Background: The domains in RNA polymerase involved in elongation slippage are unknown.
Results: We isolated E. coli RNA polymerase rpoB mutants with altered transcriptional slippage.
Conclusion: The fork domain of RNA polymerase controls slippage. Biochemical analysis of the mutants validates the genetic schemes.
Significance: Our work sheds light on the mechanism for maintenance of RNA-DNA register during transcription.
Keywords: RNA, RNA Editing, RNA Polymerase, Transcription, Transcription Regulation
Abstract
Transcription fidelity is critical for maintaining the accurate flow of genetic information. The study of transcription fidelity has been limited because the intrinsic error rate of transcription is obscured by the higher error rate of translation, making identification of phenotypes associated with transcription infidelity challenging. Slippage of elongating RNA polymerase (RNAP) on homopolymeric A/T tracts in DNA represents a special type of transcription error leading to disruption of open reading frames in Escherichia coli mRNA. However, the regions in RNAP involved in elongation slippage and its molecular mechanism are unknown. We constructed an A/T tract that is out of frame relative to a downstream lacZ gene on the chromosome to examine transcriptional slippage during elongation. Further, we developed a genetic system that enabled us for the first time to isolate and characterize E. coli RNAP mutants with altered transcriptional slippage in vivo. We identified several amino acid residues in the β subunit of RNAP that affect slippage in vivo and in vitro. Interestingly, these highly clustered residues are located near the RNA strand of the RNA-DNA hybrid in the elongation complex. Our E. coli study complements an accompanying study of slippage by yeast RNAP II and provides the basis for future studies on the mechanism of transcription fidelity.
Introduction
Escherichia coli RNA polymerase (RNAP)6 is a multisubunit enzyme that exists in two forms: holoenzyme (α2ββ′ωσ) and core enzyme (α2ββ′ω). The holoenzyme recognizes a promoter to initiate transcription. After initiation, σ is released leaving core RNAP engaged in transcription elongation and subsequent termination (1, 2). Mutational analyses of RNAP have identified important regulatory regions in the enzyme affecting different steps in the transcription cycle (3). Regions in the large subunits of bacterial RNAP, β and β′, form the active center of the enzyme and template/transcript binding sites (1). Residues in these regions are conserved in eukaryotic RNAP II (4), suggesting that the functions of RNAP are also conserved throughout evolution.
Transcription fidelity (5) is an important mechanism to ensure the accuracy of genetic information. Despite its importance, transcription fidelity is understudied, mainly because of the difficulties of identifying phenotypes associated with an increase in transcription errors (6), which are often masked by the much higher error rates in translation (7, 8). In this regard, earlier pioneering studies examining the effect of RNAP mutants on misreading of a point nonsense mutation during transcription (9, 10) have been complicated by the frequency of nontranscriptional errors induced under stress conditions (11). It becomes evident that new approaches that do not depend on the misreading of point mutations in a message are needed to study transcription fidelity. Two mutations in the largest Rpb1 subunit (the homolog of β′) of the yeast RNAP II were recently reported with a pronounced effect on transcription fidelity in vivo and in vitro (12, 13). The slow growth phenotype of the yeast Rpb1 fidelity mutants highlighted the potential importance of transcription fidelity for cell viability (12, 13). Point mutations D675Y in the β subunit and N458D/S in the β′ subunit of E. coli RNAP that reduced fidelity in vitro were also described (14, 15). Rifampicin-resistant P564L mutation in the β subunit (also known as ack-1 (9)) was reported to decrease transcription fidelity in vivo and was recently shown to increase molecular noise in E. coli cells because of transient RNA errors (16).
There are two types of transcription mistakes: misincorporation of nucleotides and transcription slippage. The mechanism of transcription slippage is unknown, but its biological consequences are well documented (17–27). Slippage constitutes a temporary loss of DNA register by RNAP leading to insertions and deletions of nucleotides in mRNA (18, 19). It is proposed that accumulation of the mistranslated proteins derived from transcription slippage may be associated with human diseases and tumorigenesis, including Alzheimer disease, Down syndrome, and colorectal cancer (24, 28–31). Bacterial RNAP makes slippage products during both initiation (25, 26, 32, 33) and elongation (20, 22, 27, 34–36). Some of these slippage events are involved in gene regulation and bacterial pathogenesis. E. coli RNAP mutants in the rpoB gene encoding the β subunit that alter initiation at promoters containing a short run of A/T base pairs have been studied (37, 38); however, RNAP mutants that alter transcriptional slippage during elongation have not been identified. High efficiencies of transcription slippage on plasmid-borne DNA templates containing homopolymeric runs of 11A/T during elongation in E. coli cells (27) reveal a potential way to study transcription infidelity genetically.
In this study, we analyzed transcription slippage during elongation in vivo using a reporter gene in the E. coli chromosome and developed a genetic system to isolate and characterize RNAP rpoB mutants with altered transcriptional slippage phenotypes. Our study took advantage of genetic results from a yeast system used for studying slippage by RNAP II as described in the accompanying article (39). We identified a cluster of amino acid residues in RNAP encapsulating the RNA-DNA hybrid region that controls transcription slippage during elongation. Biochemical analysis of representative slippery mutant RNAPs confirmed involvement of this region of the β subunit in regulation of transcription slippage.
EXPERIMENTAL PROCEDURES
Molecular Biology Reagents and Techniques
Standard molecular biology and DNA techniques are as described (40). Reagents for PCR were from Invitrogen, NTPs were from GE HealthCare, and chemicals were from Sigma-Aldrich. DNA and RNA oligonucleotides were from IDT (San Diego, CA).
Bacterial Strains and Bacterial Techniques
Bacterial strains used in this study are E. coli K-12 strain MG1655 and its derivatives (Tables 1 and 2). The bacterial techniques and media used have been described (41). The β-gal activities (Miller units) were assayed by using overnight cultures grown in LB, as described (41). To correct for potential variations in different experiments, wild type controls were always included, and at least two duplicates for each of the strains tested were assayed in independent experiments. Random mutations in a plasmid-borne rpoB gene were introduced in an E. coli mutator strain XL1-Red (XL1-Red, from Stratagene) according to the manufacturer's manual.
TABLE 1.
Strains and plasmids used in the study
| Relevant genotype/comment | Source | |
|---|---|---|
| Strain | ||
| MG1655 | Prototype E. coli K-12 strain | Carol Gross |
| CAG3002 | MG1655 btuB::Tn10 linked to rpoBC | Ref. 48 |
| CAG3246 | MG1655 recA56 srl::Tn10 | Ref. 48 |
| NY348 | rpoC-his tag fusion<>kan | Don Court |
| NC397 | lacI′<>kan<>cat sacB<>lacZYA harboring a defective λ cI857 prophage for recombineering | Ref. 44 |
| MM111 | NC397 lacI′<>spacer<>′lacZ | This work |
| MH131A | MM111 mhpR::cat<>pTac*lacI' | This work |
| Slippage strains | MG1655 mhpR::cat<>pTac*lacI'<> spacer<>′lacZ | This work |
| Plasmids | ||
| pSim19 | Provides λ Red system; curable at ≥37 °C because of temperature-sensitive plasmid replication; spectomycin resistance | Don Court |
| pDJ853 | Constitutive expression of rlpJL rpoB-his | This work |
TABLE 2.
