Background: Cruciferin represents the most abundant protein in Brassica napus seeds where its efficient storage is essential under minimized space conditions.
Results: The cruciferin complex has an octameric barrel-like structure of ∼420 kDa.
Conclusion: The barrel-like structure represents a compact building block optimized for maximal storage of amino acids.
Significance: Novel insights into structure and packing of seed storage proteins.
Keywords: Gel Electrophoresis, Plant Physiology, Protein Complexes, Protein Structure, Subcellular Organelles, Blue Native PAGE, Oil Seed Proteomics, Plant Protein Storage Vacuoles
Abstract
Globulins are an important group of seed storage proteins in dicotyledonous plants. They are synthesized during seed development, assembled into very compact protein complexes, and finally stored in protein storage vacuoles (PSVs). Here, we report a proteomic investigation on the native composition and structure of cruciferin, the 12 S globulin of Brassica napus. PSVs were directly purified from mature seeds by differential centrifugations. Upon analyses by blue native (BN) PAGE, two major types of cruciferin complexes of ∼ 300–390 kDa and of ∼470 kDa are resolved. Analyses by two-dimensional BN/SDS-PAGE revealed that both types of complexes are composed of several copies of the cruciferin α and β polypeptide chains, which are present in various isoforms. Protein analyses by two-dimensional isoelectric focusing (IEF)/SDS-PAGE not only revealed different α and β isoforms but also several further versions of the two polypeptide chains that most likely differ with respect to posttranslational modifications. Overall, more than 30 distinct forms of cruciferin were identified by mass spectrometry. To obtain insights into the structure of the cruciferin holocomplex, a native PSV fraction was analyzed by single particle electron microscopy. More than 20,000 images were collected, classified, and used for the calculation of detailed projection maps of the complex. In contrast to previous reports on globulin structure in other plant species, the cruciferin complex of Brassica napus has an octameric barrel-like structure, which represents a very compact building block optimized for maximal storage of amino acids within minimal space.
Introduction
Seed storage proteins accumulate during the seed filling process and serve as a source of nitrogen and amino acids for the germinating embryo. In dicotyledonous plants, e.g. sunflower, soybean, Arabidopsis, and rapeseed, the most abundant storage proteins are the 2 S albumins and 7 S and 12 S globulins, the latter ones representing the phylogenetically most widely distributed group of storage proteins. Mature 12S globulins consist of two polypeptide chains, which are covalently linked by a disulfide bond. Both chains stem from the same precursor molecule, which is co-translationally synthesized into the rough endoplasmic reticulum. During import, the endoplasmic reticulum signal peptide is cleaved off and a disulfide bond is formed between the N- and C-terminal parts of the polypeptide chain (1, 2). These proglobulins are then assembled into trimers (3), transported through the Golgi apparatus, and finally stored as mature hexamers in protein storage vacuoles (PSVs).4 In this process, at least a second cleavage of the proglobulin occurs, which forms the mature protein with α and β polypeptide chains. The latter step is a prerequisite for hexamer assembly (4, 5). The site of this second cleavage is between an asparagine and glycine residue and both amino acids are highly conserved among the group of the 12 S globulins (5, 6). Several proteases, so-called vacuolar processing enzymes, which belong to a novel family of cysteine proteinases, are responsible for this second cleavage of the monomers (7). Recently, it was shown that vacuolar processing enzymes as well as the corresponding storage proteins are sorted in the Golgi apparatus simultaneously in two different types of vesicles. Subsequently, these vesicles fuse to form a prevacuolar compartment, the multivesicular body in which the proteolytic process takes place (8, 9). Later, multivesicular bodies fuse into a PSV. Most members of the 12 S globulins are believed to undergo these two events of posttranslational cleavage (4, 6, 7), and the resulting polypeptide chains are referred to as the α polypeptide chain (the original N-terminal part of the precursor protein) and the β polypeptide chain (the original C-terminal part).
Also in Brassica napus, a 12 S globulin, cruciferin, is the major storage protein and accounts for ∼60% of the total seed protein (10). Five different B. napus cruciferins belong to three different families, which are listed in the reviewed UniProtKB database. The P1 family contains only one protein, CRU1, with a sequence length of 509 amino acids (AA) (α1 = 296 AA and β1 = 190 AA). In contrast, the P2 family contains three proteins: BnC1 with a sequence length of 490 AA (α2 = 277 AA and β2 = 190 AA); BnC2 with a sequence length of 496 AA (α3 = 283 AA and β3 = 190 AA) and CRU2/3 with a sequence length of 488 AA (α2/3 = 275 and β2/3 = 190 AA). The P3 family contains one protein, CRU4 with a sequence length of 465 AA (α4 = 254 AA and β4 = 189 AA). Similar to other 12S globulins, all cruciferin family members are secreted into the endoplasmic reticulum, and all exhibit the typical conserved 12S globulin cleavage site between asparagine and glycine. They are thought to be transported as trimers via the Golgi apparatus and stored as hexamers in PSVs (11–15).
Currently, little is known about the structure and organization of B. napus cruciferin within the PSV, although this most likely is important for ensuring efficient use of storage space in developing seeds. The first reports on the quaternary structure of a 12S globulin were given by Badley et al. (16) in 1975. Using electron microscopy, it was shown that the soybean 12S globulin glycinin extracted from soy flour is composed of two hexagonal rings with alternating α and β polypeptide chains. This dodecameric model was also supported by Marcone et al. (17) for amaranth globulin isolated from defatted flour. However, for rapeseed and sunflower globulin a triagonal antiprism of six subunits was suggested (18). In subsequent x-ray crystallographic studies, soybean proglycinin and rapeseed procruciferin overexpressed in Escherichia coli were reported to exhibit triangular patterns (19–21). The crystal structure of a mature soybean glycinin extracted from a mutant soybean cultivar indicates a hexamic particle formed by two face-to-face stacking trimers (22). Two conclusions can be drawn from these results: (i) quaternary structures of 12S globulins may vary between species, and (ii) the quaternary structure of 12S globulins also can differ for the same species, depending on the expression system used for globulin biosynthesis (e.g. soybean versus E. coli). Consequently, detailed structural information of native cruciferin is best obtained directly from the plant species of interest without the use of heterologous expression systems.
In this study, B. napus cruciferin complexes were directly isolated from mature seeds under native conditions. The resulting fractions were highly enriched with cruciferin. Purified proteins were used for investigations by various gel electrophoresis systems, mass spectrometry and single particle EM. A large variation with respect to isoelectric points and molecular mass was observed for the cruciferin α and β polypeptide chains. In contrast to 12S globulin complexes from other plant species, the cruciferin complex exhibits a unique octameric structure that allows packaging of the amino acids under most space-saving conditions.
EXPERIMENTAL PROCEDURES
Cultivation of B. napus
B. napus plants were cultivated in growth chambers under the following conditions: 16-h light (16 klux) at 22 °C, 8-h dark at 18 °C with a relative humidity of 55%. Seeds were harvested 53 to 60 days after pollination (DAP) and directly used for the isolation of PSVs.
