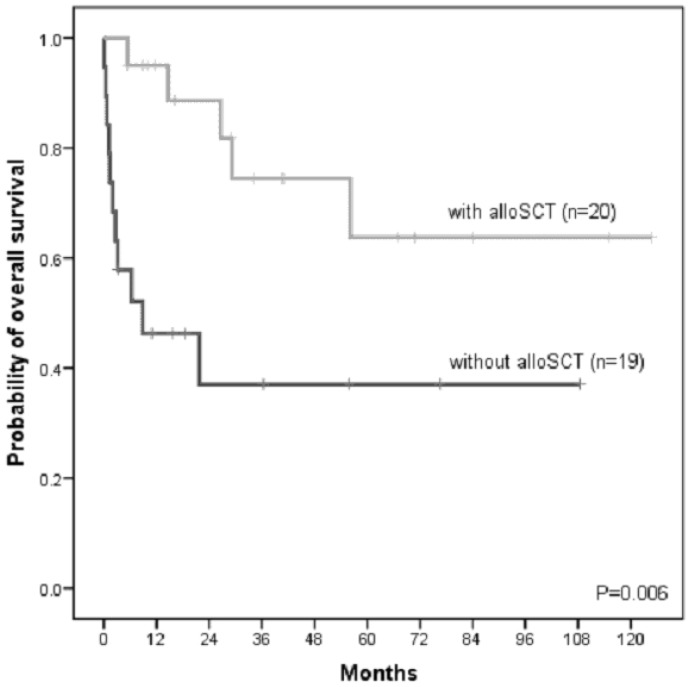
Based on the ETP-ALL specific immunophenotype, we identified pre-treatment samples of 68 newly diagnosed ETP-ALL patients. The median age was 38 years (range: 17–74 years). More patients were male (81%). Of all 68 ETP-ALL patients, follow-up data were available in 52 patients. Forty-five patients were treated according to a GMALL-like protocol, three patients to an AML-like protocol. Fifty-eight percent of patients achieved a complete remission (CR) after induction therapy (Supplementary Table S2). The cumulative 3-year OS was 60%. We further examined outcome with respect to the treatment of chemotherapy only or the allocation to allogenic stem cell transplantation (alloSCT). With the limitation of a potential selection bias for patients undergoing alloSCT, we observed for ETP-ALL patients receiving an alloSCT a favorable outcome (n = 20; 3-year OS: 74%) compared to ETP-ALL patients that were treated with chemotherapy only (n = 19; 3-year OS: 37%, P = 0.006; Figure 1). To address the potential selection bias we have also performed a landmark analysis with a time to transplant of two months. In this analyses patients undergoing alloSCT showed a favourable however not significant benefit compared to patients only receiving chemotherapy.
Figure 1. Kaplan Meier analysis of overall survival in adult ETP-ALL patients receiving chemotherapy only (without alloSCT) or undergoing alloSCT.
P-value was calculated by the Log-Rank test. Abbreviations: alloSCT, allogeneic stem cell transplantation.
Aberrant gene expression and mutational analyses in ETP-ALL compared to non-ETP T-ALL
To further characterize ETP-ALL on the molecular level, we analyzed this large ETP-ALL cohort for the expression of selected genes involved in the pathogenesis and with prognostic implications in adult acute leukemia [22]. BAALC and IGFBP7 were higher expressed in ETP-ALL compared to non-ETP T-ALL patients (BAALC, 8.6-fold, P<.001; IGFBP7, 2.5-fold, P = .009). Furthermore, expression levels of WT1 and MN1 were higher in ETP-ALL compared to non-ETP T-ALL (WT1, P<.001; MN1, 7-fold, P<.001). Additionally, expression of MEF2C, a gene associated with ETP-ALL [32], was significantly higher in ETP-ALL versus non-ETP T-ALL (2.6-fold, P = .001). As critical players in the differentiation program of T-lymphopoiesis, we explored the expression of the transcription factors GATA3, required for the development of normal ETPs [33], and BCL11B, necessary for the subsequent T-cell lineage commitment [34]. Both, GATA3 and BCL11B, were lower expressed in ETP-ALL compared to non-ETP T-ALL (1.9-fold, P = .005; and 4.9-fold, P<.001; respectively). Similarly, ETS transcription factor ERG was also significantly downregulated in ETP-ALL vs. non-ETP T-ALL (9.2-fold, P<.001; Table 1) [35].
The analysis of the TCR rearrangement status revealed that 40 ETP-ALL patients (59%) lacked clonal TCR rearrangements, while 28 patients (41%) showed a monoclonal status. In contrast, 66 (78%) of non-ETP T-ALL patients showed clonal TCR rearrangements, whereas only 18 of these patients (22%) lacked monoclonal TCR rearrangements (Table 2). In addition, differences of the NOTCH1 and FLT3 mutation status between ETP-ALL and non-ETP T-ALL cases were explored. We found a low rate of NOTCH1 mutations in the ETP-ALL subgroup (n = 10/68, 15%), whereas NOTCH1 mutations were more frequent (40%) in non-ETP T-ALL patients (P<.001). In contrast, we found a high rate of FLT3 mutations in ETP-ALL compared to non-ETP T-ALL patients: 24 of the 25 FLT3 mutations were found in the ETP-ALL group, displaying a frequency of 35.3%, whereas non-ETP T-ALL showed a FLT3 mutations frequency of only 1.2% (P<.001). Ten cases (6.5%) had TKD mutations, of which nine occurred in the ETP-ALL group (P = .003). ITD mutations were found in 15 cases, all belonging to the ETP-ALL group (P<.001, Table 2). In a multivariate analysis, NOTCH1 mutation status, low expression of BAALC, WT1, ERG, IGFBP7, and TCR rearrangement had no additional prognostic impact in the subgroup of ETP-ALL.