Chromosomal rpoB RifR mutants used in the study
| Strain | Amino acid change | Codon change | Source |
|---|---|---|---|
| CC256 | V146F | GTT → TTC | This work |
| CC257 | V146G | GTT → GGG | This work |
| CC258 | Q148H | CAG → CAC | This work |
| CC259 | Q148L | CAG → ΤΤΑ | This work |
| CC262 | Q148K | CAG → AAA | This work |
| CC261 | Q148R | CAG → CGA | This work |
| CC148 | L511P | CTG → CCC | This work |
| CC141 | S512F | TCT → TTT | This work |
| CC143 | S512Y | TCT → TAC | This work |
| rpoB3445 | Δ[G507-Q510] | Δ12 nt | Ref. 48 |
| rpoB8 | Q513P | CAG → CCG | Ref. 48 |
| rpoB101 | Q513L | CAG → CTG | Ref. 48 |
| CC264 | Q513C | CAG → TGT | This work |
| CC271 | Q513D | CAG → GAT | This work |
| rpoB113 | D516N | GAC → AAC | Ref. 48 |
| rpoB148 | D516V | GAC → GTC | Ref. 48 |
| CC144 | D516Y | GAC → TAT | This work |
| rpoB3051 | [D516Q517]X2 | [GACCAG]X2 | Ref. 48 |
| CC140 | N518D | AAC → GAT | This work |
| rpoB3595 | S522F | TCT → TTT | Ref. 48 |
| CC137 | S522Y | TCT → TAC | This work |
| CC117 | T525R | ACG → CGC | This work |
| rpoB2 | H526Y | CAC → TAC | Ref. 48 |
| CC121 | H526L | CAC → CTT | This work |
| CC122 | H526N | CAC → AAT | This work |
| rpoB3401 | R529C | CGT → TGT | Ref. 48 |
| CC253 | R529L | CGT → TTA | This work |
| CC254 | R529S | CGT → TCT | This work |
| rif180 | R529H | CGT → CAT | Ref. 61 |
| rpoB114 | S531F | TCC → TTC | Ref. 48 |
| CC124 | S531Y | TCC → TAT | This work |
| rpoB3449 | ΔΑ532 | ΔGCA | Ref. 48 |
| CC125 | A532E | GCA → GAG | This work |
| rpoB3443 | L533P | CTC → CCC | Ref. 48 |
| CC460 | L533H | CTC → CAT | This work |
| CC461 | L533R | CTC → CGA | This work |
| CC136 | G534A | GGC → GCA | This work |
| CC130 | G534S | GGC → AGT | This work |
| rpoB3370 | T563P | ACC → CCC | Ref. 48 |
| rpoB111 | P564L | CCT → CTT | Ref. 48 |
| CC276 | P564H | CCT → ATG | This work |
| CC463 | G570C | GGT → TGC | This work |
| CC464–116 | G570Q | GGT → CAA | This work |
| CC464–114 | G570M | GGT → ATG | This work |
| CC464–104 | G570A | GGT → GCA | This work |
| rpoB7 | I572F | ATC → TTC | Ref. 48 |
| CC139 | I572N | ATC → AAT | This work |
| CC267 | I572K | ATC → AAA | This work |
| CC149 | I752T | ATC → ACA | This work |
| CC134 | S574F | TCT → TTC | This work |
| rpoB3406 | R687H | CGT → CAT | Ref. 48 |
Strains and Plasmid Constructions
Various mutations in rpoB were constructed using the Lambda Red-mediated recombineering method (42, 43). To make elongation slippage fusion strains, single-stranded oligonucleotides containing a 50-nt segment of lacI that encodes amino acid residues 16–33, a 9–10-nt spacer, and a 53-nt segment of lacZ that encodes amino acid residues 15–33 were used to replace the chromosomal lacI-lacZ region of the NC397 strain (44). Then the lacI promoter region of the resulting strains was replaced with the tac (or tac*) promoter (pKK223-3; Pharmacia) using a 1-kb PCR fragment, in which a cat cassette (45) was flanked by the mhpR and lacI sequences using the following two primers: DJ24F, 5′-CTGAATCTGGTGTATATGGCGAGCGCAATGACCATTGAAACCTGTGACGGAAGATCACTTCGC-3′;and DJ24R, 5′-TGGTTTCACATTCACCACCCTGAATTGACTCTCTTCCGCCACACATTATACGAGCCGATGATTAATTGTCAACTAAGGGCACCAATAACTGCC-3′. The resulting constructs were confirmed by DNA sequencing. Those fusions were moved into MG1655 by P1 transduction, and the transductants were selected on LB plates containing chloramphenicol (15 μg/ml) and then screened for appropriate Lac phenotypes. Alternatively, similar fusions were constructed in derivatives of MG1655 harboring a plasmid-borne Lambda Red system that could be cured at ≥37 °C (46). New rpoB Rifr mutations were made by allele exchange with single-stranded oligonucleotides in strains that contained the rpoC-his tag fusion immediately linked to a kan cassette as verified by DNA sequencing. The resulting rpoB Rifr alleles were then introduced to different strains by phage P1 transduction. The transductants were selected on LB plates containing 30–50 μg/ml of kanamycin followed by screening of the Rifr phenotype.
The plasmid pDJ853, which contained the rpoB-his tag fusion, was constructed in two steps. First, a 0.9-kb SphI-SacI fragment in the wild type rpoB derivative of pXT7β (47) was replaced with a ∼110-bp SphI-SacI DNA fragment that encoded the last 24 amino acids of the β subunit and the additional GGHHHHHH His tag residues to yield plasmid pDJ2080His (pT7wt rpoB-his). Second, a 1.6-kb HindIII fragment from pDJJ11 (48) that contained the rplJL promoter and the genes at its normal location before the rpoB gene was inserted into the HindIII site of pDJ2080His, resulting in pDJ853 (pRplJL-rpoB-his) or prpoB.
RNAP Purification and Transcription in Vitro
E. coli RNAP carrying a His6 tag at the C terminus of the β′ subunit (from wild type and six rpoB mutants) was immobilized on Ni2+-NTA-agarose beads (Qiagen) directly from a crude cell lysate. The cells were grown in 8 ml of LB to an optical density at 600 nm of 0.8–1, centrifuged (Eppendorf 5804R, rotor A-4-44: 5,000 rpm, 6 min), and resuspended in 350 μl of lysis buffer (300 mm Tris acetate, pH 7.9, 100 mm potassium acetate, 10 mm MgCl2, 1 mm EDTA, 4 mm 2-mercaptoethanol). All of the steps were performed at 4 °C. The cells were sonicated using a microprobe tip and Sonicator-Ultrasonic Processor VHX 750 watt (model GEX 750; PG Scientific) by five cycles of a 5-s pulse and 45-s cooling periods. The samples were centrifuged (10,000 rpm, 10 min). The supernatant (∼300 μl) was transferred to a 1.5-ml Eppendorf tube and mixed with 100 μl of 30% suspension of Ni2+-NTA-agarose beads, which was prewashed with transcription buffer as described previously (49), and the 200 μl of premixed 3 m KCl with 30 mm of imidazole (pH 7.9) was added. The samples were shaken for 30 min (1,100 rpm) and centrifuged on the table-top centrifuge, and the supernatant was discarded. The Ni2+-NTA-agarose beads were washed with transcription buffer again, and the immobilized RNAP was used in transcription assays. The immobilized RNAP was ∼80% pure with the yield ∼1–2 μg/1 ml of the culture.
Ternary elongation complexes (TEC) were assembled with the following DNA and RNA oligonucleotides: RNA9, 5′-AUC GAG AGG-3′; RNA10, 5′-AUCGAGAGGG-3′; NDS79 (nontemplate DNA strands 79 nt long), 5′-CCT ATA GGA TAC TTA CAG CCA TCG AGA GGG ACA CGG CGA ATA GCC ATC CCA ATC CAC ACG TCC AAC GGG GCA AAC CGT A-3′; TDS76 (template DNA strand 76 nt long), 5′-GGT TTG CCC CGT TGG ACG TGT GGA TTG GGA TGG CTA TTC GCC GTG TCC CTC TCG ATG GCT GTA AGT ATC CTA TAG G-3′; NDS48, 5′-GGT ATA GGA TAC TTA CAG CCA TCG AGA GGG AAA AAA AAA CGA GGC GAA-3′; TDS48, 5′-TTC GCC TCG TTT TTT TTT CCC TCT CGA TGG CTG TAA GTA TCC TAT ACC-3′; NDS60, 5′-GGT ATA GGA TAC TTA CAG CCA TCG AGA GGG AAA AAA AAA CGA GGC GAA TAG AGA ACC CAA-3′; and TDS60, 5′-TTG GGT TCT CTA TTC GCC TCG TTT TTT TTT CCC TCT CGA TGG CTG TAA GTA TCC TAT ACC-3′. TDS62/NDS62 oligonucleotides were similar to TDS48/NDS48, except they were longer and contained 11-nt homopolymeric tracts.
Assembly of TEC9 (number indicates the RNA length in the TEC is 9 nt long, RNA9) on the beads, washing, elution, and 5′ RNA labeling were performed as described (50). For the bulk elongation assay (see Fig. 3), the 5′-labeled RNA9 and nonlabeled TDS76/NDS79 DNA oligonucleotides were used, and the reaction was performed in solution after elution of the TEC9 with imidazole. The RNA in TEC9 was elongated to 20-nt length with ATP, CTP, and GTP (10 μm each) for 1 min at room temperature. 10 μm UTP was added, at different times the reaction was stopped with gel loading buffer, and the products were subjected to PAGE as described (50). For the slippage assay measuring the rate of ATP incorporation by slippage (see Fig. 4), another TEC9 was made using nonlabeled RNA9 and TDS48/NDS48 oligonucleotides and elongated to TEC10 with 0.3 μm [α-32P]GTP (3000 Ci/mmol; PerkinElmer Life Sciences) for 4 min followed by washing with transcription buffer. The slippage reaction was performed on Ni2+-NTA beads. To initiate slippage, ATP (10 μm) was added to TEC10 for different times, and the samples were processed as described above. The assay that was established to detect insertions and deletions during elongation on the 9A (or 11A) track (see Fig. 5) was similar to the slippage assay except that the longer TDS60 template and RNA10 primer were used, and a three times larger amount of the beads and the RNA-DNA oligonucleotides were employed. Also, labeling of the RNA with [α-32P]CTP was used to replace the labeling with [α-32P]GTP. TEC10 was directly assembled on TDS60/NDS60 (for 9A tract) or TDS62/NDS62 (for 11A tract) with unlabeled RNA10 followed by the addition of a mixture of the nonlabeled ATP (10 μm), CTP (100 μm), and 0.8 μm [α-32P]CTP (3000 Ci/mmol; PerkinElmer Life Sciences) in 150 mm KCl for 7 min (TEC10 → TEC20 or TEC22 reaction). The beads were washed with transcription buffer to remove the labeled NTPs, and the samples were processed for PAGE as described above.
FIGURE 3.
General in vitro elongation properties of the rpoB mutants. A single round of transcription was performed with 10 μm NTPs and 5′-labeled RNA9 primer in two steps as indicated in the scheme at the top. TEC20 once formed was elongated with four NTPs for 10–180 s to form a runoff RNA. The arrow marks the position of the 20-nt RNA in TEC20; the arrowheads mark the 45-nt runoff RNA and the major transcription pause sites. The numbers at the bottom indicate the effects of a mutation (fold increase or decrease) on the bulk elongation rate, and NA indicates that the rate was not affected relative to wild type under the low NTP conditions. The similar results were obtained with high 200 μm NTPs (data not shown).