Isolation of Protein Storage Vacuoles
Native proteins from PSVs were isolated as described previously (23). All steps were carried out at 4 °C or on ice. 14 g of mature seeds were harvested and directly homogenized in 100 ml of glycerol (100% (v/v)) using a mortar and a pestle. The homogenate was filtered through one layer of Miracloth (20–25 μm, Calbiochem) and centrifuged for 10 min at 1100 × g. To collect PSVs, the supernatant was centrifuged for 20 min at 41,000 × g. The pellet was again resuspended in glycerol (30 ml, 100% (v/v)) and centrifuged for 20 min at 41,000 × g. This pellet contained the PSV fraction and was further resuspended in 10 ml of TE buffer (1 mm EDTA, 5 mm Tris-HCl, pH 8.5) to disrupt the PSV and loaded onto a discontinuous sucrose step gradient (30, 45, and 68% (w/v) sucrose in TE buffer) for 2 h at 78,000 × g. The fraction above the 30% sucrose layer represents the vacuolar matrix containing the storage proteins and was directly used or frozen in liquid nitrogen and stored at −80 °C.
Gel Electrophoresis Procedures
Proteins were analyzed by one-dimensional blue native polyacrylamide gel electrophoresis (BN PAGE) and by two-dimensional BN/SDS-PAGE using 100 μl of the frozen PSV matrix fraction supplemented with 5 μl of “blue loading buffer” (5% (w/v) Coomassie Blue in 750 mm aminocaproic acid) according to the protocol outlined in Wittig et al. (24). Mitochondria from Arabidopsis thaliana were prepared as outlined in Sunderhaus et al. (25) and solubilized by 5 g/g digitonin. One-dimensional SDS-PAGE was carried out as described by Schägger and von Jagow (26) using 30 μl of the frozen PSV matrix fraction. For two-dimensional isoelectric focusing/SDS-PAGE (IEF/SDS-PAGE), 50 μl of the PSV fraction were mixed with 300 μl of resuspension buffer (8 m urea, 2 m thiourea, 50 mm DTT, 2% CHAPS (w/v), 5% IPG buffer 3–11 nl (v/v), 12 μl/ml DeStreak reagent, bromphenol blue) and directly applied into a strip holder. Isoelectric focusing was carried out with the Ettan IPGphor 3 apparatus (GE Healthcare) using 3–11 nl of Immobiline DryStrip gels (18 cm). Rehydration took place at 30 V for 12 h and focusing during 4 steps at 500 V (1 h), 500–1000 V (1 h), 1000–8000 V (3 h), and 8000 V (6 h). Afterward, strips were equilibrated for 15 min in equilibration solution (6 m urea, 30% glycerol (87%, v/v), 2% SDS, 50 mm Tris-HCl, pH 8.8, bromphenol blue) with (i) 1% DTT (w/v) and (ii) 2.5% iodacetamide (w/v). IPG strips were finally transferred horizontally onto a 16.5% tricine gel, and electrophoresis was carried out for 20 h at 35 mA/mm gel layer.
Gel Staining Procedures
All polyacrylamide gels were stained with Coomassie colloidal (27, 28).
Generation of Antibodies against Cruciferin
Two different polyclonal antibodies directed against peptides of the α or β polypeptide chains of B. napus cruciferin were generated (Eurogentec S.A. Antisera Production, Seraing, Belgium). For details, see supplemental Fig. 1.
Western Blotting
Proteins separated on polyacrylamide gels were blotted onto nitrocellulose membranes for antibody staining using the Trans Blot Cell from Bio-Rad. Blotting was carried out as described in Kruft et al. (29). Immunostainings were performed using the VectaStain ABC kit (Vector Laboratories, Burlingame, CA).
MS Analyses
Tryptic digestion of proteins and peptide extraction were carried out as published in Sunderhaus et al. (25). MS analyses were performed using an EASY-nLC-system (Proxeon) coupled to a MicrOTOF-Q-II mass spectrometer (Bruker Daltonics). Identification of proteins was carried out using the MASCOT search algorithm against (i) SwissProt (ii) NCBI non-redundant protein database and (iii) The Arabidopsis Information Resource (Tair release 9).
Electron Microscopy and Image Analyses
Fractions containing cruciferin were applied to carbon-coated copper grids and negatively stained with 2% uranyl acetate. Electron microscopy was performed on a Philips CM120 electron microscope. Data were collected with a 4K slow-scan CCD camera (Gatan) at a magnification of 130,000 with a pixel size (after binning the images) of 0.23 nm at the specimen level. Single particle analysis was performed with the Groningen Image Processing (GRIP) software package on a CPU cluster as outlined by Dudkina et al. (30). The three-dimensional model of cruciferin was created with a Blender three-dimensional creation suite. X-ray structure of procruciferin from B. napus (Protein Data Bank code 3KGL (20)) was used for the superimposing on projection maps of cruciferin. The octamer of cruciferin was generated from the hexameric x-ray structure of procruciferin maintaining monomer-monomer interfaces by using the Chimera program from the University of California, San Francisco (31).
RESULTS
PSVs were isolated from freshly harvested seeds at 53 to 60 DAP under most gentle conditions using the glycerol protocol published by Jiang et al. (23). The total protein fraction was directly extracted from PSVs.
Characterization of Cruciferin Complexes from B. napus Storage Vacuoles
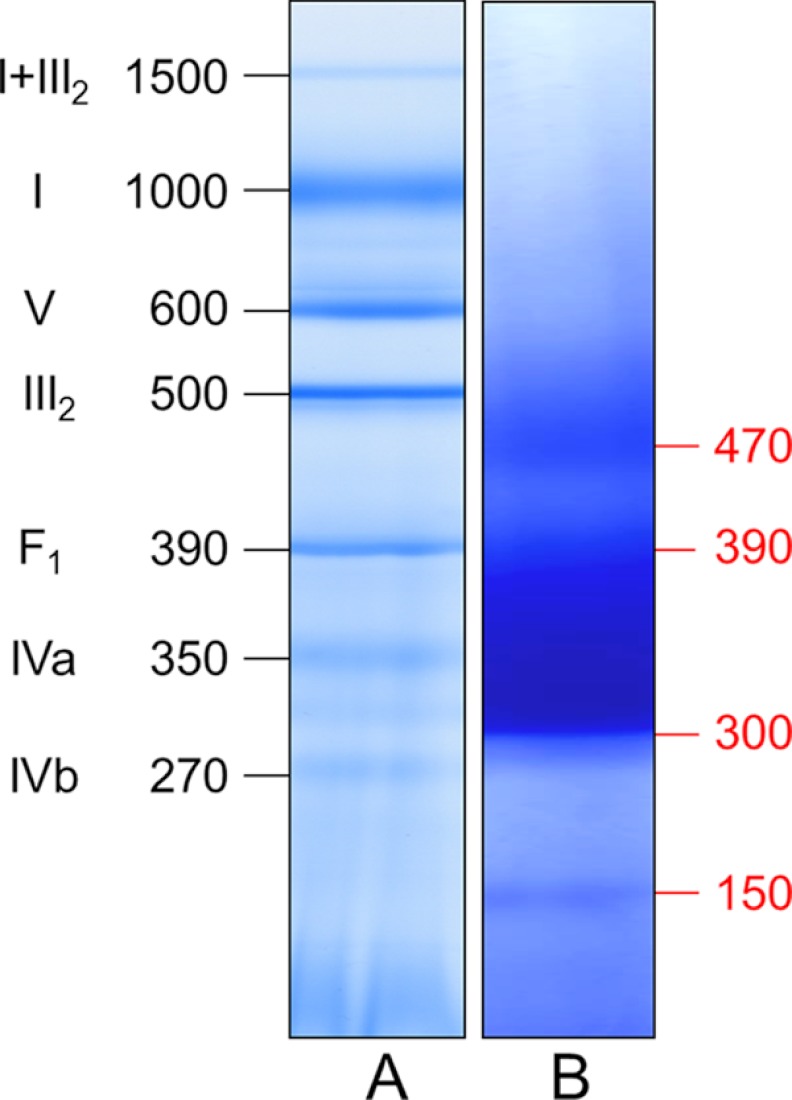
BN PAGE was originally developed for the characterization of native mitochondrial protein complexes but also proved to be a very suitable procedure for the investigation of protein complexes from other cellular compartments (24). To characterize the protein composition of the PSVs, their soluble fraction was directly supplemented with Coomassie blue and subsequently loaded onto a one-dimensional BN gel. As a control and molecular mass standard, respiratory protein complexes of A. thaliana mitochondria were solubilized by 5 g/g digitonin and separated on the same gel (Fig. 1). Three bands are visible in the lane of the PSV fraction: a central broad band sharply confined at its lower border at 300 kDa with a more diffuse upper border at ∼390 kDa is flanked by two bands of lower abundances at ∼150 and 470 kDa. Judging from the molecular masses of the bands, it can be expected that they represent multisubunit protein complexes.
