Abstract
Purpose
The constitutive androstane receptor (CAR, NR1I3) is a xenobiotic sensor governing the transcription of numerous hepatic genes associated with drug metabolism and clearance. Recent evidence suggests that CAR also modulates energy homeostasis and cancer development. Thus, identification of novel human (h) CAR activators is of both clinical importance and scientific interest.
Methods
Docking and ligand-based structure-activity models were used for virtual screening of a database containing over 2000 FDA-approved drugs. Identified lead compounds were evaluated in cell-based reporter assays to determine hCAR activation. Potential activators were further tested in human primary hepatocytes (HPHs) for the expression of the prototypical hCAR target gene CYP2B6.
Results
Nineteen lead compounds with optimal modeling parameters were selected for biological evaluation. Seven of the 19 leads exhibited moderate to potent activation of hCAR. Five out of the seven compounds translocated hCAR from the cytoplasm to the nucleus of HPHs in a concentration-dependent manner. These compounds also induce the expression of CYP2B6 in HPHs with rank-order of efficacies closely resembling that of hCAR activation.
Conclusion
These results indicate that our strategically integrated approaches are effective in the identification of novel hCAR modulators, which may function as valuable research tools or potential therapeutic molecules.
Keywords: CAR, Pharmacophore, CYP2B6, Induction, Hepatocytes
INTRODUCTION
The constitutive active/androstane receptor (CAR, NR1I3) has been established as one of the xenosensors that organize a cellular defense system against various environmental challenges by regulating hepatic genes associated with xenobiotic metabolism/detoxification and excretion, including phase I oxidation enzymes [e.g., cytochrome P450s (CYPs)], phase II conjugation enzymes (e.g., UDP-glucuronosyltransferases), and phase III efflux transport proteins such as multidrug resistance proteins (1, 2). Thus, clinical used drugs that activate CAR, such as phenobarbital (PB) and phenytoin are often associated with pharmacokinetics-related drug-drug interactions (3). In addition to these well-documented roles, recent studies suggest that CAR also mediates various hepatic functions that control diverse physiological and pathophysiological processes, including fatty acid oxidation, gluconeogenesis, insulin signaling, cell apoptosis and proliferation, tumor development, as well as the biotransformation of endogenous bile acids, bilirubin and steroid hormones (4-6). In these regards, CAR influences energy homeostasis through crosstalking with other hormone responsive regulators such as peroxisome proliferator activated receptor gamma coactivator 1 alpha and forkhead box O1, while promoting tumor progression by disturbing the expression and function of the growth arrest and DNA damage-inducible 45beta and murine double minute 2 genes (7-9). Notably, most of these discoveries were derived from studies utilizing animal models. Given the significant species differences between human (h) CAR and its rodent counterparts, direct extrapolation of these conclusions from animal to man is risky. Therefore, a need for the identification of novel hCAR activators is evident.
Consistent with its designated name, CAR is constitutively activated in immortalized cells prior to chemical activation. Moreover, CAR is characterized as a transcription factor that could be transactivated by both direct ligand binding and ligand-independent indirect mechanisms (1). These features of CAR make the identification of its activators extremely challenging, particularly towards a high-throughput format in vitro. As a result, only a limited number of CAR activators thus far have been reported in comparison to its sister receptor, pregnane X receptor (PXR), in which numerous agonistic drugs and environmental toxicants have been documented (10). Conversely, in contrast to the observations in immortalized cells, CAR is primarily retained in the cytoplasm of primary hepatocytes without activation and translocates to the nucleus upon exposure to chemical activators (11). Additionally, several laboratories including ours have established the hCAR splicing variant (hCAR3) and a hCAR chimerical construct (hCAR1+A) as chemical-responsive surrogates for hCAR with chemical specificities closely correlated with that of the reference hCAR (12-14).
Although the aforementioned biological approaches may offer valuable avenues for the identification of hCAR activators in vitro, these assays are low to media throughput thus far and are often unable to provide structure-activity insights for CAR activation. Conversely, virtual screening of chemical databases with integrated docking and ligand-based structure-activity models has been approved to be effective in identifying new compounds as modulators of nuclear receptors (15, 16). These combined models provide significant advantages over single algorithm methods since a priori knowledge of the ligand binding domain (LBD) can provide a scaffold for overlapping diverse structural elements frequently found in highly promiscuous proteins (17). Accordingly, we build our computational screening models for hCAR activation based on the crystal structure of hCAR-LBD (Protein DataBank ID: 1XV9) (18), as well as several previously published hCAR modulators (15, 19, 20).
In this report, we tested the hypothesis that integration of newly generated computational models with biological approaches can efficiently identify and evaluate novel hCAR activators. Given that activation of CAR may affect energy homeostasis, cancer development, as well as metabolism-based drug-drug interactions (DDI), identification of human CAR specific activators, in particular from drugs already on the market, is of both scientific and clinical significance. To this end, virtual screening of an FDA-approved drug database [the collaborative drug discovery (CDD) database (www.collaborativedrug.com, Berlingame, CA)] was performed with the generated pharmacophore models, as well as binding affinity to the crystal structure of hCAR-LBD. Nineteen retrieved compounds based on their pharmacophore and docking scores were subjected to further biological evaluations. Whereby seven drugs exhibit concentration-dependent hCAR activation and induction of the prototypical hCAR target gene CYP2B6. Together, these results suggest our combinatory strategy is effective in the identification of novel hCAR activators from relatively large databases.
MATERIALS AND METHODS
Chemicals and Biological Reagents
PB, 6-(4-chlorophenyl)imidazo [2,1-b][1,3] thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime (CITCO), dimethyl sulfoxide (DMSO), and ketoconazole were obtained from Sigma-Aldrich (St. Louis, MO). Ondansetron was purchased from BIOMOL Research Laboratories (Plymouth Meeting, PA). Glyburide, moexipril hydrochloride, and Telmisartan (TEL) were from AK Scientific, Inc. (Union City, CA). Tolnaftate (TOL) was acquired through Fisher Scientific (Pittsburgh, PA). Ezetimibe, octicizer (OCT), and pantoprazole were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Amiodarone (AMI), indomethacin, metolazone, nefazodone (NEF), propafenone, raloxifene, ritonavir, verapamil, carvedilol (CAL), and nicardipine hydrochloride (NIC) were generously provided by Dr. James Polli (University of Maryland School of Pharmacy, Baltimore, MD). The Dual-Luciferase Reporter Assay System was obtained through Promega (Madison, WI). β-actin antibody was acquired from Sigma-Aldrich. CYP2B6 and CYP3A4 antibodies were purchased from Millipore Co. (Billerica, MA). Matrigel, insulin, and insulin/transferrin/selenium were from BD Biosciences (Bedford, MA). All other cell culture reagents were purchased from Invitrogen (Carlsbad, CA) or Sigma-Aldrich.
