Background: O-Acetylpeptidoglycan esterase functions to release O-acetyl groups from peptidoglycan, thereby permitting the continued metabolism of this essential cell wall component.
Results: Gly-236 and Asn-268 were identified as participating at the oxyanion hole to stabilize the tetrahedral species in the reaction mechanism.
Conclusion: Ape1a functions as a classical serine esterase.
Significance: Ape represents a novel target for the development of new antibiotics.
Keywords: Enzyme Mechanisms, Enzymes, Peptidoglycan, Protein Motifs, Site-directed Mutagenesis, Acetylesterase, Oxyanion Hole, Serine Esterase, Mechanism, Oxyanion Hole
Abstract
O-Acetylpeptidoglycan esterase from Neisseria gonorrhoeae functions to release O-acetyl groups from the C-6 position of muramoyl residues in O-acetylated peptidoglycan, thereby permitting the continued metabolism of this essential cell wall heteropolymer. It has been demonstrated to be a serine esterase with sequence similarity to the family CE-3 carbohydrate esterases of the CAZy classification system. In the absence of a three-dimensional structure for any Ape, further knowledge of its structure and function relationship is dependent on modeling and kinetic studies. In this study, we predicted Neisseria gonorrhoeae Ape1a to be an SGNH hydrolase with an adopted α/β-hydrolase fold containing a central twisted four-stranded parallel β-sheet flanked by six α-helices with the putative catalytic triad, Asp-366, His-369, and Ser-80 appropriately aligned within a pocket. The role of eight invariant and highly conserved residues localized to the active site was investigated by site-directed replacements coupled with kinetic characterization and binding studies of the resultant engineered enzymes. Based on these data and theoretical considerations, Gly-236 and Asn-268 were identified as participating at the oxyanion hole to stabilize the tetrahedral species in the reaction mechanism, whereas Gly-78, Asp-79, His-81, Asn-235, Thr-267, and Val-368 are proposed to position appropriately the catalytic residues and participate in substrate binding.
Introduction
Peptidoglycan (PG)3 is an essential component of the bacterial cell wall, providing stability against osmotic stress, as well as an exoskeleton through which cellular shape is imparted. The repeating unit of the glycan backbone of this heteropolymer is composed of N-acetylglucosamine (GlcNAc)-linked β,1→4 to N-acetylmuramic acid. These glycan chains are cross-linked to each other through peptides associated with the N-acetylmuramic acid residues thus providing a covalently linked macromolecule (sacculus) that encompasses the bacterial cell. This PG sacculus can be modified by N-deacetylation of the GlcNAc or N-acetylmuramic acid residues, or by O-acetylation of the C-6 hydroxyl moiety of N-acetylmuramic acid (reviewed in Ref. 1). This latter modification is particularly significant because it both inhibits the action of many muramidases (lysozymes), major constituents of the innate immune system, and it completely precludes the activity of lytic transglycosylases, endogenous bacterial enzymes involved with cell growth and division, PG turnover, and the insertion of wall spanning structures such as flagella and secretory systems (recently reviewed in Refs. 2 and 3).
The PG sacculus is not a static structure but rather it is constantly metabolized involving the lytic transglycosylases for its biosynthesis and turnover. Given this, we hypothesized the need for (4), and later demonstrated (5), the existence of a new class of enzymes that function to de-O-acetylate PG. These O-acetyl-PG esterases (Ape) appear to be ubiquitous in Gram-negative bacteria that O-acetylate their PG (3, 4). A genomic study led to the identification of three families of enzymes, each with some amino acid sequence similarity to the family CE-3 O-acetylxylan esterases of the CAZy classification system (4). The gene encoding Neisseria gonorrhoeae Ape1a was cloned and expressed in Escherichia coli transformants. To facilitate its handling and purification, it was overproduced in a form lacking its N-terminal membrane anchor and possessing a C-terminal His6 tag, respectively (5). Efforts to delete the gene in N. gonorrhoeae were unsuccessful, suggesting that the cell does indeed require the ability to remove O-acetyl groups from PG, thereby permitting its continued metabolism. To further test this hypothesis at the enzyme level, a high-throughput screen was conducted to identify inhibitors of Ape. This search led to the identification of purpurin, a natural dye produced by the madder plant, as a competitive inhibitor of Ape1a. Moreover, purpurin was found to serve as a bacteriostatic antibacterial toward both Gram-positive and Gram-negative bacteria that produce both O-acetyl-PG and Ape (6). Thus, to further the development of Ape as a potential novel antibacterial target, knowledge of its structure and function relationship is essential. Unfortunately, exhaustive attempts by three separate research groups to produce suitable crystals of Ape1a that diffract with appropriate resolution have failed. The issue appears to be one of structural mosaicity, and neither the engineering of different forms with alternate truncations and amino acid replacements, chemical modification of surface amino acid residues, nor co-crystallization with ligands has helped to increase resolution below 3.2 Å.
In the absence of a known three-dimensional structure, any further understanding of the mechanism of action of Ape is dependent on kinetic analyses and modeling studies. A chemical modification and site-directed engineering study was conducted on Ape1a to confirm the essential role of Ser-80, His-369, and Asp-366 in the catalytic mechanism of the enzyme (7), supporting its preliminary identification as a GDS(L/H) serine esterase (5). Thus, following the established mechanism for serine esterases, we proposed that a salt bridge between Asp-366 and His-369 would facilitate the imidazolium group of the latter to abstract a proton from neighboring Ser-80, rendering it nucleophilic and suitable to function as a catalytic residue (Fig. 1). This Ser-80 nucleophile would attack the carbonyl carbon of an ester linked acetate group on the substrate (Fig. 1A), leading to the formation of a tetrahedral species (Fig. 1B), which would subsequently collapse to a covalent acyl enzyme intermediate (Fig. 1C) with the concomitant release of the product. The acetate would then be hydrolyzed from the catalytic Ser-80 (i.e. deacylation) in a reversal of this process (Fig. 1D) with water playing the role of the departed hydroxyl group, thereby returning Ser-80 to its protonated state and ready for another catalytic event.