FIGURE 1.
Separation of soluble proteins from PSVs by BN PAGE indicates presence of three major classes of protein complexes of 150, 300–390, and 470 kDa. The proteins of storage vacuoles of B. napus (lane B) and of a mitochondrial fraction of A. thaliana (molecular mass standard; lane A) were separated on a native gradient gel according to molecular mass. Molecular masses are given in kDa. Designations of A. thaliana mitochondrial protein complexes are given as follows: I+III2, supercomplex containing NADH dehydrogenase and dimeric cytochrome c reductase; I, NADH dehydrogenase; V, F0F1 ATP synthase; III2, dimeric cytochrome c reductase; F1, F1 part of the ATP synthase; IVa and IVb, larger and smaller form of cytochrome c oxidase.
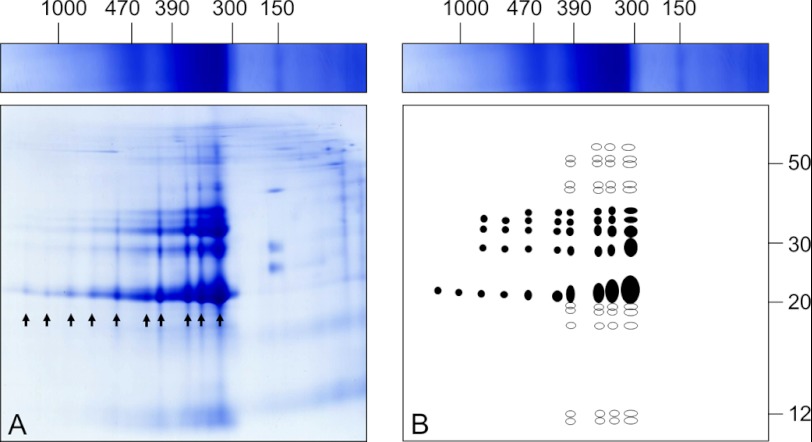
To investigate the subunit compositions of the PSV protein complexes, two-dimensional BN/SDS-PAGE was carried out for separating the subunits of the resolved complexes (Fig. 2A). Under the denaturing conditions of the second gel dimension, the smaller 150-kDa complex dissociated into three subunits of ∼40, 25, and 20 kDa. The two larger complexes dissociated into ∼15 different subunits of 60 to 12 kDa. The protein patterns of both bands indicate that the two complexes have the same composition but differ in subunit stoichiometry. In contrast to the one-dimensional BN PAGE, even larger forms of this complex are visible on the two-dimensional gel, which is schematically summarized in Fig. 2B.
FIGURE 2.
Separation of proteins isolated from PSVs by two-dimensional BN/SDS-PAGE (A) suggests specified associations of cruciferin. Arrows indicate the occurrence of 10 defined cruciferin complexes with a maximum of 15 different protein species (given on the scheme in B). Highly abundant subforms are indicated in filled circles, less abundant subforms are in open circles. Molecular masses are given in kDa above and to the right of the gel.
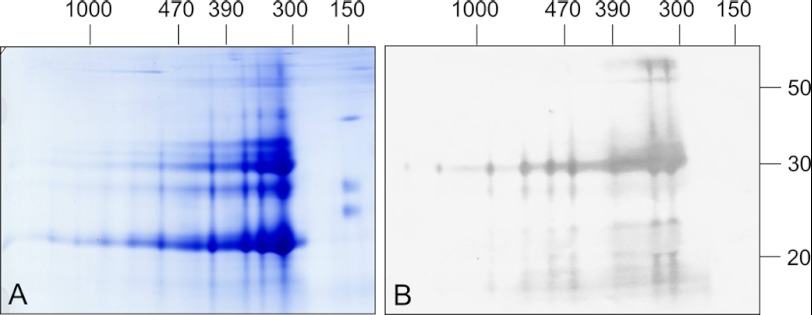
To verify that separated fractions contain cruciferin, peptide-specific antibodies were generated against the α and β polypeptide chain of the protein (see supplemental Fig. 1). Two-dimensional BN/SDS gels were then blotted onto nitrocellulose membranes and incubated with antibodies directed against both chains (Fig. 3).
FIGURE 3.
Immunoblotting analyses of a B. napus PSV fraction separated by two-dimensional BN/SDS-PAGE reveals high enrichment of cruciferin. Gels were either stained by Coomassie colloidal (A) or blotted onto nitrocellulose (B). Western blots were incubated with antibodies directed against the α chain of cruciferin. Molecular masses of standard proteins are shown above and to the right of the gel (in kDa).
The obtained results illustrate the purity of the PSV fraction. A strong signal is present in the 30-kDa range, representing α polypeptide chains from the different cruciferin families. Additionally, a strong signal is visible in the 55-kDa range indicating that few cruciferins still have an intact disulfide bond between both polypeptide chains after the β-mercaptoethanol treatment preceding the second dimension. Some weaker signals in the molecular mass range below 20 kDa most likely represent α polypeptide chain breakdown products. Blots developed with antibodies against the β polypeptide chain of cruciferin show a strong signal in the 20 to 25 kDa range as well as in the 60 kDa range (data not shown). The two-dimensional gel shows that the broad band observed in the one-dimensional gel is actually composed of several individual bands and that cruciferin is present in all those bands, also in the 470-kDa band. Additionally, cruciferin at least partially occurs in larger forms up to 1500 kDa on one-dimensional BN gels but does not form part of the 150-kDa complex.
