Background: Murine SAA1.1 is pathogenic and SAA2.2 is non-pathogenic in AA amyloidosis.
Results: SAA1.1 and SAA2.2 exhibit different biophysical properties, including fibrillation kinetics and fibril morphology.
Conclusion: The distinct biophysical properties of highly homologous SAA proteins may contribute to their different pathogenicity during chronic inflammation.
Significance: Structural and kinetic factors, more than their intrinsic amyloidogenicity, may determine the diverse pathogenicity among nearly identical SAA isoforms.
Keywords: Aggregation, Amyloid, Inflammation, Intrinsically Disordered Proteins, Protein Misfolding
Abstract
Serum amyloid A (SAA) is best known for being the main component of amyloid in the inflammation-related disease amyloid A (AA) amyloidosis. Despite the high sequence identity among different SAA isoforms, not all SAA proteins are pathogenic. In most mouse strains, the AA deposits mostly consist of SAA1.1. Conversely, the CE/J type mouse expresses a single non-pathogenic SAA2.2 protein that is 94% identical to SAA1.1. Here we show that SAA1.1 and SAA2.2 differ in their quaternary structure, fibrillation kinetics, prefibrillar oligomers, and fibril morphology. At 37 °C and inflammation-related SAA concentrations, SAA1.1 exhibits an oligomer-rich fibrillation lag phase of a few days, whereas SAA2.2 shows virtually no lag phase and forms small fibrils within a few hours. Deep UV resonance Raman, far UV-circular dichroism, atomic force microscopy, and fibrillation cross-seeding experiments suggest that SAA1.1 and SAA2.2 fibrils possess different morphology. Both the long-lived oligomers of pathogenic SAA1.1 and the fleeting prefibrillar oligomers of non-pathogenic SAA2.2, but not their respective amyloid fibrils, permeabilized synthetic bilayer membranes in vitro. This study represents the first comprehensive comparison between the biophysical properties of SAA isoforms with distinct pathogenicities, and the results suggest that structural and kinetic differences in the oligomerization-fibrillation of SAA1.1 and SAA2.2, more than their intrinsic amyloidogenicity, may contribute to their diverse pathogenicity.
Introduction
Serum amyloid A (SAA)3 belongs to a small and highly conserved family of apolipoproteins that plays a fundamental role in the acute phase response to an inflammatory stimulus, such as tissue damage, trauma, or infection (1). Although SAA has been linked to many functions related to innate immunity and lipid transport/metabolism as well as to various diseases (2, 3) including atherosclerosis, rheumatoid arthritis, Alzheimer disease and cancer, SAA is perhaps best known for being the main component of the amyloid fibrils in reactive amyloid A amyloidosis (4, 5). Amyloid A (AA) amyloidosis is a clinical manifestation that occurs in some individuals with chronic inflammatory disorder. During inflammation, the concentration of SAA in plasma rises up to 1000-fold its normal levels to a concentration of up to 1–2 mg/ml (4). Persistence of this hyperexpression is a prerequisite for AA amyloidosis (4). Although the main component of the amyloid deposits is an N-terminal 76-residue fragment of SAA (1), the presence of different fragments, including full-length SAA, suggest that proteolytic processing may be a post-deposition event (6).
Despite the high sequence identity among different SAA proteins, not all SAA isoforms are pathogenic. For example, humans have several acute phase SAAs that are equally overexpressed during inflammation, but SAA1 is the main SAA component of the amyloid fibrils. Similarly, inflammation-induced AA amyloidosis in mice results in mouse laden with amyloid deposits from only the SAA1.1 isoform (7). Furthermore, the CE/J type mouse expresses a single SAA2.2 protein that is 94% identical to SAA1.1 yet is non-pathogenic despite its overexpression during induced inflammation (8). To date, little is known about the molecular basis of AA amyloidosis and the differing pathogenicity among SAA isoforms across different vertebrates. Although it is tempting to assume that pathogenic SAAs are highly amyloidogenic and that non-pathogenic SAAs are amyloid-resistant, we have shown that the non-pathogenic murine SAA2.2 rapidly forms amyloid fibrils in vitro at 37 °C, making it one of the most inherently amyloidogenic proteins known to date (9). Therefore, other biochemical or biophysical properties of SAA proteins must account for their pathogenicity.
Here we have characterized various biophysical properties of the pathogenic murine SAA1.1 and compared them to those of non-pathogenic SAA2.2. Most notably, we show that upon incubation at 37 °C, SAA1.1 exhibits a fibrillation lag phase of several days that is highly populated with spherical oligomers. In contrast, SAA2.2 has a negligible lag phase and forms small curvilinear amyloid fibrils within a few hours. Spectroscopic, microscopy, and kinetic experiments suggest that SAA1.1 and SAA2.2 fibrils are of different morphology. Furthermore, the globular oligomers formed during the long lag phase of SAA1.1 and the fleeting prefibrillar oligomers of SAA2.2 were able to permeabilize synthetic membranes in vitro, whereas the respective fibrils did not. Overall, our results suggest possible structural and kinetic reasons that may contribute to the distinct pathogenic tendencies of SAA proteins.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant SAA with an N-terminal His tag and a linker containing a TEV protease recognition site (His-TEV-SAA) was expressed in Escherichia coli BL21(DE3)pLysS (Invitrogen). Upon induction, the cells were lysed and sonicated in Tris-urea buffer (20 mm Tris, 6 m urea, 500 mm NaCl, pH 8.0). The lysate was centrifuged, and His-tagged SAA was isolated by a 5-ml Histrap column (HP®, GE Healthcare) and gel filtration (HiLoad™ 16/60 Superdex 75, GE Healthcare). Proteolysis of His-TEV-SAA (20 °C) with TEV protease at a protease:substrate molar ratio of 1:20 yielded SAA isoforms without non-natural residues. The TEV reaction was quenched after 6 h with 5 m urea and 0.5 m NaCl, and SAA was purified by passing the mixture through a Histrap column as described above. The SAA fraction was then collected, concentrated, and further purified by gel filtration. The concentrated SAA was briefly centrifuged to remove any insoluble debris and subjected to three rounds of dialysis at 4 °C against 50–100-fold excess volume of Tris buffer over a period of 15 h. SAA was then aliquoted, and its absorbance was measured at 280 nm (Mr: SAA1.1 11,605, SAA2.2 11,670; extinction coefficient, 25440 m−1 cm−1).
Analytical Gel Filtration
The oligomeric states of SAA isoforms were monitored using an analytical gel filtration column (Superdex® 200 HR 10/300, GE Healthcare Life Sciences) coupled to a GE ÄKTA Prime® chromatography system at 4 °C. The column was pre-equilibrated with 20 mm Tris, 150 mm NaCl, pH 8.0. The sample load was 20 μg of protein, and the flow rate was 0.5 ml/min at 1 MPa. Protein elution was monitored at 280 nm. The column was precalibrated with common laboratory proteins.
Sedimentation Equilibrium (SE) and Sedimentation Velocity (SV) Analytical Ultracentrifugation (AUC)
SE-AUC experiments were performed at 4 °C using a Beckman XL-I instrument with an AN-50Ti rotor. Different SAA concentrations (0.1–0.6 mg/ml) in 20 mm Tris, pH 8.2, 150 mm NaCl buffer were loaded into 6-sector cells and spun at 10,000, 12,000, and 19,000 rpm. The absorbance at 280 nm was recorded as a function of the radial position. Equilibrium was considered attained when replicate scans 6 h apart were indistinguishable. The data were globally fitted to a model for a single species in solution as previously described (10). SV-AUC experiments were performed at 4 °C and 40,000 rpm. About 0.5 mg/ml SAA in Tris buffer was loaded in a double sector cell with a 12-mm centerpiece. The SV absorbance profiles were recorded at 280 nm (293 nm in the concentration dependence experiments) and acquired every 6 min (or 12 min in the concentration dependence experiments) for a total of ∼275 scans. They were analyzed using the SEDFIT software.
Far-UV Circular Dichroism (CD)
Far-UV CD measurements were performed using an Olis DSM® 1000 CD spectrophotometer (Bogart, Georgia) with a 2-mm path quartz cuvette (Hellma). When monitoring the loss of secondary structure, wavelength scans were performed from 200 to 260 nm with 1-nm intervals and plotted against the molar ellipticity. For denaturation experiments, measurements were averaged over 60 s at 222 nm (base line at 255 nm). For thermal denaturation, a Peltier device was used for temperature control. Urea denaturation was performed by co-dilution method, as previously described (11). For both thermal and chemical denaturation, the concentration of SAA was 0.3 mg/ml, and the sample was incubated for ∼5 min at the desired condition before collecting data.
Fibrillation Studies
Fibril formation of SAA was induced by incubating the protein at 37 °C in a dry bath incubator. The protein concentrations were 0.3 mg/ml in 20 mm Tris, pH 8.0, unless otherwise noted. For seeding assays, preformed SAA protofibrils or fibrils were added at 5% w/w to purified refolded oligomers of SAA and immediately incubated at 37 °C and allowed to aggregate.
Thioflavin T (ThT) Fluorescence
SAA (10 μg) at various conditions was added to a 400-μl working solution containing 50 mm glycine-NaOH, pH 8.5, and 12.5 μm ThT. The resulting mixture was transferred to a quartz cuvette (1-cm path length, volume 500 μl). Fibrillation of SAA was monitored at 37 °C in a dry bath. At regular intervals, a volume containing 10 μg of SAA was withdrawn and mixed with 50 μl of 100 μm ThT and enough glycine, pH 8.5, to a final volume of 400 μl. After brief resuspension/stirring, the sample was equilibrated for 30 s, and changes in ThT intensity were recorded after excitation at 440 nm. The emission maximum at 485 nm was averaged over 60 s. The slit widths were set at 10 nm for both excitation and emission.