FIGURE 1.
Proposed mechanism of action of Ape1a. The Ser-80 nucleophile attacks the carbonyl carbon of an ester linked acetate group on the substrate (A) leading to the formation of a tetrahedral species (B), which would subsequently collapse to a covalent acyl enzyme intermediate (C) and the concomitant release of the product. The acetate is then hydrolyzed from the catalytic Ser-80 in a reversal of this process (D) with water playing the role of the departed hydroxyl group. R1 and R2 denote β-(1→4)-linked GlcNAc residues, and R3 denotes stem peptides.
If Ape1a does function in this manner, the transient negatively charged oxygen atom associated with the putative tetrahedral species (Fig. 1B) would need to be stabilized by hydrogen bonds to appropriately positioned residues comprising an “oxyanion hole.” The identity of these residues for any Ape has yet to be experimentally proven, but the alignment of amino acid sequences of Ape1a and CE-3 enzymes suggests that they may be SGNH hydrolases. The SGNH hydrolases are a subfamily of the GDSL esterases that have been identified based on the presence of four invariant residues found in separate conserved motifs throughout the member proteins (reviewed in Ref. 8). Accordingly, the catalytic Ser in motif I, the amide of the Gly residue in motif II, and the Nδ2 of the Asn residue in motif III are predicted to each participate as proton donors for the formation of the oxyanion hole, whereas the His of motif VII participates as the base in the catalytic triad.
In this study, we investigated the participation of a number of highly conserved amino acids in the catalytic activity of N. gonorrhoeae Ape1a that are predicted to line its active-site pocket, including Gly-78, Asp-79, His-81, Gly-236, and Asn-268, by engineering a series of site-specific replacements of these residues. In addition, the role of Asn-235, a residue uniquely conserved among the Ape1a family of esterases, was also studied.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
DNase I, RNase A, pronase, isopropyl-β-d-1-thiogalactopyranoside, and EDTA-free protease inhibitor tablets were purchased from Roche Diagnostics Canada (Laval, PQ), whereas Fisher Scientific (Nepean, ON) provided acrylamide, glycerol, and Luria Bertani (LB) growth medium. All other growth media were purchased from Difco. Ni2+ nitrilotriacetic acid-agarose was purchased from Qiagen (Valencia, CA), and the Mono S 5/5 FPLC column was obtained from GE Healthcare. Stratagene (La Jolla, CA) provided pfu turbo DNA polymerase and DpnI. Unless otherwise stated, all other chemicals and reagents were acquired from Sigma.
Bacterial Strains and Culture Conditions
The strains of bacteria used in these studies and their genotypes are presented in supplemental Table 1. N. gonorrhoeae FA62 was grown for 24 h at 37 °C in GCBL broth supplemented with Kellogg's defined supplement (9, 10) in a humid, 5% CO2 environment as described previously (5). Plasmids constructed during these studies were all screened and maintained in E. coli DH5α. Overproduction of recombinant engineered proteins was performed with E. coli Rosetta(λDE3)pLysS cells that were always freshly transformed with the desired construct and grown in Super Broth (5 g of NaCl, 20 g of yeast extract, and 32 g of tryptone) at 37 °C with agitation. E. coli cultures were supplemented with chloramphenicol (35 mg·ml−1) and kanamycin (50 mg·ml−1) when required.
Isolation and Purification of O-Acetyl PG
O-Acetyl-PG was harvested from overnight cultures of N. gonorrhoeae FA62 grown in GCBL broth. Cells harvested by centrifugation (8,000 × g for 15 min at 4 °C) were suspended in a minimal volume of 25 mm sodium phosphate buffer, pH 6.5. Insoluble PG was isolated and purified following treatment with α-amylase, DNase, RNase, and MgSO4 as described previously (11), taking care throughout to ensure that the pH remained below 7.0 to prevent unwanted saponification of the ester-linked acetate.
Site-directed Mutagenesis
Using the appropriate primers (supplemental Table 2), the codons for Gly-78, Asp-79, His-81, Asn-235, Gly-236, Thr-267, Asn-268, and Val-368 of ape1a were individually replaced with those for Ala, Gln, Glu, Leu, Ser, or Phe, as appropriate. Mutagenesis was performed according to the QuikChange site-directed mutagenesis kitTM (Stratagene) using pACJW16 as the template (supplemental Table 1), which encodes a soluble, truncated derivative of Ape1a (5). Following PCR amplification, DpnI was added to the reaction mixture to remove the original methylated template. Reaction mixtures were then used to transform E. coli DH5α, and the resultant constructs were screened for the correct alterations by DNA sequencing (Guelph Molecular Supercenter, University of Guelph, Guelph, ON).
Production and Purification of His6-tagged Proteins
E. coli Rosetta(λDE3)pLysS cells freshly transformed with the appropriate plasmid (supplemental Table 1) were grown in Super Broth at 37 °C with aeration until mid-exponential phase (A600 ∼ 0.6). Following induction with 1 mm isopropyl-β-d-1-thiogalactopyranoside (final concentration) at 15 °C for 16 h, cells were harvested by centrifugation (5000 × g at 4 °C for 15 min). The overproduced Ape1a derivatives possessing C-terminal His6 tags were isolated and purified by a combination of affinity and cation exchange chromatographies as described previously (5–7). Briefly, crude enzyme preparations in lysis buffer (50 mm sodium phosphate, pH 8.0, and 500 mm NaCl) were added to Ni2+nitrilotriacetic acid-agarose (1 ml of resin per 15 ml of cleared lysate), and the suspensions were incubated for 1 h at 4 °C before being applied to 15-ml disposable plastic columns. Unbound proteins were removed in 10 column volumes of lysis buffer followed by 10 column volumes each of lysis buffer containing 20 mm imidazole and 30 mm imidazole, respectively. The bound Ape1a derivatives were then batch eluted in 10 ml of lysis buffer containing 150 mm imidazole. Following dialysis for 16 h against two changes of 4 liters of 25 mm sodium phosphate buffer, pH 7.0, the His6-tagged proteins were further purified by cation-exchange chromatography on Mono S. Protein was applied in dialysis buffer at a flow rate of 0.7 ml·min−1 and then a linear gradient of 0 to 1 m NaCl was applied. Under these conditions, the Ape1a derivatives eluted in buffer containing ∼75 mm NaCl.