Pharmacophore Generation
Pharmacophore modeling is a ligand-based computational technique that characterizes the structural features of molecules interacting with the same protein target and defines the three-dimensional distance constraints between minimal numbers of functional groups that can still interact with the protein. Such qualitative pharmacophore models can be used for classification between active or non-active compounds without considering activity. In this study, we used the “Common Feature” pharmacophore protocol (formerly named HIPHOP) within Catalyst™ ) embedded in Discovery Studio (DS) software suite (version 3.0; Accelrys; San Diego, CA) to generate a qualitative CAR pharmacophore based on 17 previously reported hCAR modulators (Table 1; Figure S1). The 3D molecular structures of compounds were obtained from either PubChem Compound (http://www.ncbi.nlm.nih.gov/pccompound) or by being sketched and then energy-minimized within the molecular modeling package SYBYL-X 1.2 (Tripos; St. Louis, MO). The stereochemistry of all chiral centers was thoroughly validated using published data of commercial products that are enantiomers or diastereoisomers. Hydrophobic, hydrophobic-aromatic, hydrogen bond acceptor and donor, and aromatic-ring were features used for pharmacophore generation. The molecular conformations of training set compounds were generated by the FAST algorithm with a maximum conformer number of 255 (21). CITCO and the compound number 17 in reference (15), namely CAR-17 in this paper, were defined as active model compounds, while other chemicals were moderately active compounds during common feature model generation.
Table 1.
Training set compounds that generated common feature pharmacophore models
| No. | Namea (abbreviation) |
Activityb | Reference |
|---|---|---|---|
| 1 | CITCOc | EC50 = 49 nM | J. Bio. Chem. 2003. 278, 17277-17283 |
| 2 | Clotrimazole (CLZ) | 4.34 | J. Med. Chem. 2008, 51, 7181–7192 |
| 3 | PK11195d | deactivator | Mol Pharmacol. 2008, 74:443–453 |
| 4 | Triphenyl phosphate (TPP) |
3.96 | J. Med. Chem. 2008, 51, 7181–7192 |
| 5 | CAR-1 | 6.35 | J. Med. Chem. 2008, 51, 7181–7192 |
| 6 | CAR-17 | 7.92 | J. Med. Chem. 2008, 51, 7181–7192 |
| 7 | CAR-18 | 5.22 | J. Med. Chem. 2008, 51, 7181–7192 |
| 8 | CAR-19 | 4.47 | J. Med. Chem. 2008, 51, 7181–7192 |
| 9 | CAR-2 | 5.5 | J. Med. Chem. 2008, 51, 7181–7192 |
| 10 | CAR-20 | 4.09 | J. Med. Chem. 2008, 51, 7181–7192 |
| 11 | CAR-3 | 4.88 | J. Med. Chem. 2008, 51, 7181–7192 |
| 12 | CAR-4 | 4.17 | J. Med. Chem. 2008, 51, 7181–7192 |
| 13 | CAR-5 | 3.87 | J. Med. Chem. 2008, 51, 7181–7192 |
| 14 | CAR-6 | 3.8 | J. Med. Chem. 2008, 51, 7181–7192 |
| 15 | CAR-7 | 3.28 | J. Med. Chem. 2008, 51, 7181–7192 |
| 16 | CAR-8 | 2.95 | J. Med. Chem. 2008, 51, 7181–7192 |
| 17 | CAR-9 | 2.7 | J. Med. Chem. 2008, 51, 7181–7192 |
Compounds with names starting with “CAR” are from reference (Jyrkkarinne et al., 2008). The number following “CAR” is the compound ID number shown in the paper.
Except CITCO and PK11195, the values indicate increased folds over vehicle control of CAR activation at 10 μM of each compound.
6-(4-chlorophenyl)imidazo [2,1-b][1,3] thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime (CITCO).
1-(2-chlorophenyl-N-methylpropyl)-3-isoquinolinecarboxamide (PK11195).
Preparation of CDD database structures
Collaborative Drug Discovery (CDD) database is a publicly available free-access database, containing 2815 FDA-approved drugs selected from all approved drugs since 1938 (22-24). Structures in the initial CDD database were filtered and optimized into an updated database with 2196 drugs. Firstly hydrogen atoms were added and protonation states were assigned with the program Molecular Operating Environment (MOE) (Chemical Computing Group Inc.). The structures were then converted into 3D in mol2 format and assigned with Gasteiger empirical charges with MOE. After structural minimization with three optimization methods in succession, namely, Steepest Descent, Conjugate Gradient, and Truncated Newton, molecules were assigned atomic partial charges based on CM2 charge model at the semi-empirical quantum chemical AM1 level using the program AMSOL (25, 26). Finally, the molecules were subjected to geometry optimization based on newly-calculated charges and MMFF94 force field with MOE (27). Compounds with overlapping structures, those containing metals, and those with molecular weight above 1600 were removed, creating a library of 2196 compounds. The stereochemistry, tautomerization, ionization state and racemic mixtures remained the same as from the original database. To avoid complicating data interpretation, all racemic structures and structures containing ionized salt forms that were retrieved from the screening database were excluded from further biological experiments.
Database Screening with Pharmacophore Models
The Ligand Profiler protocol in DS 3.0 can be used for rapid screening of compounds in large-scale databases against multiple pharmacophores simultaneously. Here, drugs from the CDD database were screened against four generated pharmacophore models with the most favorable fitting scores. Their conformers were generated with the FAST algorithm and their fitting algorithm was RIGID (28). The maximum number of features that were allowed to miss when mapping these ligands to the pharmacophore models was 1.
Docking of Retrieved Drugs to hCAR-LBD
The retrieved drugs obtained by virtual screening with pharmacophore models were docked to the crystal structure of hCAR-LBD (Protein DataBank ID: 1XV9) by docking algorithms FlexX (BioSolveIT, Sankt Augustin, Germany) and Surflex (Tripos, St. Louis, MO). For both docking programs, the protein was kept rigid while a ligand remained flexible during docking. Residues within 6.5 Å of a ligand were defined as the active site by FlexX. The maximum number of solutions per iteration and fragmentation was 500. The Surflex docking was based on alignment to a protomol indicating an ideal ligand comprising a cluster of molecular fragments featuring the binding pocket surface (29). The maximum number of fragment conformations was 20 and the maximum number of poses per ligand was 20.
Human Primary Hepatocytes
Human liver tissues were obtained after surgical resection by qualified pathology staff after diagnostic criteria were met, with consent forms signed by all participant patients, and prior approval from the Institutional Review Board at the University of Maryland School of Medicine was obtained. Human primary hepatocytes (HPHs) were isolated from human liver specimens by a modification of the two-step collagenase digestion method as described previously (30). HPHs for the current studies were also obtained from Life Technologies Corporation (Durham, NC). Fresh HPHs were seeded at 1.5 × 106 or 3.75 × 105 cells/well in 6-well and 24-well collagen coated plates, respectively. Hepatocytes were cultured for 36 hr at 37°C before treatment with specified compounds for another 24 or 72 hr for detection of mRNA and protein expression, respectively.
Real-Time PCR Analysis
Total RNA was isolated from treated hepatocytes using the TRIzol® reagent and reverse transcribed using a High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) following the manufacturers’ instruction. CYP2B6 mRNA expressions were normalized against that of GAPDH. Real-time PCR assays were performed in 96-well optical plates on an ABI Prism 7000 Sequence Detection System with SYBR Green PCR Master Mix. Primers used for CYP2B6 and GAPDH mRNA expression were described previously (19). Induction values were calculated using the equation: Fold = 2ΔΔCt, where ΔCt represents the differences in cycle threshold numbers between CYP2B6 and GAPDH, and ΔΔCt represents the relative change in these differences between control and treatment groups.
Transient Transfection in HepG2 Cells
HepG2 cells in 24-well plates were co-transfected with the CYP2B6-2.2kb reporter and hCAR1+A expression vectors as described previously (14, 31), using a Fugene 6 Transfection Kit (Roche Applied Science, Indianapolis, IN) following the manufacturer’s instruction. Eighteen hours after transfection, cells were treated for another 24 hours with vehicle control (0.1% DMSO), CITCO (1 μM) or each of the 19 test compounds at concentration(s) as indicated in the Figures. Subsequently, cell lysates were assayed for firefly luciferase activities normalized against the activities of cotransfected renilla luciferase using a Dual-Luciferase Kit (Promega). Data were represented as mean ± SD of three individual transfections.