FIGURE 4.
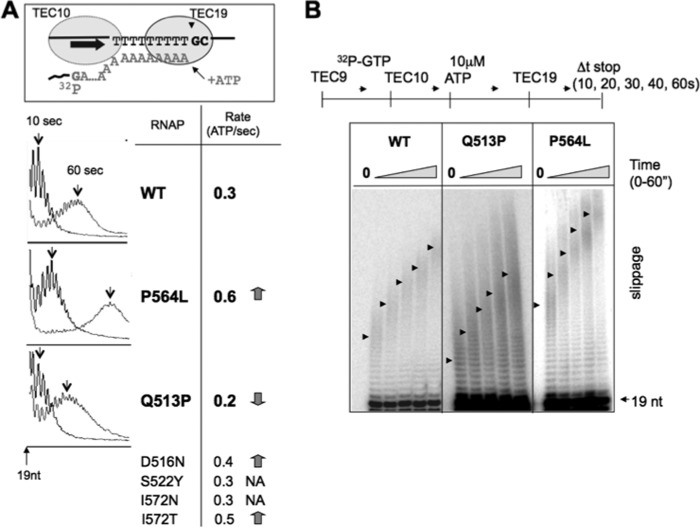
Processive slippage of RNAP halted at the end of the 9A tract. A, a cartoon at the top depicts TEC19 labeled at the +10G position of the RNA that idles at the end of the 9A tract in the presence of 10 μm ATP. The graphs show the extension of the slippage products beyond the 19-nt length by slippage after 10 s (arrows) and 60 s (arrows) of incubation with ATP (WT and β-P564L and β-Q513P mutants). The second column presents an average slippage rate for six mutants calculated as a number of ATP molecules added to the RNA/s. The decrease or increase of the ATP addition rate relative to WT is indicated by downward and upward arrows, respectively, and where NA means not affected. B, the representative autoradiogram shows time-dependent slippage in TEC19 formed by β-Q513D, Q513P, and P564L mutants. The arrows indicate the center of the peak for the distribution of the slippage products at each time point after ATP addition.
FIGURE 5.
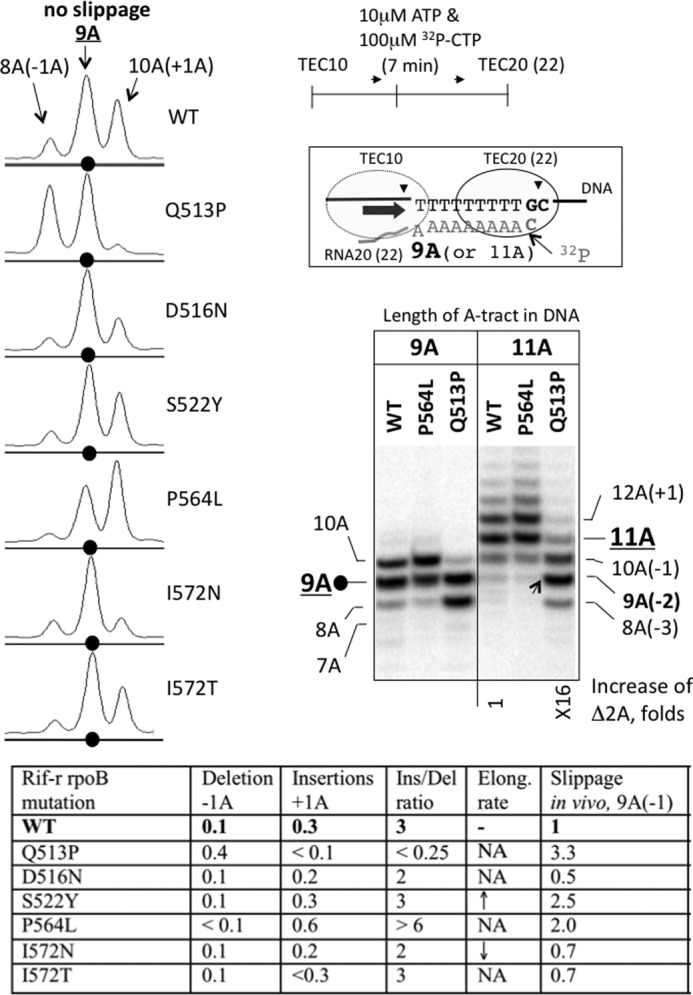
Detection of deletions and insertions in the RNA during transcription through 9A and 11A tracts in vitro. The cartoon shows the 3′-labeled TEC20 (or TEC22) halted one nucleotide downstream from the end of the 9A (or 11A) tract after elongation of unlabeled TEC10 with ATP and [α-32P]CTP. The autoradiogram depicts the slippage products (7A, 8A, and 10A RNAs for 9A tract; 8–10A and >11A RNAs for 11A tract) generated by the WT RNAP and β-P564L and β-Q513P mutants relative to the expected 9A or 11A transcript. The number under the autoradiogram shows the increase in the yield of 2A deletion in 11A tract caused by β-Q513P mutation. The phosphorimaging scans (on the left) reveal the difference in the pattern of slippage products by the WT RNAP and the six rpoB mutants. Black dots indicate the RNA product containing the 9A tract, and the slippage products are labeled according to the number of adenine residues in the tract. The table panel at the bottom summarizes slippage properties of the rpoB mutants. The numbers in the second and third columns represent the fraction of 1A deletion and 1A insertion normalized to the total amount of the CMP-labeled RNAs (include 8A, 9A, and 10A products). The fourth column shows the ratio between insertions and deletions for each mutant. The fifth column presents the effect of the mutations on elongation rate of RNAP on heteropolymeric DNA reproduced from the results of Fig. 3. The last column displays the relative slippage phenotype with the 9A(−1) reporter in vivo (from Table 4).
RESULTS AND DISCUSSION
Rationale
Identification of E. coli RNAP mutations affecting transcription slippage during elongation involved monitoring expression of a reporter gene (lacZ) carrying insertion of a homopolymeric A tract in the coding region of β-gal (27). For instance, the insertion of a 9-nt-long A tract and 1-nt deletion of the lacZ sequence adjacent to the tract generated a frameshift that inactivated the ORF encoding β-gal. The advantage of the E. coli lacZ reporter was that the change in color of colonies on indicator McConkey lactose (MacLac) plates could directly identify potential mutations in RNAP that alter transcription slippage under the conditions used. Altered expression of the lacZ reporter in the RNAP mutants could be due to nonslippage effects, such as changes in transcription initiation compared with wild type. To identify and discard those RNAP mutants in which the color phenotype derived from an effect on the promoter activity of the reporter gene, we introduced an additional lacZ reporter carrying a nonslippery in-frame 9-nt-long spacer insertion to normalize slippage activity to promoter strength. To further validate this approach, we performed biochemical analysis of the mutant RNAPs in vitro for their elongation properties and slippage at the homopolymeric A tracts.
Construction of in Vivo Slippage Reporter Strains
To avoid complications caused by plasmid copy number variations, we engineered a chromosomal-based slippage strain rather than a plasmid-borne system as previously described (27). The genome region encompassing the coding sequences for the C terminus of LacI and the promoter and N terminus of LacZ were deleted, and the two genes were joined with a spacer resulting in a lacI′-spacer-′lacZ fusion (Fig. 1A). The spacer AAAAAAAAA(−1), called 9A(−1), contained an insertion of a homopolymeric run of 9A, with the number in the parentheses indicating a nucleotide deletion in reading frame relative to the downstream fused ′lacZ gene. A 0 frame is defined as the same reading frame as ′lacZ; a −1 is out of frame by a one nucleotide deletion (27). Thus, for restoration of lacZ ORF from the 9A(−1) strain, a 1-nt insertion or a 2-nt deletion would be needed during transcription through the 9A track. We also made two nonslippage strains, one with spacer sequences of AAAATAAAA(−1) called 4AT4A(−1) as an out of frame control and another of AAAAATAAAA(0) called 5AT4A(0) as an in-frame control. These spacer variants were important for the analysis of slippage phenotypes of wild type and mutant RNAPs (see below). Hereafter these strains are named after their spacer designations.
FIGURE 1.