Structural Analysis by Circular Dichroism Spectrometry
Far UV CD spectra of proteins at concentrations of 0.1 mg·ml−1 were recorded using a Jasco J-810 spectrapolarimeter in a 0.1-cm path length cell while maintaining an internal temperature of 25 °C with a Julabo F25ME heating water bath. The spectra were recorded as an average of four data accumulations, with a scan speed of 50 nm·min−1, bandwidth of 1 nm, 4 s, data pitch of 0.2 nm, and range of 180 to 260 nm. Analysis of the spectra was performed using the DichroWeb software with the Selecon 3 program (12, 13) and protein reference set 3 (14, 15).
Kinetic Assay for Esterase Activity
Routine detection of acetyl esterase activity was performed using 2 mm p-nitrophenyl acetate as substrate at 25 °C in 50 mm sodium phosphate buffer, pH 6.5. Reactions involving a total volume of 300 μl were prepared in microtitre plates and initiated with the addition of the p-nitrophenyl acetate dissolved in ethanol (final ethanol concentration < 5% v/v). The appearance of released p-nitrophenol was monitored over 5 min by recording the increase in absorbance at 405 nm. One unit of esterase activity was defined as the amount of enzyme required to release 1 μmol of p-nitrophenol·min−1·mg of protein−1. Michaelis-Menten parameters of Ape1a and its mutagenic derivatives were determined using p-nitrophenyl acetate as a substrate at concentrations ranging from 0.1 to 6.0 mm. Analyses were performed in, at least, triplicate, and plots of initial reaction velocities as a function of substrate concentration were analyzed by non-linear regression using the Microcal Origin program (version 6.0) assuming a “one-site binding model” (5–7).
PG Binding Assay
A “pulldown” assay (16) was used to estimate the capacity of the Ape1a derivatives to bind insoluble O-acetyl-PG. The O-acetyl-PG substrate was prepared by sonicating the insoluble material in 50 mm sodium phosphate buffer, pH 6.5, for 2 min to create an evenly dispersed suspension. Purified samples (20 μg) of Ape1a and its mutagenic derivatives were incubated with 0.2 mg of purified, insoluble N. gonorrhoeae PG in 300 μl (total volume), 50 mm sodium phosphate buffer, pH 6.5 for 1 h at 4 °C. The insoluble PG was collected by ultracentrifugation (160,000 × g for 20 min at room temperature) using a Beckman Airfuge and washed free of unbound material. Bound protein was recovered by extraction of the PG pellet with 4% SDS. Each of the collected fractions was analyzed by SDS-PAGE and Western immunoblotting using an anti-His6 antibody.
Other Analytical Techniques
Amino acid sequence alignments were prepared using ClustalW (version 1.8) (17) and HHpred, whereas predictions of secondary structure were performed using the algorithms of GOR IV (18), Jpred (19), and PSIpred (20–22). Three-dimensional modeling was performed using MODELLER. Protein concentrations were determined using a bicinchoninic acid assay (Pierce). SDS-PAGE was performed by the method of Laemmli (23) using 12% acrylamide gels and Coomassie Brilliant Blue staining. Acetic acid analysis was performed by either HPLC (24) using an Aminex HPX-87H Bio-Rad column or a coupled enzymatic assay using a commercially available kit according to the manufacturer's specifications (Megazyme, International Ireland Ltd., Wicklow, Ireland). Muramic acid content of PG samples was determined by aminosugar analysis using a Beckman System Gold amino acid analyzer (25) or by HPLC (24) as described previously.
RESULTS
Identification of Potential Binding Site Residues in Ape1a
As stated earlier, the three-dimensional structure of any Ape remains unknown. However, the structures of several other members of the SGNH hydrolase superfamily have been determined, including the family CE-3 acetylxylan esterase from Clostridium thermocellum (Protein Data Bank code 2VPT) (26). Thus, the amino acid sequence of Ape1a from N. gonorrhoeae was aligned with the N-terminal catalytic module of CE-3 acetylxylan esterase from C. thermocellum (labeled CE-3 acetylxylan esterase from C. thermocellum 3-1) using the Web-based program HHpred. As we noted previously (6), reasonable alignment of the leader sequence of Ape1a involving its N-terminal 66 residues with CE-3 acetylxylan esterase from C. thermocellum 3-1 could not be made. Also, a comparable sequence of residues in Ape1a stretching from Trp-106 to Gln-208 does not appear to be present in the xylan esterase (or in any other related esterase for which structural information is available), but significant alignment resumed from Thr-209 through to the C-terminal Gln-387 of Ape1a. Consequently, a structural prediction of Ape1a was made involving its Gly-67-Gly-105 sequence and its complete C-terminal segement from Thr-209 to Gln-387 using MODELLER with CE-3 acetylxylan esterase from C. thermocellum 3-1 as the template. This prediction yielded a fidelity score of 2.2 e−20. As expected for an SGNH hydrolase, the predicted structure adopts an atypical α/β-hydrolase fold that contains a central four-stranded parallel twisted β-sheet flanked by six α-helices (Fig. 2). The putative catalytic triad, Asp-366, His-369, and Ser-80 (7) are positioned in an appropriate alignment within a pocket of the enzyme to serve as the active center. Fortunately, none of the residues of interest in this study are positioned within the absent segment (viz. Trp-106-Gln-208).
FIGURE 2.
Predicted three-dimensional structure of N. gonorrhoeae Ape1a catalytic domain. The catalytic triad residues are in orange, and the N- and C-terminal residues of the two visualized chains are also labeled (Gly-105 is hidden).
