Background: Insulin-degrading enzyme (IDE) is a highly conserved metallopeptidase initially described because of its ability to degrade insulin.
Results: (i) IDE expression is stress-inducible; (ii) IDE concentration is up-regulated in some CNS tumors; (iii) IDE down-regulation impairs SHSY5Y cell proliferation/viability.
Conclusion: IDE is a multifunctional protein.
Significance: IDE is a novel HSP with important implications in cell growth regulation.
Keywords: Brain Tumors, Heat Shock Protein, Metalloenzymes, Neuroblastoma, Proteasome, IDE
Abstract
Insulin-degrading enzyme (IDE) is a highly conserved zinc metallopeptidase that is ubiquitously distributed in human tissues, and particularly abundant in the brain, liver, and muscles. IDE activity has been historically associated with insulin and β-amyloid catabolism. However, over the last decade, several experimental findings have established that IDE is also involved in a wide variety of physiopathological processes, including ubiquitin clearance and Varicella Zoster Virus infection. In this study, we demonstrate that normal and malignant cells exposed to different stresses markedly up-regulate IDE in a heat shock protein (HSP)-like fashion. Additionally, we focused our attention on tumor cells and report that (i) IDE is overexpressed in vivo in tumors of the central nervous system (CNS); (ii) IDE-silencing inhibits neuroblastoma (SHSY5Y) cell proliferation and triggers cell death; (iii) IDE inhibition is accompanied by a decrease of the poly-ubiquitinated protein content and co-immunoprecipitates with proteasome and ubiquitin in SHSY5Y cells. In this work, we propose a novel role for IDE as a heat shock protein with implications in cell growth regulation and cancer progression, thus opening up an intriguing hypothesis of IDE as an anticancer target.
Introduction
Insulin-degrading enzyme (IDE)2 is a highly conserved zinc metallopeptidase, which is ubiquitously distributed in human tissues, particularly abundant in brain, liver, and muscles (1, 2). In these tissues, even though the relationship between its function and subcellular distribution is far from being understood, IDE shows a prevalent cytosolic localization with small amounts also found on plasma membranes, in the peroxisomes, endosomes, and mitochondria; IDE is also secreted through a non-conventional pathway (3–6).
IDE was initially described for its ability to degrade insulin and then was found to interact with several substrates, including β-amyloid, glucagon, amylin, somatostatin, and natriuretic peptide (4, 7–9). In the last decade, there have been studies that have also linked IDE to the etiology of some diseases, such as Alzheimer Disease (AD) and Type 2 Diabetes Mellitus (DM2) (4, 10–12), associating the protein to a wide range of cellular processes, such as Varicella Zoster Virus infection, steroid receptor signaling, and insulin-dependent proteasome modulation (13–17). It has also been proposed to play a role in rat muscle cell differentiation and rat development, envisaging for IDE multiple functions related to cellular growth and tissue homeostasis (18, 19). Moreover, recent findings indicate that (i) IDE appears to be involved in processing sets of antigenic peptides presented by class I MHC through a proteasome-independent pathway (20) and (ii) ubiquitin can be degraded by IDE “in vitro” (9), even though this last result is somewhat contradictory (21). Finally, preliminary experiments performed in our laboratory demonstrated that in neuronal cells infected with viruses able to induce the shut-off of cellular proteins, IDE content was nevertheless preserved or even increased with an HSP-like behavior. All these data suggest that IDE might be involved in ubiquitin-proteasome pathways and therefore in the heat shock response (HSR), prompting us to investigate this hypothesis.
HSR is an organized genetic response to diverse environmental and physiological stressors that results in the immediate induction of genes encoding molecular chaperones, proteases, and other proteins (mostly belonging to the heat shock gene superfamily) essential for protection and recovery from cellular damage associated with the accumulation of misfolded proteins. The heat shock gene superfamily is organized according to molecular size and functional class, including the Hsp100, Hsp90, Hsp70, Hsp60, Hsp40 (J-domain proteins), and small heat shock protein (sHSP) families. Many of these proteins function as molecular chaperones, inducing conformational states that are critical for the synthesis, folding, translocation, assembly, and degradation of proteins. Under stressful conditions, most HSPs stabilize denaturing proteins and refold proteins that have been already denatured. If proteins are irreversibly denatured, molecular chaperones help to hand them over to the proteolytic machineries of the cells, mainly along the ubiquitin-proteasome pathway (22–24).
In this work we demonstrate that IDE follows a heat shock protein-like behavior, and we report up-regulated levels of the enzyme in CNS tumors in vivo. Further results reveal that IDE could play a critical role in neuroblastoma cell growth, acting on the ubiquitin-proteasome system.
EXPERIMENTAL PROCEDURES
Stress Procedures
SHSY5Y, LAN-5, and ACN cells were cultivated in high glucose-DMEM whereas Jurkat, PBL, NHLF, and SK-N-BE cells were grown in RPMI 1640. Both were supplemented with 15% fetal bovine serum (FBS), l-glutamine (2 mm), penicillin (50 IU/ml), streptomycin (50 μg/ml), sodium pyruvate (1 mm), and ciprofloxacin (0.03 mm). SHSY5Y were seeded on a 96-well plate ranging from 5 × 102/well to 4 × 103/well and grown overnight. The following day, cells were exposed to different stresses, namely: (i) 48 °C for 15 and 30 min; (ii) increasing H2O2 concentrations ranging from 1 μm to 50 μm for 24 h; (iii) 60 min of serum starvation followed by 15% fetal calf serum addition for 24 h. NHLF (60 × 104/well), Jurkat and PBL (purified from the peripheral blood of healthy volunteers by Ficoll-Hypaque gradient centrifugation) cells were seeded on a 24-well plate (8 × 104/well) and the following day incubated at 48 °C for 45 min and 60 min (NHLF) and at 42 °C for 30 min and 45 min (Jurkat and PBL) or treated with increasing H2O2 concentrations ranging from 1 μm to 10 μm for 24 h or 60 min serum starvation followed by 15% fetal calf serum addition for 24 h. Lysates were collected after 24 h of recovery (unless otherwise indicated), by using a lysis buffer (50 mm NaCl, 1% Triton, 50 mm Tris-HCl) supplemented with PMSF and a protease-inhibitor mixture with broad specificity for the inhibition of serine, cysteine, aspartic, and aminopeptidase (Sigma-Aldrich). After each treatment, Trypan blue exclusion viability tests demonstrated that almost 95% of cells were still alive.