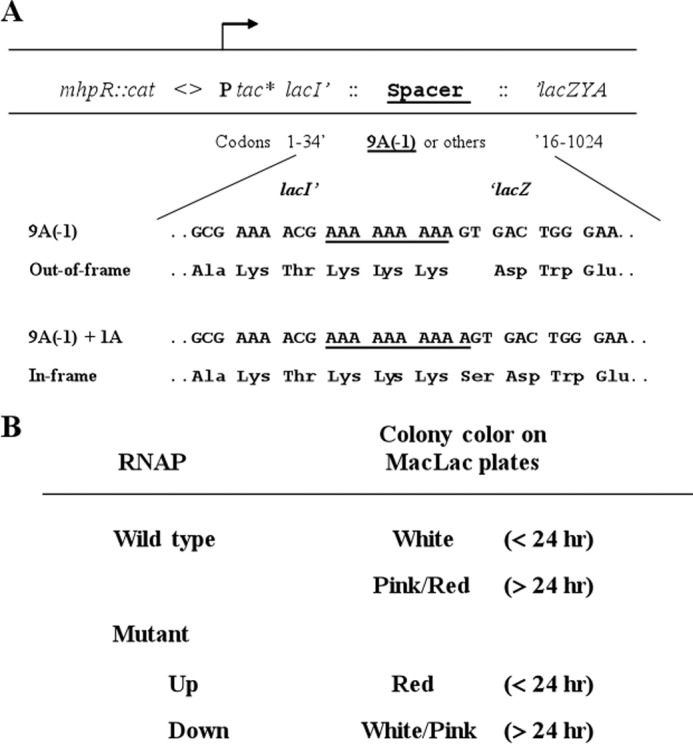
A, schematic illustration of a set of chromosomal constructs used for the isolation and characterization of RNAP slippage mutants. These reporter strains differed in the spacer sequence and the resulting reading frame relative to the fused ′lacZ gene. The numbers of residues of the N-terminal of LacI′ 1–34 and C-terminal of ′LacZ, respectively, are indicated. As examples, the flanking DNA sequences of lacI′ and ′lacZ (and their codons) that are fused with the 9A(−1) spacer are shown; amino acid residues, which are encoded by the spacer sequences (underlined), either out-of-frame or restored in-frame with the lacZ because of slippage, are indicated. The tac* promoter had a −35 mutation (TTGACA → TTGACC) compared with the wild type tac promoter. Selection for the cat cassette facilitated the introduction of the constructs into different strain backgrounds. See text for details. B, identification of potential RNAP slippage mutants using the 9A(−1) reporter. For wild type RNAP, the expression of the out of frame ′lacZ was low, and colonies were a white color on MacLac plates after an overnight incubation (<24 h) and became a pink/red color after a prolonged incubation (>24 h). RNAP mutants with increased expression of the reporter (Up) exhibited red colonies on MacLac plates after less than 24 h of incubation. However, RNAP mutants with decreased expression of the reporter (Down) formed white/pink colonies even after a prolonged incubation.
To develop a genetic scheme for the isolation of RNAP mutants that alter transcription slippage during elongation, it was critical to adjust expression of the reporter to the level enabling identification of the candidate mutant colonies by their color on indicator MacLac plates. Our initial studies found that although the lacI promoter was too weak, the tac promoter was too strong (data not shown) to be used for the identification of RNAP mutants. To tune the expression levels of the lacI′-spacer-′lacZ fusions, the chromosomal lacI promoter in each of these strains was replaced with a mutated tac promoter (ptac*) carrying a change of A for C in the “−35” region (TTGACC) compared with the (TTGACA) tac promoter. The ptac* promoter supported a level of the β-gal expression suitable for identification of slippage mutations on MacLac plates. For example, the expression level of the lacZ fusion in the 9A(−1) slippage strain was such that single colonies of the cells appeared white after ∼15 h of incubation, turning to pink/red after ≥24 h of incubation on MacLac plates at 37 °C. The color of colonies was also affected by temperature during incubation. This phenotype provided a useful screening scheme for the isolation of potential RNAP mutants by looking for color changes of colonies on the MacLac plates under the same growth conditions (Fig. 1B).
The β-gal activity of the slippage strain and nonslippage control strains were determined (Table 3). The activity produced from the in-frame nonslippage 5AT4A(0) strain was ∼655 units, whereas the out-of-frame nonslippage 4AT4A(−1) reporter produced ≤1 unit of β-gal. Importantly, the 9A(−1) slippage reporter produced ∼35 units of β-gal, indicating that wild type RNAP was undergoing slippage at homopolymeric A/T runs in vivo, as was shown for runs of 11A nt (27). The colonies of the 9A(−1) reporter strain were white to pink/red on MacLac plates, depending on the incubation time and temperature as described above.
TABLE 3.
β-Gal activities of different reporter strains
| Reporter staina | β-Gal activities |
|---|---|
| 9A(−1) | 35 ± 4 |
| 4AT4A(−1) | ∼1 |
| 5AT4A(0) | 655 ± 33 |
a The spacer sequence and the reading frame relative to the fused ′lacZ in parentheses are described in the text.
Isolation of Plasmid-borne rpoB Mutations Causing an Increased Elongation Slippage Phenotype
The rpoB gene encodes the β subunit, which forms a part of the active center in RNAP. The β subunit also encases the RNA side of the RNA-DNA hybrid in the elongation complex (1, 2). Moreover, certain rpoB mutations affect slippage during initiation (38).7 Reasoning that some RNAP mutations conferring an enhanced slippage phenotype during elongation could be lethal, we decided to isolate rpoB mutations initially from a plasmid-borne gene by looking for increased lacZ expression in the presence of a chromosomal wild type rpoB gene in the 9A(−1) reporter strain.
A new rpoB expression construct with specific features was needed for the isolation of rpoB mutants. We constructed a plasmid-borne rpoB gene (pDJ853) prpoB with the following properties: (i) the expression of the rpoB gene is constitutive at a level that was able to complement chromosomal rpoB temperature-sensitive mutants (51) without affecting the growth of wild type cells (data not shown); and (ii) the rpoB gene on the plasmid is fused to a hexahistidine tag at the C terminus of β, so that RNAP carrying the mutation can be separated from the wild type counterpart for in vitro analysis of slippage. When the prpoB plasmid was introduced into the 9A(−1) reporter strain, colonies appeared white on a MacLac+Amp plate after overnight incubation at 37 °C, similar to the strain without the plasmid.
To introduce random mutations in rpoB, the prpoB plasmid was propagated in an E. coli mutator strain (XL1-Red; Stratagene). The mutagenized prpoB pools were introduced into the 9A(−1) reporter strain by transformation. The transformants were screened on the MacLac+Amp indicator plates at different temperatures. Occasionally, fish eye (red in the center of a white colony) or red colonies appeared, indicative of increased expression of the 9A(−1) lacZ fusion. These cells exhibiting a “Lac+” phenotype harbored prpoB candidates for RNAP slippage mutants. Plasmid DNA from each of the candidates was purified and reintroduced into a ΔrecA derivative of the 9A(−1) reporter strain to prevent recombination between the chromosomal wild type rpoB gene and a plasmid-borne rpoB mutation. All of the six prpoB mutations that exhibited a Lac+ phenotype after retransformation were sequenced, and changes in the β subunit are shown in Table 4. All of these mutations conferred a slow growth phenotype on the host cells at 30 °C, and cells harboring the plasmid-borne M1273I mutation had an additional temperature-sensitive phenotype at 42 °C.
TABLE 4.
Effects of plasmid-borne rpoB mutations on elongation slippage in vivo
The values are the averages of at least two independent experiments. Each experiment was performed with minimum of two repetitions. The values of different independent experiments generally deviated less than 10% from the average value. WT indicates 35 ± 4 units of β-Gal in 9A(−1) and 655 ± 33 units in 5AT4A (see Table 3).
| RNAP mutation |
Relative β-Gal activity |
Relative slippage phenotype (9A(−1)/5AT4A(0)) | ||
|---|---|---|---|---|
| Amino acid change | Codon change | In 9A(−1) | In 5AT4A(0) | |
| Wild type | 1.0 | 1.0 | 1.0 | |
| D444G | GAT → GGT | 1.7 | 1.0 | 1.7 |
| R451C | CGT → TGT | 1.8 | 1.1 | 1.6 |
| A543V | GCA → GTA | 1.3 | 1.1 | 1.2 |
| G570D | GGT → AGT | 1.6 | 1.3 | 1.2 |
| R687C | CGT → TGT | 1.8 | 1.1 | 1.6 |
| M1273I | ATG → ATA | 1.9 | 1.3 | 1.5 |
The β-gal expression level of the lacZ fusion in the ΔrecA 9A(−1) reporter strain harboring each prpoB mutation, as well as the wild type prpoB clone, was determined (Table 4). Relative β-gal activity of each mutant was calculated by dividing the β-gal activity from cells with an rpoB mutation over that of the cells containing the wild type plasmid. Consistent with the Lac+ phenotypes on MacLac+Amp indicator plates, all of the six plasmid-borne prpoB mutations increased expression of the 9A(−1) out-of-frame lacZ fusion compared with wild type (Table 4). To correct for potential effects of the mutations on transcription initiation at the ptac* promoter, the plasmids with the six rpoB mutations and wild type were introduced into a ΔrecA derivative of the 5AT4A(0) nonslippage control strain, and the relative β-gal activity of the 5AT4A(0) cells harboring each of these mutations was calculated similarly (Table 4). A relative slippage phenotype of each RNAP mutant was estimated as a ratio of the two relative β-gal activities (Table 4). By this analysis, four of the six mutants increased elongation slippage ≥1.5-fold compared with wild type.
Isolation of Chromosomal rpoB RifR Mutants with Altered Elongation Slippage Phenotypes
We noted that all of the six plasmid-borne prpoB slippage mutations, with the exception of M1273I, are located in the elongation complex bordering the RNA strand of the 9-bp RNA-DNA hybrid region (Fig. 2); in particular, two of them (G570D and R687C) affected amino acid residues harboring other previously identified mutations that cause resistance to the antibiotic rifampicin (RifR) (48, 52). Although none of our new slippage mutants conferred a RifR phenotype, this observation prompted us to test the effects of known chromosomal rpoB RifR mutations on elongation slippage.