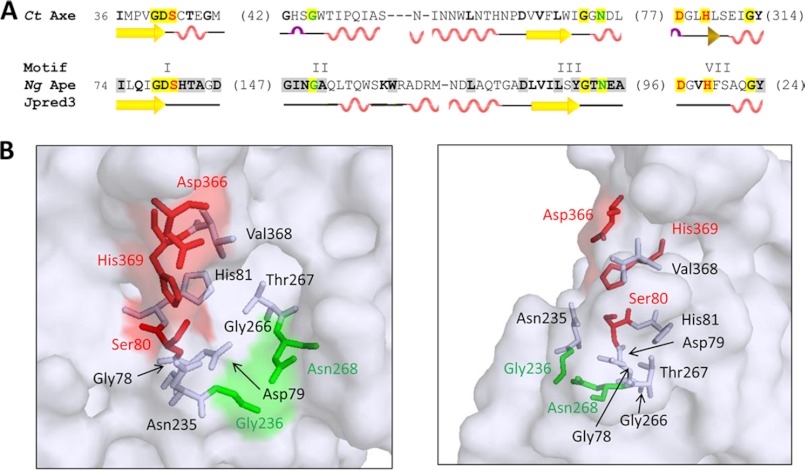
Alignment of Ape1a with the hypothetical amino acid sequences of other family 1 O-acetyl-PG esterases identified 11 invariant and many highly conserved residues (supplemental Fig. 1). Gly-236 of motif II (4), and both Gly-266 and Asn-268 of motif III are invariant among the Ape family members and, moreover, they also align with equivalents in C. thermocellum O-acetylxylan esterase and its related family homologs (Fig. 3). Each of these residues are located within the active site region of the predicted N. gonorrhoeae Ape1a structure but of the two Gly residues, only Gly-236 would appear to be positioned appropriately to potentially stabilize the tetrahedral species oxyanion of the reaction of the enzyme (Fig. 3). Hence, as both Gly-236 and Asn-268 are invariant in all Ape1 subfamilies and they comprise motifs II and III, respectively, of the consensus sequences, they were tentatively identified as forming the oxyanion hole and thus were targeted for site-specific replacements. Recognizing the neighboring position of Thr-267, a highly conserved residue in Ape subfamilies 1a and 1c (supplemental Fig. 1), in this region of the enzyme, its role, if any, was investigated by engineering its replacement as well. Similarly, Gly-78 and Asp-79 of motif I were also targeted as candidates for study given: (i) they are invariant, (ii) their immediate proximity to the catalytic Ser-80 in the primary structure of Ape1a, and (iii) their potential exposure to the putative active site pocket of Ape1a (Fig. 3). Asn-235 and Val-368 are highly conserved in the family 1a enzymes, and because they are predicted to be exposed to the active site cleft and would be proximal to catalytic residues, they too were replaced with Ala. Finally, His-81 neighbors catalytic Ser-80, and it is invariant in both Ape subfamilies 1a and 1c (supplemental Fig. 1), whereas with families 1b and 1d, it is replaced most often with Phe. Recognizing this trend, both the His-81→Phe and His-81→Ala replacements were engineered in N. gonorrhoeae Ape1a.
FIGURE 3.
Consensus sequence alignment of N. gonorrhoeae Ape1a with C. thermocellum acetylxlan esterase (Ct Axe) and predicted structure of Ape1a active site pocket. A, residues in boldface type and highlighted in gray denote greater than 50 and 80% identity, respectively, within the respective O-acetylxylan esterase and Ape families, whereas those highlighted in yellow are invariant. The catalytic triad residues are in red, and those proposed to stabilize the tetrahedral species at the oxyanion hole are in green. The known and predicted (by Jpred 3) secondary structures of O-acetylxylan esterase and Ape, respectively, are presented below their sequences. B, top and side views of the predicted structure of Ape1a active site. The catalytic triad residues are in red, the proposed oxyanion hole residues are in green, and the other residues targeted for replacement are in gray.
Site-specific Replacements
All of the amino acid replacements described in this study, G78A, D79A, H81A, H81F, N235A, G236A, G236P, T267A, N268A, N268E, N268L, N268S, and V368A were generated by site-directed mutagenesis of ape1a. With the exception of the H81A, G236P, and N268A replacements, overproduction of the engineered proteins in respective E. coli Rosetta(λDE3)pLysS transformants and their purification was monitored by SDS-PAGE and Western blot analysis using an anti-His6 antibody (data not shown)). Precaution was taken to ensure that new chromatography media were employed for the purifications of the respective enzymes to preclude any potential cross contamination of the enzyme forms with one another. In all cases, purities of >95% were achieved as determined by densitometric analysis. However, overall yields varied considerably (2–90%) compared with the isolation and purification of wild-type enzyme, which was recovered at levels of almost 16 mg per liter of culture (supplemental Table 3). The major issue confronted with the poor yields was one of solubility whereby protein would precipitate from solution despite efforts to control expression of respective genes with varying both concentrations of isopropyl-β-d-1-thiogalactopyranoside and growth temperatures, and employing buffers with different concentrations of salts and detergents. With the H81A, G236P, and N268A replacements, no expression of the encoding genes was detected despite varying the growth conditions of transformants and concentrations of the isopropyl-β-d-1-thiogalactopyranoside inducer. Nonetheless, sufficient protein was obtained with each of the other engineered enzymes for their kinetic analyses.
As a different buffer condition was used to kinetically characterize the various forms of Ape1a produced in this study compared with that reported previously for the identification of the catalytic triad residues (7), the Ser-80→Ala, Asp-366→Ala, and His-369→Ala forms of Ape1a were also produced and purified. These three enzymes were obtained in yields and levels of purity similar to those reported previously.
Circular dichroism (CD) spectroscopy was used to assess the overall secondary structure of the different forms of Ape1a purified. The spectra were analyzed using the DichroWeb server and, as found previously with the replacements of the catalytic triad residues (7), each was indistinguishable from that of the wild-type enzyme (data not shown). These analyses thus indicated that the site-specific replacements of the different amino acids did not cause any significant structural changes in the respective variant enzymes. It is recognized, however, that localized shifts of one of more residues in the vicinity of the replacements would not be detected by this analysis.