Qualitative Insulin-degrading Enzyme Analysis by Western Blotting
For the Western blotting analyses, 10 μg of total lysate was separated on a 10% SDS-PAGE gel and transferred to Hybond-ECL nitrocellulose filters (Amersham Biosciences) for 1 h at 4 °C. Unsaturated binding sites were blocked by incubating filters in a 0.01% PBS Tween (T-PBS), 5% fat-free milk solution. Membranes were then probed with an anti-IDE rabbit polyclonal antibody (BC2) (Covance, Princeton, NJ), diluted 1:3000 in 0.1% T-PBS, and incubated with a horseradish peroxidase-conjugated anti-rabbit IgG antibody (Bio-Rad), diluted 1:50,000 in 0.2% T-PBS fat-free milk. The overall content of poly-ubiquitinated proteins in lysates from IDE-silenced SHSY5Y cells and from untreated cells was analyzed by Western blotting. Immunoreactive signals were detected with an ECL Advance Western blotting Detection Kit (Amersham Biosciences).
Quantitative Real-time PCR
The total RNA from each SHSY5Y sample was isolated using an RNeasy Plus micro kit for RNA isolation according to the manufacturer's instructions (Qiagen, Hilden, Germany). cDNA of total RNA was generated in two reaction volumes of Masterscript kit using random hexamers (5PRIME, Applied Biosystems, Foster City, CA). A real-time PCR was performed by SDS7700 (5PRIME, Applied Biosystems) with a Manual SYBER ROX Master Mix. GAPDH was measured as the internal control. Primer sequences were: for IDE, GTCCTGTTGTTGGAGAGTTCCCATGTCA (Forward); GGGGAATCTTCAGAGTTTTGCAGCCAT (Reverse) and for GAPDH were: AACTTTGGCATTGTGGAAGG (Forward) and CACATTGGGGGTAGGAACAC (Reverse). Each sample measurement was carried out in triplicate. The following cycles were performed: initial denaturation cycle at 95 °C for 2 min, followed by 40 amplification cycles at 95 °C for 15 s, at 58 °C for 30 s and at 68 °C for 30 s.
IDE siRNA Transfection
SHSY5Y cells (2 × 103) were seeded on a 96-well Transwell plate, and the following day cells were transfected with 1 μm Accell IDE-siRNA and Accell siRNA control (synthesized by Dharmacon, Lafayette, CO). Lysates were collected 48 and 72 h after transfection. The rate of cell viability was determined through a Cy-Quant proliferation kit according to the manufacturer's instructions (Invitrogen).
SHSY5Y Stimulation with All Trans Retinoic Acid (ATRA)
SHSY5Y cells were seeded on a 96-well Transwell plate (2 × 103) and the following day, 1 μm ATRA and DMSO alone were added. Lysates were collected 24, 48, and 72 h after ATRA administration. Proliferation curves were determined through a Cy-Quant proliferation kit according to the manufacturer's instructions.
Immunofluorescence
Cellular expression of IDE was investigated by immunofluorescence. Sparse adhering SHSY5Y and NHLF cells were washed in PBS, fixed in cold methanol, and, after PBS rinsing, stained for 30 min with an anti-IDE rabbit polyclonal antibody (BC2, 1:100) and Hoechst (5 μg/ml in PBS) followed by a TRITC-conjugated secondary antibody (Jackson ImmunoResearch Labs, West Grove, PA). As a negative control, diluted (1:40) human IG was used.
Immunohistochemical Investigation of IDE Expression in Human Tumor Tissues
Tissue sections were obtained from the diagnostic biopsy paraffin block Archive of Anatomic Pathology of Tor Vergata University over the last five years, according to the guidelines of the local Ethical Committee and in accordance with the local Ethical Committee and Declaration of Helsinki guidelines. Tumor classification and grading was in accordance with recent WHO criteria and with the most widespread immunohistochemical panel (25). We evaluated 7 astrocytic tumors (grade III-IV), of which there were three cases of anaplastic astrocytoma and four of glioblastoma (age range 41–66 years), and three cases of olfactory neuroblastoma (age range 35–52 years). The exclusion criteria included preoperative radiation or chemotherapy and an adequate amount of tumor tissue for the correct routine processing diagnosis (at least two tissue cores). As a control for the tumors of the nervous system, we used paraffin sections of bioptic material, which contained small fragments of non-neoplastic cerebral tissue that had been removed during the surgical excision of benign tumors (n = 4). For the immunohistochemical procedure (26), serial 4-μm-thick paraffin sections were stained with hematoxylin/eosin or used for immunohistochemistry. Regarding the latter, we performed deparaffinization, blocking of endogenous peroxidase activity with 0.2% H2O2 (20 min) and incubation with normal goat serum (30 min). Sections were exposed for 1 h to a polyclonal antibody anti-IDE (BC2, Covance,1:100), Diaminobenzidine was used as final chromogen. All immunohistochemical procedures were performed at room temperature. Semi-quantitative evaluation of IDE immunoreaction was estimated at 200× magnification in at least 10 fields by two of the authors, who used a grading system in arbitrary units (a.u.) as follows: negative = 0; <25% of cells weakly positive = 0.5; diffuse weakly positive = 1; moderately positive = 2 and strongly positive = 3 (26). The results were expressed as mean number ± S.E., with an interobserver reproducibility of >95%. Differences were evaluated by a Student's t test and were considered significant at p value ≤0.05.