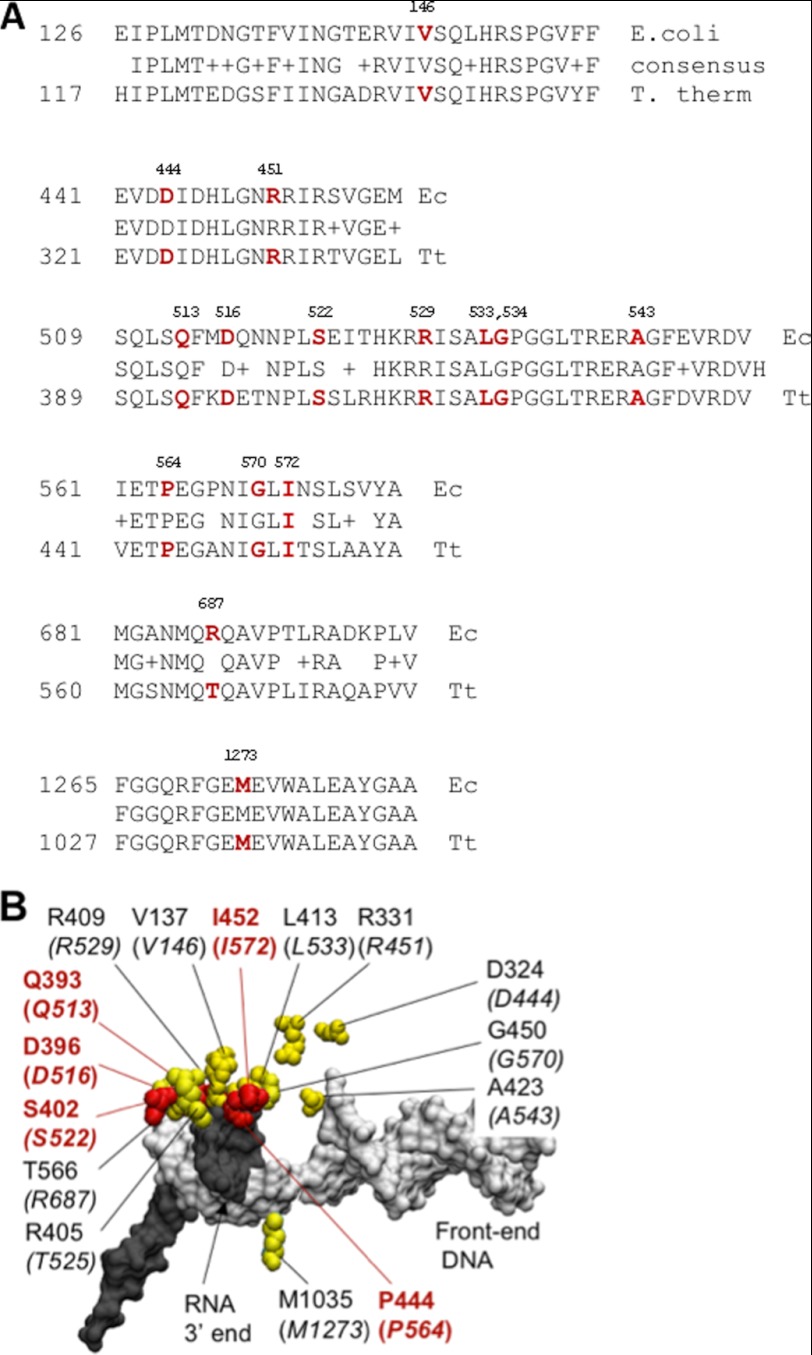
FIGURE 2.
Slippage mutations in the β subunit have close proximity to the 9-bp RNA-DNA hybrid. A, a sequence alignment of the Rif-binding region in the β subunit of T. thermophilus (Tt) and E. coli (Ec) RNAP (the fork domain). The amino acid residues harboring both the plasmid-borne and the chromosomal rpoB mutations that affect slippage on a 9A tract in vivo are shown in red. B, the amino acid residues of the β subunit involved in control of slippage are highly clustered (shown in yellow) in the structure of the T. thermophilus RNAP TEC (Protein Data Bank code 2O5J) (2). The corresponding positions in E. coli RNAP are shown in brackets. The residues carrying mutations with the confirmed slippage phenotype in vitro are highlighted in red. The RNA transcript and the template DNA are colored gray and white, respectively.
E. coli RifR mutations are exclusively located in the rpoB gene and affect 25 clustered amino acid residues of the fork domain of the β subunit (48, 52, 53). All are clustered along the RNA flank of the 9-bp RNA-DNA hybrid region near the active center of RNAP (Fig. 2; see also Fig. 6B, colored in yellow). In addition to testing all chromosomal RifR mutants from our collection (48), we also generated many other chromosomal rpoB RifR mutations (52) by Lambda Red recombineering (43, 54). All of these RifR mutations were made in the strain carrying the His-tagged rpoC gene on the chromosome. We examined more than 50 RifR mutations affecting 24 positions (Tables 2) for their effects on elongation slippage in the 9A(−1) reporter strain. Many of the RifR mutants had a Lac+ phenotype on MacLac plates. Interestingly, we also identified several RifR mutants with a reduced Lac colony phenotype because they formed white colonies after prolonged incubations (>24 h) compared with wild type under the same conditions.
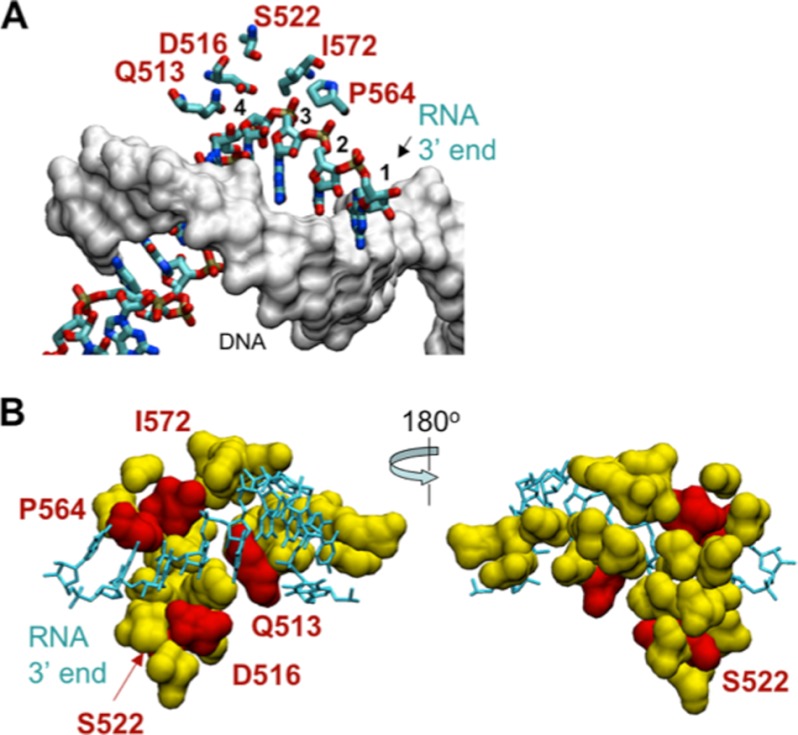
FIGURE 6.
A small segment of the fork domain in the β subunit is important for controlling transcription slippage. A, a nucleic acid scaffold in the T. thermophilus RNAP TEC (Protein Data Bank code 2O5J) (2) and location of the slippage domain. The RNA strand in the 9-bp RNA-DNA hybrid is shown as a wire frame model, and the DNA strand is shown as a surface model (gray). The arrow indicates the 3′ end of the RNA, and individual residues in the 3′-proximal part of the RNA are numbered starting from the 3′ end. Amino acid residues contacting the RNA are shown. B, amino acid residues in the β subunit involved in formation of the Rif-binding pocket (25 residues of the fork domain are shown in yellow). Red highlights five amino acid residues that when mutated affect slippage both in vivo and in vitro. The RNA strand in the hybrid is shown as a wire frame model (cyan), and the DNA strand in the hybrid is omitted.
Next, we determined the relative β-gal activities of the affected mutants in both the 9A(−1) and 5AT4A(0) reporter strains (Table 5). As described above, a relative slippage phenotype of a RNAP mutant was estimated as a ratio of the two relative β-gal activities. This analysis yielded 14 rpoB RifR mutants affecting 10 positions in the β subunit that altered elongation slippage compared with wild type. Among them, 11 RifR mutants increased elongation slippage, and three had decreased slippage. The positions of these mutations at the RNA-DNA hybrid region of RNAP are shown (Fig. 2), and the results strongly indicate that amino acid residues in this region are important for slippage.
TABLE 5.