Table 2. Mutational events in ETP-ALL compared to non-ETP T-ALL.
| Mutation Status | ETP-ALL | non-ETP T-ALL | P-value | |
| TCR rearrangement | monoclonal | 28 (41%) | 66 (79%) | <.001 |
| n = 153 | polyclonal | 40 (59%) | 18 (21%) | |
| NOTCH1 | mut | 10 (15%) | 35 (41%) | <.001 |
| n = 151 | wt | 56 (85%) | 50 (59%) | |
| FLT3 total | mut | 24 (35%) | 1 (1%) | <.001 |
| n = 153 | wt | 44 (65%) | 84 (99%) | |
| FLT3 ITD | mut | 15 (22%) | 0 (0%) | <.001 |
| wt | 53 (78%) | 85 (100%) | ||
| FLT3 D835 | mut | 9 (13%) | 1 (1%) | .003 |
| wt | 59 (87%) | 84 (99%) | ||
P values were calculated by Pearson's Chi-square test and Fisher's exact test, respectively.
Immunophenotype of FLT3mut ETP-ALL compared to FLT3wt ETP-ALL
In addition to the distinct immunophenotype, FLT3mut ETP-ALL showed specific immunophenotypic and molecular characteristics compared to FLT3wt ETP-ALL. In this study, 83% (20/24) of FLT3mut ETP-ALL patients were positive for CD117, compared to only 28% (13/44) of FLT3wt ETP-ALL cases (P<.001). Furthermore, FLT3mut ETP-ALL had a higher rate of positivity for CD2 (88% vs. 30%, P<.001) and CD13 (100% vs. 37%, P<.001) compared to FLT3wt ETP-ALL patients. FLT3wt ETP-ALL was characterized by expression of CD5 (54% vs. 4%, P<.001) and CD33 (54% vs. 4%, P<.001; Figure S1 in Supplementary Figures).
A recent study described the immunophenotype of TdT+/CD7+/CD13+/CD34+/CD117+ as highly specific for the prediction of FLT3 mutations in an unselected cohort of T-ALL [17]. In our ETP-ALL cohort, 75% (18/24) of patients with FLT3 mutations showed this immunophenotype, while only 7% (3/44) without FLT3 mutations displayed this phenotype. Another FLT3 mutation associated marker profile (sCD3-/CD117+/CD34+/CD62L+/CD56-/CD2+/CD7+/CD1a-/CD4-/CD5-/CD8-/CD13+ [36] can be adopted to 71% (17/24) of our FLT3mut ETP-ALL patients; this profile was highly specific for FLT3 mutations without a false prediction. Here we established the combination of CD2+/CD5-/CD13+/CD33-, able to detect 21 of the 24 FLT3mut ETP-ALL patients, as highly sensitive (88%) and specific (95%) algorithm (Table 3).
Table 3. Combinations of antigens as a surrogate marker for FLT3 mutations in ETP-ALL.
| FLT3mut (n = 24) | FLT3wt (n = 44) | P-value | Sensitivity | Specificity | ||
| CD117 | pos | 20 | 13 | <.001 | 83% | 70% |
| neg | 4 | 31 | ||||
| TdT+/CD7+/CD13+/CD34+/CD117+ § | pos | 18 | 3 | <.001 | 75% | 93% |
| neg | 6 | 41 | ||||
| CD117/CD34+/CD62L+/CD56/CD7+/CD2+/CD5-/CD13+ # | pos | 17 | 0 | <.001 | 71% | 100% |
| neg | 7 | 44 | ||||
| CD2+/CD5-/CD13+/CD33- & | pos | 21 | 2 | <.001 | 88% | 95% |
| neg | 3 | 42 |
Molecular characteristics of FLT3mut ETP-ALL in contrast to FLT3wt ETP-ALL
FLT3mut ETP-ALL showed a specific gene expression profile compared to FLT3wt ETP-ALL. Higher expression levels of WT1 (2.2-fold, P = .003) and lower expression of IGFBP7 (0.11-fold, P<.001) were characteristic for FLT3mut ETP-ALL. Remarkably, FLT3mut ETP-ALL had a significantly lower expression of the T-cell transcription factor GATA3 compared to FLT3wt ETP-ALL (P<.001). All except one FLT3mut ETP-ALL case had GATA3 expression levels below the median. No significant differences were found for BAALC, ERG, MN1, BCL11B, and MEF2C (Table 3). As for other lymphoblastic leukemias described [37], [38], FLT3 itself is overexpressed in ETP-ALL. In this subgroup FLT3mut ETP-ALL showed a higher expression compared to FLT3wt ETP-ALL samples (P<.01, Figure S2 in Supplementary Figures).
TCR rearrangement analysis demonstrated that FLT3mut ETP-ALL patients predominantly lacked clonal TCR rearrangements. Only 21% of the FLT3mut ETP-ALL patients in contrast to 52% of the FLT3wt ETP-ALL patients showed a TCR rearrangement (P = .01). In addition, none of the FLT3mut ETP-ALL patients showed a NOTCH1 mutation, while 23% (10/44) FLT3wt ETP-ALL had NOTCH1 mutations (Table 4).
Table 4. Molecular characteristics of FLT3mut ETP-ALL versus FLT3wt ETP-ALL patients.
| A Expression | FLT3mut (n = 24) | FLT3wt (n = 44) | P-value | |
| WT1 | median (range) | 0.78 (0.2–4.2) | 0.36 (0.0–3.4) | .003 |
| IGFBP7 | median (range) | 0.30 (0.02–4.0) | 2.52 (0.1–9.9) | <.001 |
| GATA3 | median (range) | 0.06 (0.0–5.7) | 5.82 (0.0–6.7) | <.001 |
| BAALC | median (range) | 0.44 (0.01–17.7) | 0.95 (0.0–27.1) | .41 |
| ERG | median (range) | 0.94 (0.2–18.6) | 1.56 (0.0–4.2) | .16 |
| MN1 | median (range) | 6.35 (0.3–16.8) | 7.42 (0.0–33.1) | .29 |
| BCL11B | median (range) | 0.20 (0.0–1.4) | 0.05 (0.0–1.0) | .09 |
| MEF2C | median (range) | 0.71 (0.02–2.2) | 0.46 (0.0–5.1) | .22 |
| B Mutation Status | ||||
| NOTCH1 | mut | 0 (0%) | 10 (23%) | .01 |
| wt | 24 (100%) | 34 (77%) | ||
| TCR status | monoclonal | 5 (21%) | 23 (52%) | .01 |
| polyclonal | 19 (79%) | 21(48%) | ||
A: P-values were calculated by Mann-Whitney-U-test.
