Background: N-Glycan processing and interactions with lectins regulate the quality control and endoplasmic reticulum-associated degradation (ERAD) of glycoproteins.
Results: Most of the same machinery targets nonglycosylated misfolded proteins.
Conclusion: They share some membrane and luminal ERAD machinery but not all cytosolic components.
Significance: ER glycan-interacting proteins must possess a dual specificity for glycan structure and for exposed misfolded polypeptide domains.
Keywords: Chaperone Chaperonin, ER-associated Degradation, Lectin, Protein Degradation, Protein Misfolding, EDEM1, HRD1, Igκ Light Chain
Abstract
Studies of misfolded protein targeting to endoplasmic reticulum-associated degradation (ERAD) have largely focused on glycoproteins, which include the bulk of the secretory proteins. Mechanisms of targeting of nonglycosylated proteins are less clear. Here, we studied three nonglycosylated proteins and analyzed their use of known glycoprotein quality control and ERAD components. Similar to an established glycosylated ERAD substrate, the uncleaved precursor of asialoglycoprotein receptor H2a, its nonglycosylated mutant, makes use of calnexin, EDEM1, and HRD1, but only glycosylated H2a is a substrate for the cytosolic SCFFbs2 E3 ubiquitin ligase with lectin activity. Two nonglycosylated BiP substrates, NS-1κ light chain and truncated Igγ heavy chain, interact with the ERAD complex lectins OS-9 and XTP3-B and require EDEM1 for degradation. EDEM1 associates through a region outside of its mannosidase-like domain with the nonglycosylated proteins. Similar to glycosylated substrates, proteasomal inhibition induced accumulation of the nonglycosylated proteins and ERAD machinery in the endoplasmic reticulum-derived quality control compartment. Our results suggest a shared ERAD pathway for glycosylated and nonglycosylated proteins composed of luminal lectin machinery components also capable of protein-protein interactions.
Introduction
Research on ER3 quality control and disposal of misfolded glycoproteins has attracted much attention in recent years. A pathway has been delineated, which involves lectin chaperones, glycosidases, and components of a putative retrotranslocation machinery, some of which also possess lectin activity, for delivery to the cytosolic proteasomes (1–5). The chain of events that leads a nascent glycoprotein through ER quality control and toward proper folding or ERAD involves processing of its N-glycans and their interactions. There is initially removal of the terminal glucose residue from the Glc3Man9GlcNAc2 precursor and also of the following glucose that allows binding to the chaperone/lectins calnexin and calreticulin. Removal of the third and last glucose causes dissociation from these lectins, although re-addition of a single glucose by the folding sensor UDP-Glc:glycoprotein glucosyltransferase triggers reassociation until folding is complete (6, 7). If proper folding cannot be achieved in a certain time frame, the glycoprotein molecules are targeted to ERAD by removal of more mannose residues (three or four) than occurs for folded molecules that exit to the Golgi (one or two) (8, 9). This trimming of mannose residues involves ER mannosidase I and the ER degradation-enhancing α-mannosidase-like proteins (EDEMs) and is essential for delivery to ERAD (2, 10, 11). The mannose trimming regulates misfolded glycoprotein association with the lectins OS-9 and XTP3-B at the ER quality control compartment (ERQC), a staging ground for ERAD (8, 12, 13). OS-9 and XTP3-B are in turn associated with a protein complex involved in substrate ubiquitination, retrotranslocation, and targeting to the proteasomes. This complex includes E3 ubiquitin ligases like HRD1 and gp78 and interestingly a family of cytosolic ER-associated SCF ligases that include F-box proteins (Fbs1 and Fbs2) with a lectin moiety that recognizes N-glycans of misfolded glycoproteins for ubiquitination (14). Recognition by these lectin components of the ligases requires prior retrotranslocation of the substrate glycoprotein to expose its glycan to the cytosol.
Much less is known about the processes that govern the fate of nascent nonglycosylated proteins. There is at least some distinction from the most upstream components of the glycoprotein pathway, as many of these proteins interact with the chaperone BiP instead of calnexin or calreticulin (15, 16), and at least some of these nonglycosylated ERAD substrates require the ERAD machinery-associated protein Herp (17). We recently found that, surprisingly, EDEM1, a central player in glycoprotein ERAD (10, 18, 19), can bind a glycoprotein ERAD substrate in a glycan-independent manner and that its overexpression or up-regulation during the unfolded protein response overrides the mannose trimming requirement for ERAD. This hinted to a potential role in nonglycosylated protein ERAD. Here, we set out to investigate the involvement of EDEM1 and of other lectin ERAD components for the disposal of three nonglycosylated unfolded proteins. As a glycosylated ERAD model control, we included the well characterized, uncleaved precursor of asialoglycoprotein receptor (ASGPR) H2a. This glycoprotein is expressed naturally in hepatocytes as a membrane precursor, which undergoes efficient cleavage producing a 35-kDa secreted form (20). When H2a is expressed in other cell lines, the membrane precursor is inefficiently cleaved, and the uncleaved precursor, as well as most of the cleaved fragment, is retained in the ER and degraded by the ubiquitin-proteasome system (12, 21). As a model of a naturally nonglycosylated ERAD substrate, we examined the unassembled nonsecreted NS-1κ light chain (NS-1 κLC). This protein is degraded by the 26 S proteasome (22), through an ERAD pathway that involves Herp, HRD1, and Derlin1 (17). We also analyzed a truncated Igγ heavy chain, γ V-CH1, (23), which, like NS-1 κLC, is a nonglycosylated BiP substrate and is also targeted to the Herp/HRD1 pathway (17). We also included a nonglycosylated mutant of H2a. We found involvement of several luminal components of the glycoprotein ERAD machinery but not of the cytosolically localized SCFFbs2, a glycan-dependent E3 ligase.
EXPERIMENTAL PROCEDURES
Materials
Rainbow 14C-labeled methylated protein standards were obtained from GE Healthcare. Promix cell labeling mix ([35S]Met plus [35S]Cys), >1000 Ci/mmol, was from PerkinElmer Life Sciences. Protein A-Sepharose was from Repligen (Needham, MA). Lactacystin (Lac) and kifunensin were from Cayman Chemicals (Ann Arbor, MI). MG-132 and other common reagents were from Sigma. S-protein-agarose was from Novagen. Streptavidin linked to Texas Red was from Jackson ImmunoResearch (West Grove, PA). Protein G-agarose was from Santa Cruz Biotechnology.
Plasmids and Constructs
H2a and calnexin were subcloned in pCDNA1 (Invitrogen) (12). H2aΔgly in pCDNA1 was described previously (24). Myc-H2aΔgly was subcloned in pCDNA4. BIP-HA in pXM was a kind gift of Peter Murray (Memphis, TN). H2a170 and H2a102 were created by site-directed mutagenesis, substituting asparagine to glutamine (N102Q and N170Q). In H2a305 threonine 305 was substituted by isoleucine (T305I).
The pSUPER vector carrying short hairpin RNAs (shRNA) for human ER mannosidase I or human EDEM1 and EDEM1-HA in pCMVsport2 were used previously (13, 25). EDEM1ΔCRD was described previously (26).
Mouse Fbs2 F box deletion mutant (Fbs2ΔF) and human hsHRD1 RING finger mutant were those used previously (24). S-tagged XTP3-B and OS-9.1 and OS-9.2 (27) were those used in Ref. 26. H2a G78R uncleavable mutant fused through its C terminus to monomeric red fluorescent protein (H2a-RFP) was described previously (24, 28). NS-1 κLC and HA-γ V-CH1 constructs were described previously (Refs. 16, 23, respectively).
Primers and RT-PCR
Total cell RNA was extracted with TRIzol reagent (Invitrogen). Reverse transcription was performed with a VersoTM cDNA kit (Thermo Fisher Scientific, Barrington, IL) as described previously (13).