Effects of the chromosomal rpoB RifR mutations on elongation slippage in vivo
The values are the averages of at least two independent experiments. Each experiment was performed with minimum of two repetitions. The values of different independent experiments generally deviated less than 10% from the average value.
| rpoB mutation | Relative β-Gal activity in |
Relative slippage phenotype (9A(−1)/5AT4A(0)) | |
|---|---|---|---|
| 9A(−1) | 5AT4A(0) | ||
| Wild type | 1.0 | 1.0 | 1.0 |
| V146F | 2.1 | 1.3 | 1.6 |
| V146G | 2.2 | 1.4 | 1.6 |
| Q513P | 3.0 | 0.9 | 3.3 |
| D516N | 0.6 | 1.2 | 0.5 |
| S522F | 1.8 | 1.1 | 1.6 |
| S522Y | 2.5 | 1.0 | 2.5 |
| T525R | 2.4 | 1.0 | 2.4 |
| R529H | 1.9 | 1.2 | 1.6 |
| Δ532 | 2.3 | 1.4 | 1.6 |
| P564L | 3.1 | 1.5 | 2.0 |
| G570C | 4.1 | 2.4 | 1.7 |
| G570Q | 2.5 | 1.3 | 1.9 |
| I572T | 0.8 | 1.2 | 0.7 |
| I572N | 0.8 | 1.2 | 0.7 |
Analysis of the Mutant RNAPs in Vitro
To confirm the slippage phenotypes of the rpoB mutants, we performed biochemical analysis of representative mutant RNAPs. We chose six chromosomal RifR mutants affecting five amino acid positions because they had the strongest slippage phenotypes in vivo. The rpoB mutants Q513P, S522Y, and P564L were “up” mutants, and D516N, I572N, and I572T, were “down” mutants. These strains, as indicated above, also carried a functional hexahistidine-tagged β′ subunit, which allowed the rapid pulldown and purification of RNAP from the crude lysate by immobilization on Ni2+-NTA-agarose beads (49, 55). Each of the purified enzymes was assembled into a TEC with a synthetic RNA-DNA scaffold, and the bulk elongation rate for each mutant RNAP was tested in a single-round “runoff” assay with 10 μm NTP (Fig. 3). The Q513P, D516N, P564L, and I572T mutants had an elongation rate similar to the wild type enzyme. The I572N were ∼2-fold slower, and S522Y was ∼2-fold faster than the wild type enzyme in a single round transcription. Because of the relative minor effect on the elongation rate in these bulk elongation assays, we concluded that these mutations do not cause a significant change in the regular catalytic activity of RNAP.
Next, we evaluated slippage pattern by the mutants on a RNA-DNA scaffold containing a tract of 9As in the transcript to match the 9A spacer sequence used for the in vivo screening. Slippage during unimpeded transcription of 9A tract generates both insertions and deletions, and their ratio should depend on RNAP dwell time at adenine positions within the 9A tract. For instance, a prolonged pausing at the +8 position of the tract should equally promote deletions and insertions by forward and backward shift of the RNA strand relative to the DNA, respectively. In contrast, the dwelling at the last 9 position should increase insertions only, because deletions are not allowed at the end of the tract. To produce deletions, RNAP should shift forward on DNA together with the nascent RNA, whereas insertions result from a shift in the opposite direction. Thus, RNAP halted at the end of the tract is unable to shift further forward without producing a mismatch between the 3′ end of the RNA and the DNA.
To separate slippage in the tract from that at the end of the tract, we developed two in vitro assays. The first assay aimed to determine slippage leading to insertions, and it included a prolonged halting RNAP at the end of 9A tract in the absence of all NTPs except ATP (Fig. 4). For this purpose, different TEC9 was assembled with the 9-nt RNA primer (5′-AUCGAGAGG) hybridized 1 base pair upstream from the coding region for the 9A tract. The RNA in the TEC9 was extended 1-nt by adding [α-32P]GTP to form TEC10 labeled at its 3′ end before the 9A tract. By adding ATP, the TEC10 transcribed the entire 9A tract followed by pausing at the last adenine in the tract because of the lack of the next CTP. A substantial fraction of RNAP was arrested at the last adenine of the 9A tract, which is consistent with previous findings that A/T-rich sequences cause arrest of RNAP (56, 57). In the absence of slippage, TECs halted at the end of the 9A tract generate a 19-nt RNA transcript (TEC19). In the experiment of Fig. 4, continuous slippage at the last base of 9A tract produced a distribution of slippage products longer than 19 nucleotides, which were processively elongated by slippage. The slippage rate was determined as the average number of AMP molecules added to the RNA per second (Fig. 4A). Relative to wild type, the D516N, P564L, and I572T mutants showed an increase of the slippage rate; the Q513P mutant showed a slower slippage rate, and S522Y and I572N mutants were similar to the wild type enzyme (Fig. 4, left panel). Importantly, the slippage at the 9A tract by the assembled TEC showed the same rate of AMP incorporation as that observed during transcription of the same tract initiated from a promoter (T7A1 promoter; data not shown). This result justified our employment of RNA-DNA scaffolds for testing slippage. Transcription slippage requires uninterrupted 9A track (Table 3). Accordingly, an interruption of 9A tract at the fifth position with a single U or G residue and replacement of the 9A track with 9C tract completely inhibited slippage in vitro (data not shown). Thus, the processive AMP addition in the assay of Fig. 4 was due to slippage as opposed to the multiple AMP misincorporations.
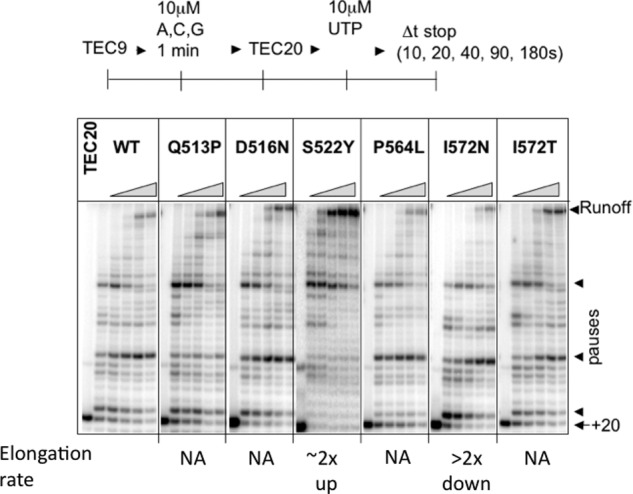
Although providing important information on the chemical rate of slippage, the assay of Fig. 4 did not allow direct comparison of the in vitro slippage with the in vivo data obtained with the 9A(−1) reporter strain. Slippage in vivo produces either insertions or deletions, whereas the assay of Fig. 4 was designed to detect insertions only. In addition, transcription through the 9A tract in vivo occurs without stopping RNAP at the end of the tract by CTP deprivation. To better match these in vivo conditions and to evaluate how the rpoB mutations affect deletions in the tract, we modified the assay of Fig. 4 by simultaneous addition of ATP and α-32P-labeled CTP that should generate a transcription complex with a 20-nt RNA if no slippage occurred (TEC20; Fig. 5). We assembled nonlabeled TEC10 with the 3′ end of its RNA located immediately upstream from the first residue of 9A tract (5′-AUCGAGAGGG) on template. By adding ATP and labeled CTP, the TEC10 proceeds to the end of the 9A tract by incorporation of nonlabeled AMP, incorporates a single labeled CMP beyond the A tract, and then stalls because of the lack of the next complementary base, GTP (Fig. 5). The modified assay served two purposes: (i) it limited any slippage to a short time period before incorporation of the [32P]CMP occurred, and (ii) it produced the 3′ end-labeled RNA containing the A tract followed by the cytosine. The 3′-labeled RNA products could easily be examined for slippage by urea-PAGE as described and summarized in the table panel of Fig. 5.
The result of Fig. 5 demonstrated that the majority of the rpoB mutants had the altered slippage pattern. Q513P mutant showed dramatic increase in deletion of 1 adenine (1A) from the 9A tract and almost complete elimination of 1A insertion. In contrast, P564L mutant significantly increased 1A insertion and decreased 1A deletion. Strikingly, the two mutants exhibited the opposite slippage directionality. D516N, I572N, and I572T mutants produced less 1A insertion than the wild type without affecting 1A deletions. Finally, S522Y mutant showed no difference in slippage from the wild type.
A bias toward insertions or deletions during slippage may depend on how rapidly RNAP transcribes through the 9A tract. As described above, the complex that has reached the end of the tract retains the ability to generate insertions but not deletions. Therefore, RNAP mutants that pause more than the wild type before the end of the 9A tract may make more deletions, whereas the mutants that induce more pausing at the end of the 9A tract may make more insertions.
Most of these results were consistent with the in vivo data. For example, the up slippage phenotypes of P564L mutant could be explained by its enhanced frequency for 1A insertion in the 9A(−1) lacZ reporter in vivo, which was designed to look primarily for 1A insertions to the 9A tract for restoration of the LacZ open reading frame (Fig. 1). Conversely, the down slippage phenotypes of D516N, I572N, and I572T could be explained by their reduced frequency for 1A insertion compared with wild type. The Q513P mutant showed the most dramatic change in slippage pattern compared with the wild type and all other rpoB mutants. Its up slippage phenotype in vivo could be explained by its strong bias toward making deletions, which would restore in-frame lacZ in the 9A(−1) reporter by the 2A deletion instead of 1A insertion. Although Q513P showed a sign of the increased 2A deletions in vitro, the sensitivity of our assay was not sufficient for reliable detection of multiple deletions in 9A tract. Therefore, we employed the longer 11A tract to test whether Q513P is capable for multiple deletions. The result of this assay was striking. The Q513P mutant showed a major ∼16-fold increase of 2A deletion in 11A tract. The minor 3A deletion was also clearly detected with the mutant (Fig. 5). The decreased 1A insertion by Q513P mutant explained its decreased rate of processive AMP addition in TEC halted at the end of 9A tract by CTP deprivation (Fig. 4). On the other hand, this mutation had no apparent effect on NTP incorporation during bulk elongation under the condition used (Fig. 3). The Q513P mutant enzyme was reported to have an increased Km for purine nucleotides during elongation (58). We suggest that this mutation primarily affected slippage as opposed to the regular elongation pathway.
Other RifR mutants, including T525R, R529H, G570C, and G570Q, did not show any effect on slippage in this in vitro assay (data not shown). The V146F and V146G mutations dramatically reduced the bulk elongation rate of RNAP during transcription of the 9A tract, which prevented us from determining the slippage pattern for these mutants.