Kinetic Properties of Ape1a and the Engineered Derivatives
The specific activity of wild-type Ape1a and each of the replacement forms was determined using 2 mm p-nitrophenyl acetate as substrate at pH 6.5 and 25 °C. Under these conditions, each had a lower activity relative to wild-type with the values ranging from 0.06 to 25% (Table 1). As expected, replacement of the catalytic Ser-80 and His-369 residues led to the greatest loss of specific activity. Consistent with its postulated role as comprising the oxyanion hole, the next greatest loss of activity was seen with the replacement of Asn-268 with Leu (0.2% residual activity). The more conservative replacement of Asn-268 with Gln resulted in a significantly more active enzyme but nonetheless it was still only 10% as active as wild-type Ape1a. Replacement of Asp-79 with Ala also resulted in an enzyme with <1% residual activity compared with wild-type, and similarly (Asp-366→Ala) Ape1a retained only 2% activity. The least disruptive replacements made to Ape1a involved His-81 and Gly-236. While still causing a substantial loss, replacement of His-81 with Phe or Gly-236 with Ala resulted in forms of the enzyme that retained 25% residual activity.
TABLE 1.
Specific activities of Ape1 and mutant derivatives
| Enzyme | Specific activitya |
|
|---|---|---|
| μmol·min−1·mg−1 | %b | |
| Ape1a wild-type | 10.4 | 100 |
| G78A | 0.473 ± 0.0653 | 4.54 |
| D79A | 0.0448 ± 0.00154 | 0.431 |
| S80A | 0.00626 ± 0.000247 | 0.0602 |
| H81F | 2.61 ± 0.0866 | 25.1 |
| N235A | 1.05 ± 0.1035 | 10.1 |
| G236A | 2.60 ± 0.0265 | 25.0 |
| T267A | 1.01 ± 0.0343 | 9.71 |
| N268L | 0.0240 ± 0.00626 | 0.231 |
| N268Q | 1.07 ± 0.0447 | 10.3 |
| N268S | 0.0866 ± 0.00619 | 0.832 |
| D366A | 0.209 ± 0.0109 | 2.01 |
| V368A | 0.965 ± 0.00738 | 9.28 |
| H369A | 0.00666 ± 0.000351 | 0.064 |
a Activity assayed in triplicate using 2 mm p-nitrophenyl acetate as substrate in 50 mm sodium phosphate buffer, pH 6.5, at 25 °C (n > 3, ± S.D).
b Values as percentage of wild-type Ape1a.
The Michaelis-Menten parameters of each Ape1a variant were determined also with p-nitrophenyl acetate as substrate using the same conditions as described above. Despite the relatively low specific activity of some of the enzymes, the kinetic constants of each were obtained with reasonable confidence as reflected by low values of S.D. (Table 2). Whereas the catalytic efficiency (viz. values of kcat/Km) of each engineered enzyme was lower than that of wild-type Ape1a, none of the amino acid replacements significantly affected Km. Thus, values ranged from between 0.39 to 2.2 mm compared with 0.6 mm for wild-type Apela. This, however, was to be expected given the relatively simple structure of the artificial ester substrate used relative to the natural O-acetyl-PG substrate of the enzyme. The complexities of quantifying an insoluble polymeric substrate and its reaction products at the molecular level though preclude its use for detailed kinetic analysis and, unfortunately, no soluble O-acetyl-PG metabolites are available in sufficient quantity.
TABLE 2.
Kinetic parameters of Ape1a and mutant variants
Reactions were conducted in triplicate in 50 mm sodium phosphate buffer, pH 6.5, at 25 °C using p-nitrophenyl acetate as substrate over a concentration range of 0.1–6 mm.
| Enzyme | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | mol−1·s−1 | |
| Wild-type | 0.60 ± 0.099 | 8.7 ± 0.40 | 14,500 ± 2480 (100%)a |
| Catalytic triad | |||
| S80A | 2.2 ± 0.57 | 0.0084 ± 0.0013 | 3.8 ± 1.14 (0.026%) |
| D366A | 0.74 ± 0.087 | 0.21 ± 0.0082 | 280 ± 34 (1.9%) |
| H369A | 1.1 ± 0.077 | 0.0078 ± 0.00019 | 7.4 ± 0.59 (0.051%) |
| Potential binding residues | |||
| G78A | 0.72 ± 0.20 | 0.43 ± 0.032 | 590 ± 160 (4.1%) |
| D79A | 0.39 ± 0.12 | 0.042 ± 0.0035 | 110 ± 24 (0.73%) |
| H81F | 0.68 ± 0.26 | 3.4 ± 0.88 | 4900 ± 1500 (34%) |
| N235A | 0.96 ± 0.27 | 1.9 ± 0.17 | 2000 ± 390 (13%) |
| G236A | 0.69 ± 0.089 | 2.5 ± 0.093 | 3600 ± 470 (25%) |
| T267A | 0.58 ± 0.094 | 0.83 ± 0.038 | 1400 ± 230 (9.8%) |
| N268L | 0.84 ± 0.12 | 0.026 ± 0.0010 | 30 ± 4.4 (0.21%) |
| N268Q | 0.42 ± 0.084 | 0.087 ± 0.0044 | 220 ± 35 (1.4%) |
| N268S | 1.7 ± 0.26 | 0.12 ± 0.0068 | 67 ± 10 (0.46%) |
| V368A | 0.91 ± 0.29 | 1.8 ± 0.18 | 2000 ± 460 (14%) |
a Percentages in parentheses were calculated relative to wild-type Ape1a.
With only modest changes to Km, the apparent decreases in overall catalytic efficiency resulted from significant decreases in kcat values. Indeed, decreases of up to four orders of magnitude were observed with the enzymes involving replacement of the catalytic triad residues, and up to three orders of magnitude for some of the others. These data thus suggest that, as expected, each of the respective amino acids influence events of the catalytic hydrolysis.