Translocation of Ad/EYFP-hCAR in Human Primary Hepatocytes
Hepatocytes cultured in 24-well collagen-coated plates were infected with 5 μl of Ad/EYFP-hCAR for 12 hr before treatment with the vehicle control (0.1% DMSO), PB (1mM), or selected compounds, AMI, TEL, NEF, TOL, CAL, NIC and OCT at indicated concentrations. Eight hours after treatment, hepatocytes were subjected to confocal microscopy analysis for the cellular localization of Ad/EYFP-hCAR. Quantitative distribution of hCAR was classified as cytosolic, nuclear, or mixed (nuclear + cytosolic); and 100 Ad/EYFP-hCAR expressing hepatocytes were counted from each group.
Western Blot Analyses
Cell homogenate proteins (20 μg) from treated HPHs were resolved on SDS polyacrylamide gels and electrophoretically transferred onto Immobilon-P polyvinylidene difluoride membranes. Subsequently, membranes were incubated with specific antibody against CYP2B6 diluted 1:5000. β-Actin was used to normalize protein loadings. Blots were washed and incubated with horseradish peroxidase goat anti-rabbit IgG antibody diluted 1:5000, and developed using ECL Western blotting detection reagent (GE Healthcare, Chalfont St. Giles, UK). Densitometry was determined using the FluorChem SP Imaging System (Alpha Innotech, San Leandro, CA).
Statistical Analysis
Experimental data are presented as a mean of triplicate determinations ± S.D. unless otherwise noted. Statistical comparisons were made by one-way analysis of variance with post-hoc Dunnett’s analysis. The statistical significance was set at p values of <0.05 (*), <0.01 (**), or 0.001 (***). Emax and EC50 values for hCAR1+A activation were estimated using the Michaelis-Menten equation (GraphPad, Prism). The Pearson coefficient was applied to evaluate the correlation between hCAR1+A activation and CYP2B6 induction.
RESULTS
Common Feature Pharmacophore Generation
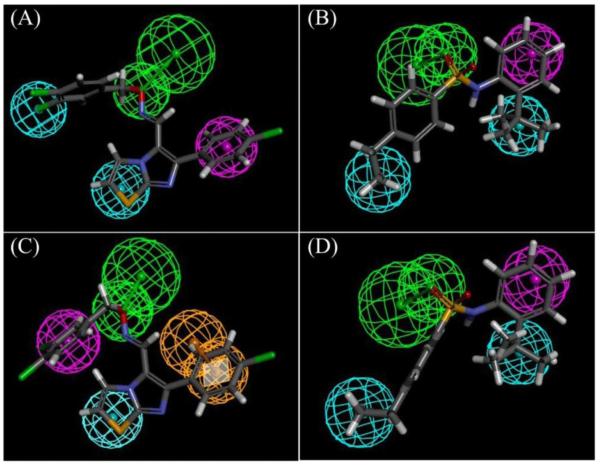
Common feature pharmacophore models were generated with 17 known hCAR modulators (Table 1; Figure S1). The top four generated pharmacophore models, rank-ordered according to their fit values with training set compounds, were used for virtual screening of the CDD database. Figure 1 illustrated the importance of the hydrophobic, hydrophobic-aromatic, hydrogen-bond acceptor, and ring-aromatic features in characterizing the pharmacophore models. The structure with best fit values to the pharmacophore models in Figure 1, A and C, is CITCO, and the one to the models in figure 1, B and D, is CAR-17.
Figure 1. Common feature pharmacophore models generated with known hCAR modulators.
The cyan, magenta, green, and orange spheres indicate hydrophobic, hydrophobic-aromatic, hydrogen-bond acceptor, and ring-aromatic features. The structures in the figures are compounds that had the best fit values to the pharmacophore models. The drug in (A) and (C) is CITCO, and the one in (B) and (D) is CAR-17.
Selection of Tested Compounds
There are 753 drugs in the CDD database matched up to one or more pharmacophore models. These drugs have fit scores to a single pharmacophore model ranging from 0 to 1, with a higher value indicating a better match. One feature is allowed to be missed when ligands were mapped to the pharmacophore models. The number of compounds that matched a pharmacophore model can be decreased by reducing the number of missed features from one to zero, and by using a cutoff of fit scores. Drugs were ranked according to their pharmacophore scores which were the summation of fit values obtained by virtual screening based on the four pharmacophore models. One hundred and six drugs with pharmacophore scores above 2.86 (average fit value of 0.71) in the CDD database were docked to the active site of the hCAR-LBD. Nineteen drugs identified for testing with cell-based experiments were selected in terms of their pharmacophore and docking scores, their category diversity and their commercial availability (Table 2; Figure S2). Drugs with positive binding energy obtained by FlexX or low binding affinity (negative docking scores) by SurfleX were removed from testing list.
Table 2.
Drugs tested with cell-based experiments
| No. | Name (abbreviation) |
CDD ID | Activitya | Pharmacophore scoreb |
FlexX scorec |
Surflex scored |
|---|---|---|---|---|---|---|
| 1 | Amiodarone (AMI) | CDD-38 | 2.9524 | 3 | −4.85 | 6.09 |
| 2 | Glyburide | CDD-308 | 1.2467 | 3.4 | −9.5 | 6.46 |
| 3 | Indomethacin | CDD-341 | 1.0042 | 2.68 | −5.42 | 5.37 |
| 4 | Ketoconazole | CDD-14714 | 1.2324 | 2.66 | −20.8 | 5.65 |
| 5 | Metolazone | CDD-440 | 1.5660 | 2.83 | −10.6 | 5.2 |
| 6 | Moexipril | CDD-46909 | 0.9642 | 3.1 | −13.5 | 4.46 |
| 7 | Nefazodone (NEF) | CDD-475 | 3.4552 | 2.73 | −1.83 | 6.71 |
| 8 | Ondansetron | CDD-16308 | 1.4337 | 2.75 | −14.1 | 5 |
| 9 | Propafenone | CDD-579 | 1.3082 | 3.1 | −15.56 | 7.82 |
| 10 | Raloxifene | CDD-598 | 1.0101 | 3.27 | −18.38 | 4.33 |
| 11 | Ritonavir | CDD-42557 | 1.5391 | 3.43 | −9.6 | −8.81 |
| 12 | Telmisartan (TEL) | CDD-46765 | 3.2181 | 3.23 | xe | 0.82 |
| 13 | Tolnaftate (TOL) | CDD-1106 | 2.7919 | 2.64 | −23.5 | 5.63 |
| 14 | Verapamil | CDD-720 | 1.2075 | 2.59 | −0.76 | 10.48 |
| 15 | Carvedilol (CAL) | CDD-105 | 3.0171 | 3.37 | −17.9 | 8.36 |
| 16 | Ezetimibe | CDD-47471 | 1.2209 | 3.12 | −17.58 | 7.74 |
| 17 | Nicardipine (NIC) | CDD-481 | 5.3983 | 3.36 | −7.1 | 6.71 |
| 18 | Octicizer (OCT) | CDD-46776 | 9.7892 | 3.51 | x | 9.52 |
| 19 | Pantoprazole | CDD-47188 | 1.4044 | 3.36 | −18.32 | 6.28 |
Values indicate increased folds over vehicle control of CAR activation at 10 μM of each compound.