Atomic Force Microscopy (AFM)
SAA isoforms incubated at specified temperatures were diluted in water to a final concentration of 0.05 mg/ml, and 20 μl of this solution was deposited on a freshly cleaved type I mica surface. After 15–30 min of incubation, the mica was gently washed with 1–2 ml of filtered Milli-Q water and then air-dried. The mica plate was mounted under the imaging head of an MFP 3D AFM (Asylum Research), and AFM scans were performed with silicon cantilevers (SSS-NCLR-10, Nanosensors) with tip radius curvature of 2 nm. Image analysis was performed with Igor Pro 5.0 image analysis software accompanying the instrument.
Antibody Dot Blot Analysis
2 μl of SAA (starting concentration 0.3 mg/ml) was spotted on nitrocellulose membranes (Hybond ECL, GE Healthcare). After adhesion of the protein and overnight drying at RT, the membrane was blocked (10% nonfat dry milk in PBS) at RT for 2 h and then washed 3 times in PBS with Tween 20 (PBST). The blots were then incubated with A11 or OC antibodies (Millipore) diluted 1:1000 (5% nonfat dry milk in PBS) at RT for 1 h. The blots were washed 3 times (PBST) and then incubated with goat-anti-rabbit horseradish peroxidase-conjugated secondary antibody at RT for 1 h. After washing, the blots were exposed to substrate (ECL Western blotting substrate, Thermo Fisher) and developed. Control experiments were performed using isotype rabbit antibody (Invitrogen).
Deep UV Resonance Raman (DUVRR) Spectroscopy
DUVRR spectra were measured at the University at Albany using a home-built Raman spectrometer as described elsewhere (12) with 199-nm excitation. A spinning NMR tube with a magnetic stirrer inside was used for sampling. GRAMS/AI 7.0 (Thermo Galactic, Salem, NH) was used for spectral data processing.
Planar Lipid Bilayer Experiments
Asolectin (Avanti Polar Lipids Inc.) bilayers were made by dissolving in n-decane at a concentration of 200 mg/ml (13, 14) and formed over a polystyrene cup with a 100-μm-diameter aperture and inserted into a thermally conductive chamber. The cup and the chamber were filled with 1 ml of 20 mm HEPES buffer (100 mm KCl, pH 7.4). Ag/AgCl leak-free electrodes (Cypress Systems) were introduced in the electrode wells containing 3 m KCl solution and connected to the cup and the chamber using salt bridges containing 3 m KCl with 2.5% agar. The chamber was then placed in a BLM-TC thermocycler (Warner) at ambient temperature. Capacitance and current were measured using a BC-535 patch clamp amplifier (Warner Instruments). A small amount of the lipid solution was then applied in the region surrounding the aperture using a fine-tipped paintbrush, and the transmembrane potential across the bilayer was set to −60 mV. The membrane quality was assessed by measuring its capacitance and conductance after stabilizing for ∼20 min. Membranes with capacitance values of ≥ 100 picofarads were used. Afterward, the SAA isoforms were allowed to aggregate by incubating the protein solution at 37 °C. At the desired time point, 20 μl of the protein solution (0.3 mg/ml) was exposed to the lipid bilayer by adding it to the chamber surrounding the bilayer. The current was then recorded with a Digidat 1440 data digitizer (Axon Instruments) and analyzed using the Clampex 10.0 software (Molecular Devices Corp). The sampling rate for collecting the data was 150 kHz, and signal filtration was achieved by using a built-in 4-pole Bessel filter at a setting of 2 kHz.
Calcein Dye Leakage Assay
Large unilamellar vesicles composed of DOPC:DOPG (dioleoylphosphatidylcholine:dioleoylphosphatidylglycerol; 80:20 w/w %) (Avanti Lipids) were prepared as previously described (15). A large unilamellar vesicle suspension containing 5 μl of diluted large unilamellar vesicles and 95 μl of TSB buffer (20 mm Tris, 150 mm NaCl, pH 8.3) was used to record base-line fluorescence, and calcein fluorescence was recorded by exciting the solution at 490 nm and monitoring the emission at 515 nm. The effect of SAA on liposome disruption was studied by mixing 5 μl of SAA obtained at different time points with 5 μl of diluted large unilamellar vesicle solution and 90 μl of TSB buffer in a black flat-bottom 96-well plate and gently shaken for 20 min. This incubation step was followed by measurement of calcein fluorescence intensity. The addition of 10 μl of 2% Triton X-100 to the assay sample further disrupted the membrane; the resultant fluorescence intensity corresponded to that due to maximum possible calcein release (Fmax). The amount of released calcein was measured by using the equation % dye leakage = ((Fs − Fb)/(Fmax − Fb)) × 100, where Fb is base-line fluorescence, and Fb is fluorescence intensity.
Cell Cytotoxicity Assay
Human embryonic kidney cells (HEK293, ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM). 200 μl of cell suspension (cell number 25 × 104 cells/ml) was added to each well of a 96-well plate (BD Biosciences) and allowed to adhere overnight at 37 °C. After this, the medium was removed, and each well of the microtiter plate was supplemented with 90 μl of fresh DMEM and 10 μl of SAA samples (starting concentration 250 μm (3 mg/ml)) or control samples. Cells were further incubated with the samples for 6, 24, or 72 h (in separate experiments) as specified. After the incubation period, cell viability was assessed using the MTS assay according to the manufacturer's instructions (Promega). SAA refolded oligomers are α-helix-rich oligomers obtained upon refolding at 4 °C. SAA misfolded oligomers are aggregates obtained after 2 h (SAA2.2) and 12 h (SAA1.1) of incubation at 37 °C. Fibrils were obtained after at least 2 weeks of incubation at 37 °C.
RESULTS
Refolding of SAA1.1 and SAA2.2 Reveals Different Oligomeric Propensities
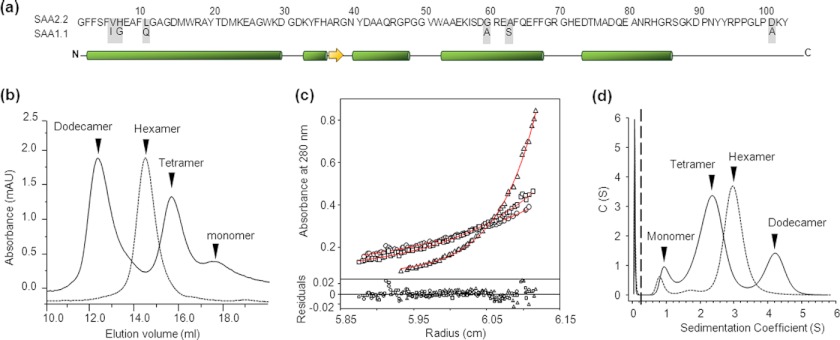
SAA1.1 and SAA2.2 share 94% sequence homology (Fig. 1a), yet they show distinct pathogenic properties. This suggests that there are significant biophysical/biochemical differences between these isoforms. To probe these inherent properties, we expressed SAA2.2 and SAA1.1 in E. coli. We previously reported that SAA2.2 refolds into a kinetically accessible octameric species that converts to a more stable hexamer (16) after a few weeks of incubation at 4 °C (10). In contrast, SAA1.1 refolded into a mixture of oligomers with retention times in size exclusion chromatography (SEC) that were consistent with dodecamer (12.4 ml), tetramer (15.7 ml), and monomer (17.9 ml) (Fig. 1b). Unlike SAA2.2, the oligomeric distribution of SAA1.1 did not change significantly over time (10). To avoid inconsistencies with the oligomeric structures, all the experiments described here were carried out with SAA2.2 and SAA1.1 samples refolded and stored at 4 °C for at least 20 days.
FIGURE 1.
The in vitro oligomeric structure of refolded SAA1.1 and SAA2.2. a, SAA1.1 and SAA2.2 share a high degree of structural similarity, including 94% sequence identity, and similar α-helix and β-sheet secondary structure according to a secondary structure prediction algorithm. b, analytical size-exclusion chromatography shows that SAA2.2 forms a hexamer (elution volume ∼14.7 ml), whereas SAA1.1 folds into a heterogeneous mixture of dodecamer (∼12.4 ml), tetramer (∼15.8 ml), and monomer (∼17.8 ml). mAU, milliabsorbance units. c, shown is equilibrium sedimentation AUC analysis of SAA1.1. Solid lines represent the global fit of representative traces at 10,000 (○), 12,000 (□), and 19,000 rpm (▵). The lower panel shows the residuals for each fit. The Mr obtained from the fit was 61.7 ± 0.9. d, sedimentation velocity AUC experiments showed at least three different sedimentation boundaries for SAA1.1 consistent with a mixture of dodecamer, tetramer, and monomer. SAA2.2 was analyzed in parallel experiments, and the data revealed one major boundary consistent with the hexameric structure observed by SEC and previous AUC experiments (8). All experiments were carried out at 4 °C.