Antibodies
Rabbit polyclonal C-terminal anti-H2 antibody was used in earlier studies (20, 29), as well as rabbit polyclonal anti-Derlin-1 (13). Mouse monoclonal anti-H2a C-terminal antibody B9 was raised in our laboratory (30). Rabbit polyclonal anti-rodent BiP antiserum was described previously (31). Goat anti-mouse κ was from SouthernBiotech (Birmingham, AL). Mouse monoclonal antibodies used were as follows: anti-FLAG (M2), anti-HA, anti-β-tubulin, anti-calnexin, rabbit anti-GM-130, anti-EDEM1, and anti-HA were from Sigma, anti-Myc was from Cell Signaling (Beverly, MA), and anti-S tag was from Novagen (Gibbstown, NJ). Goat anti-rabbit IgG antibody conjugated to Cy2, goat anti-mouse IgG DyLight549, goat anti-mouse IgG FITC, goat anti-rabbit and anti-mouse IgG conjugated to HRP, donkey anti-goat IgG linked to Cy2, and donkey anti-mouse IgG conjugated to Cy3 were from Jackson ImmunoResearch (West Grove, PA). Goat anti-rabbit IgG DyLight650 was from Abcam. Goat anti-mouse IgG conjugated to agarose was from Sigma. Biotinylated donkey anti-rabbit IgG was from IBA (Olivette, MO).
Cell Culture and Transfections
Human embryonic kidney (HEK) 293 cells were grown in DMEM plus 10% fetal calf serum (FCS) and NIH 3T3 cells in DMEM plus 10% newborn calf serum. All cells were grown at 37 °C under an atmosphere of 5% CO2.
Transient transfection of NIH 3T3 cells was performed using an MP-100 Microporator (Digital Bio) according to the manufacturer's instructions. Transient transfection of HEK 293 cells was done according to the calcium phosphate method. The experiments were performed 24–48 h after the transfection.
Metabolic Labeling, Immunoprecipitation, SDS-PAGE, and Quantitation
Subconfluent (90%) cell monolayers in 60-mm dishes were labeled with [35S]Cys, lysed, and immunoprecipitated with anti-H2 antibodies, as described previously (20, 21), or using mouse anti-H2a B9 antibody and goat anti-mouse IgG immobilized on agarose beads where indicated. The same procedure was used for κLC but labeling with 100 μCi of [35S]Met plus [35S]Cys mix, lysis with a buffer containing 0.15 m NaCl, 10 mm Tris-HCl (pH 7.5), 1% Nonidet P-40, 0.1% SDS, 0.2% deoxycholate, and immunoprecipitation with goat anti-κLC and protein G-agarose. Reducing SDS-PAGE was performed on 10% Laemmli gels if not stated otherwise. The gels were analyzed by fluorography using 20% 2,5-diphenyloxazole and were exposed to Biomax MS film using a TransScreen-LE from Eastman Kodak (Vancouver, BC). Quantitation was performed in a Fujifilm FLA 5100 phosphorimager (Japan).
Coimmunoprecipitation and Immunoblotting
Cell lysis and immunoprecipitation of H2a and related constructs were done as described previously (21). Cell lysis was performed in the presence of protease inhibitor mixture (Roche Applied Science). For immunoprecipitation from HEK 293 cells, cell lysis was done in 1% Nonidet P-40, 50 mm Tris-HCl (pH 8), 150 mm NaCl or as indicated for 30 min on ice, and debris and nuclei were pelleted in a microcentrifuge for 30 min at 4 °C. The samples were immunoprecipitated with appropriate antisera and protein A-agarose. For immunoprecipitation with anti-HA monoclonal antibody, goat anti-mouse IgG-agarose was used. After overnight precipitation, the beads were washed three times with lysis buffer diluted 1:5, followed by elution of the bound proteins by boiling with sample buffer containing β-mercaptoethanol at 100 °C for 5 min. For coimmunoprecipitation with NS-1 κLC, cells were lysed in a buffer containing 150 mm NaCl, 10 mm Tris-HCl (pH 7.5), 1% Nonidet P-40, 0.1% SDS, 0.2% deoxycholate followed by immunoprecipitation with anti-κLC and protein G-agarose (if not stated otherwise).
Coimmunoprecipitation with anti-calnexin antibody was performed as described previously (32). Briefly, after metabolic labeling, cells were lysed in HBS buffer (pH 7.5), containing 2% sodium cholate; cell lysates were immunoprecipitated with anti-CNX antibody, boiled in 1% SDS, then diluted with 10 volumes of 1% Triton X-100, 0.5% sodium deoxycholate in HBS, and reimmunoprecipitated with anti-H2a antibody.
Immunoblotting and detection by ECL were done as described previously (12), except for exposure and quantitation in a ChemiDoc XRS Imaging System (Bio-Rad).
Immunofluorescence Microscopy
The procedures employed were as described previously (12, 25) Confocal microscopy was done on a Zeiss laser scanning confocal microscope (LSM 510; Carl Zeiss, Jena, Germany) as described previously (25).
Statistical Analysis
Data are expressed as means ± S.E. Student's t test (unpaired, two-tailed) was used to compare the two groups, and the p value was calculated in GraphPad Prism 5 (GraphPad software). p < 0.05 was considered as statistically significant.
RESULTS
Components of the Glycoprotein ERAD Pathway Target a Nonglycosylated Mutant of the ERAD Substrate ASGPR H2a Precursor to the ERQC and Are Required for Its Degradation
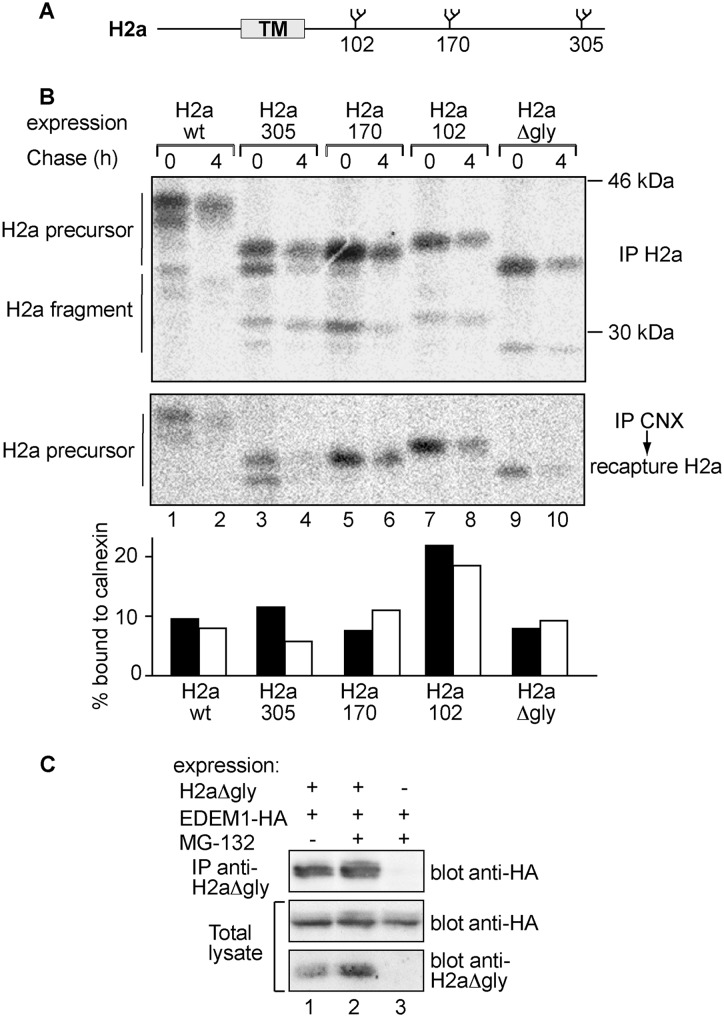
We previously reported that ASGPR H2a precursor associates after synthesis with the ER chaperone calnexin, dissociating slowly compared with its fast dissociation from the calnexin-interacting oxidoreductase ERp57 (32). We created three constructs where two alternative N-glycosylation sites of the three that exist in H2a were abrogated, with a single N-glycan remaining in either position 102, 170, or 305 (Fig. 1A). In addition, we made a construct with all glycosylation sites removed (H2aΔgly). We had seen previously that wild-type H2a is degraded soon after dissociation from calnexin (32). Therefore, in a pulse-chase analysis of H2a in the absence of proteasomal inhibition, followed by either direct immunoprecipitation or coimmunoprecipitation with anti-calnexin, a similar fraction of H2a is found associated with calnexin before or after the chase period (Fig. 1B, lanes 1 and 2, and graph). Although there were slight variations, all H2a mutants retaining a single N-glycan had similar half-lives and interacted with calnexin similarly to wild-type H2a, except for H2a102, which associated to a greater extent than WT H2a both initially and after the 4-h chase (Fig. 1B). Unexpectedly, the nonglycosylated H2aΔgly showed association with calnexin similar to WT H2a. Therefore, we focused on characterizing the involvement of other factors considered to be part of the glycoprotein ER quality control machinery on the targeting to degradation of H2aΔgly. EDEM1 showed a significant association with H2aΔgly under steady state conditions, both in the presence and absence of the proteasomal inhibitor MG-132 (Fig. 1C). We then examined the kinetics of calnexin and EDEM1 association with H2aΔgly in the presence of the proteasomal inhibitor to minimize degradation. The association of EDEM1 with H2aΔgly was more prolonged than that with calnexin (Fig. 2, A and B). However, unlike the association of EDEM1 with WT H2a (Fig. 2C) and other glycoprotein ERAD substrates (33, 34), it did not increase with time (Fig. 2B). Overexpression of EDEM1 significantly accelerated the degradation of H2aΔgly, whereas knockdown of EDEM1 inhibited its turnover (Fig. 2, D–F). We previously found that glycosylated H2a is a substrate for two E3 ubiquitin ligases, HRD1 and SCFFbs2 (13). The latter carries the substrate-recognition subunit Fbs2, which possesses a high mannose glycan binding domain (35). H2aΔgly and WT H2a showed similar association with HRD1 (Fig. 3A). In contrast, H2aΔgly showed no significant binding to Fbs2 (Fig. 3B). Consistently, overexpression of a dominant-negative RING mutant of HRD1 blocked ERAD of H2aΔgly, whereas a dominant-negative F-box mutant of Fbs2 did not (Fig. 3C). Both dominant-negative proteins were shown to block the degradation of WT H2a (13). Conversely to the interactions of Fbs2, the chaperone BiP showed negligible binding to H2a, whereas it associated significantly with H2aΔgly (Fig. 3D).