Pharmacophore score is the summation of fit values when a ligand is mapped to the four pharmacophore models. A fit value ranges between 0 and 1. Value 1 indicates a complete match to a pharmacophore model.
FlexX score is an indicator for binding energy between protein and ligand. Favorable binding is indicated by higher negative values.
Surflex score is an indicator for -logKd. Favorable binding is indicated by higher values.
Indicates a ligand fails to be docked into hCAR by FlexX.
Activation of hCAR in HepG2 cells
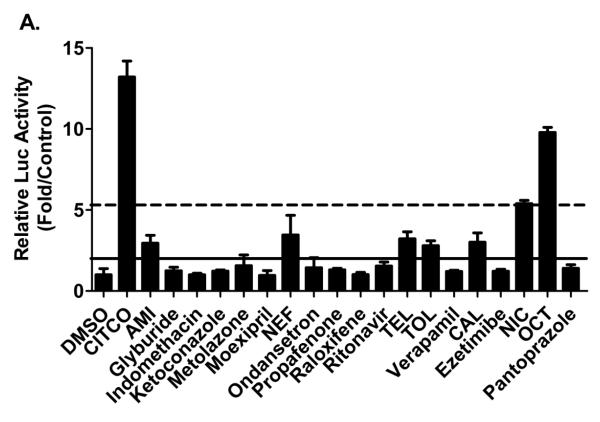
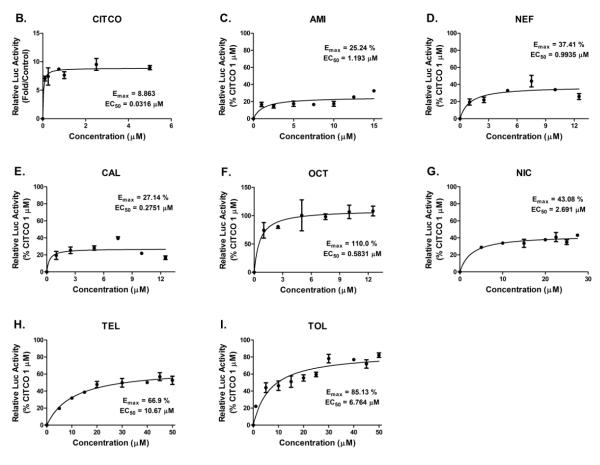
The difficulty in the investigation of hCAR activation in vitro has been substantially alleviated with the establishment of hCAR3 and hCAR1+A as the chemical responsive surrogates for the constitutively activated hCAR (13, 14). In particular, hCAR1+A exhibits chemical-mediated activation superior to that of hCAR3 and demonstrates chemical specificities closely resembling that of the reference hCAR (14). To characterize the biological activity of potential hCAR ligands identified from virtual screening of the computational models, 19 lead compounds with optimal pharmacophore parameters were evaluated in HepG2 cells co-transfected with hCAR1+A expression vector and a CYP2B6 reporter construct (CYP2B6-2.2kb) as described previously (31). As demonstrated in Figure 2A, initial validation of the 19 compounds at the concentration of 10 μM each intends to compare extents of activations relative to CITCO, the known positive control for hCAR1+A assays (14). CITCO-mediated activation of hCAR1+A reached plateau at the concentration around 1 μM (Figure 2B). Accordingly, treatment with CITCO at the concentration of 1 μM was used throughout experiments in the current study. In light of the suggested criteria in cell-based PXR reporter assays (32), five out of the 19 leads (AMI, NEF, CAL, TEL, and TOL) were categorized as moderate activators of hCAR achieving 15%-40% of CITCO-normalized hCAR activation; and two compounds (OCT and NIC) were classified as strong hCAR activators with more than 40% of CITCO-mediated activation (Figure 2A).
Figure 2. Activation of hCAR in HepG2 cells.
HepG2 cells were transfected with hCAR1+A expression and CYP2B6 reporter constructs, then treated for 24 hours with vehicle control (0.1% DMSO), positive control CITCO (1 μM), and test compounds at a dose of 10 μM each (A) or in a concentration-dependent manner (C-I). The Emax and EC50 values of CITCO were based on multi-concentration treatment (B). The solid and dotted lines represent 40% and 15% of activation attained by CITCO (1 μM). Data represent mean ± SD (n = 3). *: P<0.05, **: P<0.01, ***: P<0.001.
Subsequently, the seven compounds which fell in these benchmarks were subjected to concentration-dependent evaluation via hCAR1+A assays. Concentrations of each compound were justified based on preliminary cytotoxicity and solubility assays (supplementary table S1). Consistent with the initial validation, AMI, NEF, and CAL demonstrated moderate hCAR activation with the maximal activation (Emax) values at 25.24%, 37.41%, and 27.14 % of CITCO activation, respectively (Figure 2C-2E), while the most potent activator OCT-mediated hCAR activation (Emax = 110%) exceeded that of CITCO, and NIC displayed an Emax of 43.08% (Figure 2F and 2G). Notably, TEL and TOL exhibited lower cytotoxicity and increased hCAR activation at higher concentrations achieving Emax values of 66.9% and 85.13% of CITCO response, respectively (Figure 2H and 2I). Together, these data indicate that our computer-based virtual screening approaches are effective in the identification of hCAR activators.
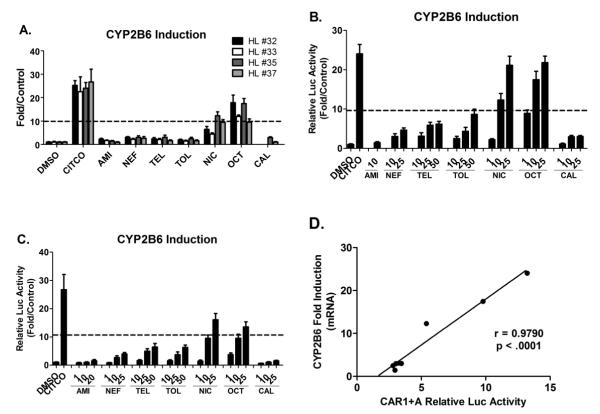
Induction of CYP2B6 Expression in Human Primary Hepatocytes
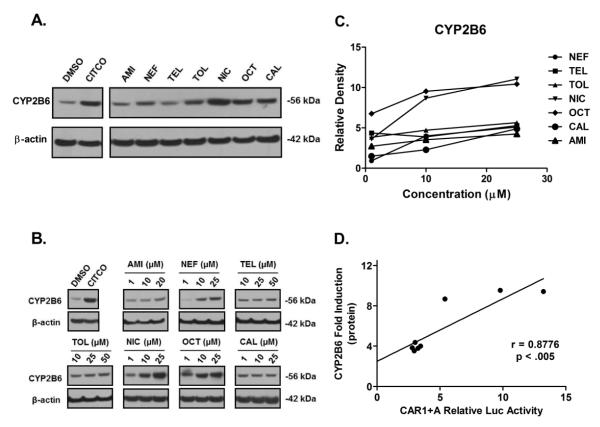
Although the nuclear receptor CAR controls the transcription of many hepatic genes, CYP2B6 symbolizes the most representative target gene of hCAR and is highly inducible upon the exposure of hCAR activators. To determine whether the predicted hCAR activators could induce the expression of endogenous CYP2B6 in a physiologically relevant in vitro system, HPHs from four donors (HL-#32, -#33, -#35, and -#37) were treated with vehicle control (0.1% DMSO), CITCO (1 μM), or AMI, NEF, CAL, TEL, TOL, NIC, and OCT at indicated concentrations. As demonstrated in Figure 3A, AMI, NEF, TEL, TOL, and CAL only marginally increased the mRNA expression of CYP2B6 at the concentration of 10 μM, while NIC and OCT resulted in strong induction at the same concentration. As expected CITCO showed robust induction of CYP2B6 gene expression. The relative CYP2B6 inducibility of all test compounds was normalized to that of CITCO; and ≥ 40% of CITCO induction was selected as the threshold for efficacious CYP2B6 inducers based partly on the similar criterion for CYP3A4 induction as recommended in the Drug Interaction Studies-Guidance for Industry (33). Under the multi-dose treatment scheme, a clear concentration-dependent increase of CYP2B6 mRNA was observed for the majority of the test compounds (Figure 3B and 3C). At the concentration of 25 μM, NIC- and OCT-mediated induction of CYP2B6 reached 90% of CITCO induction in HL-#35. Whereas the maximal induction of CYP2B6 resulted from all other test compounds didn’t attain 40% of CITCO induction.