B: P-values were calculated by Pearson's Chi-square test and Fisher's exact test, respectively.
Clinical characteristics of FLT3 mutated ETP-ALL patients
With respect to clinical characteristics, no differences were observed between the FLT3mut ETP-ALL and the FLT3wt ETP-ALL patients regarding sex and age. The response to induction therapy was similar between both groups (CR: 13/21 vs. 13/24). Three of the 24 FLT3mut ETP-ALL patients were treated with an AML protocol, but none of the patients with FLT3wt ETP-ALL (Supplementary Table S3). The overall survival rate was similar between FLT3mut ETP-ALL and FLT3wt ETP-ALL patients (3-year survival: 58% versus 61%, P = 0.86; Figure S3 in Supplementary Figures).
Sensitivity of T-ALL cell lines transfected with FLT3 expression constructs to TKI
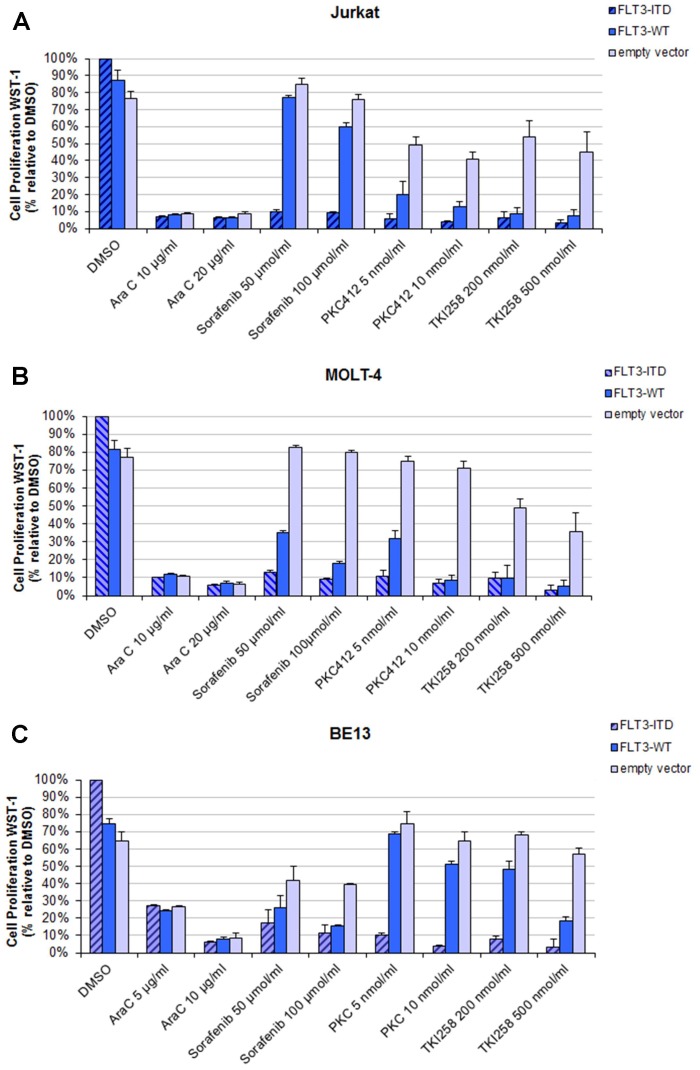
In order to assess the sensitivity of TKI in a model of T-ALL with FLT3-ITD mutations, we transfected the T-ALL cell lines Jurkat, BE13 and MOLT-4 with FLT3-ITD or FLT3-wt constructs and an empty vector as control. Transfection of FLT3 expression constructed did not alter the surface expression of myeloid (CD13, CD33) or stem cell (CD34, CD117) markers (data not shown). Cell lines transfected with FLT3-wt and FLT3-ITD constructs revealed a growth advantage compared to the empty vector transfected cells DMSO (first columns in Figure 2A–C). Treatment with TKI resulted in a selective and significant inhibition of the proliferation of FLT3-wt or FLT3-ITD transfected cells: Jurkat cells transfected with FLT3-ITD or FLT3-wt constructs showed a significant decrease in proliferation compared to empty vector transfected cells when treated with TKIs (including Sorafenib, PKC412, TKI258; Figure 2A). FLT3-ITD and FLT3-wt transfected cells were almost equally sensitive to PKC412 and TKI258, whereas empty vector transfected cells were relative insensitive. Similar results were observed for FLT3-ITD transfected MOLT-4 (Figure 2B) and BE13 cells (Figure 2C). FLT3-wt transfected cells behave different for MOLT-4 and BE13; MOLT-4 cells were more sensitive to sorafenib and BE13 cells were more resistant to PKC412 and TKI258. No differences in proliferation were observed with respect to the FLT3 status for AraC treated cells (Figure 2A–C).
Figure 2. Effects of tyrosine kinase inhibitors on proliferation in T-ALL cell lines transfected with FLT3 expression constructs (A–C).
Fourty-eight hours (hrs) after transfection, cells were seeded and cultured for additionally 48 hrs with tyrosine kinase inhibitors (PKC412, TKI258, and Sorafenib) and chemotherapy (AraC). Cell proliferation was measured using the WST-1 proliferation reagent. The mean optical density (OD) values corresponding to non-treated FLT3-ITD transfected cells were taken as 100%. The results were expressed in percentages of the OD of treated versus untreated control cells. Two experiments were performed in duplicates. For each drug two different doses were used. All results were expressed as means ±S.D. A: Jurkat cells. B: MOLT4 cells. C: BE13 cells.