FIGURE 1.
CNX and EDEM1 associate with glycosylation site mutants of ASGPR H2a. A, schematic representation of ASGPR H2a N-glycosylation sites. The gray rectangle shows the transmembrane domain (TM). B, HEK 293 cells were transfected with vectors encoding either H2a WT or its glycosylation site mutants in which two of the three H2a N-glycosylation sites were abrogated with one N-glycan remaining in the indicated positions or a mutant in which all N-glycosylation sites were canceled (H2aΔgly). The cells were pulse-labeled for 20 min with [35S]Cys and chased for the indicated times. After the pulse (0-h chase) or the chase periods, the cells were lysed; H2a was immunoprecipitated (IP) from 15% of the cell lysates, and the remainders were immunoprecipitated with anti-CNX antibody followed by elution and re-immunoprecipitation with anti-H2a, as described under “Experimental Procedures.” All immunoprecipitates were separated in 12% SDS-PAGE followed by phosphorimaging. Bands corresponding to the H2a precursor and the naturally occurring cleaved fragment are indicated on the left. For the pulse samples, two bands can be seen for the H2a precursor and also for the fragment, and the lower ones correspond to underglycosylated species (one of the glycosylation sites unoccupied). Amounts of H2a WT and glycosylation mutants coprecipitated with calnexin were plotted relative to their total amounts (IP H2a). The results of this and other experiments shown are representative of three independent experiments. C, 2 days after transfection with vectors encoding for HA-tagged EDEM1 (EDEM1-HA) and H2aΔgly, HEK-293 cells were incubated for 3 h in the absence/presence of 40 μm MG-132 and lysed in Nonidet P-40 buffer (“Experimental Procedures”). Lysates (10% of total) were run on SDS-PAGE and immunoblotted with anti-HA antibody (middle panel) or with anti-H2a (bottom panel). The rest of the lysates were immunoprecipitated with mouse monoclonal anti-H2a, subjected to SDS-PAGE, and immunoblotted with anti-HA (upper panel).
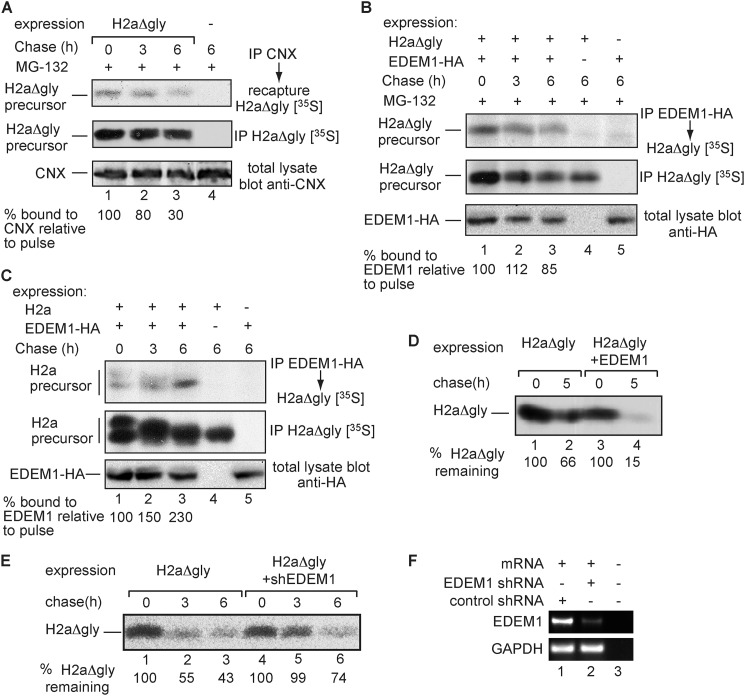
FIGURE 2.
H2aΔgly is a substrate of EDEM1. A, similar to Fig. 1B, except that cells expressing H2aΔgly were incubated with 40 μm MG-132 and 10% of the lysates were run on SDS-PAGE and immunoblotted with anti-CNX antibody (bottom panel). Phosphorimager quantitation values of H2aΔgly bound to CNX (upper panel) divided by total H2aΔgly (middle panel) are shown at the bottom relative to the pulse labeling. B, similar to A but with cells expressing H2aΔgly and EDEM1-HA and cell lysis in buffer containing 1% Triton X-100 and 0.5% sodium deoxycholate. EDEM1-HA was immunoprecipitated (IP) from 70% of the cell lysates using anti-HA and H2aΔgly from 25% of the lysates with anti-H2a. The remaining 5% of the lysates were run on SDS-PAGE and immunoblotted with anti-HA (bottom panel). C, similar to B but with WT H2a instead of H2aΔgly. Note that the fully glycosylated band is shifted to a faster migration due to trimming of mannose residues, whereas the underglycosylated lower species is degraded (8). D, similar to B (middle panel) but with cells expressing H2aΔgly together with control GFP or with EDEM1-HA and without MG-132. Quantitations of the percent of H2aΔgly remaining after the chase relative to the pulse are shown at the bottom. E, similar to D but with cells expressing H2aΔgly together with control anti-LacZ shRNA or anti-EDEM1 shRNA. F, in parallel with E, RNA was extracted from the cells and used for RT-PCR with primers for EDEM1 mRNA (upper panel) compared with GAPDH (lower panel). No RNA template was added in lane 3.
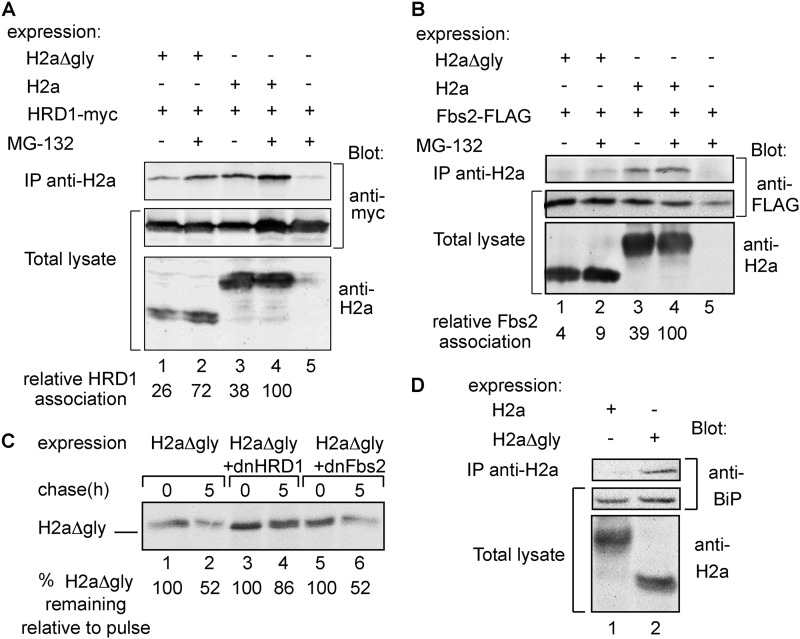
FIGURE 3.