Figure 3. Quantitative PCR Analysis of CYP2B6 mRNA induction.
HPHs isolated from 4 donors (HL-#32, -#33, -#35, and -#37) were cultured in Williams’ E medium and treated with vehicle control (0.1% DMSO), CITCO (1 μM), and test compounds at 10 μM each (A) or multiple concentrations (B and C) for 24 hr. Real-time PCR was employed to detect CYP2B6 induction as described in Materials and Methods. CYP2B6 inducibility of each compound was normalized to that of CITCO. Correlation between hCAR1+A activation and CYP2B6 induction was depicted in D. Data are expressed as mean ± SD (n=3). Dashed line indicates 40% of CITCO-mediated CYP2B6 induction.
Homogenates prepared from HPHs treated with aforementioned test compounds at the same concentrations were subjected to immunoblotting analysis for CYP2B6 protein expression. As demonstrated in Figure 4, NIC and OCT showed robust induction of CYP2B6 protein in a concentration-dependent manner with the maximal induction challenging that of CITCO, while AMI, NEF, CAL, TEL, and TOL exhibited relatively moderate induction of CYP2B6 through multiple concentrations. Collectively, these observations demonstrate that the inductive expression of CYP2B6 gene at the mRNA and protein levels correlates well with each other, and the two potent hCAR1+A activators, OCT and NIC, also exhibit the strongest induction of CYP2B6 expression. Further statistical analysis of the current data revealed that the potency of hCAR1+A activation in cell-based reporter assays is highly correlated with the induction of CYP2B6 mRNA and protein in HPHs with R2 values of 0.9790 and 0.8776, respectively (Figure 3D and 4D).
Figure 4. Induction of CYP2B6 protein in human primary hepatocytes.
Cultured HPHs were treated with vehicle control (0.1% DMSO), CITCO (1 μM), and test compounds at 10 μM each (A) or multiple concentrations (B) for 72 hr. After harvesting, cell homogenates (20 μg/each) were subjected to CYP2B6 protein Immunoblotting analysis as described in Materials and Methods. Densitometry analysis was performed using FluorChem SP Imaging System (C). Correlation between hCAR1+A activation and induction of CYP2B6 protein was demonstrated in D.
Nuclear Translocation of hCAR in Human Primary Hepatocytes
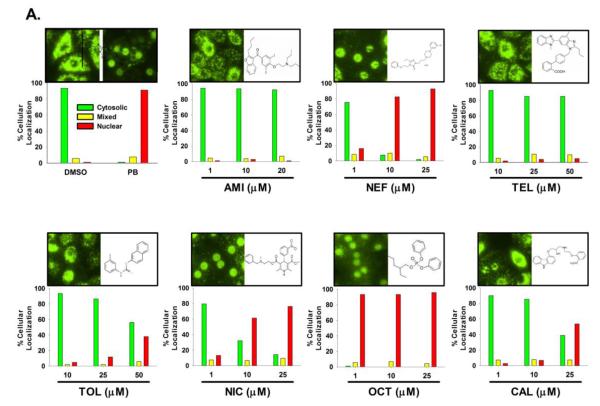
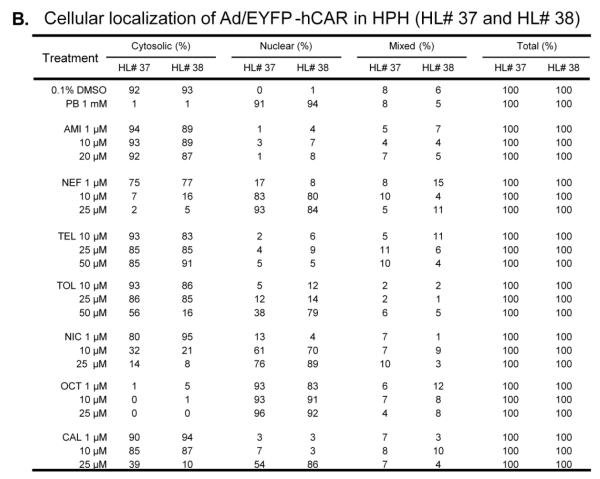
CAR activation is a multi-step process with nuclear translocation as the important initial step (34). In HPHs, CAR is primarily localized in the cytoplasm prior to activation and translocates to the nucleus only after exposure to hCAR modulators, which contrasts to the spontaneous nuclear accumulation observed in immortalized cells (11). To further define the capacity of hCAR activation, compounds tested in hCAR1+A activation and CYP2B6 induction experiments, were subjected to a hCAR nuclear translocation experiment in cultured HPHs infected with the Ad/EYFP-hCAR, which has demonstrated exceptional efficacy in transducing HPHs (35). As outlined in Materials and Methods, transfected HPHs were treated with controls or test compounds for eight hours prior to confocal microscopy analysis. As expected, our results showed that without activation, over 90% Ad/EYFP-hCAR was expressed in the cytoplasm of infected hepatocytes, while approximately 90% Ad/EYFP-hCAR translocated to the nucleus after being treated with the prototypical CAR activator PB (1mM) (Figure 5A). In agreement with hCAR1+A and CYP2B6 induction data, treatment of HPHs with AMI, CAL, TEL and TOL resulted only in negligible to moderate nuclear translocation of Ad/EYFP-hCAR, whereas NIC and OCT were associated with approximately 80% and over 90% nuclear accumulation, respectively (Figure 5A). Unexpectedly, HPHs treated with NEF, a moderate hCAR activator and weak CYP2B6 inducer, displayed robust and concentration-dependent translocation of hCAR achieving 17%, 83%, and 93% nuclear distribution after the treatment at 1, 10, and 25 μM, respectively. Together, these results indicate that chemical-mediated hCAR nuclear accumulation is well-correlated with hCAR1+A activation and CYP2B6 induction in general.
Figure 5. Nuclear translocation of hCAR in human primary hepatocytes.
HPHs from donor (HL-#37, and -#38) were infected with Ad/EYFP-hCAR and then subjected to eight hours of treatment with vehicle control (0.1% DMSO), the positive control, PB (1 mM), or each test compound at three indicated concentrations. Cellular localization of Ad/EYFP-hCAR was analyzed under confocal microscopy as outlined in Materials and Methods. Representative cellular distribution and chemical-mediated translocation were demonstrated (A). After treatment, one-hundred Ad/EYFP-hCAR expressing hepatocytes from donors (HL-#37, and -#38) were categorized into one of three groups: cytoplasmic, nuclear, or mixed (cytoplasmic + nuclear) (B).