Co-immunoprecipitation
A anti-IDE monoclonal antibody (Abcam, Cambridge, UK) was coupled to M-270 epoxy Dynabeads (Invitrogen) using an optimized version of the protocol suggested by the manufacturer (5 μg of Ab/1,5 mg of beads). Magnetic beads, coated with anti-IDE Ab and stored at 4 °C in PBS, 0.02% NaN3, were washed three times with lysis buffer, and added to the soluble fraction of cell lysates. Total protein extracts were obtained by rinsing the cells twice with ice-cold PBS followed by the addition of ice-cold lysis buffer (Co-IP), supplemented with 1 mm DTT, 50 mm NaCl, and a protease-inhibitor mixture with broad specificity for the inhibition of serine, cysteine, aspartic, and aminopeptidase (Sigma-Aldrich). Harvested cells were washed once in PBS, and the pellet was resuspended in 1:9 ratio of cell mass to extraction buffer supplemented with a protease inhibitors mixture, according to manufacturer's instruction (Invitrogen). Cells were incubated for 15 min in ice and centrifuged at 2600 × g for 5 min at 4 °C. The extract was used immediately for co-immunoprecipitation. Purification was achieved by slow mixing at 4 °C. The isolated protein complex was eluted from the beads for 20 min at room temperature in a fresh aqueous solution. WB analysis was performed by using anti-IDE monoclonal antibody, anti-ubiquitin monoclonal antibody, and P26S monoclonal antibody (Abcam).
Statistical Analysis
One-way analysis of variance (ANOVA) was used to determine significant differences among groups. Tukey's significant difference post hoc test was used for pairwise comparisons after the analysis of variance.
RESULTS
IDE Induction upon Stress Exposure
Cells respond to environmental stresses by triggering the synthesis of heat shock proteins (HSPs) to restore protein homeostasis and to guarantee cell survival (22). Here, we investigated the stress-dependent modulation of IDE expression in different human cell lines, namely: SHSY5Y cells (human neuroblastoma line), NHLF cells (human fibroblast derived cell line), Jurkat cells (human lymphoblastic-like cell line), and PBL (human peripheral blood lymphocytes).
SHSY5Y cells were exposed to: (i) heat shock at 48 °C for 15 and 30 min; (ii) increasing concentrations of H2O2, and (iii) serum starvation for 60 min. On the other hand, for heat shock NHLF cells were incubated at 48 °C for 45 and 60 min and PBL and Jurkat cells at 42 °C for 30 and 45 min. Incubation times and temperature were set for each line to obtain the maximal IDE induction: incubation time of NHLF was longer than SHSY5Y because setting experiments demonstrated that for these cells a shorter incubation time is not sufficient for an acceptable stress induction, and Jurkat cells and PBL were stressed at a lower temperature since cells die quickly at temperatures above 42 °C. NHLF and Jurkat cells underwent H2O2 and serum starvation experiments following the same conditions described for SHSY5Y. Preliminary time-course analysis revealed that IDE concentration in SHSY5Y (see supplemental Fig. S2) and NHLF is not modulated within 12 h of recovery, maximum modulation becoming evident only after 24 h, while for Jurkat cells and human PBL the increase occurs earlier, evident after 6 h of recovery. These findings are consistent with the notion that HSPs have a cell- and time-specific pattern of induction and transcription (24).
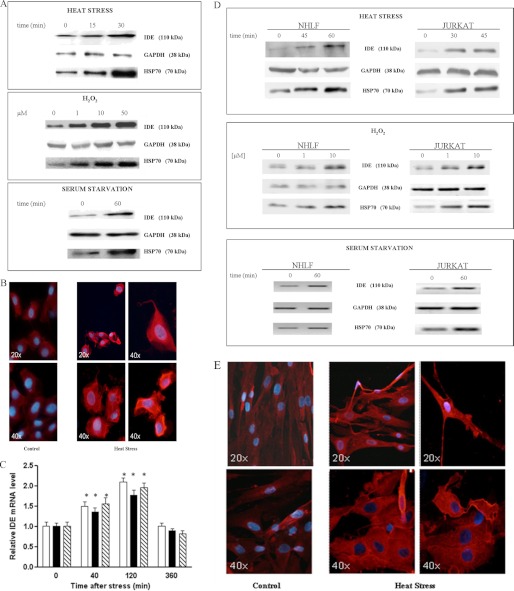
As shown in Fig. 1A for SHSY5Y, under each experimental condition, IDE increases in a similar way to HSP70, while the housekeeping gene GAPDH levels remain unaltered. The fluorescence microscopy analysis of heat-stressed cells after 24 h of recovery strengthened this evidence because it revealed a widespread cytoplasmic localization (Fig. 1B). IDE up-regulation was also validated at the transcriptional level by quantifying IDE mRNA after 40, 120, 300 min of recovery. In fact, we found that IDE mRNA increases 120 min after stress and returns to a basal level within 5 h (Fig. 1C).
FIGURE 1.
IDE up-regulation in stress-exposed SHSY5Y, NHLF, and Jurkat cells. A, Western blotting analyses of IDE, HSP70, and GAPDH in lysates from heat-stressed SHSY5Y cells after 24 h of recovery. Three different stresses were used: heat stress for 15 and 30 min at 48 °C, oxidative stress using 1, 10, 50 μm H2O2 for 24 h, and nutrient deprivation for 60 min. A representative immunoblot of five independent experiments is shown. See supplemental Fig. S1 for the densitometric analysis. B, immunofluorescence analysis of heat-stressed SHSY5Y cells after 24 h of recovery: IDE content dramatically increases in the cytoplasm of SHSY5Y cells. Images were captured at different magnification (×20 and ×40). C, RT-PCR quantification of IDE mRNA in SHSY5Y-stressed cells: Heat stress for 30 min at 48 °C (white column); oxidative stress using 50 μm H2O2 (black column). IDE mRNA increases within 2 h of recovery and returns to basal levels within 5 h. Results are the means ± S.E. of five independent experiments and are displayed for treated cells as the relative expression of untreated controls. *, significantly different from control (p < 0.05, one-way ANOVA, followed by Tukey's test, n = 15). D, Western blotting analyses of IDE level in lysates from NHLF and Jurkat cells, respectively, incubated at 48 °C for 45 min and 60 min and at 42 °C for 30 min and 45 min, with increasing H2O2 concentrations ranging from 1 μm to 10 μm for 24 h and 60 min of serum starvation. See supplemental Fig. S3 for Western blotting analyses of IDE levels in lysates from PBL. The reported immunoblot represents five independent experiments. See supplemental Fig. S1 for the densitometric analysis. E, immunofluorescence analysis of heat-stressed NHLF cells after 24 h of recovery: the intracellular IDE content consistently increases. Images were captured at different magnifications (×20 and ×40).