H2aΔgly associates with BiP and is a substrate of HRD1 but not of SCFFbs2. A, experiment similar to that in Fig. 1C but with coexpression of Myc-tagged HRD1 (HRD1-myc) with H2a or H2aΔgly and immunoblotting with anti-Myc or anti-H2a. Quantitations of HRD1 association with H2a or H2aΔgly are shown at the bottom relative to association with H2a upon MG-132 treatment. Nonspecific coprecipitation of HRD1 (lane 5) was subtracted from the rest. The results were normalized by dividing the intensity of the HRD1 band in the top panel by that of the corresponding bands in the middle and bottom panels. IP, immunoprecipitation. B, similar to A, except that FLAG-tagged Fbs2 (Fbs2-FLAG) was used instead of HRD1-myc and blotting was with anti-FLAG. C, experiment similar to that in Fig. 2D but with cells expressing H2aΔgly with control GFP or with dominant-negative mutants of either HRD1 (dnHRD1) or Fbs2 (dnFbs2). D, similar to A but without HRD1 expression and immunoblotting with anti-H2a or of endogenous BiP with rabbit anti-BiP.
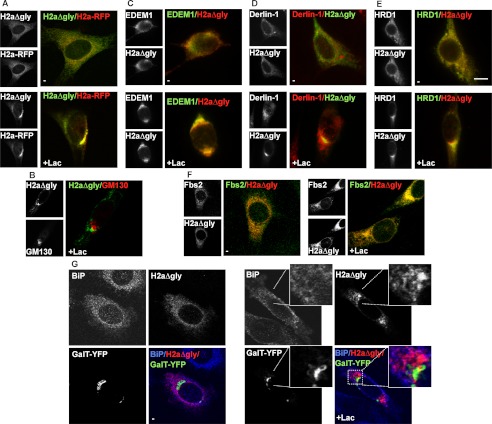
We next determined whether H2aΔgly accumulates like WT H2a and other glycoprotein substrates in the juxtanuclear ERQC (8, 12, 28). Indeed, proteasomal inhibition caused accumulation of H2aΔgly from an initial dispersed ER pattern to the ERQC, where it colocalized with the glycoprotein ERAD substrate H2a linked to a monomeric red fluorescent protein (H2a-RFP) (Fig. 4A) and not with a Golgi marker, GM130 (Fig. 4B). Accumulated H2aΔgly also colocalized to a fair extent with EDEM1, Derlin-1, HRD1, and Fbs2. (Fig. 4, C–F). We had seen previously that these ERAD machinery components are recruited to the ERQC upon proteasomal inhibition or glycoprotein substrate overexpression (13, 28). Although H2aΔgly does not appear to bind to or require SCFFbs2 for its degradation (Fig. 3), both proteins are still recruited to the ERQC, SCFFbs2 apparently from the cytosolic side and H2aΔgly on the membrane. This suggests that the ERQC may be a staging ground for ERAD of glycoproteins (13, 28) as well as nonglycoproteins and that SCFFbs2 is recruited either independent of the glycan nature of the substrate or that sufficient amounts of endogeneous glycoproteins exist to recruit it. H2aΔgly remained separate from a Golgi marker, β1,3-galactosyltransferase linked to YFP (GalT-YFP) and also from most of the abundant ER chaperone BiP, which is not concentrated in the ERQC (Fig. 4G). This result is similar to the segregation of BiP from overexpressed ERQC-localized glycosylated H2a (12, 28).
FIGURE 4.
Upon proteasomal inhibition H2aΔgly colocalizes with EDEM1 and ERAD factors at the ERQC. A, plasmids encoding for H2a-RFP and myc-tagged H2aΔgly were cotransfected in NIH 3T3 cells. One day after transfection, cells were incubated for 3 h without (upper panels) or with Lac (25 μm) (lower panels), fixed, permeabilized, and incubated with mouse anti-Myc and FITC-conjugated goat anti-mouse IgG. Representative confocal optical slices are shown. B, similar to A but with endogenous GM130 detected with rabbit anti-GM130 and Cy3-conjugated goat anti-rabbit IgG. C, cells cotransfected with HA-tagged EDEM1 and untagged H2aΔgly and incubated with rabbit anti-H2a and Cy3-conjugated goat anti-rabbit IgG and with mouse anti-HA and FITC-conjugated goat anti-mouse IgG. D, similar to B; endogenous Derlin-1 was visualized using rabbit anti-Derlin-1 and Cy3-conjugated goat anti-rabbit IgG. E, similar to C but with Myc-tagged HRD1 instead of EDEM1-HA. Mouse anti-Myc antibodies and FITC-conjugated goat anti-mouse IgG were used to visualize HRD1. Bar, 10 μm. F, similar to C with cells expressing H2aΔgly and FLAG-tagged Fbs2, detected with mouse anti-FLAG and FITC-conjugated goat-anti mouse IgG. G, cells transfected with Myc-tagged H2aΔgly and GalT-YFP, incubated with mouse anti-Myc and goat-anti mouse Dylight549 IgG. Endogenous Bip was visualized using rabbit anti-Bip and goat anti-rabbit DyLight650 IgG, pseudocolored blue. A section was enlarged for better detail of the lack of colocalization.
Altogether, the results show a similar routing and requirement of ERAD pathway components for H2aΔgly as compared with WT H2a, including calnexin, EDEM1, and HRD1. Notable exceptions are BiP, which binds strongly to nonglycosylated H2aΔgly but not to the glycoprotein, H2a, and SCFFbs2, which targets H2a but is not required for degradation of H2aΔgly.
Glycan-independent Targeting of the Nonglycosylated Substrate by a Mutant EDEM1 Lacking Its Carbohydrate Recognition Domain
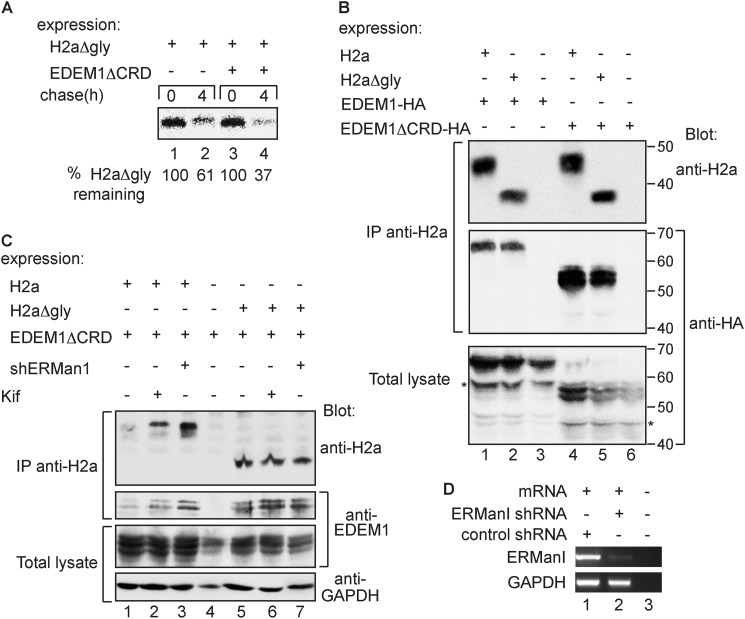
We had shown that when EDEM1 is overexpressed or up-regulated by the UPR it bypasses the glycan dependence for glycoprotein ERAD. In these conditions, the carbohydrate recognition domain of EDEM1 was not required for it to target WT H2a (26). We tested whether a mutant EDEM1 (EDEM1ΔCRD), lacking most of its carbohydrate-recognition domain, which corresponds to the catalytic portion of homologous mannosidases (26), would target H2aΔgly for degradation. In a pulse-chase analysis, overexpression of EDEM1ΔCRD significantly increased the degradation of H2aΔgly (Fig. 5A). EDEM1ΔCRD associated to a similar degree with both WT H2a and H2aΔgly, and even more strongly than WT EDEM1 (Fig. 5B). Trimming of mannose residues of the substrate, in the case of WT H2a, or of any putative intermediate glycoprotein in the case of an indirect interaction was not required for the association of EDEM1ΔCRD with WT H2a or with H2aΔgly (Fig. 5C). Consistently, neither the expression nor activity of ER mannosidase I was necessary for this association (Fig. 5, C and D).
FIGURE 5.