Summary of Slippage Effects
The slippage mutants analyzed in this work fell into four categories: (i) mutations that increase slippage efficiency (the combined yield of insertions and deletions) and additionally increased the intrinsic tendency of the wild type enzyme to make more insertions than deletions (defined here as slippage directionality) (P564L); (ii) mutations that change slippage directionality but not slippage efficiency (Q513P); (iii) mutations that decreased slippage efficiency; most of these mutations also affect slippage directionality by inhibiting insertions more than deletions (D516N, I572N, and I572T); and (iv) mutations that affect slippage in vivo but not in vitro (the chromosomal mutants: β-S522Y, T525R, R529H, G570C, and G570Q; and the plasmid-borne mutants: β-D444G, R451C, A543V, G570D, and R687C) (data not shown). The plasmid-borne β-M1273I mutant exhibited an extremely slow elongation rate in vitro, causing its complete arrest within the 9A tract, which did not allow its further analysis. The difference between the in vitro and in vivo results on several mutants could be attributed to the design of the in vitro assay involving a nonphysiologically low ATP concentration (cellular ATP concentration in the millimolar range rather than 10 μm used in vitro).
Fig. 6 shows the location of the mutated amino acid residues in the crystal structure of the Thermus thermophilus RNAP (TEC) (2). The majority of the mutations identified in our work affect residues encapsulating the 9-bp RNA-DNA hybrid at the bottom of the catalytic cleft in RNAP. In particular, β-Gln-513, Asp-516, Pro-564, and Ile-572 are sufficiently close to the 9-bp RNA-DNA hybrid to form hydrogen bonds with the RNA strand at nucleotide positions 3 and 4 upstream from the 3′ end of the transcript. The Rif-binding pocket in the β subunit is part of the fork region (residues 500–690), which undergoes a major conformational change during the switch of RNAP from initiation to elongation mode (2, 59). The fork region is an interface between the β and β′ subunits; the conformational change during TEC formation is proposed to promote closure of the β′ protein clamp with the β subunit around the RNA-DNA hybrid (2). After testing mutations within the 25 amino acid residues forming an extended binding pocket for rifampicin, we identified a small protein path forming an interface between the RNA strand of the hybrid and the fork domain of the β subunit involved in slippage both in vivo and in vitro. We suggest that mutations in this region weaken the DNA clamp in the elongation complex, which facilitates spontaneous melting of the RNA-DNA hybrid leading to slippage. The present work analyzed the effect on slippage of mutations in a small region of the β subunit adjacent to the RNA-DNA hybrid. The approach used in this study will be useful to further identify other positions (non-RifR sites) in the same region, as well as other regions in the β and β′ subunits that are important for transcription slippage.
A significant fraction of the mutations identified for increased slippage in vivo did not show effects on slippage in vitro. These mutations were primarily localized to the area of the protein around the DNA duplex downstream of the RNA-DNA hybrid in the elongation complex. Although several mutations from this class were localized near the RNA-DNA hybrid, the majority of them were not directly involved in interactions with the RNA strand in the hybrid. This class of mutations is likely to affect the interaction of RNAP with the front end DNA duplex in the elongation complex. They also may affect interaction of RNAP with transacting factors, which were not present in our “minimal” in vitro system. Alternatively, an increased slippage phenotype exhibited by these mutants in vivo may require a different concentration of ATP substrates or some other cellular conditions, which we cannot fully recapitulate in vitro. Transcription elongation in bacteria is coupled to translation. Therefore, some of the RNAP mutations might affect slippage indirectly by promoting translational frameshifts at the 9A tract in the lacZ mRNA (60). This group of mutants deserves further investigation.
Summary Remarks
We have developed a simple genetic system to isolate RNAP mutants that alter transcription slippage during elongation, complementary to an accompanying paper on slippage in yeast (39). We have identified the fork region in RNAP near the RNA-DNA hybrid as being important for slippage. Our successful isolation of the highly clustered fork mutants that decrease or increase slippage indicated that wild type bacterial RNAP is evolved for some slippage, which may be regulated according to the cellular conditions. Identification of several mutations that change slippage directionality argues that elongation intermediates leading to deletions and insertions include distinct conformations of RNAP. This work provides a basis for further elucidation of the elongation slippage mechanism responsible for out of DNA register errors in transcription.
Acknowledgments
We acknowledge the highly interactive environment at the Gene Regulation and Chromosome Biology Laboratory and Zachary Burton for critical reading of the manuscript.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program. This work was also supported by the National Cancer Institute and the Center for Cancer Research.
D. J. Jin, unpublished results.
- RNAP
- RNA polymerase
- nt
- nucleotide(s)
- NTA
- nitrilotriacetic acid
- TEC
- ternary elongation complex(es)
- MacLac
- McConkey lactose.
REFERENCES
- 1. Vassylyev D. G., Vassylyeva M. N., Zhang J., Palangat M., Artsimovitch I., Landick R. (2007) Structural basis for substrate loading in bacterial RNA polymerase. Nature 448, 163–168 [DOI] [PubMed] [Google Scholar]
- 2. Vassylyev D. G., Vassylyeva M. N., Perederina A., Tahirov T. H., Artsimovitch I. (2007) Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448, 157–162 [DOI] [PubMed] [Google Scholar]
- 3. Trinh V., Langelier M. F., Archambault J., Coulombe B. (2006) Structural perspective on mutations affecting the function of multisubunit RNA polymerases. Microbiol. Mol. Biol. Rev. 70, 12–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ebright R. H. (2000) RNA polymerase. Structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. 304, 687–698 [DOI] [PubMed] [Google Scholar]
- 5. Springgate C. F., Loeb L. A. (1975) On the fidelity of transcription by Escherichia coli ribonucleic acid polymerase. J. Mol. Biol. 97, 577–591 [DOI] [PubMed] [Google Scholar]
- 6. Strathern J. N., Jin D. J., Court D. L., Kashlev M. (2012) Isolation and characterization of transcription fidelity mutants. Biochim. Biophys. Acta 1819, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shaw R. J., Bonawitz N. D., Reines D. (2002) Use of an in vivo reporter assay to test for transcriptional and translational fidelity in yeast. J. Biol. Chem. 277, 24420–24426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reynolds N. M., Lazazzera B. A., Ibba M. (2010) Cellular mechanisms that control mistranslation. Nat. Rev. Microbiol. 8, 849–856 [DOI] [PubMed] [Google Scholar]
- 9. Libby R. T., Gallant J. A. (1991) The role of RNA polymerase in transcriptional fidelity. Mol. Microbiol. 5, 999–1004 [DOI] [PubMed] [Google Scholar]
- 10. Libby R. T., Nelson J. L., Calvo J. M., Gallant J. A. (1989) Transcriptional proofreading in Escherichia coli. EMBO J. 8, 3153–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foster P. L. (2007) Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42, 373–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kireeva M. L., Nedialkov Y. A., Cremona G. H., Purtov Y. A., Lubkowska L., Malagon F., Burton Z. F., Strathern J. N., Kashlev M. (2008) Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol. Cell 30, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koyama H., Ueda T., Ito T., Sekimizu K. (2010) Novel RNA polymerase II mutation suppresses transcriptional fidelity and oxidative stress sensitivity in rpb9Delta yeast. Genes Cells 15, 151–159 [DOI] [PubMed] [Google Scholar]
- 14. Holmes S. F., Santangelo T. J., Cunningham C. K., Roberts J. W., Erie D. A. (2006) Kinetic investigation of Escherichia coli RNA polymerase mutants that influence nucleotide discrimination and transcription fidelity. J. Biol. Chem. 281, 18677–18683 [DOI] [PubMed] [Google Scholar]
- 15. Svetlov V., Vassylyev D. G., Artsimovitch I. (2004) Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J. Biol. Chem. 279, 38087–38090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gordon A. J., Halliday J. A., Blankschien M. D., Burns P. A., Yatagai F., Herman C. (2009) Transcriptional infidelity promotes heritable phenotypic change in a bistable gene network. PLoS Biol. 7, e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ba Y., Tonoki H., Tada M., Nakata D., Hamada J., Moriuchi T. (2000) Transcriptional slippage of p53 gene enhanced by cellular damage in rat liver. Monitoring the slippage by a yeast functional assay. Mutat. Res. 447, 209–220 [DOI] [PubMed] [Google Scholar]
- 18. Baranov P. V., Hammer A. W., Zhou J., Gesteland R. F., Atkins J. F. (2005) Transcriptional slippage in bacteria. Distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 6, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anikin M., Molodtsov D., Temiakov D., McAllister (2010) Transcript Slippage and Recoding, in Recoding: Expansion of Decoding Rules Enriches Gene Expression (Atkins J. F., Gesteland Raymond F., eds) pp. 409–434, Springer, New York [Google Scholar]