To further characterize the oligomeric states of SAA1.1, we performed sedimentation equilibrium analytical ultracentrifugation (SE-AUC). Recent SE-AUC studies of SAA2.2 yielded a Mr of 70, which is consistent with a hexamer Mr and is in excellent agreement with SEC results (10). The SE-AUC traces for SAA1.1 shown in Fig. 1c were globally fitted, yielding a Mr of 61.7 ± 0.9. This averaged Mr is not consistent with hexamer (69.6 kDa) or tetramer (46.4 kDa) but supports the SEC data, suggesting that SAA1.1 exists as a mixture of oligomeric species. We carried out SV-AUC to probe further the structure of SAA1.1 and SAA2.2 (Fig. 1d). The SAA2.2 data showed a major peak with a sedimentation coefficient of 3 S, consistent with a hexameric protein, whereas for SAA1.1 it showed two major peaks with maxima at 2.5 S and 4.3 S and a minor peak at ∼1 S. By comparison with the SEC results, we conclude that the 4.3, 2.5, and 1 S peaks represent the dodecamer, tetrameric, and monomeric species, respectively. However, in contrast to the SEC data, the tetramer was the major species seen in SV-AUC, suggesting that the dodecamer might have dissociated to the tetramer during ultracentrifugation. Interestingly, when we calculated the area under the curve, the SV-AUC peaks corresponding to SAA1.1 yielded an area distribution of ∼71% tetramer, ∼21% dodecamer, and ∼7% monomer, corresponding to an average Mr of 64.1 kDa. This Mr value is within 5% of the Mr (61.7 kDa) obtained by SEC.
SAA1.1 is Less Stable Than SAA2.2 Despite Similar Secondary and Tertiary Structure
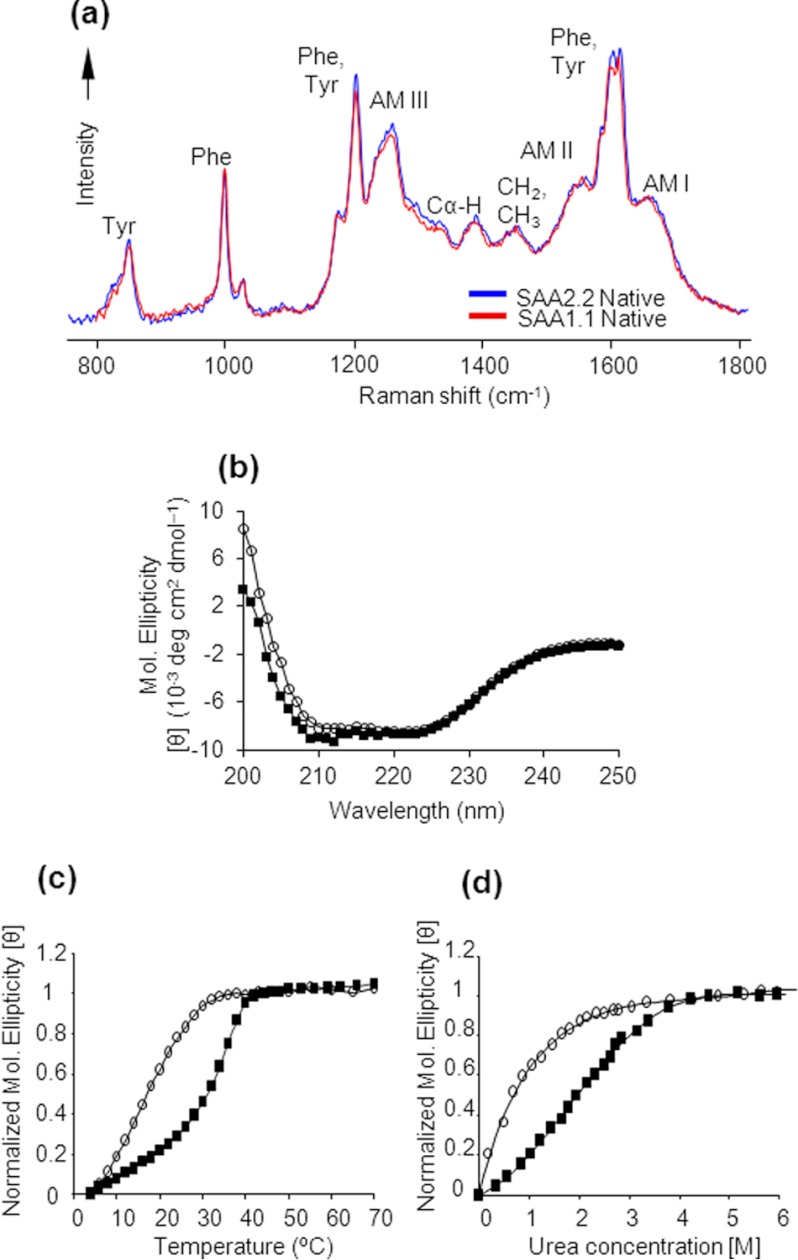
The differences in quaternary structure between SAA1.1 and SAA2.2 suggest that there might be differences in their secondary and tertiary structures. Therefore, we characterized the structure of the isoforms by DUVRR spectroscopy, a powerful tool for structural characterization of proteins in both refolded and misfolded oligomeric forms (17, 18). A high sensitivity of DUVRR spectra to the protein secondary structure is based on the dependence of amide vibrational modes on ψ and φ dihedral angles determining the three-dimensional conformation of the polypeptide backbone (17, 18). DUVRR spectra are most sensitive to β-sheet and unordered protein conformations in contrast to CD, which is most sensitive to α-helix. Amide I vibrational mode is dominated by C O stretching, with a small contribution from C—N stretching and N—H bending. Amide II and amide III bands involve significant C—N stretching, N—H bending, and C—C stretching. The Cα-H bending vibration mode involves Cα—H symmetric bending and C—Cα stretching (17).
O stretching, with a small contribution from C—N stretching and N—H bending. Amide II and amide III bands involve significant C—N stretching, N—H bending, and C—C stretching. The Cα-H bending vibration mode involves Cα—H symmetric bending and C—Cα stretching (17).
We found that oligomers obtained after refolding both SAA isoforms exhibit identical positions and intensities of amide bands I, II, and III, indicating that their secondary structure compositions are nearly identical (Fig. 2a). The maximum of the amide I band at ∼1645 cm−1 and the relatively weak Cα—H peak suggest that both SAAs have a predominantly α-helical secondary structure, consistent with their far UV-CD spectra (Fig. 2b) (16). In addition to similar amide bands, the vibrational modes of phenylalanine and tyrosine in the DUVRR spectra of SAA1.1 and SAA2.2 have almost the same intensity, indicating that the local environment of these residues is the same in both proteins. Thus, despite different oligomeric structures, SAA1.1 and SAA2.2 appear to have similar secondary and tertiary structure, as expected from their 94% sequence identity.
FIGURE 2.
Comparison of the secondary structure and stability of SAA1.1 (○) and SAA2.2 (■). a, SAA1.1 and SAA2.2 at 4 °C exhibited nearly identical DUVRR spectra, and the amide I (AM I) band maximum at ∼1645 cm−1 indicates the presence of largely α-helical secondary structure. b, far UV-CD spectra of SAA isoforms at 4 °C indicate similar and predominantly α helical secondary structure. c, the thermal denaturation of SAA was monitored by far-UV CD at 222 nm. d, the urea-induced denaturation of SAA at 4 °C was monitored by CD spectroscopy. The protein concentrations in a–d) were 0.3 mg/ml (20 mm Tris buffer, pH 8.0).
SAA2.2 is marginally stable in vitro and is intrinsically disordered at physiological temperature (11). Therefore, we compared the sensitivity of SAA1.1 and SAA2.2 to thermal and urea denaturation by monitoring their far-UV CD signal at 222 nm. Thermal denaturation experiments showed that SAA1.1 has a lower thermal stability than SAA2.2 (Fig. 2b). The melting curve of SAA1.1 lacked a pretransition base line and yielded a midpoint (Tm) of ∼18 °C, which is significantly lower than that of SAA2.2 (∼32 °C; Ref. 11) (Fig. 2b). The Tm values obtained from experiments are for comparison purposes only, as the thermal unfolding of SAA1.1 and SAA2.2 is largely irreversible. We then proceeded to probe the urea-induced denaturation of SAA1.1. We previously showed that upon exposure to low concentrations of urea, SAA2.2 undergoes a hexamer to monomer transition (11). Because the thermal denaturation data showed that SAA1.1 is mostly unfolded at RT, we performed the urea experiments at 4 °C. The cooperative urea-induced denaturation of SAA1.1 yielded an apparent transition mid-point (Cm) of ∼ 0.5 m, significantly lower than the ∼2.0 m value obtained for SAA2.2 (Fig. 2b). Thus, SAA1.1 is less stable than SAA2.2 and also appears to be an intrinsically disordered protein under physiological conditions.