Even with NHLF cells, (which express appreciable amounts of IDE under basal conditions), with Jurkat cells and normal PBL, we confirm that this enzyme is stress-inducible. Western blotting analyses on cell lysates revealed that, in NHLF, in Jurkat cells, IDE intracellular content under stress conditions is significantly increased in a way similar to HSP70 (Fig. 1D and for WB on PBL results (see supplemental Fig. S3). Similar to SHSY5Y, NHLF cells were further analyzed by fluorescence microscopy after 24 h of recovery. Even in this case, a broad cytoplasmic distribution of the enzyme was observed (Fig. 1E).
It is worth pointing out that other neuroblastoma cell lines (namely LAN-5, CAN, and SK-N-BE) have been tested for IDE levels in our laboratory under both normal and stressed conditions showing: (a) high IDE concentrations in unstressed cell lines and (b) a pattern of stress induction of IDE clearly overlapping with that of SHSY5Y in all cell lines (data not shown).
Histological and Immunohistochemical Findings
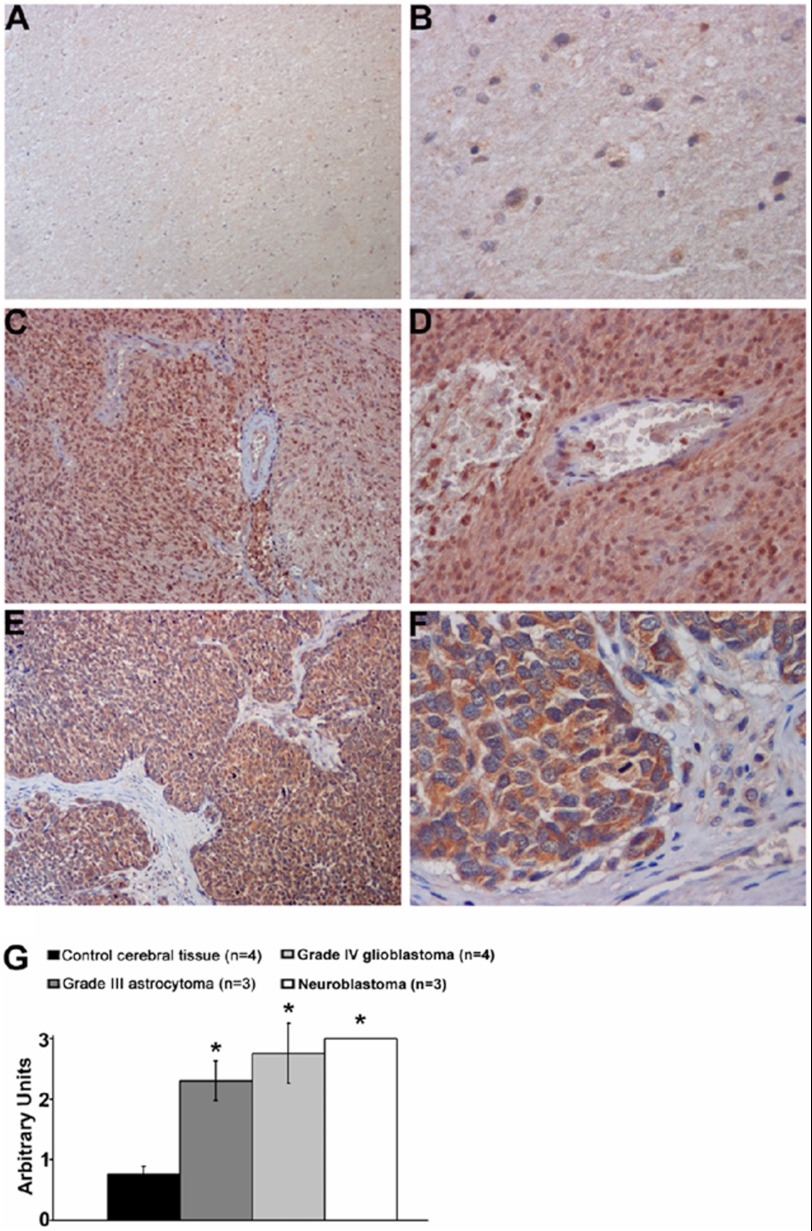
Based on the reported evidence of IDE constitutive expression in the brain (27), IDE distribution was investigated in tumor bioptic tissues of the central nervous system (Fig. 2, A–F). Semiquantitative evaluation (Fig. 2G) revealed that IDE immunostaining is almost similarly high in grade III and IV malignant glioma cells and olfactory neuroblastoma tumor cells, while it was significantly lower in normal nerve cells (p ≪ 0.001). These findings strongly support the hypothesis that IDE overexpression is associated with a more aggressive behavior and progression of tumors.
FIGURE 2.
IDE expression in human tumors by immunohistochemistry. At different magnification, representative images of (A, B) normal cerebral tissue showing a weak IDE immunoreactivity of nerve cells, whereas (C, D) glioblastoma and (E, F) olfactory neuroblastoma tumor cells display a strong IDE immunostaining. G, bar graph showing the semiquantitative evaluation of IDE immunoreactivity, expressed as arbitrary units + S.E. Diaminobenzidine is used as chromogen and hematoxylin as counterstaining. Original magnification: A, C, E, ×150; B, F, ×400; D, ×250; *, p < 0.01.