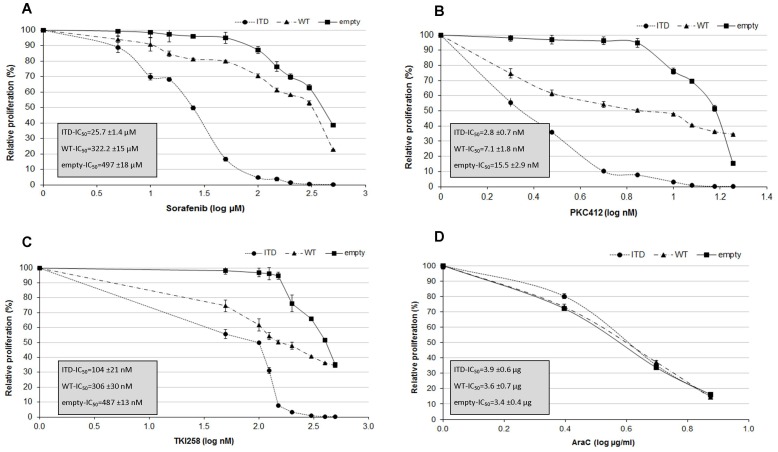
We further examined the TKI mediated apoptosis in Jurkat cells transfected with FLT3 expressing constructs. All TKIs induced enhanced apoptosis in cells transfected with FLT3 expressing constructs compared to empty vector controls (Figure S4 in Supplementary Figures). Cells treated with Sorafenib revealed a 3-fold and 2-fold increase in apoptosis in FLT3-ITD and FLT3-wt transfected cells, respectively. Similar results were observed for PKC412 and TKI258 treated cells, whereas no significant changes in apoptosis were observed for AraC (Figure S4 in Supplementary Figures). Finally, we defined the concentration (IC50) of Sorafenib, PKC412, TKI258 and AraC that induced 50% growth inhibition of Jurkat cells (Figure 3A–D). The IC50 for Sorafenib was 25.7 µM in FLT3-ITD transfected cells, compared to to 305.5 µM and 486.5 µM in FLT3-wt, and empty vector transfected cells, respectively (Figure 3A). The IC50 for PKC412 was 2.8 nM in FLT3-ITD transfected cells, compared to 7.1 nM and 15.5 nM in FLT3-wt and empty vector transfected cells, respectively (Figure 3B). Similar growth inhibitory effects were observed for TKI258 (Figure 3C). No differences in the IC50 between the different transfected cells were seen for AraC (Figure 3D).
Figure 3. Growth inhibition of Jurkat cells transfected with FLT3 expression constructs (FLT3-ITD, FLT3-wt, and empty vector) and treated with Sorafenib, PKC412, TKI258 and AraC.
IC50 was determined by WST-1 assay with different concentrations.
Discussion
In the past decades the molecular characterization of T-ALL broadly expanded and unraveled key events that drive malignant transformation. These genetic alterations may in future lead to the development and the implementation of targeted therapy. Coustan Smith et al. first identified ETP-ALL as a high risk subgroup of pediatric T-ALL characterized by a specific immature immunophenotype and a distinct gene expression profile [4]. Most recently, the genetic heterogeneity of pediatric ETP-ALL was further assessed by whole genome sequencing and next generation sequencing [6]. While various novel somatic mutations were identified, no single alteration could be detected pointing to the heterogeneous genetic background of ETP-ALL, despite the apparently common clinical and immunophenotype features. However, features shared with myeloid leukemias were present as well as mutations in genes of cytokine receptor and RAS signaling and genes involved in histone-modification were frequently observed [6]. In this work, we now focused on genes with already established prognostic and pathogenic value in AML and/or T-ALL.
We have previously characterized ETP-ALL as a high risk subgroup of early T-ALL in adults [22]. To further delineate the molecular pathomechanisms for this distinct T-ALL subgroup with stem cell like and myeloid features, we examined molecular alterations and clinical outcome in a large cohort of adult ETP-ALL patients (n = 68).
ETP-ALL is defined by a specific immunophenotype as described [4]. Recently, an additional score based on the immunophenotype was suggested to define ETP-ALL [39]. In our ETP-ALL cohort, 96% (65/68) of patients had a score greater than 6, which we used as the defining cut-off. In addition to this distinct phenotype, expression analyses of candidate genes revealed significant higher expression of stem cell associated genes and genes with adverse prognostic significance in ETP-ALL versus non-ETP T-ALL. High expression of BAALC and IGFBP7, associated with an immature high risk leukemic phenotype in adult T-ALL and AML [26], [40], underscores the immature nature of ETP-ALL. Similarly, IGFBP7, like MEF2C, were also found to be significantly upregulated in pediatric ETP-ALL [4]. In addition, MN1 identified to be associated with ETP-ALL [32], and WT1, a gene known to be of unfavorable prognosis in AML as well as in T-ALL in the presence of a mutation [25], [41], were also overexpressed in the cohort of ETP-ALL.
We further observed distinct differences in the mutational profile: compared to non-ETP T-ALL, ETP-ALL patients showed less frequent NOTCH1 mutations (15%). On the other hand, a high rate of FLT3 mutations was observed in ETP-ALL (35%), contrasting the mutational profile of non-ETP T-ALL. FLT3 mutations are one of the most frequent genetic alterations in AML [18], whereas FLT3 is only infrequently mutated in leukemic lymphoblasts [42]. This underscores to some extent the association of ETP-ALL with early myeloid differentiation. In pediatric patients, a similar low rate of NOTCH1 mutations (16%) was seen in ETP-ALL. FLT3 mutations, although in a lower frequency (14%), occurred exclusively in ETP-ALL [6]. In addition, we analyzed the TCR rearrangement status in these ETP-ALL patients. In normal human T-cell development, TCR rearrangements are rare in prothymocytes, but are commonly found at the prethymocyte stage [43], [44]. The frequent absence of TCR rearrangement in our cohort confirms the immaturity of ETP-ALL. Together, these data further indicate that ETP-ALL represents a distinct leukemic subtype.