Analyses of Cruciferin α and β Polypeptide Chains at Reducing/Non-reducing Conditions
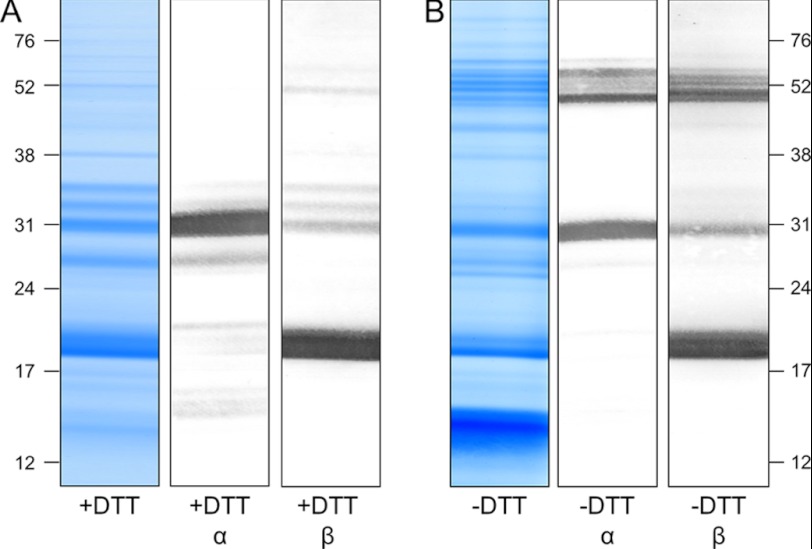
To investigate the size of the monomeric cruciferin complex as well as its α and β polypeptide chains, cruciferin isolated from PSVs was dissolved in either denaturating buffer containing DTT to reduce the disulfide bond between the N and the C terminus or in buffer without DTT to keep the protein in its native monomeric form. Fractions were separated using one-dimensional SDS gels and blotted onto nitrocellulose membranes, which were incubated with antibodies against either the α or the β chain (Fig. 4).
FIGURE 4.
Immunoblotting analyses of a B. napus PSV fraction separated by one-dimensional SDS-PAGE reveals cruciferin complexes of ∼48 to 58 kDa under non-reducing conditions. PSV fractions were separated under reducing conditions (A; +DTT) and non-reducing conditions (B; −DTT). Gels were either stained Coomassie colloidal or blotted onto nitrocellulose membranes and incubated with antibodies directed against the α or the β chains of cruciferin as indicated. Molecular masses of standard proteins are shown to the right and to the left in kDa.
Under denaturing conditions, antibodies directed against the α polypeptide chain produced a strong signal visible in the 30-kDa range. Some additional but much weaker signals are observable in the range of 27 and 33 kDa and in the low molecular mass range between 14 and 20 kDa. No signals could be detected above 35 kDa, indicating that all cruciferin is cleaved into α and β polypeptide chains by this treatment. In addition, incubation with antibodies directed against the β polypeptide chain produced a strong signal at 18 to 20 kDa as well as some minor bands in the range of 30–35 and at ∼50 kDa (Fig. 4A). Under non-reducing conditions both antibodies gave the same signals in the 27–33 kDa and 18–20 kDa range, respectively, but with notably reduced intensities. Additionally, a number of new signals are visible in the range of ∼52 kDa (Fig. 4B). Because these proteins are detected by the antibodies directed against the α and the β polypeptide chains, these bands most likely represent cruciferins with intact disulfide bonds.
Because isoforms of cruciferin precursor proteins as well as their respective α and β polypeptide chains cannot be separated on one-dimensional SDS gels due to similar molecular masses, the PSV fraction was analyzed by two-dimensional IEF/SDS-PAGE (Fig. 5). IEF is often capable of separating proteins with the same or similar molecular masses due to different occurrences of ionizable amino acids within isoforms.
FIGURE 5.
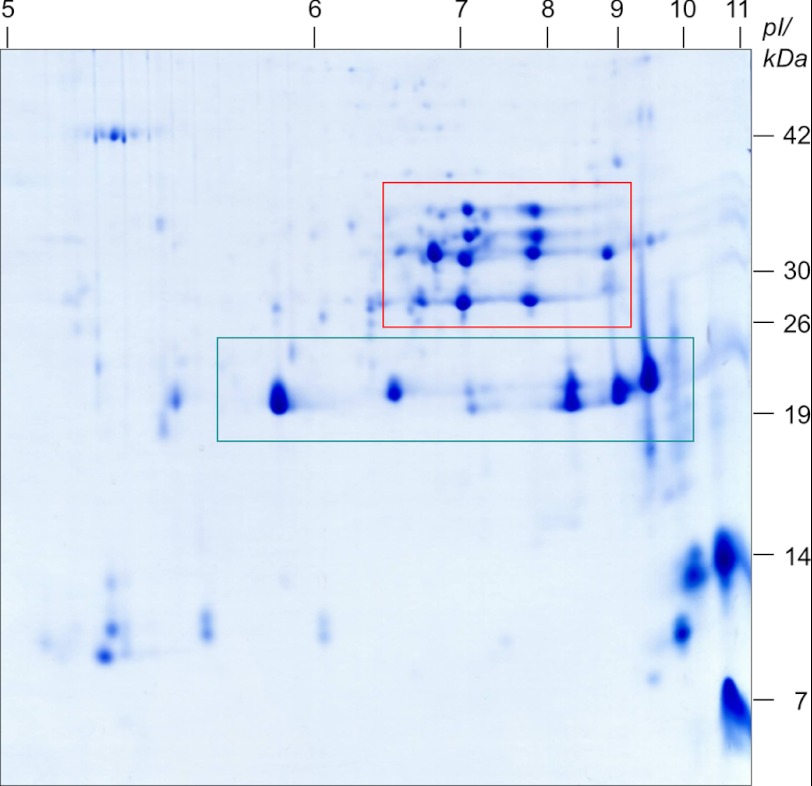
Resolving a PSV fraction according to isoelectric point and molecular mass reveals existence of cruciferin α chains in a pH range from 6. 7 to 8.8 (red box) and β-chains in a pH range from 5.9 to 9.5 (green box). Molecular masses of standard proteins as well as the pI range are given on the right and above the gel, respectively.
As shown in Fig. 5 and by Western blotting (data not shown), α polypeptide chains are separated in a range between 27 and 36 kDa and isoelectric points between 6.7 and 8.8 (red box). The corresponding β polypeptide chains are separated in a range between 19 and 21 kDa and isoelectric points between 5.9 and 9.5 (green box).
For assigning cruciferin polypeptide chains from the three individual cruciferin families to each detected spot/band on our one-dimensional BN (Fig. 1), two-dimensional BN/SDS (Fig. 3), one-dimensional SDS (Fig. 4), and two-dimensional IEF/SDS (Fig. 5) gels, protein identifications by mass spectrometry were carried out. A total number of 95 spots, representing the most abundant proteins, were picked from the four gel systems and analyzed by LC-ESI-Q-TOF-MS/MS (supplemental Fig. 2 and supplemental Table 1). Members of all five cruciferin families were detected as well as the 2 S storage protein Napin. The latter one is also known to accumulate during the seed filling process in B. napus PSVs. Furthermore, five proteins were identified, which do not belong to the family of storage proteins.