Effect of IDE Silencing on Cell Proliferation and Death
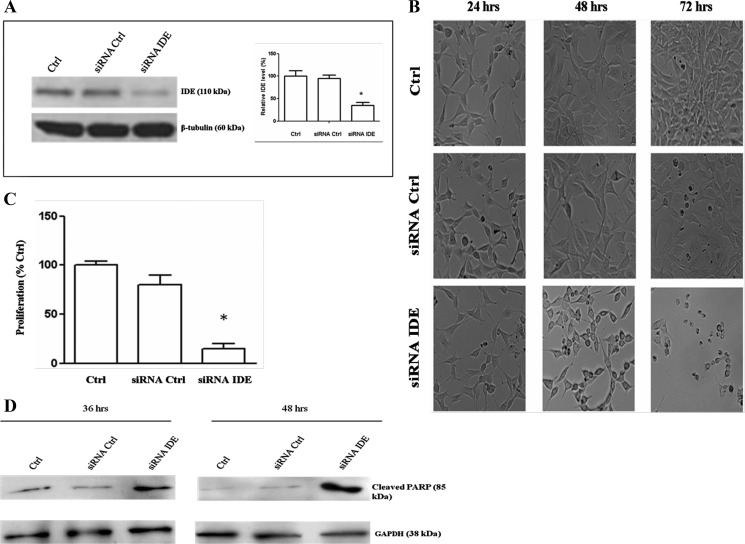
The effect of IDE down-regulation on SHSY5Y cells has been investigated by inhibiting IDE expression with a specific siRNA. The reduction in IDE content of 70 ± 5.7% (Fig. 3A) correlates with a significant decrease in SHSY5Y proliferation rate and precedes SHSY5Y apoptotic death, which occurs 72–84 h after IDE silencing (Fig. 3, B and C). In fact, since a burst of cleaved PARP occurs in the early stages of apoptosis (28), Western blotting analysis of lysates at 48 h after treatment highlighted a consistent increase of cleaved PARP in IDE-silenced cells (Fig. 3D).
FIGURE 3.
Effect of IDE silencing on neuroblastoma proliferation and viability. A, Western blotting analysis of IDE concentration in cell SHSY5Y lysates collected 48 h after stimulation either with an IDE-specific siRNA or a nontargeting siRNA pool. A representative immunoblot of six independent experiments is shown. Results presented are the means ± S.E. of six independent experiments. *, significantly different from control (p < 0.05, one way ANOVA, followed by Tukey's test, n = 18). B, after 48 h of transfection, SHSY5Y cells treated with the anti-IDE siRNA show a marked decrease of cell proliferation rate. Extensive cell death occurs 24 h later. C, proliferation rate is displayed for treated cells as the relative expression of untreated controls at 72 h after IDE-silencing. The results presented are the means ± S.E. of six independent experiments. *, significantly different from control (p < 0.05, one way ANOVA, followed by Tukey's test, n = 18). D, in the case of IDE-silenced cells at 48 h after silencing an increment of cleaved PARP-level is detected in the early stage of apoptosis.
As expected, control cells grown in Accell media display an optimal proliferation rate, whereas cells transfected with an siRNA-specific pool after 72–84 h of treatment showed only a slight reduction in proliferation and viability rates (Fig. 3B).
ATRA Treatment Down-regulates IDE Expression in Neuroblastoma Cells
All trans-retinoic acid (ATRA) is a vitamin A precursor used in the clinical treatment of neuroblastoma. It reduces the malignancy by switching cells toward a less invasive and distinctly neuronal phenotype characterized by a slow division rate and neurite outgrowth (29, 30).
Western blotting analysis of SHSY5Y cell lysates, 24, 48, and 72 h after 1 μm ATRA administration (Fig. 4A) clearly indicates that IDE concentration is progressively down-regulated. Notably, in accordance with previously reported data on IDE stress inducibility, cells incubated with DMSO alone (ATRA vector) display a transient increase of IDE enzymatic concentration. As expected, a significant effect in SHSY5Y cell differentiation (data not shown), and a reduction in proliferation rate was observed (Fig. 4B).
FIGURE 4.
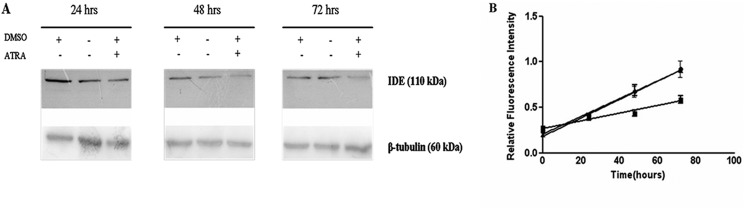
ATRA effect on IDE expression. A, Western blotting analysis of IDE in SHSY5Y cells treated with ATRA. Lysates collected 24 h after the addition of 1 μm ATRA indicate a significant down-regulation of IDE expression compared with untreated cells and to DMSO (ATRA vector)-treated cells. A representative immunoblot of four independent experiments is shown. See supplemental Fig. S4 for the densitometric analysis. B, proliferation curve in arbitrary units (relative fluorescence units) in the absence and in the presence of ATRA is shown. Control (●); DMSO (▴); ATRA(■). The results presented are the mean ± S.E. of four independent experiments.
IDE Effect on the Ubiquitin-Proteasome System Cascade
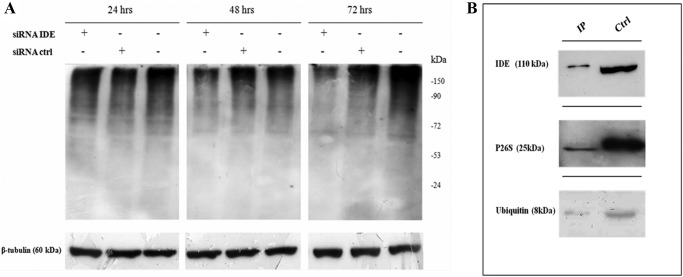
To evaluate the effect of IDE silencing on the proteasome-ubiquitin system, we have compared the overall content of poly-ubiquitinated proteins of IDE-silenced SHSY5Y cells versus control and/or scrambled cells. Interestingly, we found that the quantity of poly-ubiquitinated proteins is dramatically reduced in SHSY5Y IDE-silenced cells (Fig. 5A), an observation in accord with previous reports that show that inhibition of the ubiquitin system stimulates apoptosis in some neural cells (31). It is equally important to underscore that the poly-ubiquitinated protein reduction occurs 36–48 h after the transfection, when IDE intracellular content is still 50–60% of the final rate.