EDEM1 mutant devoid of its carbohydrate recognition domain (CRD) can interact with substrates and target them to degradation. A, similar to Fig. 2D but with EDEM1 mutant with most of its CRD deleted (EDEM1ΔCRD) instead of EDEM1 WT. B, same procedure as in Fig. 1C was performed, but with cells expressing either HA-tagged EDEM1 WT or EDEM1ΔCRD, together with either H2a or H2aΔgly. Asterisks indicate C-terminal (HA-tagged) cleavage products of EDEM1 and EDEM1ΔCRD, observed in total lysates but almost absent from the coimmunoprecipitation. Molecular mass markers are indicated on the right in kDa. IP, immunoprecipitation. C, similar to B but with cotransfection with or without an anti-ER mannosidase I (ERManI) shRNA encoding p-SUPER plasmid as indicated. Two days post-transfection, cells were incubated in the presence or absence of 10 μm kifunensine (Kif) for 6 h. Immunoblots were with anti-H2a, anti-EDEM1, and anti-GAPDH. D, RNA was extracted from cells transfected with the plasmids encoding anti-ER mannosidase I or control anti-LacZ shRNA, and used for RT-PCR with primers for ER mannosidase I mRNA (upper panel) compared with GAPDH (lower panel). Lane 3 shows a sample with no RNA template.
Targeting of a Naturally Nonglycosylated Substrate by EDEM1 and Routing to the ERQC
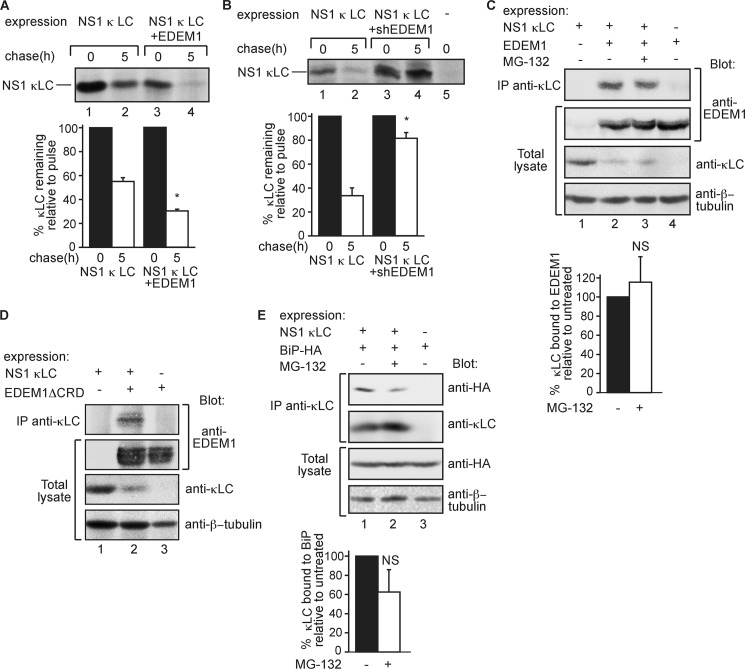
As the above experiments were done on a nonglycosylated mutant of a glycoprotein, we wondered whether a naturally nonglycosylated ERAD substrate would behave similarly. Therefore, we analyzed a nonsecreted Ig κ light chain (NS-1 κLC), which utilizes several components of the ERAD machinery, Derlin-1, Herp, HRD1, and p97 (17), but is recognized by the ER chaperone BiP instead of calnexin (15, 16). The degradation of NS-1 κLC was accelerated by overexpression of EDEM1 and strongly inhibited by knockdown of EDEM1 (Fig. 6, A and B). In keeping with this finding, EDEM1 coimmunoprecipitated substantially with NS-1 κLC (Fig. 6C). This association did not change significantly upon inhibition of its degradation. Similar to H2aΔgly, NS-1 κLC also coimmunoprecipitated with EDEM1ΔCRD, the overexpression of which decreased NS-1 κLC levels (Fig. 6D).
FIGURE 6.
ERAD of nonsecreted κ light chain involves EDEM1. A and B, experiments similar to those in Fig. 2, D and E, respectively, but with nonglycosylated nonsecreted κ light chain (NS-1 κLC) instead of H2aΔgly. Cell lysis and immunoprecipitation (IP) with anti-NS-1 κLC antibody were as described under “Experimental Procedures.” Note that in A transfections were done with 6 μg of plasmid carrying NS-1 κLC and in B with 4 μg, to better observe the effect of the knockdown. Results were quantitated and normalized as in Fig. 2D; values are averages of three independent experiments; error bars are standard deviations. p values between chase time with control and with EDEM1 or shEDEM1 expression: 0.02, marked with asterisks. C, similar to Fig. 1C but with expression of NS-1 κLC instead of H2aΔgly and lysis and immunoprecipitation with goat anti-κLC as explained under “Experimental Procedures.” Immunoblotting was with anti-EDEM1, anti-NS-1 κLC, and anti-β-tubulin for loading control. p value >0.05. D, similar to C but with EDEM1ΔCRD instead of EDEM1. E, similar to C but with BiP-HA but instead of EDEM1-HA. BiP-HA was detected with anti-HA. p value >0.05. NS, not significant.
NS-1 κLC coimmunoprecipitated with BiP, as seen previously (16), and in this case the association decreased upon proteasomal inhibition (Fig. 6E), suggesting release from BiP at late ERAD stages. However, it was not a significant decrease.
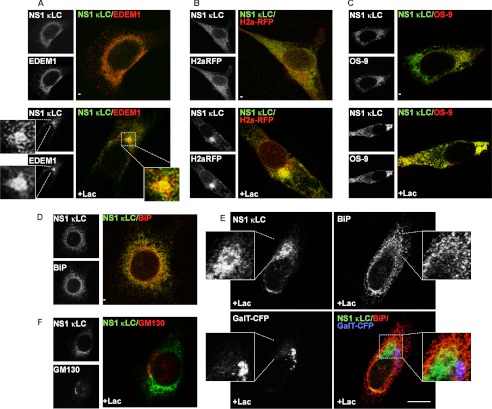
In untreated cells, NS-1 κLC appeared in a diffuse ER pattern, and upon proteasomal inhibition it accumulated and colocalized significantly with EDEM1 at the ERQC (Fig. 7A), where a substantial fraction of NS-1 κLC also colocalized with H2a-RFP (Fig. 7B). The HRD1/SEL1L-associated lectin OS-9 (27) concentrated in the ERQC, as seen previously (26), and upon proteasomal inhibition, it colocalized with accumulated NS-1 κLC (Fig. 7C, lower panels).
FIGURE 7.
Routing of nonsecreted κ light chain to the ERQC. A, NIH 3T3 cells cotransfected with EDEM1-HA together with a plasmid encoding for NS-1 κLC, treated and processed as in Fig. 4, and incubated with goat anti-κLC and Cy2-conjugated donkey anti-goat IgG and with rabbit anti-HA, biotinylated donkey anti-rabbit IgG, and streptavidin linked to Texas Red. A section was enlarged for better detail of the colocalization. B, similar to A but with cells expressing H2a-RFP and NS-1 κLC. C, similar to B but with of a mixture of S-tagged OS-9.1 and OS-9.2 (OS-9) instead of H2a-RFP. Mouse anti-S tag and donkey anti-mouse IgG-Cy3 were used to visualize OS-9.1/2. D, similar to C but transfection was only with NS-1 κLC. Endogenous BiP was stained with rabbit anti-BiP, biotinylated donkey anti-rabbit IgG, and streptavidin linked to Texas Red. E, similar to D but with cotransfection of GalT-CFP (Golgi marker) and cell incubation with Lac. Bar, 10 μm. F, similar to D, except that endogenous GM130 was detected with rabbit anti-GM130, biotinylated donkey anti-rabbit IgG, and streptavidin linked to Texas Red.
We had seen that even in the absence of expression of an ERAD substrate, calnexin accumulates in the ERQC upon proteasomal inhibition, whereas BiP does not (12, 32). We wondered whether upon expression of NS-1 κLC, a protein that associates strongly with BiP, BiP would now appear in the ERQC. As expected, in the absence of proteasomal inhibitors, NS-1 κLC colocalized with BiP in a disperse ER pattern (Fig. 7D). However, upon proteasomal inhibition, although NS-1 κLC is now concentrated in the ERQC, BiP remained mostly secluded from this region and neither protein colocalized with a Golgi marker (Fig. 7, E and F), similar to what we observed upon coexpression of H2aΔgly (Fig. 4G).