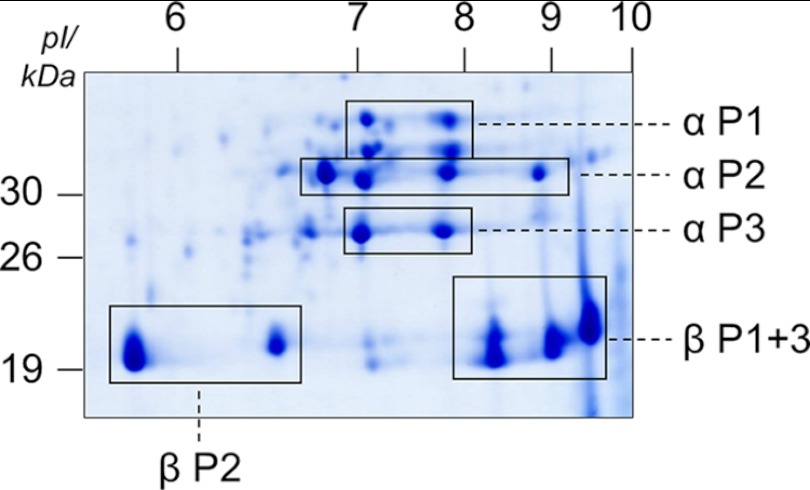
Altogether, mass spectrometry enabled the assignment of spots on two-dimensional IEF/SDS gels to distinct cruciferin families and polypeptide chains (Fig. 6). With respect to α polypeptide chains, four spots in the range of 34-kDa spots were identified as αCRU1 (P1); another four spots in the range of 32 kDa were assigned to αBnC1, αBnC2, and αCRU2/3 (P2); and two spots in the range of 27 kDa represent αCRU4 (P3). All six spots in the range of 19 to 21 kDa were distinguishable exclusively by their isoelectric points, and they all represent β polypeptide chains. Four spots exhibiting isoelectric points of ∼8 to 9.5 were identified as βCRU1 and βCRU4 (P1 and P3), and two spots exhibiting isoelectric points from ∼5.8 to 6.8 were assigned to βBnC1, βBnC2, and βCRU2/3 (P2).
FIGURE 6.
Analyses of cruciferin subunits by LC-MS/MS allows assigning clusters of spots on two-dimensional IEF/SDS gels to the three distinct cruciferin families. Names of families and polypeptide chain attribution are given to the right and below the gel. Molecular masses and pIs are given to the left and above the gel, respectively. P1, family 1 containing cruciferin Cru 1; P2, family 2 containing cruciferin BnC1, BnC2, and Cru 2/3; P3, family 3 containing cruciferin Cru 4.
The Cruciferins of Rapeseed Are Phosphorylated
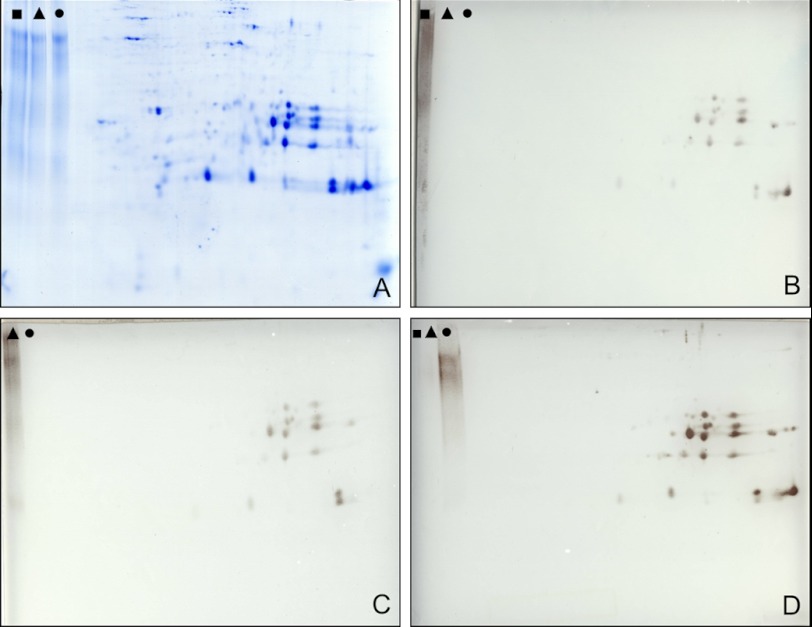
Phosphorylation of cruciferins was investigated using antibodies directed against phosphoserine, phosphothreonine, and phosphotyrosine. For this approach, storage vacuole protein fractions were separated by two-dimensional IEF/SDS-PAGE and blotted onto filter membranes (Fig. 7). Several cruciferin forms belonging to both the α and the β chains are recognized by all three sera. Overall, the α chain of cruciferin reacts stronger with the sera than the β chain. The strongest immune signals were obtained with the serum directed against phosphoserine (Fig. 7D).
FIGURE 7.
Immunological analysis of cruciferin phosphorylation. Protein storage vacuole fractions were resolved by two-dimensional IEF/SDS-PAGE and blotted onto membranes, and blots were developed using antibodies directed against three different phosphorylation sites. As a control, phosphorylated BSA (phosphorylated at tyrosine (■); phosphorylated at threonine (▴); phosphorylated at serine (●)) was separated within the same gel. A, Coomassie-stained reference gel. B, immunoblot developed with an IgG directed against phosphorylated tyrosine; C, immunoblot developed with an IgG directed against phosphorylated threonine; D, immunoblot developed with an IgG directed against phosphorylated serine. Note that the phosphotyrosine-BSA control on C is not indicated because it partially was beyond the limit of the blot. However, the specificity of the IgG was verified by independent experiments (not shown).
Structure of the Native Cruciferin Storage Form in Mature PSVs
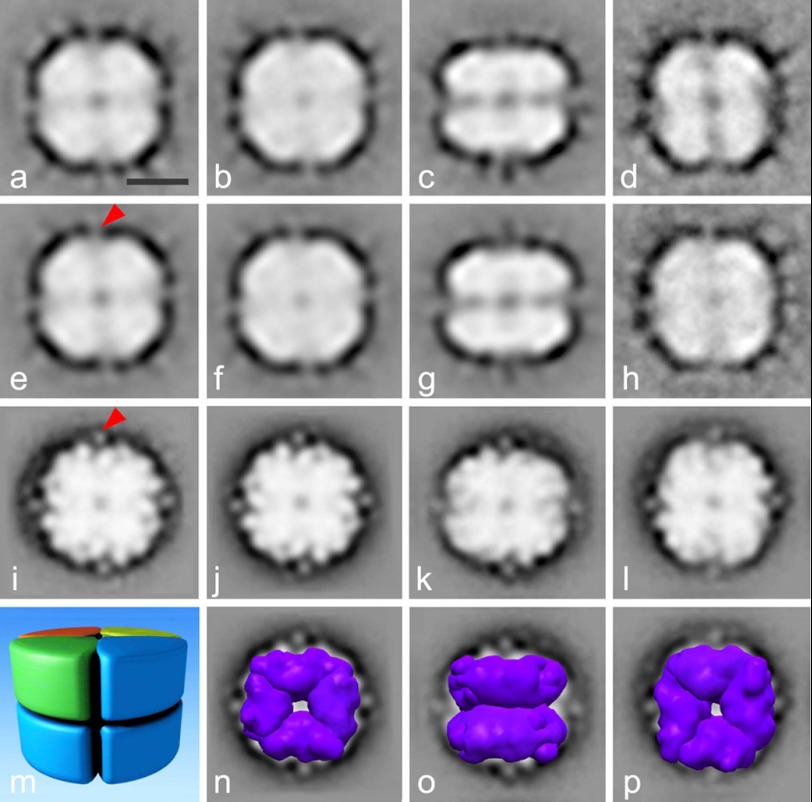
For a structural analysis of the native cruciferin oligomers, PSV fractions purified from developing (53 DAP) or mature seeds (60 DAP) were examined and analyzed by single particle EM (Fig. 8). Altogether, 23,000 protein projections were analyzed to obtain final averages at ∼20 Å resolution after several iterations of alignment and classification. As can be seen from class averages, the majority of molecules appeared to be oriented in a top or bottom view positions (Fig. 8, a, b, d, e, f, h, i, j, and l), whereas only a smaller group of projections represented side views (Fig. 8, c, g, and k). The top and bottom views clearly demonstrate a 4-fold symmetry. By combining the information obtained on the top and side views, it can be concluded that the structure is composed of two layers, each consisting of four identical subunits in a tetrameric configuration enclosing a pore in the center of the complex. A groove divides the side-view projection into two halves, each of which appears to be mirror the other one in the projection. This indicates that the two sets of tetramers are facing each other in antiparallel positions.