FIGURE 5.
IDE effect on ubiquitin system. A, Western blotting of the overall poly-ubiquitinated proteins content in lysates from IDE-silenced SHSY5Y. IDE inhibition is accompanied by a significant reduction of the poly-ubiquitinated proteins compared with untreated or scrambled cells. A representative immunoblot of six independent experiments is shown. B, Western blotting analysis of co-immunoprecipitation sample in SHSY5Y. Immunoaffinity purification shows that IDE interacts with ubiquitin and 26S proteasome in SHSY5Y neuroblastoma cells. (Ctrl indicates the whole lysate.)
Since IDE seems involved in the degradation of amyloidogenic proteins (7), we performed Congo Red staining on neuroblastoma cells treated with IDE siRNA, to investigate whether IDE silencing leads to an alteration of the amount of amyloidogenic proteins. However, no difference was observed between treated and normal cells after 24 h and 48 h of silencing, respectively (data not shown). It is important to state that we cannot rule out an alteration of the amount of misfolded protein forms that are not positive to Congo Red staining.
Additionally, in accord with previous reports that show that IDE immunoprecipitates with proteasome in rat skeletal muscle extracts and interacts with ubiquitin (9, 21, 31), we show that also in SHSY5Y neuroblastoma cells, IDE co-immunoprecipitates with 26S proteasome and ubiquitin (Fig. 5B). These data clearly suggest a role for IDE in the control of the proteasome-ubiquitin pathway.
DISCUSSION
Cells are physiologically exposed to noxious stimuli (i.e. heat stress, oxidative stress, nutrient deprivation, pathogens) with profound pathological consequences, including protein denaturation, lipid oxidation, and DNA damage. The HSR is the strategy that cells have developed to restore protein homeostasis and to promote cell survival. In this context, two alternative fates await non-native and damaged proteins, which is either (a) destruction through the error checking ubiquitin-proteasome system or (b) refolding back to the native state helped by HSPs (22, 32). Although the mechanism for addressing a damaged protein toward either one of the two pathways is poorly known, HSPs play a key role in the “protein triage” decision, by interacting with the ubiquitin-proteasome machinery (23). In addition, a great deal of data have recently indicated that, besides their contribution to the repair mechanisms after stress exposure, there is a direct involvement of several HSPs in the modulation of cell proliferation and apoptosis, which is confirmed by the fact that almost all human tumors display a specific combined alteration of different HSPs (in particular HSP70, HSP90, and HSP27) (33, 34). Hence, HSPs have become potential therapeutic targets and their quantification could become a putative diagnostic marker (35–38).
In this report, three major and closely correlated findings are reported, namely (i) IDE expression is stress-inducible in malignant and not-malignant cells; (ii) IDE concentration is in vivo markedly up-regulated in some tumors of the central nervous system; (iii) IDE down-regulation impairs SHSY5Y cell proliferation and viability, being accompanied by a decrease in the overall content of poly-ubiquitinated proteins, likely reflecting an influence of IDE on the ubiquitin-proteasome system.
Finding (i) is clearly demonstrated following the expression of the enzyme in different human cell lines (i.e. SHSY5Y, NHLF cells, Jurkat cells, and human peripheral blood lymphocytes, PBL are discussed in detail, see above). Our results clearly show that both malignant and not-malignant cells, once exposed to various stressful conditions, trigger IDE gene transcription, strengthening the hypothesis for a role of IDE in the HSR.
A bioinformatic analysis of the IDE gene promoter (GenBankTM database, accession number: NG_013012)(−4799/−1 bp upstream of the first ATG) seems to suggest the presence of multiple heat shock elements (HSE) in the region between 4010/−4700 bp. More specifically, TESS and TF search software (which can be found at: www.cbrc.jp/research/db/TFSEARCH.html) make it possible to identify several 5′-nGAAn-3′ sequences, known as high affinity binding sites for the heat shock factor (HSF). The suggestion of IDE as HSP-like is also strengthened by the recent demonstration of Aβ trapping in the IDE chamber, which seems to prevent the formation of soluble Aβ aggregates, leading us to the intriguing perspective of IDE as a “dead-end chaperone” (39).
The HSP-like IDE behavior prompted us to analyze its distribution in tissues where it is usually expressed, such as cells from the nervous system, and to investigate whether IDE modulation is important in tumor growth (which represents finding (ii), see above).
Although IDE is already known to be overexpressed in some human malignancies, such as in ovarian or breast cancer (27), to date there is no information regarding IDE expression in nervous system tumors.
Here, we report that IDE immunostaining was marked in malignant tumors, such as olfactory neuroblastoma and high grade glioma, suggesting that IDE overexpression in these tumors is generally linked to a more aggressive behavior and progression. Further studies are needed to clarify the possible molecular pathways through which IDE overexpression influences carcinogenesis in vivo. In any case, this evidence raises the intriguing scenario that elevated intracellular levels of IDE might be of vital importance for neuronal tumors cells.
On the basis of the above-reported findings the effect of IDE down-regulation on SHSY5Y neuroblastoma cells has been investigated (Fig. 3). Results demonstrated that when a IDE-specific siRNA is administered to SHSY5Y cells, extensive apoptotic cell death occurs; these data confirm that the reduction of IDE content is not compatible with proliferation and viability of these tumor cells, where the maintenance of IDE high levels seems to be a pre-requisite for tumor viability (corresponding to finding (iii)), see above).
The importance of high IDE levels in SHSY5Y for cell malignancy is also indirectly confirmed by the observation that a significant decrease in SHSY5Y IDE intracellular content is induced by ATRA, a vitamin A precursor used in the clinical treatment of neuroblastoma (Fig. 4) (29, 30). Since preliminary results obtained on other neuroblastoma cell lines indicate that IDE inhibition, under conditions identical to those used for SHSY5Y, does not unequivocally correlate with apoptosis triggering (data not shown), we cannot state whether the ATRA effects on proliferation rates of neuroblastoma cells are due only to the reduction of IDE cell content. However, this side effect must be also taken into consideration.