Binding of OCT and NIC with the hCAR-LBD
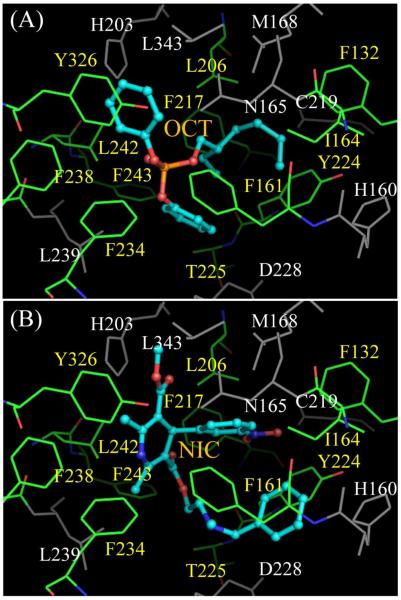
The interaction between the LBD of hCAR and the newly identified potent hCAR activators are demonstrated by docking program Surflex. Figure 6 shows that two compounds, OCT and NIC, are able to favorably bind to the LBD of hCAR. Through computer simulation, the crystal structure of hCAR-LBD was kept rigid and the two compounds were allowed to have some flexibility to be able to fit correctly into the LBD. Unlike PXR ligands that may interact with different sites in its large binding pocket (16, 36), both compounds overlap in the same area having contacts with a hydrophobic pocket bounded by L206, F217, F243, L242, Y326, F238, F234, F161, I164, F132, Y224, and T225. Although neither compound has specific hydrogen bond with the receptor, the CH3OC(O) group of NIC does have electrostatic interaction with residue H203. The binding modes are similar to that between hCAR and ligand CITCO or 5β-pregnanedione suggested by Xu et al (18), where a ligand interacts with hCAR mostly by hydrophobic contacts, with or without obvious hydrogen bond or electrostatic interaction. The suggested binding modes between hCAR and its ligands can be helpful for future design of hCAR modulators by structure-based lead compound optimization.
Figure 6. Docking of new hCAR activators to hCAR-LBD with Surflex.
The ligands (OCT, A; NIC, B) are represented with ball-and-stick, while the protein residues are shown with lines. The ligands interact with hCAR mostly by hydrophobic interaction, without obvious hydrogen bond formation. Residues L206, F217, F243, L242, Y326, F238, F234, F161, I164, F132, Y224, and T225 (names shown in yellow) that form a hydrophobic pocket in the binding site are colored by elements, with carbon, nitrogen and oxygen atoms in green, blue, and red, respectively. The corresponding atoms in ligands are colored with cyan, blue, and red. Other residues belonging to the binding site are in grey.
DISCUSSION
Mounting evidence indicates that CAR plays pivotal roles in the regulation of various liver functions from xenobiotic defense to endocrine homeostasis, and can serve as a predictive biomarker for metabolism-associated attenuated drug efficacy, or increased toxicity, and as a potential therapeutic target for metabolic disorders (6, 37). Thus, there is an escalating need for efficient screening of hCAR activators at the earliest stages of drug development as well as among drugs on the market. In the present study, we have demonstrated that a combined approach strategically integrating computational methods and biological experiments is effective in the identification of novel hCAR activators from marketed drugs.
The CDD drug database, containing over 2000 FDA-approved drugs, was screened initially with four pharmacophore models generated using 17 training compounds recently reported as hCAR modulators such as CITCO. Drugs with favorable pharmacophore scores in the screening were subsequently docked to the active site of the hCAR-LBD with FlexX and Surflex. The selection of drugs for biological evaluation was based on their pharmacophore and docking scores, market availability, and structural diversity. To date, a number of computational models built upon the crystal structures of PXR-, VDR-, and hCAR-LBD, as well as ligand-based pharmacophores have been applied to characterize the molecular interaction between hCAR and its modulators (15, 38). However, to our knowledge, this study represents the first to explore a large set of commercial FDA-approved drugs for hCAR activation and CYP2B6 induction by integrated computational and biological approaches.
Structure- and ligand-based pharmacophore models are rapid and effective approaches for the discovery of new lead compounds as potential hCAR ligands. Nevertheless, outstanding issues associated with this strategy include the limitation in distinguishing agonists from antagonists and non-activators. Determination of the physiological roles of lead compounds from computational models relies ultimately on their biological CAR activation and target gene modulation. Notably, unlike most nuclear receptors, CAR exhibits unique subcellular distribution and activation patterns in that CAR is constitutively activated in immortalized cells independent of chemical activation (1). As a result, no pharmaceutical company at present has an assay available to monitor the activation of CAR in vitro on a routine basis (32). Intriguingly, several lines of evidence indicate that a number of hCAR splicing variants such as hCAR2 and hCAR3, converts the constitutive nature of CAR to chemical-responsive in immortalized cells (13, 39). Moreover, a hCAR3-based chimeric mutant, hCAR1+A, was established in this laboratory as a robust chemical-responsive surrogate of the reference hCAR (14). In the current study, nineteen drugs with optimal pharmacophore and docking parameters were validated in cell-based hCAR1+A assays, where seven of which significantly enhanced the transactivation of hCAR1+A over that of vehicle control (Figure 1A). At present, there is no established criterion to rank order hCAR activators and their clinical DDI potentials. In contrast, criteria defining the transactivation of hPXR, the closest nuclear receptor member of hCAR, has been purported by previous reports (32, 40), in which chemical-mediated activation responses were normalized to that of rifampicin (10 μM), where >40% for high potential, between 15 and 40% for moderate potential, and <15% for low potential. Although arbitrary, we have adopted this threshold in categorizing hCAR1+A activators, by which OCT and NIC were predicted as potent activators while AMI, NEF, TEL, OCT, and CAL were seen as moderate modulators.
Transcriptional up-regulation of CYP2B6 gene expression in the liver represents a hallmark of hCAR activation. To date, accumulated evidence indicates that cross-talk between CAR and PXR leads to the transactivation of many shared target genes including CYP2Bs and CYP3As. In human, however, asymmetrical cross-regulation of CYP2B6 and CYP3A4 by hCAR but not hPXR was reported; hCAR exhibited preferential induction of CYP2B6 over CYP3A4 (41). In the current investigation, seven suspected hCAR activators identified from cell-based reporter assays were further evaluated for their inducibility of CYP2B6 expression in HPHs. It is noteworthy that similar to that of hCAR1+A assays, OCT and NIC displayed the most potent induction of CYP2B6 at both mRNA and protein levels; and the other five moderate hCAR1+A activators exhibited concentration-dependent but only moderate CYP2B6 induction. Previously, we have reported that activation of CAR1+A is closely correlated with that of the reference hCAR (14). The current findings further reveal that the hCAR1+A assay appears to be able to quantitatively predict hCAR activators in their capacity of target gene induction.