SAA1.1 Exhibits an Oligomer-rich Long Fibrillation Lag Phase
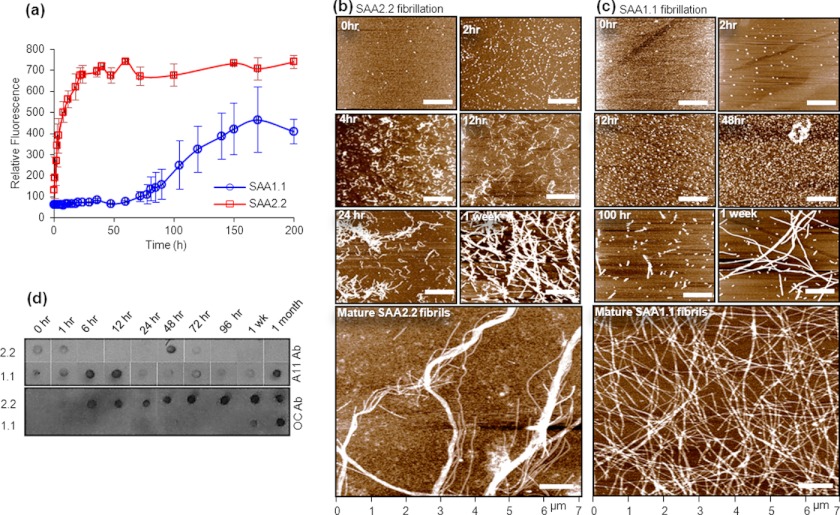
SAA2.2 spontaneously forms amyloid fibrils in vitro upon incubation at 37 °C (9), and therefore, it was expected that SAA1.1 would be even more amyloidogenic due to its pathogenic nature. The kinetics of SAA fibrillation at 37 °C was investigated using the ThT binding assay. SAA2.2 (0.3 mg/ml) spontaneously aggregated and formed cross-β-rich aggregates within a few hours, whereas SAA1.1 showed no increase in ThT intensity until about 3 days (Fig. 3a). Interestingly, after 70–100 h at 37 °C, the ThT intensity of SAA1.1 samples increased and reached a maximum in about a week. AFM was used in parallel to monitor the species formed during amyloid formation at 37 °C. SAA2.2, freshly taken from cold storage and spotted on mica, showed small spherical structures that are thought to be refolded hexamer (Fig. 3b, 0 h). SAA2.2 formed prefibrillar oligomers rapidly at 37 °C and assembled within a few hours into short curvilinear fibrils that further grew into longer fibrils and then into mature linear fibrils by 1 week (Fig. 3b). Refolded SAA1.1 (as shown in Fig. 1b) at 4 °C showed similar spherical structures as observed for SAA2.2, suggesting refolded assemblies (Fig. 3c, 0 h). When incubated at 37 °C, SAA1.1 assembled into globular oligomers as early as 2 h (Fig. 3c). Further incubation for 2–3 days resulted in more and larger oligomers (i.e. late-stage prefibrillar oligomers). Rod-shaped prefibrillar structures were observed as early as ∼70 h and eventually assembled into long fibrils of similar length and height as that of SAA2.2 mature fibrils.
FIGURE 3.
Kinetics of SAA aggregation at 37 °C probed by ThT fluorescence, AFM, and immunoblotting. a, upon incubating SAA1.1 and SAA2.2 at 37 °C, their fibrillation kinetics was monitored by ThT fluorescence at 485 nm. The error bars represent the normalized S.D. from three independent experiments. b and c, the time course of SAA aggregation induced by incubation at 37 °C was monitored by AFM. The images are shown as height traces, and the scale bar corresponds to 1 μm. d, shown is a time-dependent dot blot analysis of SAA1.1 and SAA2.2 fibrillation using the conformation-specific A11 and OC antibody. The starting protein concentrations in a–d were 0.3 mg/ml (20 mm Tris buffer, pH 8.0).
We utilized the fibril-specific (OC) and oligomer-specific (A11) antibody to confirm the slow and fast fibrillation rates of SAA1.1 and SAA2.2, respectively. OC is a conformation-specific antibody that recognizes generic amyloid fibrils from a wide range of proteins but does not bind to monomeric or non-fibrillar oligomeric forms(19). A11 antibodies have been shown to bind generic epitopes specific to prefibrillar oligomers (19). Dot blot experiments performed on SAA2.2 and its aggregates showed moderate OC binding to SAA2.2 oligomers obtained as early as ∼1 h (Fig. 3d) and moderate to strong OC antibody binding thereafter, confirming the fast fibrillation kinetics of SAA2.2. In contrast, OC antibody showed no binding to SAA1.1 oligomers until about 1 week (Fig. 3d), when moderate binding to OC was observed. Strong OC binding was observed only for SAA1.1 fibrils obtained after ∼1 month of SAA1.1 incubation at 37 °C. The A11 data did not show the consistent trend observed for OC binding. A11 bound to SAA1.1 aggregates throughout its fibrillation pathway (Fig. 3d), suggesting the coexistence of prefibrillar oligomeric intermediates and fibrils. A11 binding to SAA2.2 varied somewhat from sample to sample, but the overall trend was consistent with weak or no binding after 1 h of incubation at 37 °C. We occasionally observed binding at later time points, as shown in the representative data in Fig. 3d, perhaps due to conformational heterogeneity and the marginal stability of SAA2.2 fibrils (20). These dot blot results are mostly consistent with the ThT and AFM data and together demonstrate that SAA1.1 and SAA2.2 have very different fibrillation kinetics.
Structural Differences between the Refolded Protein and Mature Fibrils of SAA1.1 and SAA2.2
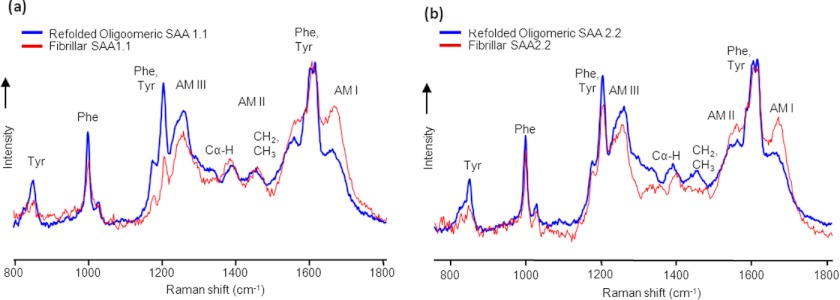
DUVRR spectroscopy is uniquely capable of identifying differences in β-sheet secondary structures between globular and fibrillar structures (21) and, therefore, was used to probe the structure of SAA1.1 and SAA2.2 mature fibrils. DUVRR spectra of mature SAA fibrils are superimposed (Fig. 4, a and b) with Raman spectra of the corresponding refolded proteins (Fig. 2a). Absolute normalization of Raman spectra acquired for large protein aggregates is a challenging task. Herein, DUVRR spectra are normalized using Tyr and Phe bands (∼1600 cm−1) for comparison.
FIGURE 4.
Comparing the structural differences between refolded and fibrillar SAA using Raman spectroscopy. a and b, shown are superimposed DUVRR spectra of the refolded oligomer(s) and the mature fibrils of SAA1.1 (a) and SAA2.2 (b). The mature fibrils were formed after incubating SAA for 1 month at 37 °C. To allow comparison, DUVRR spectra were normalized using the Tyr and Phe bands (∼1600 cm−1).
Raman spectra of SAA1.1 and SAA2.2 fibrils showed an increase in the intensity of the amide I and II bands compared with those of refolded proteins, consistent with increase in β-sheet structure. Changes in the shape and intensity of amide III, CH2/CH3, and Cα—H bands were observed in the spectra of SAA2.2 fibrils compared with the refolded protein, suggesting that SAA2.2 undergoes substantial a change in polypeptide backbone conformation upon fibril formation (Fig. 4b). In contrast, changes in the shape and intensity of CH2/CH3 and Cα-H bands were less evident for SAA1.1, suggesting that the formation of SAA1.1 fibrils involves much less perturbation of the protein secondary structure. In the case of the Phe and Tyr bands at 1000 cm−1 and 856 −1, respectively, fibrillation of SAA1.1 was accompanied by a decrease in their intensity, indicating substantial changes in the local environments of these amino acid residues upon SAA1.1 fibrillation. In the case of SAA2.2, although a decrease in the Tyr band intensity occurred upon SAA2.2 fibrillation, there were almost no changes in the intensity of the Phe band at 1000 cm−1. The latter suggests that, in contrast to SAA1.1, the local environment of Phe residues does not change significantly upon SAA2.2 fibrillation. The conclusions above are further supported by the spectral changes in the region of 1200–1300 cm−1 upon fibrillation (Fig. 4). Although the overlap of Phe, Tyr, and amide III bands in this region complicates the analysis and definitive interpretation, more significant change in both 1200-cm−1 aromatic amino acid band and amide III band on fibrillation is evident for SAA1.1. In summary, the DUVRR data suggest that SAA1.1 and SAA2.2 may aggregate via different aggregation mechanisms.
Prefibrillar Species and Fibrils of SAA1.1 and SAA2.2 Exhibit Differences in Structure and Morphology
The differences between SAA1.1 and SAA2.2 in their fibrillation kinetics, AFM images of their early stages of fibrillation, and fibrillar structure as determined by DUVRR suggest differences in their fibril morphology. Therefore, we further compared the AFM data of the prefibrillar oligomers/early fibrils and mature fibrils of SAA1.1 and SAA2.2. AFM data show that SAA2.2 forms spherical oligomers that rapidly assembled into short curvilinear fibrils (Fig. 5a and Fig. 3b, 12 h). In contrast, after a long oligomer-rich lag phase, SAA1.1 formed straight rigid-looking protofibrillar (very short fibrils) structures (Fig. 5b and Fig. 3c, 100 h). The fibril assembly units of SAA1.1 and SAA2.2 were clearly different, with oligomer heights of 2.0 and 1.0 nm, respectively (Fig. 5c). It appears that the curvilinear protofibrillar units of SAA2.2 result in flexible fibrils that can braid together into bundles (Fig. 3b, Mature SAA2.2). Such flexibility was absent in mature SAA1.1 fibrils, which due to their rod-like protofibrils, seem to mostly self-interact via lateral associations (Fig. 3b). Thus, although at first glance the AFM images of mature fibrils from SAA1.1 and SAA2.2 appear to be similar (Figs. 3, b and c, and 5d), the differences at the prefibrillar and early fibrillar levels are consistent with mature fibrils with significant structural differences.
FIGURE 5.