Interestingly, within the ETP-ALL subgroup FLT3mut ETP-ALL define a new molecular stem cell entity as these cases show a specific immunophenotype and molecular characteristics compared to FLT3wt ETP-ALL. We observed a high expression of CD2, the myeloid antigen CD13, and CD117 in the FLT3mut ETP-ALL. CD117, encoded by the c-KIT protooncogene, is highly expressed at the early stages of hematopoietic development [45], and in acute leukemia the highest frequency of CD117 expression is found in AML [46]–[48]. Expression of CD117 has been associated with FLT3 mutations in rare cases of T-ALL [15], [16]. Here, in this yet largest cohort of FLT3mut T-ALL cases, only four FLT3mut patients lacked CD117 expression. Recently, combinations of surface markers were suggested as surrogate marker for FLT3 mutations in T-ALL [17], [36]. However, while these combinations yield a high sensitivity, none could detect all of the ETP-ALL cases with FLT3mut. In our study, a combination of CD2+/CD5-/CD13+/CD33- resulted in the highest sensitivity for the presence of FLT3 mutations in ETP-ALL with a high specificity. For the routinely performed diagnostic flow cytometry, these combinations may help to identify ETP-ALL patients that should be tested for FLT3 mutations.
We further observed that FLT3mut ETP-ALL predominantly lacked clonal TCR-rearrangements pointing to a leukemic transformation before the prothymocyte stage of T-cell development. The absence of TCR rearrangements had already been linked to early treatment failure in children with T-ALL [49], providing an indirect support for the poor prognosis of ETP-ALL. The early developmental arrest of FLT3mut ETP-ALL is also emphasized by the low GATA3 expression. In normal T-cell development, GATA3 plays a definite role in the early T-lineage specification as it is required for the transformation of the ETP/DN1 to the DN2a stage [34]. Thus the leukemic transformation in FLT3mut ETP-ALL lacking GATA3 expression might occur at a stem cell pluripotent prothymic stage before GATA3 expression is induced. These data in combination with the absence of activating NOTCH1 mutations reflect an even more immature nature of the FLT3mut ETP-ALL within the ETP-ALL subgroup.
ETP-ALL as a subgroup of early T-ALL reflects a high risk entity with an overall survival of approximately 50% in adults [22]. Based on the findings of the GMALL study group [50], an alloSCT should be planned in first complete remission for early T-ALL patients. Even though the selection for patients undergoing alloSCT is biased due to various confounding parameters, ETP-ALL patients receiving an alloSCT showed a remarkable favorable outcome in our cohort, whereas the outcome for ETP-ALL patients receiving chemotherapy was relatively poor. The poor response to lymphoid cell-directed ALL chemotherapy only, as already reported for pediatric ETP-ALL [4], might be due to the immature nature and myeloid characteristic of the ETP-ALL. Thus, to further improve outcome for these high risk patients, in addition to alloSCT the implementation of targeted therapies should be considered. Due to the high frequency of FLT3 mutations in ETP-ALL, TKIs already studied in FLT3 mutated AML [51], [52] would be an attractive treatment option. We assessed the sensitivity of T-ALL cell lines transfected with FLT3-ITD and FLT3-wt expression constructs and observed that FLT3 transfected T-ALL cells, despite of their enhanced proliferation, were particular sensitive to TKIs similar to results in AML [31]. Although the transfection of FLT3 expression constructs in T-ALL cell lines remains an in vitro system, the distinct sensitivity to TKIs together with the positive experience in AML support the rational for the clinical use of TKIs in FLT3mut ETP-ALL. In this work, TKI side effects and the impact of TKI on the FLT3 D835Y mutation were not evaluated. However, in analogy to AML it would be expected that the tested TKI are also able to target TKD mutations. Regarding side effects in the clinical use of TKI, the experience in AML have shown that chemotherapy backbone in combination TKIs have to be carefully chosen.
Herein, we describe that ETP-ALL patients represent a distinct molecular subgroup of adult T-ALL patients with a low frequency of NOTCH1 mutations and a high rate of FLT3 mutations. Moreover, we characterize FLT3mut ETP-ALL as a new subgroup of ETP-ALL with unique immunophenotypical and molecular features pointing to a stem cell leukemia. To further improve outcome of this high risk leukemia, targeted therapies with TKIs as well as the allocation to alloSCT should further explored.
Supporting Information
Expression of surface antigens comparing FLT3 mut ETP-ALL patients and FLT3 wt ETP-ALL patients. Median and quartiles of the percentage of positive cells in the flow cytometry are pictured. Abbreviations: * statistically significant; ns, not significant.
(DOC)
FLT3 mRNA expression in 68 adult ETP-ALL samples measured by quantitative RT-PCR. The FLT3 expression was significantly higher in FLT3mut ETP-ALL (n = 21) compared to FLT3wt ETP-ALL (n = 37) (p<.01).
(DOC)
Clinical outcome of FLT3 mut ETP-ALL versus FLT3 wt ETP-ALL patients. The plot shown is the Kaplan Meier analysis of overall survival. P-value was calculated by the Log-Rank test.
(DOC)
Effects of tyrosine kinase inhibitors on apoptosis in Jurkat cells transfected with FLT3 expression constructs. Fourty-eight hrs after transfection the cells were cultured with tyrosine kinase inhibitors (A: Sorafenib, B: PKC412, and C: TKI258) or D: AraC. Apoptosis assay was performed by Annexin V/7AAD labeling of the cells. The results are expressed in percentage of apoptotic cells. Experiments were performed in duplicates. All results were expressed as means ±S.D.
(DOC)
Immunphenotype used for the classification of the 68 ETP-ALL patients.
(DOCX)
Clinical characteristics of ETP-ALL patients.
(DOCX)
Clinical characteristics of FLT3 mut ETP-ALL versus FLT3 wt ETP-ALL patients.
(DOCX)
Acknowledgments
We thank Ouidad Benlasfer for excellent technical assistance.