In contrast to the spontaneous nuclear localization of CAR in immortalized cells, CAR is primarily compartmented in the cytoplasm of primary hepatocytes and intact liver in vivo prior to activation; and often chemical-mediated nuclear translocation alone is sufficient to confer CAR activation. Although the exact molecular mechanisms behind this process remain unclear, particularly direct and indirect activators of CAR may rely upon different mechanisms, chemical-mediated nuclear translocation of hCAR in HPHs can be used as an alternative model to identify physiologically relevant hCAR activators. Recent results from our laboratory demonstrated that an adenovirus construct expressing enhanced yellow fluorescent protein tagged hCAR (Ad/EYFP-hCAR) infects HPHs with high efficiency and displays the cellular localization and chemical-stimulated translocation of hCAR in a physiologically relevant manner (35). Utilizing this system, we showed that OCT and NIC significantly translocated hCAR from the cytoplasm to the nucleus in HPHs, while AMI, TEL, CAL, and TOL only resulted in moderate but concentration-dependent nuclear accumulation. These observations are in agreement with the results from hCAR1+A activation and CYP2B6 induction assays, supporting the rank order of these novel hCAR activators in general. Interestingly, we observed that NEF treatment ellicited marked nuclear accumulation of hCAR in HPHs but only exhibited relatively moderate hCAR1+A activation and CYP2B6 induction. Although we cannot entirely explain this observed discrepancy, it’s not surprising that different assay formats may produce discrepant results that warrant interrogation when multi-endpoint data are interpreted. Future in vitro to in vivo correlations will reveal the most important changes and dominant pathways controlling CAR activation.
CONCLUSION
In summary, this study reveals that integration of structure- and ligand-based computational models with biological experiments is effective in the identification and evaluation of novel hCAR activators. Importantly, our results revealed that chemical-mediated activations of hCAR1+A in vitro are quantitatively associated with induction of endogenous CYP2B6 by the same panel of xenobiotics. Forty percent of CITCO response in hCAR1+A assays appears to be a reasonable cut-off threshold for initial categorizing of efficacious hCAR activators, and may serve as a preliminary criterion in evaluating hCAR activation in vitro. OCT, the most potent hCAR activator according to our investigation, is used commonly as a flame retarding plasticizer and periodically as a pharmaceutical aid. The current realization of OCT as a potent hCAR activator and CYP2B6 inducer warrants further toxicological assessment. Notably, the optimal occupancy of the hCAR-LBD by OCT and NIC will facilitate retuning of our generated models in assisting future virtual screening for hCAR activators. In the meantime, we do realize that our current models cannot be applied to identify indirect CAR activators, which exhibit their activities through less defined signaling pathways. Whereas, in the absence of other suitable in vitro models, the current strategy that combines the computational models and biological approaches may offer powerful tools for identification of hCAR activators in a relatively high-throughput manner.
Supplementary Material
Figure S1. Chemical structures of training compounds. Seventeen known human CAR ligands extracted from literature were used as training compounds to generate the common features of pharmacophore model in this study. The chemical structures of these compounds were drawn using ChemDraw Ultra 10.
Figure S2. Chemical structures of tested lead compounds. Nineteen lead compounds with optimal pharmacophore parameters from virtual screening of the FDA-approved drug data base were selected for biological assessment of CAR activation. The chemical structures of these compounds were drawn using ChemDraw Ultra 10.
ACKNOWLEDGEMENTS
The authors thank Dr. James Polli (The University of Maryland School of Pharmacy) for kindly offering multiple compounds and Dr. Alex MacKerell (The University of Maryland School of Pharmacy) for making the Discovery Studio available for this study. We also thank Dr. Sean Ekins (Collaborations in Chemistry, Jenkintown, PA) for offering initial CDD database and Dr. Taiji Oashi, a previous lab member from Dr. MacKerell’s lab for database optimization. The authors appreciatively acknowledge The University of Maryland Medical Center and Life Technologies (Durham, NC) for providing the human hepatocytes used in this study. The research is supported in part by the National Institutes of Health Grants DK061652 (H.W) and DK061425 (P.S).
ABBREVIATIONS
- (AMI)
amiodarone
- (CAL)
carvedilol
- (CAR)
constitutive androstane receptor
- (Ad/EYFP-hCAR)
adenovirus expressing enhanced yellow fluorescent protein-tagged human CAR
- (CITCO)
6-(4-chlorophenyl) imidazo[2,1-b][1,3]-thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl)oxime
- (CYP)
cytochrome P450
- (DMSO)
dimethyl sulfoxide
- (GAPDH)
glyceraldehyde-3-phosphate dehydrogenase
- (HPH)
human primary hepatocytes
- (LBD)
ligand-binding domain
- (NEF)
nefazodone
- (NIC)
nicardipine
- (OCT)
octicizer
- (PB)
phenobarbital
- (PXR)
pregnane X receptor
- (RT-PCR)
reverse transcription-polymerase chain reaction
- (TEL)
telmisartan
- (TOL)
tolnaftate
REFERENCES
- 1.Honkakoski P, Sueyoshi T, Negishi M. Drug-activated nuclear receptors CAR and PXR. Ann Med. 2003;35:172–182. doi: 10.1080/07853890310008224. [DOI] [PubMed] [Google Scholar]
- 2.Qatananiand M, Moore DD. CAR, the continuously advancing receptor, in drug metabolism and disease. Curr Drug Metab. 2005;6:329–339. doi: 10.2174/1389200054633899. [DOI] [PubMed] [Google Scholar]
- 3.Yap KY, Chui WK, Chan A. Drug interactions between chemotherapeutic regimens and antiepileptics. Clin Ther. 2008;30:1385–1407. doi: 10.1016/j.clinthera.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Maglich JM, Watson J, McMillen PJ, Goodwin B, Willson TM, Moore JT. The nuclear receptor CAR is a regulator of thyroid hormone metabolism during caloric restriction. J Biol Chem. 2004;279:19832–19838. doi: 10.1074/jbc.M313601200. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004;64:7197–7200. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
- 6.Gao J, He J, Zhai Y, Wada T, Xie W. The constitutive androstane receptor is an anti-obesity nuclear receptor that improves insulin sensitivity. J Biol Chem. 2009;284:25984–25992. doi: 10.1074/jbc.M109.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kodama S, Koike C, Negishi M, Yamamoto Y. Nuclear Receptors CAR and PXR Cross Talk with FOXO1 To Regulate Genes That Encode Drug-Metabolizing and Gluconeogenic Enzymes. 2004;Vol. 24:7931–7940. doi: 10.1128/MCB.24.18.7931-7940.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, Moore DD. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol Endocrinol. 2005;19:1646–1653. doi: 10.1210/me.2004-0520. [DOI] [PubMed] [Google Scholar]