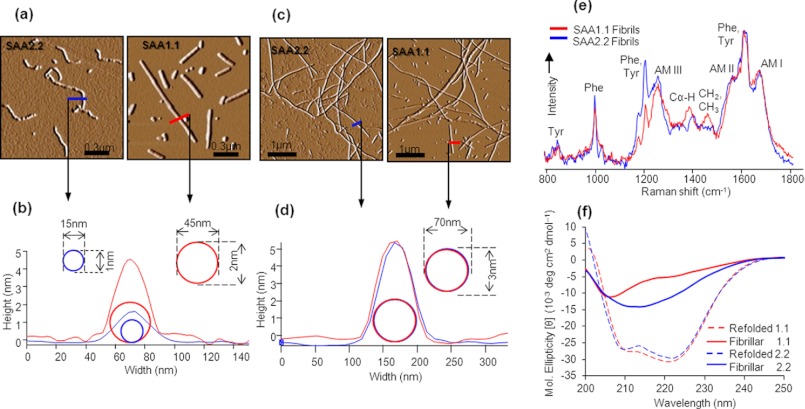
SAA1.1 and SAA2. 2 self-assemble into amyloid fibrils of different morphologies. a, AFM amplitude traces of early aggregates show clear differences in protofibrillar shape and size between SAA1.1 (70 h) and SAA2.2 (10 h). b, a plot of the protofibril cross-section of SAA1.1 and SAA2.2 was obtained from the corresponding height traces in a. c, shown are AFM images of full-length fibrils formed by SAA1.1 (150 h) and SAA2.2 (50 h). d, a plot of the fibril cross-section was obtained from the corresponding height traces in c. e and f, the mature fibrils formed by the two isoforms were analyzed by DUVRR (e) and far UV-CD (f) showing significant structural difference. The DUVRR spectra are the same as shown in Fig. 4. AM, amide.
To further compare the structural differences between the fibrils of SAA1.1 and SAA2.2, the DUVRR and far-UV CD spectra were superimposed (Fig. 5, d and e). The DUVRR spectra (Fig. 5d) were normalized using the Tyr and Phe bands at ∼1600 cm−1. The differences of Phe bands at 1000 cm−1 indicate that Tyr and Phe aromatic amino acids have a different local environment in these two types of fibrils. Also, there is a large difference in the frequency of Cα—H bending mode, indicating a significant change in the polypeptide backbone conformation. Cα-H peak has a maximum at 1385 cm−1 in the case of SAA 1.1 fibrils, indicating a substantial presence of disordered protein conformation. In contrast, the Cα-H band maximum at 1400 cm−1 for the SAA2.2 fibrils is indicative of β-sheet structure. The far UV-CD spectrum of SAA2.2 shows a prominent minimum near 215 nm, suggesting a high β-sheet content (Fig. 5e). SAA1.1 fibrils show two observable minima at 222 and 205 nm. The presence of a more prominent extrema at 205 nm (a red shift of ∼10 nm from the typical random coil minima at ∼195 nm) suggests the presence of significant amounts of unordered secondary content. Thus, altogether, the AFM, Raman, and CD data suggest that SAA2.2 and SAA1.1 aggregate via different mechanism, leading to the formation of amyloid fibrils with significant structural differences.
Cross-seeding Experiments Suggest That SAA1.1 and SAA2.2 Form Amyloid Fibrils via Different Pathways
Amyloid formation can usually be accelerated by the addition of a small amount of preformed amyloid from the same protein that acts as a nucleation seed (22). To further probe whether SAA1.1 and SAA2.2 were forming fibrils via distinct pathways, we self-seeded and cross-seeded refolded-SAA with a 5% w/w aliquot of SAA protofibrils and fibrils. The AFM images of the SAA samples used for seeding experiments are shown in Fig. 6a. The addition of SAA1.1 protofibrils to SAA1.1 resulted in a mild seeding effect, but the addition of SAA2.2 protofibrils had no effect on SAA1.1 fibrillation (Fig. 6b). In contrast, seeding SAA1.1 with SAA1.1 fibrils resulted in a strong seeding effect, whereas SAA2.2 fibrils had no seeding activity on SAA1.1 (Fig. 6c).
FIGURE 6.
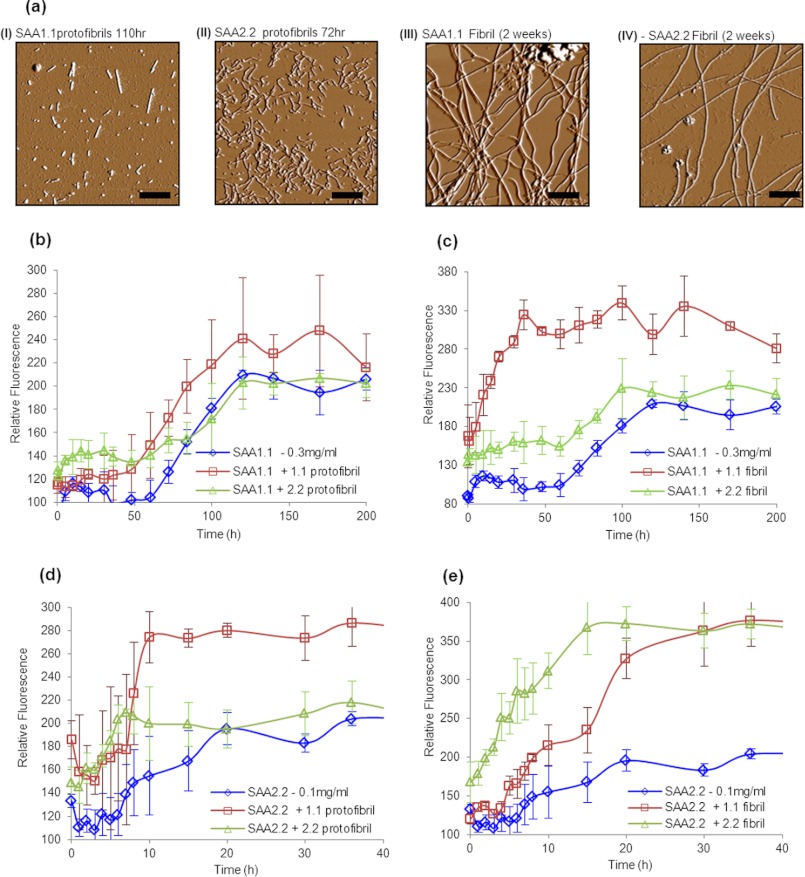
Self- and cross-seeding effects of preformed SAA aggregates on the kinetics of SAA aggregation. a, amplitude AFM traces of SAA1.1 and SAA2.2 aggregates (I, 1.1 protofibrils (3–4 days); II, 2.2 protofibrils (3 days); III, 1.1 fibrils (2 weeks); IV, 2.2 fibrils (2 weeks)) were preformed by allowing aggregation at 37 °C for the specified time and added (5% v/v) to refolded oligomers of SAA isoforms 1.1 and 2.2 as amyloid seeding agents. Self- and cross-seeding effects of protofibrils (b) and fibrils (c) on SAA1.1 fibrillation are shown. Self- and cross-seeding effects of protofibrils (d) and fibrils (e) on SAA2.2 fibril formation are shown.
For the seeding experiments of SAA2.2 fibrillation, we used 0.1 instead of 0.3 mg/ml protein to slow down the aggregation rate and better evaluate the seeding effects. Interestingly, both SAA2.2 and SAA1.1 protofibrils were able to seed the fibrillation of SAA2.2, with the former exhibiting a much higher ThT fluorescence intensity (Fig. 6d). When mature fibrils were used to seed SAA2.2 fibrillation, the SAA2.2 fibrils exhibited a much stronger seeding effect than the SAA1.1 fibrils (Fig. 6e). Although the seeding kinetic effect of the SAA1.1 fibrils was modest, the increase in ThT fluorescence was much higher than in the absence of seeds. All in all, these results show that SAA1.1 and SAA2.2 differ in their ability to cross-seed each other's fibrillation, suggesting that they inherently fibrillate via different pathways.