After replacement of the catalytic Ser-80 and His-369 residues, replacement of Asn-268 resulted in the greatest decrease in kcat and, consequently, catalytic efficiency of Ape1a (Table 2). Interestingly, the impact of this replacement on kcat and catalytic efficiency was greater than that resulting from replacement of Asp-366, the third residue of the catalytic triad. This suggests an essential role for Asn-268 in the mechanism of action of Ape1a, and it is consistent with its earlier identification as a potential residue comprising the oxyanion hole. Neighboring Thr-267 is predicted to be slightly buried behind the active site pocket of Ape1a, but its replacement with Ala still caused a 10-fold decrease in kcat, presumably from affecting the appropriate alignment of Asn-268. That a precise alignment of the Asn amide is required for full activity is further supported by the observation that the relatively conservative replacement of Asn-268 with Gln was nearly as deleterious as replacing it with either Leu or Ser. Thus, while still providing the same functional group for hydrogen bonding, the extra methylene group of the Gln side chain would not permit its precise localization required at the active center of the enzyme. In contrast, the relatively high catalytic activity retained by (Asn-235→Ala)Ape1a excludes the possibility that this highly conserved residue functions at the oxyanion hole.
The second residue proposed to comprise the oxyanion hole was Gly-236. Contrary to initial expectations, however, its replacement with Ala while still deleterious had much less impact on kcat and the consequent catalytic efficiency of the enzyme which, in fact, was even higher than that of (Asn-235→Ala)Ape1a.
Of the replacements involving residues that neighbor the catalytic Ser-80, not surprisingly the greatest effect was seen with (Asp-79→Ala)Ape1a, suggesting that the Asp residue serves to appropriately align Ser-80. A similar effect was seen with replacement of Val-368, which presumably influences the positioning of catalytic His-369. Replacement of Gly-78 with Ala led to an enzyme with a kcat value one order of magnitude greater than that of (Asp-79→Ala)Ape1a, whereas that of the His-81→Phe enzyme was yet another order of magnitude greater, suggesting that N. gonorrhoeae Ape1a can accommodate this substitution with Phe that is present at this position in the subfamily 1b and 1c enzymes.
Substrate Binding to Ape1a and Derivatives
Although the insolubility of the natural O-acetylated PG substrate precludes its use for rigorous kinetic analyses, it was used in a pulldown affinity assay to provide a preliminary characterization of the binding properties of the respective Ape1a forms. PG from N. gonorrhoeae FA62 was used for this experiment, and compositional analysis indicated that the material isolated from stationary growth phase cells was O-acetylated to an extent of 54 ± 3.6% relative to muramic acid content. Following incubation, enzymes remaining bound to the insoluble PG were recovered by centrifugation and then subjected to SDS-PAGE analysis after their elution in SDS. As evident from the preliminary data presented in Table 3, the most deleterious replacements for binding to this natural substrate involved Gly-78, Asp-79, Thr-267, and His-369, followed closely by Ser-80, and Asn-268 when replaced with Leu. Replacing Asn-268 with Gln, however, substantially increased the apparent binding capacity of the enzyme, as did replacing Asn-235 or Val-368 with Ala. As with the effect on Michaelis-Menten parameters, replacement of Gly-236 with Ala had little impact on the binding of O-acetyl-PG to Ape1a.
TABLE 3.
Binding affinity of wild-type Ape1 and its mutant variants for insoluble O-acetyl-PG
Enzymes (20 μg) were incubated on ice with 200 μg of insoluble O-acetyl-PG suspended in 50 mm sodium phosphate buffer, pH 6.5, for 1 h prior to recovery of enzyme-bound ligand by ultracentrifugation.
| Enzyme | % Bounda | % Changeb |
|---|---|---|
| Ape1a wild type | 45.6 | |
| Catalytic triad | ||
| S80A | 1.64 | −96.4 |
| D366A | 14.1 | −69.1 |
| H369A | 0 | −100 |
| Potential binding residues | ||
| G78A | 0 | −100 |
| D79A | 0 | −100 |
| H81F | 27.7 | −39.3 |
| N235A | 73.1 | +160 |
| G236A | 37.4 | −18.0 |
| T267A | 0 | −100 |
| N268L | 1.95 | −95.7 |
| N268Q | 75.5 | +166 |
| N268S | 12.2 | −73.2 |
| V368A | 68.6 | +150 |
a Representative data calculated based on the amount remaining in the supernatant following incubation with ligand.
b % Change in PG binding ability of mutant derivatives relative to wild-type enzyme.
DISCUSSION
There are many structural and sequence alignment studies that identify the roles of specific residues in the GDSL hydrolase superfamily, but few enzymatic studies have been performed to confirm these roles (8). In the current study, we attempted to specifically fill this void, at least for Ape and related CE-3 esterases, by characterizing the replacement of eight residues in N. gonorrhoeae Ape1a. These residues were identified through alignment to be highly conserved, if not invariant, and three-dimensional modeling predicted them to occupy a position within the active site pocket and near to the catalytic triad residues. Of these, Gly-236 and Asn-268 were predicted specifically to participate in the oxyanion hole. Our results would thus appear to provide the first experimental evidence supporting this role proposed for Asn-268. Whereas we were unable to generate (Asn-268→Ala)Ape1a, replacement of Asn-268 with Leu, a residue of similar size but lacking a hydrogen bond donor/acceptor, was possible and caused a 335-fold decrease in kcat. This assignment is further supported by the observation that only minimal efficiency is retained when the conservative replacement of Asn-268 was made with Gln, despite providing the same opportunities for appropriate hydrogen bonding with the amide side chain. Presumably, the extra methylene of the Gln side chain compromises the precise positioning of the functional group required at the active center. Likewise, replacement of Thr-267, a residue predicted to be buried behind the active site pocket of Ape1a, with Ala would likely alter the positioning of Asn-268, thus accounting for the observed 90% decrease in catalytic efficiency. Unequivocal interpretation of these findings, however, would require the determination of the three-dimensional structure of Ape1a in complex with ligand. Unfortunately, such has not been determined for Ape nor any related CE-3 enzyme, but the need for precise positioning of binding and catalytic groups has been observed with many other enzymes, including those that act on PG. For example, the single oxidation of a residue that comprises binding subsite B of hen egg-white lysozyme, Trp-62, to oxindole alanine only causes a rotation of the residue but totally precludes productive binding of PG and consequently its hydrolysis (27). Hence, we suggest that, as proposed for C. thermocellum acetylxylan esterase (26) and other SGNH esterases, such as rhamnogalactuonan acetylesterase from Aspergillus aculeatus (28) and E. coli thioesterase I/protease I/lysophospholipase L1 (29), our modeling and experimental data support the assignment of Asn-268 as comprising the oxyanion hole of Ape1a to stabilize the negative charge of the tetrahedral species of the reaction pathway.