FIGURE 8.
Structural analyses of the cruciferin complex by single particle electron microscopy. Top view (a, b, d–f, and h) and side view (c and g) projection maps of negatively stained cruciferin complexes isolated 53 DAP and top view (i, j, l) and side view (k) projections of cruciferin isolated 60 DAP. Panels e, f, g, and j demonstrate the views of a, b, c, and i correspondingly after symmetrization. a represents a sum of 705 projections; b, 582 projections; c, 372 projections, d, 575 projections; h projections, 335 projections; i, 5000 projections; k, 5000 projections; l, 5000 projections. m shows the model of the octameric cruciferin complex based on the negatively stained projection maps. n–p demonstrate the superimposing of procruciferin density maps (purple) on projection maps from j–l correspondingly. The bar is 5 nm. The red arrows point to additional mass in every monomer in our projections, which is not visible in the monomers of the hexameric cruciferin complex solved x-ray diffraction.
From the single particle EM data shown in Fig. 8, we generated a model of the cruciferin storage complex (Fig. 8m). It is composed of eight subunits arranged in an octameric configuration rather than the hexamer, which was proposed by other studies for globulins from other organisms (11–15, 20, 22). From the dimensions observed in the projection maps, we estimated the mass of the octamer to be ∼420 kDa. Estimation was done by using a formula for the cylinder-shaped complex as m = ΠR2h/ρ, where m is the mass of the protein, R and h are the radius and the height of the cylinder, and ρ is the average protein density, which is 1.5 g/cm3 (32). Thus, each monomer, composed of an α and β polypeptide chain, should have a molecular mass of ∼50 kDa. The obtained model is in agreement with the observations from electrophoretic analyses showing average molecular masses of α and β chains of ∼32 kDa and 20 kDa, respectively. To interpret the projection maps, we generated the octamer of cruciferin from the x-ray structure of procruciferin and superimposed the density map of the octamer on our projection maps (Fig. 8, n–p). Importantly, by fitting the crystal structure, we gain a resolution by a factor of 5 (33). Based on the close fit, we conclude that the analyzed projections indeed represent the top and side views of the cruciferin complex in an octameric configuration. There is an additional mass in every monomer in our projections, seen as spikes (Fig. 8, e and i, red arrows), which do not belong to the monomers of the hexameric cruciferin complex solved by x-ray diffraction. This could be a result of incompleteness of the hexamer structure, which has missing residues as seen from the deposited atomic coordinates. Alternatively, crystallization conditions might cause changes in the protein configuration or subunit arrangements.
DISCUSSION
In this study, the structural properties of cruciferin proteins as well as the supramolecular structure of the complex formed by cruciferin subunits is investigated. In previous studies, storage protein complexes were analyzed e.g. by the expression of the proteins in a heterologous expression system, which allowed purification of the complexes to a level of purity rendering them amenable for crystallographic structural analysis. Here, we chose a different approach. Protein complexes were purified directly from freshly harvested seeds and analyzed using gel electrophoretic approaches, mass spectrometry, and single particle electron microscopy. Although the latter method is compromised in the resolution obtained at the structural level, it allows investigations of the protein from native environment. Differences in the results obtained by both approaches and their implications are discussed below.
Suitability of the Vacuolar Fraction for Biochemical and Structural Analyses
The purity of the isolated cruciferin fraction was demonstrated by the different gel electrophoresis procedures in combination with antibodies directed against cruciferin α and β polypeptide chains (Figs. 3 and 4) as well as by mass spectrometry. Results from the 95 analyzed spots/bands revealed that only five proteins do not belong to the family of storage proteins. These are as follows: glyceraldehyde-3-phosphate dehydrogenase, elongation factor 1-α, myrosinase binding protein, protein disulfide isomerase, and oleosin (supplemental Fig. 2 and supplemental Table 1). The first three proteins most likely represent cytosolic constituents, whereas the protein disulfide isomerase is reported to be an integral part of protein storage vacuoles (34). Oleosin is a component of oil bodies in B. napus seeds (35). The purity of the vacuolar fraction is additionally confirmed by electron microscopy. Negatively stained specimens were found to exclusively contain oligomeric Cruciferin, either in side or in top views. Therefore, the PSV fractions obtained are well suited for a detailed analysis of native B. napus cruciferin.
Heterogeneity of B. napus Cruciferin
The analyses of cruciferin and its polypeptide chains were carried out using different electrophoretic approaches. The results obtained by one-dimensional BN, one-dimensional SDS, two-dimensional BN/SDS, and two-dimensional IEF/SDS/PAGE, respective immunoblots, and mass spectrometry analyses suggest a strong diversity in physicochemical properties of the cruciferin polypeptide chains, proteins, and protein complexes (Table 1). In all gel systems, α polypeptide chains show molecular masses between 27 and 36 and β polypeptide chains between 19 and 21 kDa. Partially, molecular masses differ from the calculated ones in a range of 6 kDa, especially for α chains. Isoelectric points of some cruciferin polypeptides differ significantly compared with their calculated pIs in databases. In general, the β polypeptide chains show a very diverse pI range on IEF/SDS gels between 5.9 and 9.5. This is also reflected by the calculated pIs (6.1 and 8.6). Hence, β polypeptide chains can be considered to be rather basic. Surprisingly, α chains, which previously were observed to have an acidic pI, are found at higher pH values (36, 37). Their apparent isolelectric points on IEF/SDS gels are all between pH 6.8 and 8.6. This clearly indicates a neutral rather than an acidic pI (Table 1 and Fig. 6).
TABLE 1.