- 20. Penno C., Hachani A., Biskri L., Sansonetti P., Allaoui A., Parsot C. (2006) Transcriptional slippage controls production of type III secretion apparatus components in Shigella flexneri. Mol. Microbiol. 62, 1460–1468 [DOI] [PubMed] [Google Scholar]
- 21. Ratinier M., Boulant S., Combet C., Targett-Adams P., McLauchlan J., Lavergne J. P. (2008) Transcriptional slippage prompts recoding in alternate reading frames in the hepatitis C virus (HCV) core sequence from strain HCV-1. J. Gen. Virol. 89, 1569–1578 [DOI] [PubMed] [Google Scholar]
- 22. Linton M. F., Raabe M., Pierotti V., Young S. G. (1997) Reading-frame restoration by transcriptional slippage at long stretches of adenine residues in mammalian cells. J. Biol. Chem. 272, 14127–14132 [DOI] [PubMed] [Google Scholar]
- 23. van Leeuwen F. W., Kros J. M., Kamphorst W., van Schravendijk C., de Vos R. A. (2006) Molecular misreading. The occurrence of frameshift proteins in different diseases. Biochem. Soc. Trans. 34, 738–742 [DOI] [PubMed] [Google Scholar]
- 24. van Leeuwen F. W., Fischer D. F., Benne R., Hol E. M. (2000) Molecular misreading. A new type of transcript mutation in gerontology. Ann. N.Y. Acad. Sci. 908, 267–281 [DOI] [PubMed] [Google Scholar]
- 25. Turnbough C. L., Jr. (2008) Regulation of bacterial gene expression by the NTP substrates of transcription initiation. Mol. Microbiol. 69, 10–14 [DOI] [PubMed] [Google Scholar]
- 26. Turnbough C. L., Jr., Switzer R. L. (2008) Regulation of pyrimidine biosynthetic gene expression in bacteria. Repression without repressors. Microbiol Mol. Biol. Rev. 72, 266–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wagner L. A., Weiss R. B., Driscoll R., Dunn D. S., Gesteland R. F. (1990) Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 18, 3529–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Leeuwen F. W., de Kleijn D. P., van den Hurk H. H., Neubauer A., Sonnemans M. A., Sluijs J. A., Köycü S., Ramdjielal R. D., Salehi A., Martens G. J., Grosveld F. G., Peter J., Burbach H., Hol E. M. (1998) Frameshift mutants of β amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science 279, 242–247 [DOI] [PubMed] [Google Scholar]
- 29. Raabe M., Linton M. F., Young S. G. (1998) Long runs of adenines and human mutations. Am. J. Med. Genet. 76, 101–102 [DOI] [PubMed] [Google Scholar]
- 30. van Leeuwen F. W., Fischer D. F., Kamel D., Sluijs J. A., Sonnemans M. A., Benne R., Swaab D. F., Salehi A., Hol E. M. (2000) Molecular misreading. A new type of transcript mutation expressed during aging. Neurobiol. Aging 21, 879–891 [DOI] [PubMed] [Google Scholar]
- 31. van Leeuwen F. W., van Tijn P., Sonnemans M. A., Hobo B., Mann D. M., Van Broeckhoven C., Kumar-Singh S., Cras P., Leuba G., Savioz A., Maat-Schieman M. L., Yamaguchi H., Kros J. M., Kamphorst W., Hol E. M., de Vos R. A., Fischer D. F. (2006) Frameshift proteins in autosomal dominant forms of Alzheimer disease and other tauopathies. Neurology 66, S86–92 [DOI] [PubMed] [Google Scholar]
- 32. Liu C., Heath L. S., Turnbough C. L., Jr. (1994) Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 8, 2904–2912 [DOI] [PubMed] [Google Scholar]
- 33. Jin D. J. (1994) Slippage synthesis at the galP2 promoter of Escherichia coli and its regulation by UTP concentration and cAMP·cAMP receptor protein. J. Biol. Chem. 269, 17221–17227 [PubMed] [Google Scholar]
- 34. de Vries N., Duinsbergen D., Kuipers E. J., Pot R. G., Wiesenekker P., Penn C. W., van Vliet A. H., Vandenbroucke-Grauls C. M., Kusters J. G. (2002) Transcriptional phase variation of a type III restriction-modification system in Helicobacter pylori. J. Bacteriol. 184, 6615–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Larsen B., Wills N. M., Nelson C., Atkins J. F., Gesteland R. F. (2000) Nonlinearity in genetic decoding. Homologous DNA replicase genes use alternatives of transcriptional slippage or translational frameshifting. Proc. Natl. Acad. Sci. U.S.A. 97, 1683–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Penno C., Sansonetti P., Parsot C. (2005) Frameshifting by transcriptional slippage is involved in production of MxiE, the transcription activator regulated by the activity of the type III secretion apparatus in Shigella flexneri. Mol. Microbiol. 56, 204–214 [DOI] [PubMed] [Google Scholar]
- 37. Jin D. J., Turnbough C. L., Jr. (1994) An Escherichia coli RNA polymerase defective in transcription due to its overproduction of abortive initiation products. J. Mol. Biol. 236, 72–80 [DOI] [PubMed] [Google Scholar]
- 38. Jin D. J. (1996) A mutant RNA polymerase reveals a kinetic mechanisms for the switch between nonproductive stuttering synthesis and productive initiation during promoter clearance. J. Biol. Chem. 271, 11659–11667 [PubMed] [Google Scholar]
- 39. Strathern J. N., Malagon F., Irvin J., Gotte D., Shafer B., Kireeva M., Lubkowska L., Jin D. J., Kashlev M. (2013) J. Biol. Chem. 288, 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 41. Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory, Plainview, NY [Google Scholar]
- 42. Sawitzke J. A., Thomason L. C., Costantino N., Bubunenko M., Datta S., Court D. L. (2007) Recombineering. In vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 421, 171–199 [DOI] [PubMed] [Google Scholar]
- 43. Sawitzke J. A., Costantino N., Li X. T., Thomason L. C., Bubunenko M., Court C., Court D. L. (2011) Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J. Mol. Biol. 407, 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Svenningsen S. L., Costantino N., Court D. L., Adhya S. (2005) On the role of Cro in lambda prophage induction. Proc. Natl. Acad. Sci. U.S.A. 102, 4465–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alton N. K., Vapnek D. (1979) Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature 282, 864–869 [DOI] [PubMed] [Google Scholar]
- 46. Datta S., Costantino N., Court D. L. (2006) A set of recombineering plasmids for gram-negative bacteria. Gene 379, 109–115 [DOI] [PubMed] [Google Scholar]
- 47. Zalenskaya K., Lee J., Gujuluva C. N., Shin Y. K., Slutsky M., Goldfarb A. (1990) Recombinant RNA polymerase. Inducible overexpression, purification and assembly of Escherichia coli rpo gene products. Gene 89, 7–12 [DOI] [PubMed] [Google Scholar]
- 48. Jin D. J., Gross C. A. (1988) Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 202, 45–58 [DOI] [PubMed] [Google Scholar]
- 49. Kireeva M. L., Lubkowska L., Komissarova N., Kashlev M. (2003) Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae. Methods Enzymol. 370, 138–155 [DOI] [PubMed] [Google Scholar]
- 50. Kireeva M. L., Kashlev M. (2009) Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 106, 8900–8905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jin D. J., Gross C. A. (1989) Characterization of the pleiotropic phenotypes of rifampin-resistant rpoB mutants of Escherichia coli. J. Bacteriol. 171, 5229–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Garibyan L., Huang T., Kim M., Wolff E., Nguyen A., Nguyen T., Diep A., Hu K., Iverson A., Yang H., Miller J. H. (2003) Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair 2, 593–608 [DOI] [PubMed] [Google Scholar]
- 53. Lisitsyn N. A., Sverdlov E. D., Moiseyeva E. P., Danilevskaya O. N., Nikiforov V. G. (1984) Mutation to rifampicin resistance at the beginning of the RNA polymerase beta subunit gene in Escherichia coli. Mol. Gen. Genet. 196, 173–174 [DOI] [PubMed] [Google Scholar]
- 54. Thomason L., Court D. L., Bubunenko M., Costantino N., Wilson H., Datta S., Oppenheim A. (2007) Current Protocols in Molecular Biology (Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., eds) Chapter 1, Unit 1.16 [DOI] [PubMed] [Google Scholar]
- 55. Kireeva M., Nedialkov Y. A., Gong X. Q., Zhang C., Xiong Y., Moon W., Burton Z. F., Kashlev M. (2009) Millisecond phase kinetic analysis of elongation catalyzed by human, yeast, and Escherichia coli RNA polymerase. Methods 48, 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Samkurashvili I., Luse D. S. (1996) Translocation and transcriptional arrest during transcript elongation by RNA polymerase II. J. Biol. Chem. 271, 23495–23505 [DOI] [PubMed] [Google Scholar]
- 57. Komissarova N., Becker J., Solter S., Kireeva M., Kashlev M. (2002) Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol. Cell 10, 1151–1162 [DOI] [PubMed] [Google Scholar]
- 58. Jin D. J., Gross C. A. (1991) RpoB8, a rifampicin-resistant termination-proficient RNA polymerase, has an increased Km for purine nucleotides during transcription elongation. J. Biol. Chem. 266, 14478–14485 [PubMed] [Google Scholar]
- 59. Vassylyev D. G., Sekine S., Laptenko O., Lee J., Vassylyeva M. N., Borukhov S., Yokoyama S. (2002) Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 417, 712–719 [DOI] [PubMed] [Google Scholar]
- 60. Atkins J. F., Björk G. R. (2009) A gripping tale of ribosomal frameshifting. Extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol. Mol. Biol. Rev. 73, 178–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chakrabarti S. L., Gorini L. (1977) Interaction between mutations of ribosomes and RNA polymerase. A pair of strA and rif mutants individually temperature-insensitive but temperature-sensitive in combination. Proc. Natl. Acad. Sci. U.S.A. 74, 1157–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]