Nonglycosylated Truncated Igγ Heavy Chain Is Also a Substrate of EDEM1
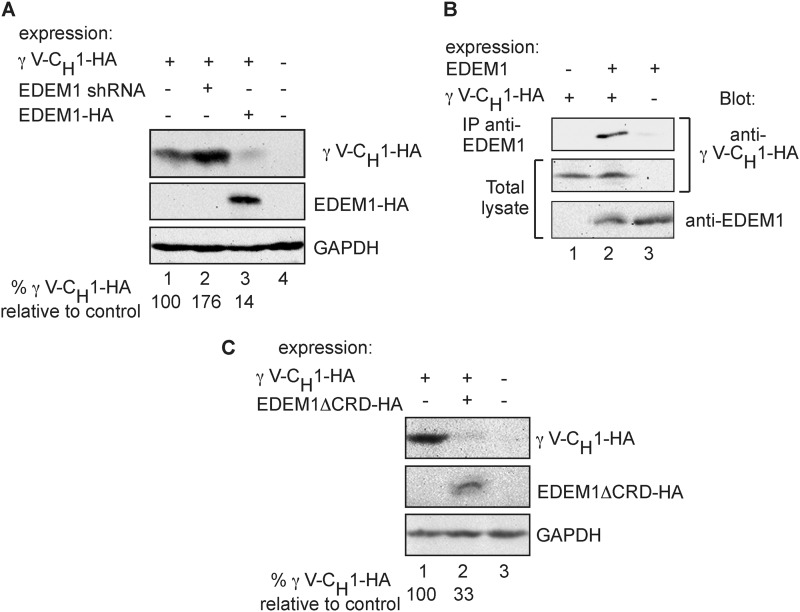
We looked at another nonglycosylated BiP substrate, truncated Igγ heavy chain (HA-γ V-CH1), also an ERAD substrate (17). Once again, we found that EDEM1 knockdown caused accumulation of HA-γ V-CH1, and EDEM1 overexpression caused a significant reduction of its level (Fig. 8A). HA-γ V-CH1 coimmunoprecipitated specifically with EDEM1 (Fig. 8B). EDEM1ΔCRD also accelerated HA-γ V-CH1 degradation (Fig. 8C).
FIGURE 8.
EDEM1 involvement in ERAD of a truncated Igγ heavy chain. A, HEK 293 cells transiently coexpressing HA-tagged truncated Igγ heavy chain (γ V-CH1-HA) with either GFP (lane 1), anti-EDEM1 shRNA (lane 2), EDEM1-HA (lane 3), or a mock-transfected control (lane 4) were lysed in Nonidet P-40 buffer (“Experimental Procedures”)); γ V-CH1-HA and EDEM1-HA were immunoprecipitated from 90% of the cell lysates with anti-HA. The immunoprecipitates and the remaining 10% of the lysates were run on SDS-PAGE and immunoblotted with anti-HA (upper and middle panels) and anti-GAPDH (bottom panel). Values at the bottom show quantitation of γ V-CH1-HA levels relative to the sample without EDEM1 overexpression or knockdown (lane 1). B, similar to A but with expression of γ V-CH1-HA and EDEM1-HA (transfected with only 4 μg of plasmid instead of 15 μg present in A, to reduce the accelerated γ V-CH1-HA degradation), with controls missing either vector; 90% of the lysates were immunoprecipitated with rabbit anti-EDEM1 and protein A-Sepharose. The immunoprecipitates and the rest of the lysates were run on SDS-PAGE and immunoblotted with anti-HA for the detection of γ V-CH1-HA (upper and middle panels). The lysates were also immunoblotted with anti-EDEM1 (bottom panel). C, similar to A but with expression of EDEM1ΔCRD-HA instead of EDEM1-HA.
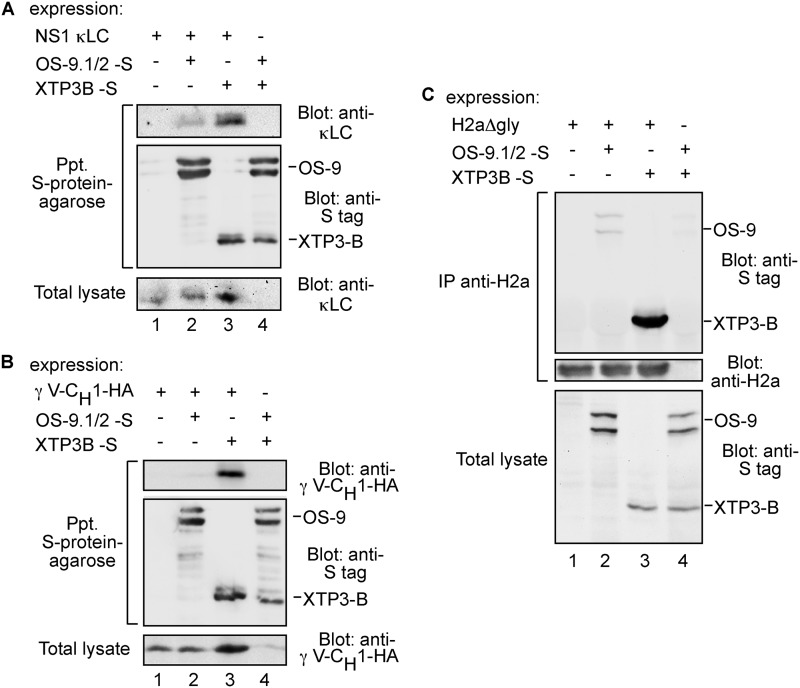
Nonglycosylated Substrates Associate with XTP3-B and to a Lesser Extent with OS-9
We then explored whether the nonglycosylated substrates associate with the lectins OS-9 and its functional homolog XTP3-B. Coimmunoprecipitation experiments showed specific and significant interaction of both NS-1 κLC and HA-γ V-CH1 with XTP3-B (Fig. 9, A and B). However, OS-9 associated only weakly with NS-1 κLC, and almost no interaction could be detected with HA-γ V-CH1 (Fig. 9, A and B). Similarly, H2aΔgly associated strongly with XTP3-B, and in a much weaker fashion with OS-9 (Fig. 9C).
FIGURE 9.
Association of XTP3-B and OS-9 with nonglycosylated proteins. A, HEK 293 cells coexpressing NS-1 κLC and either S-tagged XTP3-B or a mixture of S-tagged OS-9.1 and OS-9.2 (OS-9.1/2-S) or a control without NS-1 κLC (lane 4) were lysed in buffer containing 1% Triton X-100 and 0.5% sodium deoxycholate. S-tagged XTP3-B and OS-9.1/2 were affinity-precipitated from 80% of the cell lysates using S-protein-agarose, and the precipitates were separated in 12% SDS-PAGE and immunoblotted with anti-κLC (top panel) and anti-S-tag antibodies (middle panel). The rest of the lysates were run on 12% SDS-PAGE and immunoblotted with anti-κLC (bottom panel). B, similar to A but with γ V-CH1-HA instead of κLC. Cells were lysed in Nonidet P-40 buffer and immunoblotting was with anti-HA. C, similar to B but with H2aΔgly. In this case H2aΔgly was immunoprecipitated (IP) from 80% of the cell lysates with anti-H2a antibody. Immunoprecipitates were separated in SDS-PAGE and immunoblotted with anti-S-tag and anti-H2a antibodies. The rest of the lysates were run on SDS-PAGE and immunoblotted with anti-S-tag (bottom panel).
DISCUSSION
Our results suggest that nonglycosylated misfolded secretory proteins are targeted by much of the so-called glycoprotein ER quality control and ERAD machinery. Indeed, calnexin has previously been implicated in the quality control of some nonglycosylated substrate proteins (36, 37). Here, we show that EDEM1 can interact in a glycan-independent manner with misfolded nonglycosylated H2aΔgly, which associates with both calnexin and BiP. In addition, we show that EDEM1 can bind to two nonglycosylated proteins, NS-1 κLC and γ V-CH1, which associate with BiP instead of calnexin (15, 16). This is consistent with a suggested chaperone function of EDEM1 (38, 39), which could bind to the polypeptide moiety of misfolded glycoproteins (26). A possibility that should be evaluated in future studies is that the association of EDEM1 with the nonglycosylated substrates might occur through interaction of EDEM1 with ERdj5 (40), which also binds to BiP.