Characteristics of B. napus cruciferins and their α and β polypeptide chains
| No AA αa | No AA βa | No AA α+βa | MM cal α+βb | MM cal αb | MM app αc | MM cal βb | MM app βc | pI cal αb | pI app αc | pI cal βb | pI app βc | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cru 1 | 296 | 190 | 486 | 54.1 | 32.9 | 31.0–36.0 | 21.2 | 19.0–21.0 | 7.17 | 6.8–7.8 | 6.94 | 8.3–9.5 |
| BnC1 | 254 | 190 | 477 | 51.4 | 30.6 | 27.0–36.0 | 20.8 | 19.0–21.0 | 6.78 | 6.8–7.8 | 6.16 | 5.9–7.0 |
| BnC2 | 283 | 190 | 473 | 51.8 | 30.8 | 31.0–32.0 | 21.0 | 19.0–21.0 | 7.95 | 7.1–7.8 | 8.6 | 8.3–8.9 |
| Cru 2/3 | 275 | 190 | 465 | 51.3 | 30.5 | 27.0–32.0 | 20.8 | 19.0–21.0 | 6.78 | 7.1 | 6.16 | 5.9–7.0 |
| Cru 4 | 254 | 189 | 443 | 48.9 | 28.1 | 27.0–32.0 | 20.9 | 19.0–21.0 | 6.66 | 6.7–7.0 | 8.6 | 8.3–9.5 |
a Number of amino acids, excluding the signal peptide, derived from the corresponding UniProt database entry.
b Calculated isoelectric points and calculated molecular masses, excluding the signal peptide, using the ExPASy MM/pI tool.
c Apparent isoelectric points and molecular masses upon analyses by two-dimensional IEF/SDS PAGE (Fig. 5).
A certain level of heterogeneity was also observed on the level of the cruciferin holo-complex. The main band for cruciferin complexes on BN gels smeared over a range of nearly 100 kDa. This band includes protein complexes ranging from 300 to 390 kDa (Fig. 1), whereas the estimated mass of the cruciferin octamer based on EM analyses is ∼420 kDa. The difference between the estimated mass of the complex and the one observed on BN gels is most likely caused by the barrel-like structure of the octamer. Indeed, the three-dimensional structure of cruciferin is considerably compact. In contrast, the structures of the respiratory complexes of A. thaliana mitochondria are much more irregular (complex I, for instance, has an l-like shape). Because the BN gel was calibrated by the masses of the respiratory complexes, the apparent molecular mass of the cruciferin complex is very likely to be underestimated. Apart from that, calculation of the mass from the EM projections does not take into account the presence of the central pore and space between the monomers, thereby providing slight overestimation for the mass of cruciferin.
Phosphorylation of Cruciferin
To explain pI variations on two-dimensional IEF/SDS gels, an immune blotting approach was carried out (Fig. 7). Western blots developed with antibodies directed against phosphoserine, phosphothreonine, and phosphotyrosine indicate stable phosphorylations on several but not all cruciferin forms separated by IEF/SDS-PAGE (Fig. 7). Similar results were previously reported for B. napus and A. thaliana (39–41). In fact, it was shown that cruciferin is one of the most phosphorylated proteins in A. thaliana seeds. In contrast, our MS analyses did not reveal direct evidence for cruciferin phosphorylation in B. napus. This negative result probably is due to the fact that phosphopeptides were not enriched during sample preparation for MS (e.g. by Immobilized metal ion affinity chromatography (IMAC) or TiO2 affinity chromatography). Most likely, phosphorylation sites in cruciferins are substoichiometrically phosphorylated. As a consequence, non-phosphorylated peptides dominated the mass spectra.
The Supramolecular Structure of Directly Isolated Cruciferin
By combining a mild purification procedure with analyses by BN PAGE and single particle EM, we were able to structurally characterize cruciferin oligomers from mature seeds. Because proteins for crystallization experiments often are produced by overexpression in heterologous systems, mainly E. coli, several factors ensuring a correct assembly may be missing, e.g. signal peptide cleavage, the proteolytic cleavage of the α and β polypeptide chains and their assembly in the endoplasmic reticulum as well as the following formation into oligomers. Furthermore, many results on seed storage proteins are obtained using defatted flours for industrial purposes or NaCl extraction, which may strongly disturb the native conformation of the proteins (17, 18, 42–44). Therefore, further analyses should be carried out to verify hexameric globulin structure in seeds as reported before for other plants. Finally, globulin complexes from additional species should be characterized to obtain more general insights into their oligomeric organization.
Acknowledgment
We thank Dr. Holger Eubel for critically reading the manuscript.

This article contains supplemental Table 1 and Figs. 1 and 2.
- PSV
- protein storage vacuole
- BN
- blue native
- AA
- amino acid(s)
- DAP
- days after pollination
- BN PAGE
- blue native PAGE
- IEF
- isoelectric focusing.
REFERENCES
- 1. Ereken-Tumer N., Richter J. D., Nielsen N. C. (1982) Structural characterization of the glycinin precursors. J. Biol. Chem. 257, 4016–4018 [PubMed] [Google Scholar]
- 2. Sengupta C., Deluca V., Bailey D. S., Verma D. P. (1981) Post-translational processing of 7S and 11S components of soybean storage proteins. Plant Mol. Biol. 1, 19–34 [DOI] [PubMed] [Google Scholar]
- 3. Chrispeels M. J., Higgins T. J., Spencer D. (1982) Assembly of storage protein oligomers in the endoplasmic reticulum and processing of the polypeptides in the protein bodies of developing pea cotyledons. J. Cell Biol. 93, 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dickinson C. D., Hussein E. H., Nielsen N. C. (1989) Role of posttranslational cleavage in glycinin assembly. Plant Cell 1, 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jung R., Scott M. P., Nam Y. W., Beaman T. W., Bassüner R., Saalbach I., Müntz K., Nielsen N. C. (1998) The role of proteolysis in the processing and assembly of 11S seed globulins. Plant Cell 10, 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Müntz K. (1996) Proteases and proteolytic cleavage of storage proteins in developing and germinating dicotyledonous seeds. J. Exp. Bot. 47, 605–622 [Google Scholar]
- 7. Shimada T., Yamada K., Kataoka M., Nakaune S., Koumoto Y., Kuroyanagi M., Tabata S., Kato T., Shinozaki K., Seki M., Kobayashi M., Kondo M., Nishimura M., Hara-Nishimura I. (2003) Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J. Biol. Chem. 278, 32292–32299 [DOI] [PubMed] [Google Scholar]
- 8. Otegui M. S., Herder R., Schulze J., Jung R., Staehelin L. A. (2006) The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18, 2567–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vitale A., Hinz G. (2005) Sorting of proteins to storage vacuoles: how many mechanisms? Trends Plant Sci. 10, 316–323 [DOI] [PubMed] [Google Scholar]
- 10. Crouch M. L., Sussex I. M. (1981) Development and storage-protein synthesis in Brassica napus L. embryos in vivo and in vitro. Planta (Berl.) 153, 64–74 [DOI] [PubMed] [Google Scholar]