Prefibrillar Oligomers of SAA1.1 and SAA2.2 Can Permeabilize Synthetic Lipid Bilayers
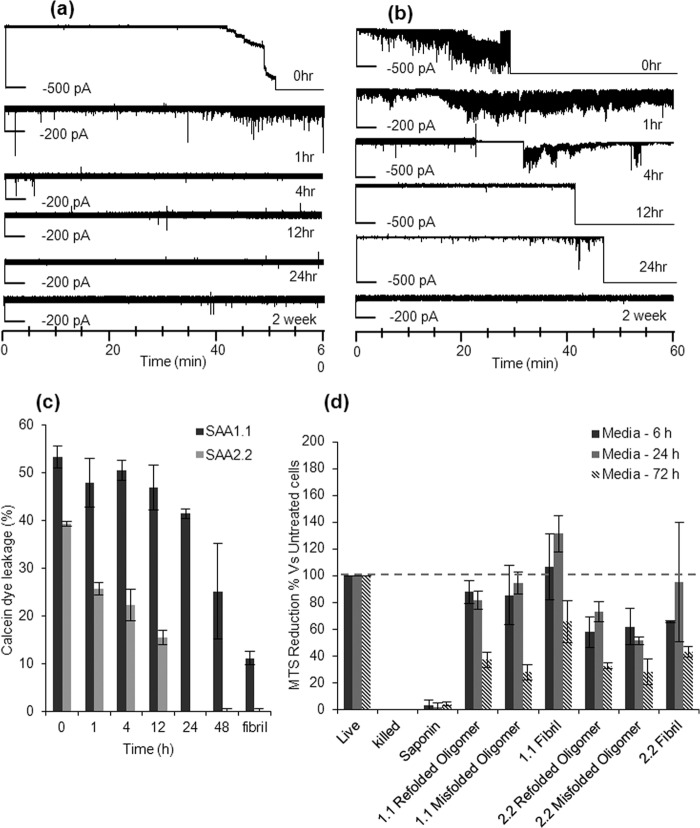
It has been shown that amyloid-related oligomers obtained from a variety of proteins possess membrane permeabilization activity (23). Therefore, we decided to probe whether the long-lived SAA1.1 oligomers could disrupt model membranes in vitro. Asolectin bilayers are good mimics of biological membranes(14, 24). Hence, we used them to screen for the ability of various SAA aggregates to destabilize or permeabilize lipid bilayers. Specifically, aggregates formed during SAA fibrillation were exposed to lipid bilayers, and we measured the conductance of the bilayer. Formation of discrete pores in the bilayer or membrane “leakage” due to interaction with the aggregates can be probed by measuring the current flowing from one chamber to another. The addition of refolded SAA2.2 at micromolar concentrations (0 h) to the solution surrounding the bilayer led to an increase of membrane conductance in all the experiments (Fig. 7a), suggesting irreparable bilayer rupture due to formation of pores. Similarly, bilayer leakage, but not disruption, was observed when the bilayer was exposed to samples of SAA2.2 preincubated at 37 °C for 1 h. Interestingly, no rupture or interaction with the bilayer was observed when the SAA2.2 samples were incubated at 37 °C for 4 h, 24 h, or 2 weeks (Fig. 7a) before exposure to the lipid bilayer. These results suggest that the early stage prefibrillar oligomers of SAA2.2, but not the short curvilinear fibrils or longer fibrils, have the ability to permeabilize the asolectin bilayers. We then followed the propensity of SAA1.1 aggregates formed upon incubation at 37 °C for different time periods to cause membrane permeabilization. Surprisingly, either bilayer leakage or bilayer disruption was observed for SAA1.1 oligomers/aggregates formed after ∼24 h (Fig. 7b). No bilayer disruption was observed for samples incubated at 37 °C for ∼2 weeks, suggesting that only the early-stage prefibrillar oligomers of SAA1.1 and not the longer fibrils were capable of membrane permeabilization.
FIGURE 7.
Lipid membrane permeabilization caused by SAA prefibrillar oligomers. a, membrane leakage or disruption by SAA aggregates was probed by measuring the conductance across the lipid bilayer. SAA2.2 oligomers interacted with the lipid bilayer and caused membrane disruption at 0 h and membrane leakage at 1 h, but no membrane interaction was observed with older aggregates. b, SAA1.1 oligomers formed within 24 h caused membrane leakage or complete membrane disruption, but no leakage was observed for the 2-week sample. c, kinetics of SAA2.2 and SAA1.1 membrane permeabilization were probed using a calcein-based liposome leakage assay. SAA1.1 oligomers showed a higher propensity to cause liposome disruption and for a longer period of time relative to SAA2.2 oligomers. Mature amyloid fibrils of both SAA1.1 and SAA2.2 caused lower membrane leakage relative to their respective oligomers. d, the effect of various SAA aggregates on HEK293 cell was monitored using cell viability (MTS reduction) assay. SAA aggregates were added to cell cultures at a final concentration of 0.3 mg/ml and incubated at 37 °C for 6, 24, and 72 h before determining cell viability. Cell viability is expressed as the percentage reduction of MTS in treated cells compared with cells not exposed to any treatment (Live). Killed cells are those wells where media were replaced by sterile water. The error bars in represent the S.D. from two independent experiments.
Membrane disruption effect of various SAA aggregates were further studied by exposing them to DOPC:DOPG (dioleoylphosphatidylcholine:dioleoylphosphatidylglycerol; 80:20 w/w%) vesicles loaded with calcein. Aggregates capable of causing membrane permeabilization would result in the release of calcein from the calcein-loaded vesicles. The amount of calcein released can be measured by using fluorometric techniques and can be correlated to provide information about the extent and kinetics of membrane disruption caused by the aggregates. About 50% leakage was obtained in the presence of aggregate at ∼1 μm concentration of SAA1.1 throughout the first 24 h of incubation at 37 °C (Fig. 7c). Subsequent aggregates showed relatively less calcein leakage with the lowest leakage corresponding to fibrillar samples. Early aggregates of SAA2.2 showed calcein leakage and gradually subsided over the first 12 h of aggregation at 37 °C. By 24 h no calcein leakage was observed for SAA2.2 aggregates. These results indicate that amyloid fibrils of SAA2.2 and SAA1.1 did not disrupt biomimic membranes, but rather, membrane permeabilization coincides with the presence of prefibrillar oligomers present during the fibrillation lag phase.
To probe the ability of SAA to interact with cellular membranes, we examined the effect of various SAA aggregates on HEK293 cell using the MTS assay (see “Experimental Procedures”) to monitor cell viability. SAA species (refolded-oligomer, misfolded oligomers/protofibrils, and mature fibrils) were concentrated to 3 mg/ml and added to cell cultures, resulting in a 10-fold dilution of SAA to a concentration (0.3 mg/ml) relevant to that present during inflammation. The cells were incubated with SAA in serum-free media for 6, 24, and 72 h before determining cell viability (Fig. 7d). Treatment with SAA1.1 for 6 and 24 h showed mild loss of viability, but significant cytotoxicity was observed at 72 h. SAA2.2 species caused cytotoxicity at all incubation times, but similar to SAA1.1, the greatest effect was observed at 72 h. Interestingly, the SAA2.2 species were more toxic than their SAA1.1 counterparts when incubated with the HEK cells for 6 and 24 h.
DISCUSSION
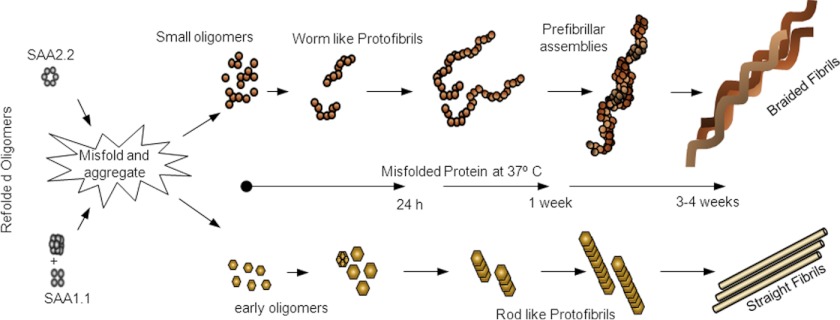
We show here that SAA1.1 and SAA2.2 form different oligomeric structures upon folding in vitro. When incubated at 37 °C, these refolding oligomers of SAA unfold, “misfold,” and assemble into larger oligomers with distinct structure and amyloidogenicity. A preliminary model to illustrate the putative in vitro fibril assembly pathway for SAA1.1 and SAA2.2 is shown in Fig. 8. The prefibrillar oligomers of SAA2.2 assemble quickly into protofibrils and curvilinear fibrils, whereas the larger SAA1.1 prefibrillar oligomers accumulate, as if lacking a conformational change that would allow them to fibrillate. Eventually, SAA1.1 begins to assemble into straight fibrils with a height similar to that of the oligomers that populate the lag phase. Thus, the six residues (Fig. 1a) that differ between SAA1.1 and SAA2.2 have a major effect on their oligomerization and fibrillation in vitro, although their specific role is not yet known. Although the pathological relevance of these findings are unclear, the potential implications are intriguing.
FIGURE 8.
Preliminary model of SAA1.1 and SAA2.2 fibrillation based on AFM data. Misfolded SAA2.2 aggregates into transient oligomers that undergo linear assembly into curvilinear early fibrils. These curvilinear fibrils further elongate and intertwine into mature fibrils capable of self-assembling further into braided bundles. In contrast, SAA1.1 forms spherical oligomers that are larger than those formed by SAA2.2 and give rise to rod-like protofibrils/early fibrils that assemble into mature straight fibrils. These SAA1.1 fibrils seem more rigid than those formed by SAA2.2, as they do not intertwine but rather appear able to interact laterally with each other.
What Is the Oligomeric Structure of Native SAA in Vivo?
The high degree of sequence homology suggests that SAA1.1 and SAA2.2 likely have a similar monomeric structure. Far UV-CD and DUVRR spectra suggest that they further share similar secondary structures and likely comparable tertiary folds, yet SAA2.2 can refold in vitro into an octamer and a hexamer, whereas SAA1.1 forms tetrameric and dodecameric oligomers. The physiological relevance of these oligomers is not yet clear, especially as our thermal denaturation results suggest that SAA2.2 and SAA1.1 may be disordered in vivo unless stabilized by ligands (25). However, the possibility that SAA may exist in different oligomeric forms in vivo is also supported by other studies showing that SAA is able to oligomerize in vitro (26–29) and in vivo (30, 31). In particular, recent studies have shown the presence of various SAA oligomeric forms similar in size to those we report here in murine spleen tissue (30) and purified exosomes (31). In fact, in the former study (30), freshly isolated SAA oligomers were α-helix-rich and in the molecular weight ranges comparable to the refolded oligomers reported in this study.
In serum, SAA is usually found associated to the third fraction of HDL (32). However, we have previously shown that SAA2.2 hexamer does not bind to HDL, suggesting that the N terminus containing the HDL binding site is buried within the hexamer, preventing interaction with lipid (33). Because it has been shown that SAA can also exist in vivo in lipid-free form, it is plausible that at high SAA concentrations (up to 1–2 mg/ml (4)) present during inflammation, lipid-free SAA might exist in an oligomeric non-HDL binding state for functional reasons (34). This reasoning is consistent with a study showing that after an acute phase reaction in mice, SAA was found to be circulating free in serum in addition to being HDL-bound (35, 36). Thus, the structural malleability of SAA1.1 and SAA2.2 we observed in vitro may reflect the in vivo properties of SAA proteins to allow the regulation of their many putative functions related to cholesterol metabolism and the inflammatory responses (34). Furthermore, it would help explain how a small (103–104 residues) and marginally stable protein like SAA is able to interact in vivo with many binding partners, including HDL, cholesterol (37), outer membrane protein (38), heparin (39), extracellular matrix glycoproteins (40), and platelets (41).