EDEM1 is involved in trimming of mannose residues from precursor N-glycans during glycoprotein ER quality control (26, 39, 41). However, this trimming is not necessary for EDEM1 binding to its substrates when EDEM1 is exogenously overexpressed (13, 42) or when EDEM1 is up-regulated during the unfolded protein response (26).
EDEM1 had been implicated in the targeting of the nonglycoprotein ricin toxin for retrotranslocation (43), although this toxin is not an ERAD substrate. Here, we show for a nonglycosylated mutant of a glycoprotein ERAD substrate and also for bona fide nonglycoprotein ERAD substrates that their degradation is dependent on EDEM1. In keeping with this, the interaction and targeting of H2aΔgly for degradation by EDEM1 does not require EDEM1's carbohydrate recognition (mannosidase-like) domain (Fig. 5). A recent study of EDEM1 association with tyrosinase showed that the N terminus of EDEM1 interacts with this substrate (44). Consistently, C-terminal EDEM1 fragments showed almost no interaction with H2a or H2aΔgly (Fig. 5B). Interestingly, the N-terminal portion (after the signal peptide) is absent in the EDEM1 homolog in Saccharomyces cerevisiae Mnl1 (Htm1). Consistent with this difference, nonglycosylated yeast ERAD substrates do not seem to be targeted by Mnl1 (45–47).
Other components of “glycoprotein ERAD” that target nonglycosylated proteins for degradation are the lectins OS-9 and XTP3-B. We detected significant binding of XTP3-B but weak association of OS-9 with H2aΔgly, NS-1 κLC, and γ V-CH1. It is unclear if XTP3-B binds directly to these substrates independently of its lectin activity or if it coprecipitates as part of a complex.
Similar to the nonglycosylated substrates that we have studied, nonglycosylated mutant amyloidogenic transthyretin was previously found to associate with XTP3-B but not OS-9 (27). However, a nonglycosylated mutant of another glycoprotein substrate, α1-antitrypsin NHK associated with both lectins (48, 49), suggesting that these two lectins may have different sets of preferred substrates. In S. cerevisiae, Yos9 was shown to be involved in the nonglycosylated protein ERAD (50, 51), underscoring the nonglycoprotein binding potential for a number of these components that are often considered to be specific for the turnover of glycosylated proteins.
The concentration of NS-1 κLC and H2aΔgly in the ERQC and relative depletion of BiP from this compartment upon proteasomal inhibition (Figs. 4 and 7) suggest that at advanced stages of targeting to ERAD, these substrates might dissociate from BiP and bind to EDEM1 and/or XTP3-B. Indeed, there was a relative decrease in the association of NS-1 κLC with BiP upon proteasomal inhibition (Fig. 6E). However, the effect on the association was not significant. This suggests that BiP is in large excess of the substrate, and upon proteasomal inhibition, BiP molecules in the peripheral ER not bound to NS-1 κLC would far exceed the amount of BiP in association with NS-1 κLC in the ERQC. This is consistent with a model that we had proposed, where binding of misfolded proteins to the relatively low amounts of BiP present in the ERQC would facilitate its displacement from the UPR sensors IRE1 and PERK, which accumulate in this compartment, leading to their activation (1, 28).
The F-box proteins Fbs2 and the similar but neural-specific Fbs1 are lectin subunits of the Skp, Cullin, F-box containing (SCF) E3 ligase, which target high mannose glycans of misfolded glycoproteins once they reach the cytosol (14). Their role is to possibly prevent premature deglycosylation of the substrates by peptide:N-glycosidase before ubiquitination (52). They do not seem to target a nonglycosylated protein in vitro (14) nor in cells in vivo as we observed here (Fig. 3). A deubiquitination step was reported as a requirement for dislocation of a substrate to the cytosol, which would imply a second ubiquitination step for the substrate to be recognized by the proteasome (53). We can speculate that this second ubiquitination step might be provided by SCFFbs2 for glycoproteins, and in the case of nonglycosylated substrates by E3 ligases involved in ubiquitination of cytosolic nonglycosylated proteins. Except for this cytosolic step in the targeting of glycoprotein ERAD substrates to the proteasomes, glycosylated and nonglycosylated substrates seem to use a similar luminal and membrane ERAD machinery. As most secretory proteins are glycosylated, with nonglycosylated proteins being a minority, it stands to reason that they would utilize at least some components of a shared machinery. It is important to emphasize that this machinery is used not only in the case of glycoprotein substrates with their glycosylation sites artificially abrogated but also by a naturally nonglycosylated ERAD substrate, NS-1 κLC.
Acknowledgment
We thank Maya Mouler Rechtman for technical assistance at the initial stages of the work.
This work was supported in part by Grant 1070/10 from the Israel Science Foundation and German-Israeli Project Cooperation (Deutsch-Israelische Projektkooperation K 5-1).
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic reticulum-associated degradation
- ERQC
- ER-derived quality control compartment
- EDEM1
- ER degradation-enhancing mannose-like protein 1
- Lac
- lactacystin
- ASGPR
- asialoglycoprotein receptor
- CNX
- calnexin.
REFERENCES
- 1. Benyair R., Ron E., Lederkremer G. Z. (2011) Protein quality control, retention, and degradation at the endoplasmic reticulum. Int. Rev. Cell Mol. Biol. 292, 197–280 [DOI] [PubMed] [Google Scholar]
- 2. Lederkremer G. Z. (2009) Glycoprotein folding, quality control, and ER-associated degradation. Curr. Opin. Struct. Biol. 19, 515–523 [DOI] [PubMed] [Google Scholar]
- 3. Brodsky J. L., Skach W. R. (2011) Protein folding and quality control in the endoplasmic reticulum. Recent lessons from yeast and mammalian cell systems. Curr. Opin. Cell Biol. 23, 464–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernasconi R., Molinari M. (2011) ERAD and ERAD tuning: disposal of cargo and of ERAD regulators from the mammalian ER. Curr. Opin. Cell Biol. 23, 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith M. H., Ploegh H. L., Weissman J. S. (2011) Road to ruin. Targeting proteins for degradation in the endoplasmic reticulum. Science 334, 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. D'Alessio C., Caramelo J. J., Parodi A. J. (2010) UDP-Glc:glycoprotein glucosyltransferase-glucosidase II, the ying-yang of the ER quality control. Semin. Cell Dev. Biol. 21, 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Määttänen P., Gehring K., Bergeron J. J., Thomas D. Y. (2010) Protein quality control in the ER. The recognition of misfolded proteins. Semin. Cell Dev. Biol. 21, 500–511 [DOI] [PubMed] [Google Scholar]
- 8. Frenkel Z., Gregory W., Kornfeld S., Lederkremer G. Z. (2003) Endoplasmic reticulum-associated degradation of mammalian glycoproteins involves sugar chain trimming to Man6–5GlcNAc2. J. Biol. Chem. 278, 34119–34124 [DOI] [PubMed] [Google Scholar]
- 9. Lederkremer G. Z., Glickman M. H. (2005) A window of opportunity. Timing protein degradation by trimming of sugars and ubiquitins. Trends Biochem. Sci. 30, 297–303 [DOI] [PubMed] [Google Scholar]
- 10. Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) N-Glycan structures. Recognition and processing in the ER. Trends Biochem. Sci. 35, 74–82 [DOI] [PubMed] [Google Scholar]
- 11. Hebert D. N., Bernasconi R., Molinari M. (2010) ERAD substrates. Which way out? Semin. Cell Dev. Biol. 21, 526–532 [DOI] [PubMed] [Google Scholar]
- 12. Kamhi-Nesher S., Shenkman M., Tolchinsky S., Fromm S. V., Ehrlich R., Lederkremer G. Z. (2001) A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell 12, 1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Groisman B., Shenkman M., Ron E., Lederkremer G. Z. (2011) Mannose trimming is required for delivery of a glycoprotein from EDEM1 to XTP3-B and to late endoplasmic reticulum-associated degradation steps. J. Biol. Chem. 286, 1292–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoshida Y., Adachi E., Fukiya K., Iwai K., Tanaka K. (2005) Glycoprotein-specific ubiquitin ligases recognize N-glycans in unfolded substrates. EMBO Rep. 6, 239–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma J., Kearney J. F., Hendershot L. M. (1990) Association of transport-defective light chains with immunoglobulin heavy chain-binding protein. Mol. Immunol. 27, 623–630 [DOI] [PubMed] [Google Scholar]