The authors thank the following institutions for kindly providing clinical data: Aachen: Medizinische Klinik IV - Hämatologie und Onkologie (Prof. Dr. med. Tim H. Brümmendorf); Bad Saarow: Klinik für Innere Medizin III Hämatologie, Onkologie und Palliativmedizin - Sarkomzentrum Berlin-Brandenburg (PD Dr. med. Peter Reichardt); Bonn: Medizinische Klinik III für Hämatologie und Onkologie Universitätsklinikum Bonn (Prof. Dr. med. Peter Brossart); Bremen: Klinikum Bremen-Mitte gGmbH Medizinische Klinik I (Prof. Dr. med. Bernd Hertenstein); Cottbus: Carl-Thiem-Klinikum Cottbus, II. Medizinische Klinik (Prof. Dr. med. Hjalmar B. Steinhauer); Duisburg: Med. Klinik II St. Johannes-Hospital (Prof. Dr. med. C. Aul); Essen: Evangelisches Krankenhaus Essen-Werden gGmbH Klinik für Hämatologie, Onkologie und Stammzelltransplantation (PD Dr. med. Peter Reimer); Frankfurt (Oder): Medizinische Klinik I (Prof. Dr. med. Michael Kiehl); Freiburg: Medizinische Universitätsklinik Abt. Innere Medizin I (Prof. Dr. Dr. h.c. R. Mertelsmann); Göttingen: Medizinische Universitäts-Klinik Abteilung für Hämatologie/Onkologie (Prof. Dr. med. Lorenz Trümper); Hagen: Kath. Krankenhaus Hagen gem. GmbH St.-Marien-Hospital Klinik für Hämatologie und Onkologie (Dr. med. Hans-Walter Lindemann); Hamburg: Asklepios Klinik St. Georg Hämatologische Abteilung (Prof. Dr. med. N. Schmitz); Hamburg: Asklepios Klinik Altona II. Medizinische Abteilung (Dr. med. D. Braumann); Hamm: Med. Klinik Abteilung für Hämatologie-Onkologie Evangelisches Krankenhaus Hamm (Prof. Dr. med. Jörg Schubert); Hannover: Universitätsklinikum, Hämatologie/Onkologie (Prof. Dr. A. Ganser); Homburg/Saar: Universitätsklinikum des Saarlandes Klinik für Innere Medizin I - Onkologie, Hämatologie (Prof. Dr. med. Michael Pfreundschuh); Jena: Universitätsklinikum Jena Klinik für Innere Medizin II Abteilung Hämatologie und Internistische Onkologie (Prof. Dr. med. Andreas Hochhaus); Kaiserslautern: Medizinische Klinik I, Westpfalz-Klinikum GmbH, Standort I Kaiserslautern (Prof. Dr. med. Hartmut Link); Karlsruhe: Städt. Klinikum Karlsruhe, Medizinische Klinik III, Schwerpunkt Onkologie, Hämatologie (Prof. Dr. med. Martin Bentz); Kiel: Universitätsklinikum Schleswig-Holstein Campus Kiel, II. Med. Klinik u. Poliklinik (Prof. Dr. Dr. M. Kneba/M. Brüggemann); Magdeburg: Universitätsklinikum Magdeburg A.ö.R Zentrum für Innere Medizin Klinik für Hämatologie/Onkologie (Prof. Dr. med. Th. Fischer); Mainz: III. Medizinische Klinik und Poliklinik Universitätsmedizin der Johannes Gutenberg-Universität (Prof. Dr. med. Matthias Theobald); Meschede: St. Walburga-Krankenhaus Meschede GmbH (PD Dr. med. M. Schwonzen); Münster: Universität Münster, Medizinische Klinik A (Prof. Dr. W.E. Berdel); Nürnberg: Klinikum Nürnberg Nord Medizinische Klinik 5 (Prof. Dr. med. M. Wilhelm); Oldenburg: Klinikum Oldenburg Innere Medizin II (Prof. Dr. med. C.-H. Köhne); Potsdam: Klinikum Ernst von Bergmann Medizinische Klinik (Prof. Dr. med. G. Maschmeyer); Stuttgart: Robert Bosch-Krankenhaus Abt. Hämatologie/Onkologie (Prof. Dr. med. W. Aulitzky); Tübingen: Medizinische Klinik und Poliklinik Abteilung 2 Hämatologie, Onkologie, Immunologie und Rheumatologie Ambulanz (Prof. Dr. med. L. Kanz); Wiesbaden: Horst-Schmidt-Kliniken, Innere Medizin III, Hämatologie/Onkologie (Prof. Dr. med. Norbert Frickhofen); Wuppertal: HELIOS Klinikum Wuppertal Med. Klinik 1 (PD Dr. med. A. Raghavachar).
Funding Statement
This study was supported by research funding from Berliner Krebsgesellschaft and Deutsche Krebshilfe to C.D.B. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, et al. (2002) Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 1: 75–87 S1535610802000181 [pii]. [DOI] [PubMed] [Google Scholar]
- 2. Soulier J, Clappier E, Cayuela JM, Regnault A, Garcia-Peydro M, et al. (2005) HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood 106: 274–286 2004-10-3900 [pii];10.1182/blood-2004-10-3900 [doi]. [DOI] [PubMed] [Google Scholar]
- 3. van Vlierberghe P, van GM, Tchinda J, Lee C, Beverloo HB, et al. (2008) The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood 111: 4668–4680 blood-2007-09-111872 [pii];10.1182/blood-2007-09-111872 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, et al. (2009) Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol 10: 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dadi S, Le NS, Payet-Bornet D, Lhermitte L, Zacarias-Cabeza J, et al. (2012) TLX Homeodomain Oncogenes Mediate T Cell Maturation Arrest in T-ALL via Interaction with ETS1 and Suppression of TCRalpha Gene Expression. Cancer Cell 21: 563–576 S1535-6108(12)00077-3 [pii];10.1016/j.ccr.2012.02.013 [doi]. [DOI] [PubMed] [Google Scholar]
- 6. Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, et al. (2012) The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481: 157–163 nature10725 [pii];10.1038/nature10725 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paganin M, Ferrando A (2010) Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Grotel M, Meijerink JP, van Wering ER, Langerak AW, Beverloo HB, et al. (2008) Prognostic significance of molecular-cytogenetic abnormalities in pediatric T-ALL is not explained by immunophenotypic differences. Leukemia 22: 124–131 2404957 [pii];10.1038/sj.leu.2404957 [doi]. [DOI] [PubMed] [Google Scholar]
- 9. Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, et al. (2004) Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306: 269–271. [DOI] [PubMed] [Google Scholar]
- 10. Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, et al. (2006) Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood 108: 1151–1157. [DOI] [PubMed] [Google Scholar]