Long-lived SAA Prefibrillar Oligomers and Implications for AA Amyloidosis
Many studies support the paradigm that the toxic species in amyloid/misfolding diseases comprise misfolded oligomers and not the amyloid fibrils (23). Amyloid-related oligomers obtained from a variety of proteins have been shown to cause membrane permeabilization and disruption (23). Although the mechanism behind this interaction is unclear, misfolded oligomeric species appear to behave like amphipathic cell-penetrating peptides. Our results (Fig. 7) similarly show that SAA1.1 and SAA2.2 oligomers are able to permeabilize model membranes in vitro and are in agreement with previous studies demonstrating that SAA can form ion channels in vitro (27). Furthermore, our observation of SAA-induced toxicity in HEK293 cells, especially during 72 h of exposure, suggests that aggregated SAA species are able to adversely interact with cellular membranes. Although the synthetic lipid membranes and cell viability results did not show a direct correlation, this is not surprising when considering the different experimental conditions. Specifically, the synthetic membrane experiments are quick and carried out at room temperature, thereby allowing us to better probe the effect of the different SAA species (refolded oligomers, misfolded oligomers/protofibrils, and amyloid fibrils). In contrast, the cellular assays are slow, resulting in continuous SAA aggregation, especially for SAA2.2 during the long incubation period at 37 °C. Therefore, the cell viability experiments are not able to identify the toxic species. Nevertheless, our results suggest that the aberrant interaction of putatively toxic SAA species with the cell membrane may involve a complex interplay of the structure/stability/aggregation kinetics of oligomers, the physiochemical feature of the cell membrane, and interaction with other proteins (42).
There is substantial evidence that SAA oligomers play a pathological role in AA amyloidosis. AA amyloidosis can be induced in mice by administering extracts of amyloid-laden tissue that act as an amyloid enhancing factor (43, 44), and the disease has been shown to propagate via a prion-like mechanism (45). More recently, SAA oligomers have been identified as the species that triggers and transmits AA amyloidosis in mice (30, 31). Our in vitro results show that the long fibrillation lag phase of pathogenic SAA1.1, in contrast to the virtually spontaneous fibrillation of non-pathogenic SAA2.2, accounts for the persistent population of SAA1.1 prefibrillar oligomers. It should be noted that to assess the pathological implications of the kinetics of SAA oligomerization and fibrillation, further studies would be required. Nevertheless, if this in vitro property of SAA1.1 and SAA2.2 were to reflect their in vivo behavior during chronic inflammation, this may imply that the oligomerization-fibrillation kinetics of SAA isoforms, and not their inherent amyloidogenicity, could partly account for their diverse pathogenicity in AA amyloidosis. More broadly, it seems likely that the rate at which toxic misfolded oligomers form and further aggregate into less toxic species may be a general pathogenic factor in amyloid diseases.
Pathological Implications of the Distinct Structure and Morphologies of Prefibrillar and Fibrillar SAA Species
Unlike small globular proteins that usually exist in a single native conformation, fibrils may exhibit a variety of morphologies (46, 47). More recently, simulation studies have shown that amyloid polymorphism is under kinetic control and that by varying the relative stabilities of an amyloid-competent and amyloid-resistant species, morphological differentiation can be observed (48, 49). Despite the similar sequence of SAA2.2 and SAA1.1, they formed different oligomeric species that led to the formation of fibrils of different structure (Fig. 5). The pathological implications of this finding are not clear, but recent studies have shown that distinct fibril morphologies can be linked to different biological activities such as phenotypic expression in yeast prions (50), toxicity to neuronal cells (51), and deposition patterns (52). Interestingly, amyloid tissue from patients with AA amyloidosis were shown to contain significant structural disorder and variability, suggesting that polymorphism is an intrinsic property of SAA fibrils (53). The high concentration of SAA during inflammation along with the presence of other in vivo ligands might modulate the kinetics of SAA aggregation, favoring certain oligomers and morphologies over others.
Overall, the results of our study suggest that factors affecting the kinetics of assembly, the pathway of aggregation, and the stability of the various SAA species on the amyloid pathway are likely to play an important role in the diverse pathogenicity of different SAA isoforms across species and might also be relevant to other amyloid diseases.
Acknowledgments
The AUC experiments were performed in the Analytical Biochemistry core facility of the Rensselaer Center for Biotechnology and Interdisciplinary Studies.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AG028158 (to W. C.) and R01 AG033719 (to I. K. L.).
- SAA
- serum amyloid A (AA)
- AFM
- atomic force microscopy
- AUC
- analytical ultracentrifugation
- RT
- room temperature
- SE
- sedimentation equilibrium
- SV
- sedimentation velocity
- SEC
- size exclusion chromatography
- ThT
- thioflavin T
- DUVRR
- deep UV resonance Raman
- TEV
- tobacco etch virus
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium.
REFERENCES
- 1. Uhlar C. M., Whitehead A. S. (1999) Serum amyloid A, the major vertebrate acute-phase reactant. Eur. J. Biochem. 265, 501–523 [DOI] [PubMed] [Google Scholar]
- 2. Malle E., Sodin-Semrl S., Kovacevic A. (2009) Serum amyloid A. An acute-phase protein involved in tumour pathogenesis. Cell. Mol. Life Sci. 66, 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cunnane G., Whitehead A. S. (1999) Amyloid precursors and amyloidosis in rheumatoid arthritis. Baillieres Best Pract. Res. Clin. Rheumatol. 13, 615–628 [DOI] [PubMed] [Google Scholar]
- 4. Gillmore J. D., Lovat L. B., Persey M. R., Pepys M. B., Hawkins P. N. (2001) Amyloid load and clinical outcome in AA amyloidosis in relation to circulating concentration of serum amyloid A protein. Lancet 358, 24–29 [DOI] [PubMed] [Google Scholar]
- 5. Röcken C., Shakespeare A. (2002) Pathology, diagnosis, and pathogenesis of AA amyloidosis. Virchows Archiv. 440, 111–122 [DOI] [PubMed] [Google Scholar]
- 6. Westermark P., Sletten K. (1982) A serum AA-like protein as a common constituent of secondary amyloid fibrils. Clin. Exp. Immunol. 49, 725–731 [PMC free article] [PubMed] [Google Scholar]
- 7. Yu J., Zhu H., Guo J. T., de Beer F. C., Kindy M. S. (2000) Expression of mouse apolipoprotein SAA1.1 in CE/J mice. Isoform-specific effects on amyloidogenesis. Lab. Invest. 80, 1797–1806 [DOI] [PubMed] [Google Scholar]
- 8. Sipe J. D., Carreras I., Gonnerman W. A., Cathcart E. S., de Beer M. C., de Beer F. C. (1993) Characterization of the inbred CE/J mouse strain as amyloid resistant. Am. J. Pathol. 143, 1480–1485 [PMC free article] [PubMed] [Google Scholar]
- 9. Wang L., Lashuel H. A., Colón W. (2005) From hexamer to amyloid. Marginal stability of apolipoprotein SAA2.2 leads to in vitro fibril formation at physiological temperature. Amyloid 12, 139–148 [DOI] [PubMed] [Google Scholar]
- 10. Wang Y., Srinivasan S., Ye Z., Javier Aguilera J., Lopez M. M., Colón W. (2011) Serum amyloid A 2.2 refolds into a octameric oligomer that slowly converts to a more stable hexamer. Biochem. Biophys. Res. Commun. 407, 725–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang L., Colón W. (2005) Urea-induced denaturation of apolipoprotein serum amyloid A reveals marginal stability of hexamer. Protein Sci. 14, 1811–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lednev I. K., Ermolenkov V. V., He W., Xu M. (2005) Deep-UV Raman spectrometer tunable between 193 and 205 nm for structural characterization of proteins. Anal. Bioanal. Chem. 381, 431–437 [DOI] [PubMed] [Google Scholar]
- 13. Herce H. D., Garcia A. E., Litt J., Kane R. S., Martin P., Enrique N., Rebolledo A., Milesi V. (2009) Arginine-rich peptides destabilize the plasma membrane, consistent with a pore formation translocation mechanism of cell-penetrating peptides. Biophys. J. 97, 1917–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ladiwala A. R., Litt J., Kane R. S., Aucoin D. S., Smith S. O., Ranjan S., Davis J., Van Nostrand W. E., Tessier P. M. (2012) Conformational differences between two amyloid β oligomers of similar size and dissimilar toxicity. J. Biol. Chem. 287, 24765–24773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nielsen S. B., Otzen D. E. (2010) Impact of the antimicrobial peptide Novicidin on membrane structure and integrity. J. Colloid Interface Sci. 345, 248–256 [DOI] [PubMed] [Google Scholar]