The evidence regarding the stress-dependent inducibility and the role of IDE in some tumor cells, laid out in this report, strengthen the idea that IDE could be a multifunctional protein, rendering of great interest the identification of unrevealed aspects of the IDE biological role.
In this context, intriguingly, we report that IDE-silenced SHSY5Y cells display a decreased proliferation rate and viability in association with an early reduction of the overall poly-ubiquitinated protein content (Fig. 5) (9, 40). Finally, we show that IDE immunoprecipitates with ubiquitin and proteasome in neuroblastoma cells, suggesting a direct role for IDE in the modulation of this pathway.
This observation would seem to indicate that there is a precise mechanism of IDE activity regulation and/or the existence of yet-to-be-identified IDE natural inhibitors, which could be misregulated, at least in this tumor. In this sense, it has been reported that monomeric Ub degradation by IDE appears inhibited by nestin, a type VI intermediate filament protein expressed by many cell types during development and at high levels by tumor cells, such as neuroblastoma and glioma cells (9, 37).
Therefore, a 2-fold role could be envisaged for IDE, namely the control of the ubiquitin-proteasome degradation system, acting on ubiquitin homeostasis and/or directly on the proteasome enzymatic activity. The physiological relevance of IDE up-regulation upon stress exposure could therefore rely on the modulation of the ubiquitin-proteasome pathway, thus facilitating the damaged protein “triage,” even though it is not possible to rule out other roles in the regulation of protein homeostasis. In conclusion, these results suggest a novel role for IDE as a heat shock-like protein and highlight the IDE involvement in the oncogenesis of the central nervous system.
This work was supported in part by the Italian Ministry for the University and Research (MiUR PRIN 2008L57JXW 002 (to S. M.) and FIRB RBNE03PX83 (to M. C.).

This article contains supplemental Figs. S1–S4.
- IDE
- insulin-degrading enzyme
- HSP
- heat shock protein
- HSR
- heat shock response
- CNS
- central nervous system
- ATRA
- all trans retinoic acid.
REFERENCES
- 1. Kirschner R. J., Goldberg A. L. (1983) A high molecular weight metalloendoprotease from the cytosol of mammalian cells. J. Biol. Chem. 258, 967–976 [PubMed] [Google Scholar]
- 2. Duckworth W. C., Bennett R. G., Hamel F. G. (1998) Insulin degradation: progress and potential. Endocr. Rev. 19, 608–624 [DOI] [PubMed] [Google Scholar]
- 3. Qiu W. Q., Walsh D. M., Ye Z., Vekrellis K., Zhang J. (1998) Insulin-degrading enzyme regulates extracellular levels of amyloid beta-protein by degradation. J. Biol. Chem. 273, 32730–32738 [DOI] [PubMed] [Google Scholar]
- 4. Hersh L. B. (2006) The insulysin (insulin degrading enzyme) enigma. Cell Mol. Life. Sci. 63, 2432–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao J., Li L., Leissring M. A. (2009) Insulin-degrading enzyme is exported via an unconventional protein secretion pathway. Mol. Neurodegener. 4, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glebov K., Schütze S., Walter J. (2011) Functional Relevance of a Novel SlyX Motif in Non-conventional Secretion of Insulin-degrading Enzyme. J. Biol. Chem. 286, 22711–22715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kurochkin I. V. (2001) Insulin-degrading enzyme: embarking on amyloid destruction. Trends Biochem. Sci. 26, 421–425 [DOI] [PubMed] [Google Scholar]
- 8. Ciaccio C., Tundo G. R., Grasso G., Spoto G., Marasco D., Ruvo M., Gioia M., Rizzarelli E., Coletta M. (2009) Somatostatin: a novel substrate and a modulator of insulin-degrading enzyme activity. J. Mol. Biol. 385, 1556–1567 [DOI] [PubMed] [Google Scholar]
- 9. Ralat L. A., Kalas V., Zheng Z., Goldman R. D., Sosnick T. R., Tang W. J. (2011) Ubiquitin is a novel substrate for human insulin-degrading enzyme. J. Mol. Biol. 406, 454–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rudovich N., Pivovarova O., Fisher E., Fischer-Rosinsky A., Spranger J., Möhlig M., Schulze M. B., Boeing H., Pfeiffer A. F. (2009) Polymorphisms within insulin-degrading enzyme (IDE) gene determine insulin metabolism and risk of type 2 diabetes. J. Mol. Med. 87, 1145–1151 [DOI] [PubMed] [Google Scholar]
- 11. Abdul-Hay S. O., Kang D., McBride M., Li. L, Zhao J., Leissring M. A. (2011) Deletion of insulin-degrading enzyme elicits antipodal, age-dependent effects on glucose and insulin tolerance. PLoS One 6, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tundo G., Ciaccio C., Sbardella D., Boraso M. S., Viviani B., Coletta M., Marini S. (2012) Somatostatin modulates insulin-degrading-enzyme metabolism: implications for the regulation of microglia activity in AD. PLoS One 7, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kupfer S. R., Wilson E. M., French F. S. (1994) Androgen and glucocorticoid receptors interact with insulin degrading enzyme. J. Biol. Chem. 269, 20622–20638 [PubMed] [Google Scholar]
- 14. Bennett R. G., Fawcett J., Kruer M. C., Duckworth W. C., Hamel F. G. (2003) Insulin inhibition of the proteasome is dependent on degradation of insulin by insulin-degrading enzyme. J. Endocrinol. 177, 399–405 [DOI] [PubMed] [Google Scholar]
- 15. Udrisar D. P., Wanderley M. I., Porto R. C., Cardoso C. L., Barbosa M. C., Camberos M. C., Cresto J. C. (2005) Androgen- and estrogen-dependent regulation of insulin-degrading enzyme in subcellular fractions of rat prostate and uterus. Exp. Biol. Med. 230, 479–486 [DOI] [PubMed] [Google Scholar]