Gly-236 was predicted to also comprise the oxyanion hole based both on its hypothetical location within the active site pocket of Ape1a and its invariance in all Apes. Indeed, the backbone amide of its equivalent was observed to serve as an H-bond donor at the oxyanion hole of E. coli thioesterase I/protease I/lysophospholipase L1 in complex with its substrate (29). Its replacement in Ape1a with Ala though did not result in the large decrease in kcat that, perhaps, may have been expected. However, by lacking a side chain the only opportunities for hydrogen bonding that a Gly residue provides would involve its backbone amino and carboxyl groups. These would still be available by its replacement with the Ala residue, and any decrease in activity would presumably result from any slight structural adjustments that would be necessary to accommodate the methyl group side chain. The only replacement that could be made to experimentally test this assignment would be with Pro, but unfortunately, expression of the gene encoding the (Gly-236→Pro)Ape1a was not observed under any condition tested.
Gly-78 and Asp-79 represent secondary structure breakers. The increasing influence of their replacement, respectively, on the catalytic efficiency of Ape1a supports their importance for the appropriate positioning of the Ser-80 nucleophile. Indeed, the equivalent of Gly-78 in other SGNH esterases has been implicated in allowing the nucleophilic Ser to adopt an unstrained conformation (8, 28). Also, residues equivalent to Asp-79 have been proposed to participate in substrate binding in addition to appropriately positioning the Ser nucleophile (8, 28, 30). Our preliminary PG binding data support this additional role for Asp-79. Interestingly, replacement of His-81 with Phe did not have the same profound effect on kcat as the Gly-78 and Asp-79 replacements despite what would intuitively be considered a significant alteration in the proximity of Ser-80. However, Phe is observed to replace His at position 81 in many of the subfamily 1b and 1d enzymes, and it appears that Ape1a can accommodate this also, at least with the N. gonorrhoeae enzyme.
Unfortunately, without knowledge of the three-dimensional structure of Ape1a, it is not possible for us to completely rationalize the PG-binding data with the kinetic data obtained using the simple substrate model compound of p-nitrophenyl acetate given the extensive structural differences between the two ligands. Whereas the nitrophenyl ring of p-nitrophenyl acetate might appropriately mimic the sugar ring of muramoyl residues, the much smaller substrate lacks all of the other H-bonding possibilities provided by the N-acetyl group at C-2 and the lactyl ether at C-3 of muramoyl residues, together with the stem peptide that extends from it. Furthermore, it is likely that the binding site of Ape1a is comprised of subsites that accommodate the neighboring GlcNAc residues of the O-acetylated muramoyl residue at the active center. Indeed, at least three substrate- binding subsites have been observed in the PG N-deacetylase from Streptococcus pneumoniae (31), whereas four have been characterized in the chitin N-deacetylase from Colletotrichum lindemuthianum (32).
As an esterase with specificity for O-acetyl-PG, Ape is required for the continued biosynthesis and maintenance of the PG sacculus by facilitating its localized lysis involving the lytic transglycosylases (reviewed in Refs. 2 and 3). The lytic transglycosylases form complexes with the penicillin-binding proteins and other proteins associated with the metabolism of PG and their activity are required to provide new sites within the existing sacculus for the insertion of wall precursors (2). Given that their mode of action results in the production of 1,6-anhydromuramoyl reaction products, the lytic transglycosylases are dependent on Ape to remove any blocking O-acetyl groups. Consequently, Apes are proposed to play a major role in controlling PG metabolism (3).
Recently, purpurin was identified as an effective competitive inhibitor of Ape1a and moreover, selectively inhibited bacteria, both Gram-positive and Gram-negative, that produce both O-acetylated PG and the esterase (6). This study thus provided the first experimental evidence indicating that Ape is essential for cell growth and division and thereby supported the principle that it presents a novel antibacterial target. That Ape is localized external to the cytoplasmic membrane of bacteria makes it all the more attractive as an antibacterial target by minimizing uptake and efflux issues. To further this effort, knowledge of its structure and function relationship would be very helpful, if not essential. However, without a three-dimensional structure, we are dependent on modeling and kinetic information such as that provided in this study, which has confirmed Ape to function as an SGNH esterase. This understanding will greatly facilitate the search and/or design of potent irreversible inhibitors, which may prove to serve as leads for new antibacterials.
Acknowledgments
We thank Chelsea Clarke, Sarah Shamiya, and Simone ten Kortenaar for expert technical assistance.
This work was supported by Operating Grant MOP 81223 (to A. J. C.) from the Canadian Institutes of Health Research and postgraduate scholarships (PGSD; to J. M. P. and J. T. W.) from the Natural Sciences and Engineering Research Council of Canada.

This article contains supplemental Tables 1–3 and Fig. 1.
- PG
- peptidoglycan
- GlcNAc
- N-acetylglucosamine
- Ape
- O-acetylpeptidoglycan esterase.