- 16. Wang L., Lashuel H. A., Walz T., Colon W. (2002) Murine apolipoprotein serum amyloid A in solution forms a hexamer containing a central channel. Proc. Natl. Acad. Sci. U.S.A. 99, 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oladepo S. A., Xiong K., Hong Z., Asher S. A., Handen J., Lednev I. K. (2012) UV resonance Raman investigations of peptide and protein structure and dynamics. Chem. Rev. 112, 2604–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lednev I. K. (2007) in Protein Structures, Methods in Protein Structures and Stability Analysis. Part B. Vibrational Spectroscopy (Uversky V. N., Permyakov E. A., eds). Nova Science Publishers, Inc., New York [Google Scholar]
- 19. Kayed R., Head E., Sarsoza F., Saing T., Cotman C. W., Necula M., Margol L., Wu J., Breydo L., Thompson J. L., Rasool S., Gurlo T., Butler P., Glabe C. G. (2007) Fibril-specific, conformation-dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ye Z., Bayron Poueymiroy D., Aguilera J. J., Srinivasan S., Wang Y., Serpell L. C., Colón W. (2011) Inflammation protein SAA2.2 spontaneously forms marginally stable amyloid fibrils at physiological temperature. Biochemistry 50, 9184–9191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shashilov V. A., Sikirzhytski V., Popova L. A., Lednev I. K. (2010) Quantitative methods for structural characterization of proteins based on deep UV resonance Raman spectroscopy. Methods 52, 23–37 [DOI] [PubMed] [Google Scholar]
- 22. Harper J. D., Lansbury P. T. (1997) Models of amyloid seeding in Alzheimier's disease and scrapie. Mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 66, 385–407 [DOI] [PubMed] [Google Scholar]
- 23. Kayed R., Sokolov Y., Edmonds B., McIntire T. M., Milton S. C., Hall J. E., Glabe C. G. (2004) Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 279, 46363–46366 [DOI] [PubMed] [Google Scholar]
- 24. Lin M. C., Kagan B. L. (2002) Electrophysiologic properties of channels induced by A β 25–35 in planar lipid bilayers. Peptides 23, 1215–1228 [DOI] [PubMed] [Google Scholar]
- 25. Wang L., Colón W. (2007) Effect of zinc, copper, and calcium on the structure and stability of serum amyloid A. Biochemistry 46, 5562–5569 [DOI] [PubMed] [Google Scholar]
- 26. Strachan A. F., Shephard E. G., Bellstedt D. U., Coetzee G. A., van der Westhuyzen D. R., de Beer F. C. (1989) Human serum amyloid A protein behavior in aqueous and urea-containing solutions and antibody production. Biochem. J. 263, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hirakura Y., Carreras I., Sipe J. D., Kagan B. L. (2002) Channel formation by serum amyloid A. A potential mechanism for amyloid pathogenesis and host defense. Amyloid 9, 13–23 [DOI] [PubMed] [Google Scholar]
- 28. Lu J., Du Clos T., Mold C., Sun P. (2010) Immunology 2010 Meeting, Baltimore, MD, May 7–11, 2010, Abstract no. 135–34, J. Immunol. 184 [Google Scholar]
- 29. Lakota K., Resnik N., Mrak-Poljsak K., Sodin-Semrl S., Veranic P. (2011) Colocalization of serum amyloid a with microtubules in human coronary artery endothelial cells. J. Biomed. Biotech. 2011, 528276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Senthilkumar S., Chang E., Jayakumar R. (2008) Diffusible amyloid oligomers trigger systemic amyloidosis in mice. Biochem. J. 415, 207–215 [DOI] [PubMed] [Google Scholar]
- 31. Tasaki M., Ueda M., Ochiai S., Tanabe Y., Murata S., Misumi Y., Su Y., Sun X., Shinriki S., Jono H., Shono M., Obayashi K., Ando Y. (2010) Transmission of circulating cell-free AA amyloid oligomers in exosomes vectors via a prion-like mechanism. Biochem. Biophys. Res. Commun. 400, 559–562 [DOI] [PubMed] [Google Scholar]
- 32. Benditt E. P., Eriksen N. (1977) Amyloid protein SAA is associated with high density lipoprotein from human serum (apolipoproteins). Proc. Natl. Acad. Sci. U.S.A. 74, 4025–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang L., Colón W. (2004) The interaction between apolipoprotein serum amyloid A and high density lipoprotein. Biochem. Biophys. Res. Commun. 317, 157–161 [DOI] [PubMed] [Google Scholar]
- 34. Urieli-Shoval S., Linke R. P., Matzner Y. (2000) Expression and function of serum amyloid A, a major acute-phase protein, in normal and disease states. Curr. Opin. Hematol. 7, 64–69 [DOI] [PubMed] [Google Scholar]
- 35. Hajri T., Elliott-Bryant R., Sipe J. D., Liang J. S., Hayes K. C., Cathcart E. S. (1998) The acute phase response in apolipoprotein A-1 knockout mice. Apolipoprotein serum amyloid A and lipid distribution in plasma high density lipoproteins. Biochim. Biophys. Acta 1394, 209–218 [DOI] [PubMed] [Google Scholar]
- 36. Webb N. R., de Beer M. C., van der Westhuyzen D. R., Kindy M. S., Banka C. L., Tsukamoto K., Rader D. L., de Beer F. C. (1997) Adenoviral vector-mediated overexpression of serum amyloid A in apoA-I-deficient mice. J. Lipid Res. 38, 1583–1590 [PubMed] [Google Scholar]
- 37. Liang J. S., Sipe J. D. (1995) Recombinant human serum amyloid-A (apoSAA37p) binds cholesterol and modulates cholesterol flux. J. Lipid Res. 36, 37–46 [PubMed] [Google Scholar]
- 38. Hari-Dass R., Shah C., Meyer D. J., Raynes J. G. (2005) Serum amyloid A protein binds to outer membrane protein A of Gram-negative bacteria. J. Biol. Chem. 280, 18562–18567 [DOI] [PubMed] [Google Scholar]
- 39. Ancsin J. B., Kisilevsky R. (1999) The heparin heparan sulfate-binding site on apo serum amyloid A. Implications for the therapeutic intervention of amyloidosis. J. Biol. Chem. 274, 7172–7181 [DOI] [PubMed] [Google Scholar]
- 40. Preciado-Patt L., Hershkoviz R., Fridkin M., Lider O. (1996) Serum amyloid a binds specific extracellular matrix glycoproteins and induces the adhesion of resting CD4(+) T cells. J. Immunol. 156, 1189–1195 [PubMed] [Google Scholar]
- 41. Urieli-Shoval S., Shubinsky G., Linke R. P., Fridkin M., Tabi I., Matzner Y. (2002) Adhesion of human platelets to serum amyloid A. Blood 99, 1124–1229 [DOI] [PubMed] [Google Scholar]
- 42. Evangelisti E., Cecchi C., Cascella R., Sgromo C., Becatti M., Dobson C. M., Chiti F., Stefani M. (2012) Membrane lipid composition and its physicochemical properties define cell vulnerability to aberrant protein oligomers. J. Cell Sci. 125, 2416–2427 [DOI] [PubMed] [Google Scholar]
- 43. Cui D., Kawano H., Hoshii Y., Liu Y., Ishihara T. (2008) Acceleration of murine AA amyloid deposition by bovine amyloid fibrils and tissue homogenates. Amyloid 15, 77–83 [DOI] [PubMed] [Google Scholar]
- 44. Lundmark K., Westermark G. T., Olsén A., Westermark P. (2005) Protein fibrils in nature can enhance amyloid protein A amyloidosis in mice. Cross-seeding as a disease mechanism. Proc. Natl. Acad. Sci. U.S.A. 102, 6098–6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Westermark G. T., Westermark P. (2009) Serum amyloid A and protein AA. Molecular mechanisms of a transmissible amyloidosis. FEBS Lett. 583, 2685–2690 [DOI] [PubMed] [Google Scholar]
- 46. Kodali R., Wetzel R. (2007) Polymorphism in the intermediates and products of amyloid assembly. Curr. Opin. Struct. Biol. 17, 48–57 [DOI] [PubMed] [Google Scholar]
- 47. Jahn T. R., Radford S. E. (2008) Folding versus aggregation. Polypeptide conformations on competing pathways. Arch. Biochem. Biophys. 469, 100–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pellarin R., Schuetz P., Guarnera E., Caflisch A. (2010) Amyloid fibril polymorphism is under kinetic control. J. Am. Chem. Soc. 132, 14960–14970 [DOI] [PubMed] [Google Scholar]
- 49. Stpkowski D., Bienia J. (2012) Nature of cross-seeding barriers of amyloidogenesis. Acta Biochim. Pol. 59, 307–312 [PubMed] [Google Scholar]
- 50. Tanaka M., Collins S. R., Toyama B. H., Weissman J. S. (2006) The physical basis of how prion conformations determine strain phenotypes. Nature 442, 585–589 [DOI] [PubMed] [Google Scholar]
- 51. Seilheimer B., Bohrmann B., Bondolfi L., Müller F., Stüber D., Döbeli H. (1997) The toxicity of the Alzheimer's β-amyloid peptide correlates with a distinct fiber morphology. J. Struct. Biol. 119, 59–71 [DOI] [PubMed] [Google Scholar]
- 52. Meyer-Luehmann M., Coomaraswamy J., Bolmont T., Kaeser S., Schaefer C., Kilger E., Neuenschwander A., Abramowski D., Frey P., Jaton A. L., Vigouret J.-M., Paganetti P., Walsh D. M., Mathews P. M., Ghiso J., Staufenbiel M., Walker L. C., Jucker M. (2006) Exogenous induction of cerebral β-amyloidogenesis is governed by agent and host. Science 313, 1781–1784 [DOI] [PubMed] [Google Scholar]
- 53. Jiménez J. L., Tennent G., Pepys M., Saibil H. R. (2001) Structural diversity of ex vivo amyloid fibrils studied by cryo-electron microscopy. J. Mol. Biol. 311, 241–247 [DOI] [PubMed] [Google Scholar]