- 16. Li Q., Ali M. A., Cohen J. I. (2006) Insulin degrading enzyme is a cellular receptor mediating varicella-zoster virus infection and cell-to-cell spread. Cell 127, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fernández-Gamba A., Leal M. C., Morelli L., Castaño E. M. (2009) Insulin-degrading enzyme: structure-function relationship and its possible roles in health and disease. Curr. Pharm. Des. 15, 3644–3655 [DOI] [PubMed] [Google Scholar]
- 18. Kayalar C., Wong W. T. (1989) Metalloendoprotease inhibitors which block the differentiation of L6 myoblasts inhibit insulin degradation by the endogenous insulin-degrading enzyme. J. Biol. Chem. 264, 8928–89234 [PubMed] [Google Scholar]
- 19. Kuo W. L., Montag A. G., Rosner M. R. (1993) Insulin-degrading enzyme is differentially expressed and developmentally regulated in various rat tissues. Endocrinology 132, 604–611 [DOI] [PubMed] [Google Scholar]
- 20. Parmentier N., Stroobant V., Colau D., de Diesbach P., Morel S., Chapiro J., van Endert P., Van den Eynde B. J. (2010) Production of an antigenic peptide by insulin-degrading enzyme. Nat. Immunol. 11, 449–454 [DOI] [PubMed] [Google Scholar]
- 21. Grasso G., Rizzarelli E., Spoto G. (2008) How the binding and degrading capabilities of insulin degrading enzyme are affected by ubiquitin. Biochim. Biophys. Acta. 1784, 1122–1126 [DOI] [PubMed] [Google Scholar]
- 22. Jolly C., Morimoto R. I. (2000) Role of the heat shock response and molecular chaperones in oncogenesis and cell death. Journal of the National Cancer Institute 92, 1564–1572 [DOI] [PubMed] [Google Scholar]
- 23. Garrido C., Solary E. (2003) A role of HSPs in apoptosis through “protein triage”? Cell Death Differ. 10, 619–620 [DOI] [PubMed] [Google Scholar]
- 24. De Maio A. (2011) Extracellular heat shock proteins, cellular export vesicles, and the Stress Observation System: a form of communication during injury, infection, and cell damage. It is never known how far a controversial finding will go! Dedicated to Ferruccio Ritossa. Cell Stress Chaperones. 16, 235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Louis D. N., Ohgaki H., Wiestler O. D., Cavenee W. K., Burger P. C., Jouvet A., Scheithauer B. W., Kleihues P. (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Orlandi A., Ferlosio A., Ciucci A., Francesconi A., Lifschitz-Mercer B., Gabbiani G., Spagnoli L. G., Czernobilsky B. (2006) Cellular retinol binding protein-1 expression in endometrial hyperplasia and carcinoma: diagnostic and possible therapeutic implications. Mod. Pathol. 19, 797–803 [DOI] [PubMed] [Google Scholar]
- 27. Yfanti C., Mengele K., Gkazepis A., Weirich G., Giersig C., Kuo W. L., Tang W. J., Rosner M., Schmitt M. (2008) Expression of metalloprotease insulin-degrading enzyme insulysin in normal and malignant human tissues. Int. J. Mol. Med. 4, 421–431 [PMC free article] [PubMed] [Google Scholar]
- 28. Simbulan-Rosenthal C. M., Rosenthal D. S., Iyer S., Boulares H., Smulson M. E. (1999) Involvement of PARP and poly(ADP-ribosyl)ation in the early stages of apoptosis and DNA Replication. Mol. Cell Biochem. 193, 137–148 [PubMed] [Google Scholar]
- 29. Wu G., Fang Y., Lu Z. H., Ledeen R. W. (1998) Induction of axon-like and dendrite-like processes in neuroblastoma cells. J. Neurocytol. 27, 1–14 [DOI] [PubMed] [Google Scholar]
- 30. Lopes F. M., Schröder R., da Frota M. L., Jr., Zanotto-Filho A., Müller C. B., Pires A. S., Meurer R. T., Colpo G. D., Gelain D. P., Kapczinski F., Moreira J. C., Fernandes Mda C., Klamt F. (2010) Comparison between proliferative and neuron-like SH-SY5Y cells as an in vitro model for Parkinson disease studies. Brain Res. 1337, 85–94 [DOI] [PubMed] [Google Scholar]
- 31. Bennett R. G., Hamel F. G., Duckworth W. C. (2000) Insulin inhibits the ubiquitin-dependent degrading activity of the 26S proteasome. Endocrinology 141, 2508–2517 [DOI] [PubMed] [Google Scholar]
- 32. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 33. Calderwood S. K., Khaleque M. A., Sawyer D. B., Ciocca D. R. (2006) Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem. Sci. 31, 164–172 [DOI] [PubMed] [Google Scholar]
- 34. Ciocca D. R., Calderwood S. K. (2005) Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 10, 86–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Evan G., Littlewood T. (1998) A matter of life and cell death. Science 281, 1317–1322 [DOI] [PubMed] [Google Scholar]
- 36. Karapanagiotou E. M., Syrigos K., Saif M. W. (2009) Heat shock protein inhibitors and vaccines as new agents in cancer treatment. Expert Opin. Investig. Drugs 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 37. Joly N., Engl C., Jovanovic G., Huvet M., Toni T. (2010) Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34, 797–827 [DOI] [PubMed] [Google Scholar]
- 38. Khalil A. A., Kabapy N. F., Deraz S. F., Smith C. (2011) Heat shock proteins in oncology: Diagnostic biomarkers or therapeutic targets? Biochim. Biophys. Acta 2, 89–104 [DOI] [PubMed] [Google Scholar]
- 39. De Tullio M. B., Morelli L., Castaño E. M. (2008) The irreversible binding of amyloid peptide substrates to insulin-degrading enzyme: a biological perspective. Prion 2, 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thomas S. K., Messam C. A., Spengler B. A., Biedler J. L., Ross R. A. (2004) Nestin is a potential mediator of malignancy in human neuroblastoma cells. J. Biol. Chem. 279, 27994–27999 [DOI] [PubMed] [Google Scholar]