- 16. Skowronek M. H., Hendershot L. M., Haas I. G. (1998) The variable domain of nonassembled Ig light chains determines both their half-life and binding to the chaperone BiP. Proc. Natl. Acad. Sci. U.S.A. 95, 1574–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Okuda-Shimizu Y., Hendershot L. M. (2007) Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol. Cell 28, 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Olivari S., Molinari M. (2007) Glycoprotein folding and the role of EDEM1, EDEM2, and EDEM3 in degradation of folding-defective glycoproteins. FEBS Lett. 581, 3658–3664 [DOI] [PubMed] [Google Scholar]
- 19. Tamura T., Cormier J. H., Hebert D. N. (2011) Characterization of early EDEM1 protein maturation events and their functional implications. J. Biol. Chem. 286, 24906–24915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tolchinsky S., Yuk M. H., Ayalon M., Lodish H. F., Lederkremer G. Z. (1996) Membrane-bound versus secreted forms of human asialoglycoprotein receptor subunits. Role of a juxtamembrane pentapeptide. J. Biol. Chem. 271, 14496–14503 [DOI] [PubMed] [Google Scholar]
- 21. Shenkman M., Ayalon M., Lederkremer G. Z. (1997) Endoplasmic reticulum quality control of asialoglycoprotein receptor H2a involves a determinant for retention and not retrieval. Proc. Natl. Acad. Sci. U.S.A. 94, 11363–11368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chillarón J., Haas I. G. (2000) Dissociation from BiP and retrotranslocation of unassembled immunoglobulin light chains are tightly coupled to proteasome activity. Mol. Biol. Cell 11, 217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee Y. K., Brewer J. W., Hellman R., Hendershot L. M. (1999) BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol. Biol. Cell 10, 2209–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Groisman B., Avezov E., Lederkremer G. Z. (2006) The E3 ubiquitin ligases HRD1 and SCFFbs2 recognize the protein moiety and sugar chains, respectively, of an ER-associated degradation substrate. Isr. J. Chem. 46, 189–196 [Google Scholar]
- 25. Avezov E., Frenkel Z., Ehrlich M., Herscovics A., Lederkremer G. Z. (2008) Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5–6GlcNAc2 in glycoprotein ER-associated degradation. Mol. Biol. Cell 19, 216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ron E., Shenkman M., Groisman B., Izenshtein Y., Leitman J., Lederkremer G. Z. (2011) Bypass of glycan-dependent glycoprotein delivery to ERAD by up-regulated EDEM1. Mol. Biol. Cell 22, 3945–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. (2008) OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kondratyev M., Avezov E., Shenkman M., Groisman B., Lederkremer G. Z. (2007) PERK-dependent compartmentalization of ERAD and unfolded protein response machineries during ER stress. Exp. Cell Res. 313, 3395–3407 [DOI] [PubMed] [Google Scholar]
- 29. Shenkman M., Ehrlich M., Lederkremer G. Z. (2000) Masking of an endoplasmic reticulum retention signal by its presence in the two subunits of the asialoglycoprotein receptor. J. Biol. Chem. 275, 2845–2851 [DOI] [PubMed] [Google Scholar]
- 30. Benyair R., Kondratyev M., Veselkin E., Tolchinsky S., Shenkman M., Lurie Y., Lederkremer G. Z. (2011) Constant serum levels of secreted asialoglycoprotein receptor sH2a and decrease with cirrhosis. World J. Gastroenterol. 17, 5305–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hendershot L. M., Wei J. Y., Gaut J. R., Lawson B., Freiden P. J., Murti K. G. (1995) In vivo expression of mammalian BiP ATPase mutants causes disruption of the endoplasmic reticulum. Mol. Biol. Cell 6, 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frenkel Z., Shenkman M., Kondratyev M., Lederkremer G. Z. (2004) Separate roles and different routing of calnexin and ERp57 in endoplasmic reticulum quality control revealed by interactions with asialoglycoprotein receptor chains. Mol. Biol. Cell 15, 2133–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oda Y., Hosokawa N., Wada I., Nagata K. (2003) EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 299, 1394–1397 [DOI] [PubMed] [Google Scholar]
- 34. Hagiwara M., Maegawa K., Suzuki M., Ushioda R., Araki K., Matsumoto Y., Hoseki J., Nagata K., Inaba K. (2011) Structural basis of an ERAD pathway mediated by the ER-resident protein-disulfide reductase ERdj5. Mol. Cell 41, 432–444 [DOI] [PubMed] [Google Scholar]
- 35. Yoshida Y., Tokunaga F., Chiba T., Iwai K., Tanaka K., Tai T. (2003) Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. J. Biol. Chem. 278, 43877–43884 [DOI] [PubMed] [Google Scholar]
- 36. Swanton E., High S., Woodman P. (2003) Role of calnexin in the glycan-independent quality control of proteolipid protein. EMBO J. 22, 2948–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leach M. R., Williams D. B. (2004) Lectin-deficient calnexin is capable of binding class I histocompatibility molecules in vivo and preventing their degradation. J. Biol. Chem. 279, 9072–9079 [DOI] [PubMed] [Google Scholar]
- 38. Hosokawa N., Wada I., Natsuka Y., Nagata K. (2006) EDEM accelerates ERAD by preventing aberrant dimer formation of misfolded α1-antitrypsin. Genes Cells 11, 465–476 [DOI] [PubMed] [Google Scholar]
- 39. Olivari S., Cali T., Salo K. E., Paganetti P., Ruddock L. W., Molinari M. (2006) EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun. 349, 1278–1284 [DOI] [PubMed] [Google Scholar]
- 40. Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., Nagata K. (2008) ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321, 569–572 [DOI] [PubMed] [Google Scholar]
- 41. Hosokawa N., Tremblay L. O., Sleno B., Kamiya Y., Wada I., Nagata K., Kato K., Herscovics A. (2010) EDEM1 accelerates the trimming of α1,2-linked mannose on the C branch of N-glycans. Glycobiology 20, 567–575 [DOI] [PubMed] [Google Scholar]
- 42. Cormier J. H., Tamura T., Sunryd J. C., Hebert D. N. (2009) EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Mol. Cell 34, 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Slominska-Wojewodzka M., Gregers T. F., Wälchli S., Sandvig K. (2006) EDEM is involved in retrotranslocation of ricin from the endoplasmic reticulum to the cytosol. Mol. Biol. Cell 17, 1664–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marin M. B., Ghenea S., Spiridon L. N., Chiritoiu G. N., Petrescu A. J., Petrescu S. M. (2012) Tyrosinase degradation is prevented when EDEM1 lacks the intrinsically disordered region. PLoS ONE 7, e42998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakatsukasa K., Nishikawa S., Hosokawa N., Nagata K., Endo T. (2001) Mnl1p, an α-mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J. Biol. Chem. 276, 8635–8638 [DOI] [PubMed] [Google Scholar]
- 46. Jakob C. A., Bodmer D., Spirig U., Battig P., Marcil A., Dignard D., Bergeron J. J., Thomas D. Y., Aebi M. (2001) Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kostova Z., Wolf D. H. (2005) Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J. Cell Sci. 118, 1485–1492 [DOI] [PubMed] [Google Scholar]
- 48. Bernasconi R., Pertel T., Luban J., Molinari M. (2008) A dual task for the Xbp1-responsive OS-9 variants in the mammalian endoplasmic reticulum. Inhibiting secretion of misfolded protein conformers and enhancing their disposal. J. Biol. Chem. 283, 16446–16454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. (2008) Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 283, 20914–20924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Izawa T., Nagai H., Endo T., Nishikawa S. (2012) Yos9p and Hrd1p mediate ER retention of misfolded proteins for ER-associated degradation. Mol. Biol. Cell 23, 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jaenicke L. A., Brendebach H., Selbach M., Hirsch C. (2011) Yos9p assists in the degradation of certain nonglycosylated proteins from the endoplasmic reticulum. Mol. Biol. Cell 22, 2937–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamaguchi Y., Hirao T., Sakata E., Kamiya Y., Kurimoto E., Yoshida Y., Suzuki T., Tanaka K., Kato K. (2007) Fbs1 protects the malfolded glycoproteins from the attack of peptide:N-glycanase. Biochem. Biophys. Res. Commun. 362, 712–716 [DOI] [PubMed] [Google Scholar]
- 53. Claessen J. H., Kundrat L., Ploegh H. L. (2012) Protein quality control in the ER. Balancing the ubiquitin checkbook. Trends Cell Biol. 22, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]