- 11. Asnafi V, Buzyn A, Le Noir S, Baleydier F, Simon A, et al. (2009) NOTCH1/FBXW7 mutation identifies a large subgroup with favorable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) study. Blood 113: 3918–3924. [DOI] [PubMed] [Google Scholar]
- 12. Zuurbier L, Petricoin EF, Vuerhard MJ, Calvert V, Kooi C, et al. (2012) The significance of PTEN and AKT aberrations in pediatric T-cell acute lymphoblastic leukemia. Haematologica haematol.2011.059030 [pii];10.3324/haematol.2011.059030 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grabher C, von Boehmer H, Look AT (2006) Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer 6: 347–359. [DOI] [PubMed] [Google Scholar]
- 14. Silva A, Jotta PY, Silveira AB, Ribeiro D, Brandalise SR, et al. (2010) Regulation of PTEN by CK2 and Notch1 in primary T-cell acute lymphoblastic leukemia: rationale for combined use of CK2- and gamma-secretase inhibitors. Haematologica 95: 674–678 haematol.2009.011999 [pii];10.3324/haematol.2009.011999 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paietta E, Ferrando AA, Neuberg D, Bennett JM, Racevskis J, et al. (2004) Activating FLT3 mutations in CD117/KIT(+) T-cell acute lymphoblastic leukemias. Blood 104: 558–560. [DOI] [PubMed] [Google Scholar]
- 16. van Vlierberghe P, Meijerink JP, Stam RW, van der Smissen W, van Wering ER, et al. (2005) Activating FLT3 mutations in CD4+/CD8- pediatric T-cell acute lymphoblastic leukemias. Blood 106: 4414–4415 106/13/4414 [pii];10.1182/blood-2005-06-2267 [doi]. [DOI] [PubMed] [Google Scholar]
- 17. Hoehn D, Medeiros LJ, Chen SS, Tian T, Jorgensen JL, et al. (2012) CD117 expression is a sensitive but nonspecific predictor of FLT3 mutation in T acute lymphoblastic leukemia and T/myeloid acute leukemia. Am J Clin Pathol 137: 213–219 137/2/213 [pii];10.1309/AJCPR3N3JMSYLPFG [doi]. [DOI] [PubMed] [Google Scholar]
- 18. Levis M, Small D (2003) FLT3: ITDoes matter in leukemia. Leukemia 17: 1738–1752 10.1038/sj.leu.2403099 [doi];2403099 [pii]. [DOI] [PubMed] [Google Scholar]
- 19. Marcucci G, Haferlach T, Dohner H (2011) Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol 29: 475–486 JCO.2010.30.2554 [pii];10.1200/JCO.2010.30.2554 [doi]. [DOI] [PubMed] [Google Scholar]
- 20. Sanz M, Burnett A, Lo-Coco F, Lowenberg B (2009) FLT3 inhibition as a targeted therapy for acute myeloid leukemia. Curr Opin Oncol 21: 594–600. [DOI] [PubMed] [Google Scholar]
- 21. Pemmaraju N, Kantarjian H, Ravandi F, Cortes J (2011) FLT3 inhibitors in the treatment of acute myeloid leukemia: the start of an era? Cancer 117: 3293–3304 10.1002/cncr.25908 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neumann M, Heesch S, Gökbuget N, Schwartz S, Schlee C, et al. (2012) Clinical and molecular characterization of early T-cell precursor leukemia: a high-risk subgroup in adult T-ALL with a high frequency of FLT3 mutations. Blood Cancer Journal e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bruggemann M, Raff T, Flohr T, Gokbuget N, Nakao M, et al. (2006) Clinical significance of minimal residual disease quantification in adult patients with standard-risk acute lymphoblastic leukemia. Blood 107: 1116–1123. [DOI] [PubMed] [Google Scholar]
- 24. Baldus CD, Martus P, Burmeister T, Schwartz S, Gokbuget N, et al. (2007) Low ERG and BAALC expression identifies a new subgroup of adult acute T-lymphoblastic leukemia with a highly favorable outcome. J Clin Oncol 25: 3739–3745. [DOI] [PubMed] [Google Scholar]
- 25. Heesch S, Goekbuget N, Stroux A, Tanchez JO, Schlee C, et al. (2010) Prognostic implications of mutations and expression of the Wilms tumor 1 (WT1) gene in adult acute T-lymphoblastic leukemia. Haematologica 95: 942–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heesch S, Schlee C, Neumann M, Stroux A, Kuhnl A, et al. (2010) BAALC-associated gene expression profiles define IGFBP7 as a novel molecular marker in acute leukemia. Leukemia 24: 1429–1436. [DOI] [PubMed] [Google Scholar]
- 27. Heuser M, Beutel G, Krauter J, Dohner K, von Neuhoff N, et al. (2006) High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood 108: 3898–3905. [DOI] [PubMed] [Google Scholar]
- 28. Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, et al. (2003) Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol 23: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baldus CD, Thibaut J, Goekbuget N, Stroux A, Schlee C, et al. (2009) Prognostic implications of NOTCH1 and FBXW7 mutations in adult acute T-lymphoblastic leukemia. Haematologica 94: 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kalender AZ, De KK, Gianfelici V, Geerdens E, Vandepoel R, et al. (2012) High accuracy mutation detection in leukemia on a selected panel of cancer genes. PLoS One 7: e38463 10.1371/journal.pone.0038463 [doi];PONE-D-12-00005 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brandts CH, Sargin B, Rode M, Biermann C, Lindtner B, et al. (2005) Constitutive activation of Akt by Flt3 internal tandem duplications is necessary for increased survival, proliferation, and myeloid transformation. Cancer Res 65: 9643–9650 65/21/9643 [pii];10.1158/0008-5472.CAN-05-0422 [doi]. [DOI] [PubMed] [Google Scholar]
- 32. Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, et al. (2011) Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell 19: 484–497. [DOI] [PubMed] [Google Scholar]
- 33. Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, et al. (2009) GATA-3 is required for early T lineage progenitor development. J Exp Med 206: 2987–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rothenberg EV (2012) Transcriptional drivers of the T-cell lineage program. Curr Opin Immunol 24: 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV (1999) Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development 126: 3131–3148. [DOI] [PubMed] [Google Scholar]