REFERENCES
- 1. Vollmer W. (2008) Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32, 287–306 [DOI] [PubMed] [Google Scholar]
- 2. Scheurwater E., Reid C. W., Clarke A. J. (2008) Lytic transglycosylases: Bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 40, 586–591 [DOI] [PubMed] [Google Scholar]
- 3. Moynihan P. J., Clarke A. J. (2011) O-Acetylated peptidoglycan: Controlling the activity of bacterial autolysins and lytic enzymes of innate immune systems. Int. J. Biochem. Cell Biol. 43, 1655–1659 [DOI] [PubMed] [Google Scholar]
- 4. Weadge J. T., Pfeffer J. M., Clarke A. J. (2005) Identification of a new family of enzymes with potential O-acetylpeptidoglycan esterase activity in both Gram-positive and Gram-negative bacteria. BMC Microbiol. 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weadge J. T., Clarke A. J. (2006) Identification and characterization of O-acetylpeptidoglycan esterase: a novel enzyme discovered in Neisseria gonorrhoeae. Biochemistry 45, 839–851 [DOI] [PubMed] [Google Scholar]
- 6. Pfeffer J. M., Clarke A. J. (2012) Identification of first-known inhibitors of O-acetylpeptidoglycan esterase: A potential new antibacterial target. ChemBioChem. 13, 722–731 [DOI] [PubMed] [Google Scholar]
- 7. Weadge J. T., Clarke A. J. (2007) Neisseria gonorrhoeae O-acetylpeptidoglycan esterase, a serine esterase with a Ser-His-Asp catalytic triad. Biochemistry 46, 4932–4941 [DOI] [PubMed] [Google Scholar]
- 8. Akoh C. C., Lee G. C., Liaw Y. C., Huang T. H., Shaw J. F. (2004) GDSL family of serine esterases/lipases. Prog. Lipid Res. 43, 534–552 [DOI] [PubMed] [Google Scholar]
- 9. Kellogg D. S., Jr., Peacock W. L., Jr., Deacon W. E., Brown L., Pirkle D. I. (1963) Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85, 1274–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pagotto F. J., Salimnia H., Totten P. A., Dillon J. R. (2000) Stable shuttle vectors for Neisseria gonorrhoeae, Haemophilus spp., and other bacteria based on a single origin of replication. Gene 244, 13–19 [DOI] [PubMed] [Google Scholar]
- 11. Dupont C., Clarke A. J. (1991) Dependence of lysozyme-catalysed solubilization of Proteus mirabilis peptidoglycan on the extent of O-acetylation. Eur. J. Biochem. 195, 763–769 [DOI] [PubMed] [Google Scholar]
- 12. Sreerama N., Woody R. W. (1993) A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209, 32–44 [DOI] [PubMed] [Google Scholar]
- 13. Sreerama N., Venyaminov S. Y., Woody R. W. (1999) Estimation of the number of α-helical and β-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 8, 370–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sreerama N., Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 15. Sreerama N., Venyaminov S. Y., Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: inclusion of denatured proteins with native proteins in the analysis. Anal. Biochem. 287, 243–251 [DOI] [PubMed] [Google Scholar]
- 16. Scheurwater E. M., Clarke A. J. (2008) The C-terminal domain of Escherichia coli YfhD functions as a lytic transglycosylase. J. Biol. Chem. 283, 8363–8373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garnier J., Gibrat J. F., Robson B. (1996) GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266, 540–553 [DOI] [PubMed] [Google Scholar]
- 19. Cuff J. A., Clamp M. E., Siddiqui A. S., Finlay M., Barton G. J. (1998) JPred: a consensus secondary structure prediction server. Bioinformatics 14, 892–893 [DOI] [PubMed] [Google Scholar]
- 20. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 21. McGuffin L. J., Bryson K., Jones D. T. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 22. Bryson K., McGuffin L. J., Marsden R. L., Ward J. J., Sodhi J. S., Jones D. T. (2005) Protein structure prediction servers at University College London. Nucleic Acids Res. 33, W36–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 24. Clarke A. J. (1993) Extent of peptidoglycan O acetylation in the tribe Proteeae. J. Bacteriol. 175, 4550–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clarke A. J. (1993) Compositional analysis of peptidoglycan by high performance anion-exchange chromatography. Anal. Biochem. 212, 344–350 [DOI] [PubMed] [Google Scholar]
- 26. Correia M. A., Prates J. A., Brás J., Fontes C. M., Newman J. A., Lewis R. J., Gilbert H. J., Flint J. E. (2008) Crystal structure of a cellulosomal family 3 carbohydrate esterase from Clostridium thermocellum provides insights into the mechanism of substrate recognition. J. Mol. Biol. 379, 64–72 [DOI] [PubMed] [Google Scholar]
- 27. Blake C. C., Cassels R., Dobson C. M., Poulsen F. M., Williams R. J., Wilson K. S. (1981) Structure and binding properties of hen lysozyme modified at tryptophan 62. J. Mol. Biol. 147, 73–95 [DOI] [PubMed] [Google Scholar]
- 28. Mølgaard A., Kauppinen S., Larsen S. (2000) Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure 8, 373–383 [DOI] [PubMed] [Google Scholar]
- 29. Lo Y. C., Lin S. C., Shaw J. F., Liaw Y. C. (2003) Crystal structure of Escherichia coli thioesterase I/protease I/lysophospholipase L1: consensus sequence blocks constitute the catalytic center of SGNH-hydrolases through a conserved hydrogen bond network. J. Mol. Biol. 330, 539–551 [DOI] [PubMed] [Google Scholar]
- 30. Chang R. C., Chen J. C., Shaw J. F. (1996) Site-directed mutagenesis of a novel serine arylesterase from Vibrio mimicus identifies residues essential for catalysis. Biochem. Biophys. Res. Commun. 221, 477–483 [DOI] [PubMed] [Google Scholar]
- 31. Blair D. E., Schüttelkopf A. W., MacRae J. I., van Aalten D. M. (2005) Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl. Acad. Sci. U.S.A. 102, 15429–15434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hekmat O., Tokuyasu K., Withers S. G. (2003) Subsite structure of the endo-type chitin deacetylase from a deuteromycete, Colletotrichum lindemuthianum: an investigation using steady-state kinetic analysis and MS. Biochem. J. 374, 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]