- 9.Phillips JM, Burgoon LD, Goodman JI. Phenobarbital elicits unique, early changes in the expression of hepatic genes that affect critical pathways in tumor-prone B6C3F1 mice. Toxicol Sci. 2009;109:193–205. doi: 10.1093/toxsci/kfp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kliewer SA. The nuclear pregnane X receptor regulates xenobiotic detoxification. J Nutr. 2003;133:2444S–2447S. doi: 10.1093/jn/133.7.2444S. [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omiecinski CJ, Coslo DM, Chen T, Laurenzana EM, Peffer RC. Multi-species Analyses of Direct Activators of the Constitutive Androstane Receptor. Toxicol Sci. 2011 doi: 10.1093/toxsci/kfr191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auerbach SS, Stoner MA, Su S, Omiecinski CJ. Retinoid X receptor-alpha-dependent transactivation by a naturally occurring structural variant of human constitutive androstane receptor (NR1I3) Mol Pharmacol. 2005;68:1239–1253. doi: 10.1124/mol.105.013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen T, Tompkins LM, Li L, Li H, Kim G, Zheng Y, Wang H. A single amino acid controls the functional switch of human constitutive androstane receptor (CAR) 1 to the xenobiotic-sensitive splicing variant CAR3. J Pharmacol Exp Ther. 2010;332:106–115. doi: 10.1124/jpet.109.159210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jyrkkarinne J, Windshugel B, Ronkko T, Tervo AJ, Kublbeck J, Lahtela-Kakkonen M, Sippl W, Poso A, Honkakoski P. Insights into ligand-elicited activation of human constitutive androstane receptor based on novel agonists and three-dimensional quantitative structure-activity relationship. J Med Chem. 2008;51:7181–7192. doi: 10.1021/jm800731b. [DOI] [PubMed] [Google Scholar]
- 16.Pan Y, Li L, Kim G, Ekins S, Wang H, Swaan PW. Identification and Validation of Novel Human Pregnane X Receptor Activators among Prescribed Drugs via Ligand-Based Virtual Screening. 2011;Vol. 39:337–344. doi: 10.1124/dmd.110.035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekins S, Chang C, Mani S, Krasowski MD, Reschly EJ, Iyer M, Kholodovych V, Ai N, Welsh WJ, Sinz M, Swaan PW, Patel R, Bachmann K. Human pregnane X receptor antagonists and agonists define molecular requirements for different binding sites. Mol Pharmacol. 2007;72:592–603. doi: 10.1124/mol.107.038398. [DOI] [PubMed] [Google Scholar]
- 18.Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, Moore JT. A structural basis for constitutive activity in the human CAR/RXRalpha heterodimer. Mol Cell. 2004;16:919–928. doi: 10.1016/j.molcel.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Chen T, Stanton JD, Sueyoshi T, Negishi M, Wang H. The Peripheral Benzodiazepine Receptor Ligand 1-(2-Chlorophenyl-methylpropyl)-3-isoquinoline-carboxamide Is a Novel Antagonist of Human Constitutive Androstane Receptor. 2008;Vol. 74:443–453. doi: 10.1124/mol.108.046656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, Moore JT. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278:17277–17283. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- 21.Kirchmair J, Laggner C, Wolber G, Langer T. Comparative analysis of protein-bound ligand conformations with respect to catalyst’s conformational space subsampling algorithms. Journal of Chemical Information and Modeling. 2005;45:422–430. doi: 10.1021/ci049753l. [DOI] [PubMed] [Google Scholar]
- 22.Hohman M, Gregory K, Chibale K, Smith PJ, Ekins S, Bunin B. Novel web-based tools combining chemistry informatics, biology and social networks for drug discovery. Drug discovery today. 2009;14:261–270. doi: 10.1016/j.drudis.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Ekinsand S, Williams AJ. Finding promiscuous old drugs for new uses. Pharmaceutical research. 2011;28:1785–1791. doi: 10.1007/s11095-011-0486-6. [DOI] [PubMed] [Google Scholar]
- 24.Ekins S, Williams AJ, Krasowski MD, Freundlich JS. In silico repositioning of approved drugs for rare and neglected diseases. Drug discovery today. 2011;16:298–310. doi: 10.1016/j.drudis.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Chambers CC, Hawkins GD, Cramer CJ, Truhlar DG. Model for aqueous solvation based on class IV atomic charges and first solvation shell effects. Journal of Physical Chemistry. 1996;100:16385–16398. [Google Scholar]
- 26.Li JB, Zhu TH, Cramer CJ, Truhlar DG. New class IV charge model for extracting accurate partial charges from wave functions. Journal of Physical Chemistry A. 1998;102:1820–1831. [Google Scholar]
- 27.Kaminskiand G, Jorgensen WL. Performance of the AMBER94, MMFF94, and OPLS-AA force fields for modeling organic liquids. Journal of Physical Chemistry. 1996;100:18010–18013. [Google Scholar]
- 28.Chang C, Bahadduri PM, Polli JE, Swaan PW, Ekins S. Rapid identification of P-glycoprotein substrates and inhibitors. Drug Metabolism and Disposition. 2006;34:1976–1984. doi: 10.1124/dmd.106.012351. [DOI] [PubMed] [Google Scholar]
- 29.Ruppert J, Welch W, Jain AN. Automatic identification and representation of protein binding sites for molecular docking. Protein Sci. 1997;6:524–533. doi: 10.1002/pro.5560060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeCluyse EL, Alexandre E, Hamilton GA, Viollon-Abadie C, Coon DJ, Jolley S, Richert L. Isolation and culture of primary human hepatocytes. Methods Mol Biol. 2005;290:207–229. doi: 10.1385/1-59259-838-2:207. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, LeCluyse EL. A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem. 2003;278:14146–14152. doi: 10.1074/jbc.M212482200. [DOI] [PubMed] [Google Scholar]
- 32.Chu V, Einolf HJ, Evers R, Kumar G, Moore D, Ripp S, Silva J, Sinha V, Sinz M, Skerjanec A. In vitro and in vivo induction of cytochrome p450: a survey of the current practices and recommendations: a pharmaceutical research and manufacturers of america perspective. Drug Metab Dispos. 2009;37:1339–1354. doi: 10.1124/dmd.109.027029. [DOI] [PubMed] [Google Scholar]
- 33.FDA Drug Interaction Studies-Study Design, Data Analysis, and Applications for Dosing and Labeling. FDA Guidance. 2006 [Google Scholar]
- 34.Liand H, Wang H. Activation of xenobiotic receptors: driving into the nucleus. 2010;Vol. 6:409–426. doi: 10.1517/17425251003598886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Chen T, Cottrell J, Wang H. Nuclear Translocation of Adenoviral-Enhanced Yellow Fluorescent Protein-Tagged-Human Constitutive Androstane Receptor (hCAR): A Novel Tool for Screening hCAR Activators in Human Primary Hepatocytes. 2009;Vol. 37:1098–1106. doi: 10.1124/dmd.108.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, Redinbo MR. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–2333. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- 37.Dong B, Saha PK, Huang W, Chen W, Abu-Elheiga LA, Wakil SJ, Stevens RD, Ilkayeva O, Newgard CB, Chan L, Moore DD. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. 2009;Vol. 106:18831–18836. doi: 10.1073/pnas.0909731106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kublbeck J, Laitinen T, Jyrkkarinne J, Rousu T, Tolonen A, Abel T, Kortelainen T, Uusitalo J, Korjamo T, Honkakoski P, Molnar F. Use of comprehensive screening methods to detect selective human CAR activators. Biochemical pharmacology. 2011 doi: 10.1016/j.bcp.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 39.DeKeyser JG, Stagliano MC, Auerbach SS, Prabhu KS, Jones AD, Omiecinski CJ. Di(2-ethylhexyl) phthalate is a highly potent agonist for the human constitutive androstane receptor splice variant CAR2. Mol Pharmacol. 2009;75:1005–1013. doi: 10.1124/mol.108.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sinz M, Kim S, Zhu Z, Chen T, Anthony M, Dickinson K, Rodrigues AD. Evaluation of 170 xenobiotics as transactivators of human pregnane X receptor (hPXR) and correlation to known CYP3A4 drug interactions. Curr Drug Metab. 2006;7:375–388. doi: 10.2174/138920006776873535. [DOI] [PubMed] [Google Scholar]
- 41.Faucette SR, Sueyoshi T, Smith CM, Negishi M, Lecluyse EL, Wang H. Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther. 2006;317:1200–1209. doi: 10.1124/jpet.105.098160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Chemical structures of training compounds. Seventeen known human CAR ligands extracted from literature were used as training compounds to generate the common features of pharmacophore model in this study. The chemical structures of these compounds were drawn using ChemDraw Ultra 10.
Figure S2. Chemical structures of tested lead compounds. Nineteen lead compounds with optimal pharmacophore parameters from virtual screening of the FDA-approved drug data base were selected for biological assessment of CAR activation. The chemical structures of these compounds were drawn using ChemDraw Ultra 10.