- 11. Breen J. P., Crouch M. L. (1992) Molecular analysis of a cruciferin storage protein gene family of Brassica napus. Plant Mol. Biol. 19, 1049–1055 [DOI] [PubMed] [Google Scholar]
- 12. Inquello V., Raymond J., Azanza J. L. (1993) Disulfide interchange reactions in 11S globulin subunits of Cruciferae seeds. Relationships to gene families. Eur. J. Biochem. 217, 891–895 [DOI] [PubMed] [Google Scholar]
- 13. Rödin J., Ericson M. L., Josefsson L. G., Rask L. (1990) Characterization of a cDNA clone encoding a Brassica napus 12 S protein (cruciferin) subunit. Relationship between precursors and mature chains. J. Biol. Chem. 265, 2720–2723 [PubMed] [Google Scholar]
- 14. Rödin J., Sjödahl S., Josefsson L. G., Rask L. (1992) Characterization of a Brassica napus gene encoding a cruciferin subunit: estimation of sizes of cruciferin gene families. Plant Mol. Biol. 20, 559–563 [DOI] [PubMed] [Google Scholar]
- 15. Sjödahl S., Rödin J., Rask L. (1991) Characterization of the 12S globulin complex of Brassica napus. Evolutionary relationship to other 11–12S storage globulins. Eur. J. Biochem. 196, 617–621 [DOI] [PubMed] [Google Scholar]
- 16. Badley R. A., Atkinson D., Hauser H., Oldani D., Green J. P., Stubb J. M. (1975) The structure, physical and chemical properties of the soy bean protein glycinin. Biochim. Biophys. Acta 412, 214–228 [DOI] [PubMed] [Google Scholar]
- 17. Marcone M. F., Beniac D. R., Harauz G., Yada R. Y. (1994) Quaternary structure and model for the oligomeric seed globulin from Amaranthus hypochondriacus K343. J. Agric. Food Chem. 42, 2675–2678 [Google Scholar]
- 18. Plietz P., Damaschun G., Müller J. J., Schwenke K. D. (1983) The structure of 11-S globulins from sunflower and rape seed. A small-angle X-ray scattering study. Eur. J. Biochem. 130, 315–320 [DOI] [PubMed] [Google Scholar]
- 19. Adachi M., Takenaka Y., Gidamis A. B., Mikami B., Utsumi S. (2001) Crystal structure of soybean proglycinin A1aB1b homotrimer. J. Mol. Biol. 305, 291–305 [DOI] [PubMed] [Google Scholar]
- 20. Tandang-Silvas M. R., Fukuda T., Fukuda C., Prak K., Cabanos C., Kimura A., Itoh T., Mikami B., Utsumi S., Maruyama N. (2010) Conservation and divergence on plant seed 11S globulins based on crystal structures. Biochim. Biophys. Acta 1804, 1432–1442 [DOI] [PubMed] [Google Scholar]
- 21. Tandang M. R., Adachi M., Utsumi S. (2004) Cloning and expression of rapeseed procruciferin in Escherichia coli and crystallization of the purified recombinant protein. Biotechnol. Lett. 26, 385–391 [DOI] [PubMed] [Google Scholar]
- 22. Adachi M., Kanamori J., Masuda T., Yagasaki K., Kitamura K., Mikami B., Utsumi S. (2003) Crystal structure of soybean 11S globulin: glycinin A3B4 homohexamer. Proc. Natl. Acad. Sci. U.S.A. 100, 7395–7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang L., Phillips T. E., Rogers S. W., Rogers J. C. (2000) Biogenesis of the protein storage vacuole crystalloid. J. Cell Biol. 150, 755–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wittig I., Braun H. P., Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 25. Sunderhaus S., Klodmann J., Lenz C., Braun H. P. (2010) Supramolecular structure of the OXPHOS system in highly thermogenic tissue of Arum maculatum. Plant Physiol. Biochem. 48, 265–272 [DOI] [PubMed] [Google Scholar]
- 26. Schägger H., von Jagow G.(1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 27. Neuhoff V., Stamm R., Pardowitz I., Arold N., Ehrhardt W., Taube D. (1990) Essential problems in quantification of proteins following colloidal staining with coomassie brilliant blue dyes in polyacrylamide gels, and their solution. Electrophoresis 11, 101–117 [DOI] [PubMed] [Google Scholar]
- 28. Neuhoff V., Stamm R., Eibl H. (1985) Clear background and highly sensitive protein staining with Coomassie Blue dyes in polyacrylamide gels: A systematic analysis. Electrophoresis 6, 427–448 [Google Scholar]
- 29. Kruft V., Eubel H., Jänsch L., Werhahn W., Braun H. P. (2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127, 1694–1710 [PMC free article] [PubMed] [Google Scholar]
- 30. Dudkina N. V., Eubel H., Keegstra W., Boekema E. J., Braun H. P. (2005) Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl. Acad. Sci. U.S.A. 102, 3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 32. Quillin M. L., Matthews B. W. (2000) Accurate calculation of the density of proteins. Acta Crystallogr. D Biol. Crystallogr. 56, 791–794 [DOI] [PubMed] [Google Scholar]
- 33. Rossmann M. G. (2000) Fitting atomic models into electron-microscopy maps. Acta Crystallogr. D Biol. Crystallogr. 56, 1341–1349 [DOI] [PubMed] [Google Scholar]
- 34. Andème Ondzighi C., Christopher D. A., Cho E. J., Chang S. C., Staehelin L. A. (2008) Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell 20, 2205–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang A. H. (1996) Oleosins and oil bodies in seeds and other organs. Plant Physiol. 110, 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moreira M. A., Hermodson M. A., Larkins B. A., Nielsen N. C. (1979) Partial characterization of the acidic and basic polypeptides of glycinin. J. Biol. Chem. 254, 9921–9926 [PubMed] [Google Scholar]
- 37. Shewry P. R., Napier J. A., Tatham A. S. (1995) Seed storage proteins: structures and biosynthesis. Plant Cell 7, 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jagow G., Schägger H. (2003) Membrane Protein Purification and Crystallization: A Practical Guide, 2nd Ed., pp. 105–130 Academic Press [Google Scholar]
- 39. Agrawal G. K., Thelen J. J. (2006) Large scale identification and quantitative profiling of phosphoproteins expressed during seed filling in oilseed rape. Mol. Cell Proteomics 5, 2044–2059 [DOI] [PubMed] [Google Scholar]
- 40. Wan L., Ross A. R., Yang J., Hegedus D. D., Kermode A. R. (2007) Phosphorylation of the 12 S globulin cruciferin in wild-type and abi1–1 mutant Arabidopsis thaliana (thale cress) seeds. Biochem. J. 404, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meyer L. J., Gao J., Xu D., Thelen J. J. (2012) Phosphoproteomic analysis of seed maturation in Arabidopsis, rapeseed, and soybean. Plant Physiol. 159, 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bhatty R. S., McKenzie S. L., Finlayson A. J. (1968) The proteins of rapeseed (Brassica napus L.) soluble in salt solutions. Can. J. Biochem. 46, 1191–1197 [DOI] [PubMed] [Google Scholar]
- 43. Lawrence M. C., Suzuki E., Varghese J. N., Davis P. C., Van Donkelaar A., Tulloch P. A., Colman P. M. (1990) The three-dimensional structure of the seed storage protein phaseolin at 3 A resolution. EMBO J. 9, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwenke K. D., Raab B., Plietz P., Damaschun G. (1983) The structure of the 12 S globulin from rapeseed (Brassica napus L.). NAHRUNG 27, 165–175 [Google Scholar]