- 36. Paietta E (2010) Surrogate marker profiles for genetic lesions in acute leukemias. Best Pract Res Clin Haematol 23: 359–368 S1521-6926(10)00058-7 [pii];10.1016/j.beha.2010.08.001 [doi]. [DOI] [PubMed] [Google Scholar]
- 37. Carow CE, Levenstein M, Kaufmann SH, Chen J, Amin S, et al. (1996) Expression of the hematopoietic growth factor receptor FLT3 (STK-1/Flk2) in human leukemias. Blood 87: 1089–1096. [PubMed] [Google Scholar]
- 38. Chillon MC, Gomez-Casares MT, Lopez-Jorge CE, Rodriguez-Medina C, Molines A, et al. (2012) Prognostic significance of FLT3 mutational status and expression levels in MLL-AF4+ and MLL-germline acute lymphoblastic leukemia. Leukemia leu2012161 [pii];10.1038/leu.2012.161 [doi]. [DOI] [PubMed] [Google Scholar]
- 39. Inukai T, Kiyokawa N, Campana D, Coustan-Smith E, Kikuchi A, et al. (2012) Clinical significance of early T-cell precursor acute lymphoblastic leukaemia: results of the Tokyo Children's Cancer Study Group Study L99-15. Br J Haematol 156: 358–365 10.1111/j.1365-2141.2011.08955.x [doi]. [DOI] [PubMed] [Google Scholar]
- 40. Baldus CD, Tanner SM, Ruppert AS, Whitman SP, Archer KJ, et al. (2003) BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: a Cancer and Leukemia Group B Study. Blood 102: 1613–1618. [DOI] [PubMed] [Google Scholar]
- 41. Paschka P, Marcucci G, Ruppert AS, Whitman SP, Mrozek K, et al. (2008) Wilms' tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol 26: 4595–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, et al. (2001) Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 97: 2434–2439. [DOI] [PubMed] [Google Scholar]
- 43. Blom B, Verschuren MC, Heemskerk MH, Bakker AQ, van Gastel-Mol EJ, et al. (1999) TCR gene rearrangements and expression of the pre-T cell receptor complex during human T-cell differentiation. Blood 93: 3033–3043. [PubMed] [Google Scholar]
- 44. Dik WA, Pike-Overzet K, Weerkamp F, de RD, de Haas EF, et al. (2005) New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J Exp Med 201: 1715–1723 jem.20042524 [pii];10.1084/jem.20042524 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gunji Y, Nakamura M, Osawa H, Nagayoshi K, Nakauchi H, et al. (1993) Human primitive hematopoietic progenitor cells are more enriched in KITlow cells than in KIThigh cells. Blood 82: 3283–3289. [PubMed] [Google Scholar]
- 46. Cascavilla N, Musto P, D'Arena G, Melillo L, Carella AM, et al. (1998) CD117 (c-kit) is a restricted antigen of acute myeloid leukemia and characterizes early differentiative levels of M5 FAB subtype. Haematologica 83: 392–397. [PubMed] [Google Scholar]
- 47. Ikeda H, Kanakura Y, Tamaki T, Kuriu A, Kitayama H, et al. (1991) Expression and functional role of the proto-oncogene c-kit in acute myeloblastic leukemia cells. Blood 78: 2962–2968. [PubMed] [Google Scholar]
- 48. Reuss-Borst MA, Buhring HJ, Schmidt H, Muller CA (1994) AML: immunophenotypic heterogeneity and prognostic significance of c-kit expression. Leukemia 8: 258–263. [PubMed] [Google Scholar]
- 49. Gutierrez A, Dahlberg SE, Neuberg DS, Zhang J, Grebliunaite R, et al. (2010) Absence of biallelic TCRgamma deletion predicts early treatment failure in pediatric T-cell acute lymphoblastic leukemia. J Clin Oncol 28: 3816–3823 JCO.2010.28.3390 [pii];10.1200/JCO.2010.28.3390 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gokbuget N, Hoelzer D (2006) Treatment of adult acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program 133–141. [DOI] [PubMed] [Google Scholar]
- 51. Ravandi F, Cortes JE, Jones D, Faderl S, Garcia-Manero G, et al. (2010) Phase I/II study of combination therapy with sorafenib, idarubicin, and cytarabine in younger patients with acute myeloid leukemia. J Clin Oncol 28: 1856–1862 JCO.2009.25.4888 [pii];10.1200/JCO.2009.25.4888 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pratz KW, Sato T, Murphy KM, Stine A, Rajkhowa T, et al. (2010) FLT3-mutant allelic burden and clinical status are predictive of response to FLT3 inhibitors in AML. Blood 115: 1425–1432 blood-2009-09-242859 [pii];10.1182/blood-2009-09-242859 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of surface antigens comparing FLT3 mut ETP-ALL patients and FLT3 wt ETP-ALL patients. Median and quartiles of the percentage of positive cells in the flow cytometry are pictured. Abbreviations: * statistically significant; ns, not significant.
(DOC)
FLT3 mRNA expression in 68 adult ETP-ALL samples measured by quantitative RT-PCR. The FLT3 expression was significantly higher in FLT3mut ETP-ALL (n = 21) compared to FLT3wt ETP-ALL (n = 37) (p<.01).
(DOC)
Clinical outcome of FLT3 mut ETP-ALL versus FLT3 wt ETP-ALL patients. The plot shown is the Kaplan Meier analysis of overall survival. P-value was calculated by the Log-Rank test.
(DOC)
Effects of tyrosine kinase inhibitors on apoptosis in Jurkat cells transfected with FLT3 expression constructs. Fourty-eight hrs after transfection the cells were cultured with tyrosine kinase inhibitors (A: Sorafenib, B: PKC412, and C: TKI258) or D: AraC. Apoptosis assay was performed by Annexin V/7AAD labeling of the cells. The results are expressed in percentage of apoptotic cells. Experiments were performed in duplicates. All results were expressed as means ±S.D.
(DOC)
Immunphenotype used for the classification of the 68 ETP-ALL patients.
(DOCX)
Clinical characteristics of ETP-ALL patients.
(DOCX)
Clinical characteristics of FLT3 mut ETP-ALL versus FLT3 wt ETP-ALL patients.
(DOCX)