Background: Physcomitrella PpORS is an ancient member of the plant type III polyketide synthase (PKS) family.
Results: PpORS, produced in nonprotonemal moss cells, synthesizes pentaketide 2′-oxoalkylresorcinols using a unique substrate binding site.
Conclusion: PpORS is a novel very long chain 2′-oxoalkylresorcinol synthase.
Significance: This is the first step toward understanding the co-evolution of the type III PKS family and land plants.
Keywords: Biosynthesis, Enzymes, Natural Product Biosynthesis, Plant Biochemistry, Polyketides, Protein Evolution, 2′-Oxoalkylresorcinol Synthase, Physcomitrella, Type III Polyketide Synthase, Very Long Chain Fatty Acyl-CoA
Abstract
The plant type III polyketide synthases (PKSs), which produce diverse secondary metabolites with different biological activities, have successfully co-evolved with land plants. To gain insight into the roles that ancestral type III PKSs played during the early evolution of land plants, we cloned and characterized PpORS from the moss Physcomitrella. PpORS has been proposed to closely resemble the most recent common ancestor of the plant type III PKSs. PpORS condenses a very long chain fatty acyl-CoA with four molecules of malonyl-CoA and catalyzes decarboxylative aldol cyclization to yield the pentaketide 2′-oxoalkylresorcinol. Therefore, PpORS is a 2′-oxoalkylresorcinol synthase. Structure modeling and sequence alignments identified a unique set of amino acid residues (Gln218, Val277, and Ala286) at the putative PpORS active site. Substitution of the Ala286 to Phe apparently constricted the active site cavity, and the A286F mutant instead produced triketide alkylpyrones from fatty acyl-CoA substrates with shorter chain lengths. Phylogenetic analysis and comparison of the active sites of PpORS and alkylresorcinol synthases from sorghum and rice suggested that the gramineous enzymes evolved independently from PpORS to have similar functions but with distinct active site architecture. Microarray analysis revealed that PpORS is exclusively expressed in nonprotonemal moss cells. The in planta function of PpORS, therefore, is probably related to a nonprotonemal structure, such as the cuticle.
Introduction
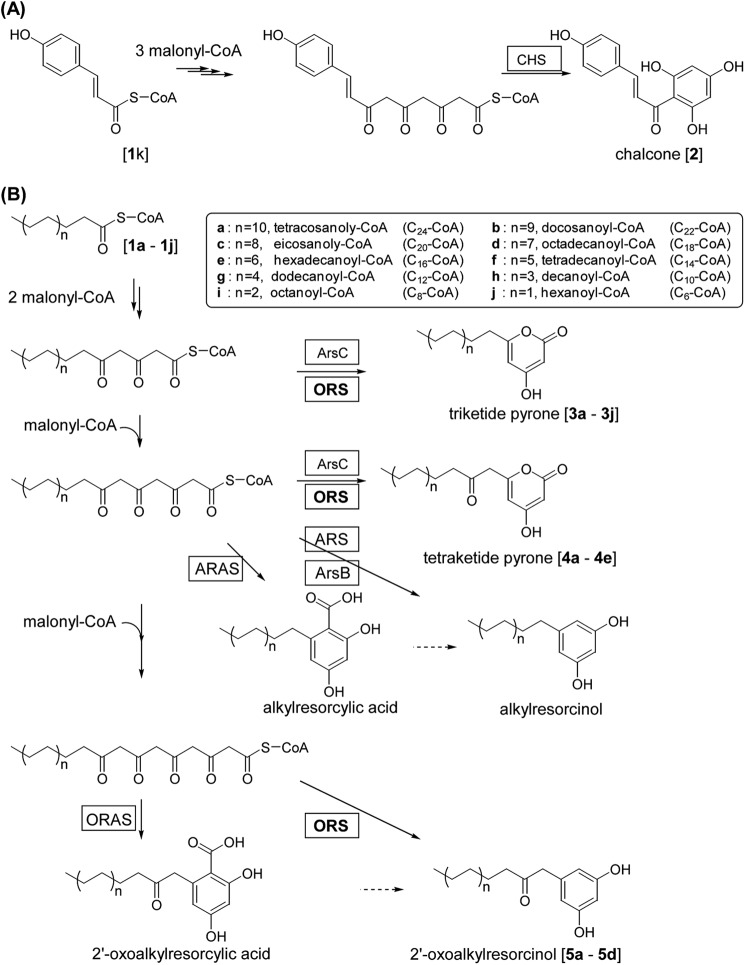
Type III polyketide synthases (PKSs),2 traditionally described as enzymes of the chalcone synthase (CHS) family, produce a variety of polyketide natural products in plants and microorganisms (1). Type III PKSs are homodimers of 42–45-kDa subunits, and each subunit has its own active site that is shaped by highly conserved residues including the signature Cys-His-Asn catalytic triad. These enzymes iteratively condense a starter CoA substrate with acetate units derived from malonyl-CoA and cyclize the linear polyketide intermediates to produce compounds with unique ring structures. Different type III PKSs produce a diverse range of products. This diversity is the result of differences in the selection of the starter CoA substrate, the number of condensation steps, and the cyclization mechanism. For example, CHS, a plant-specific type III PKS that catalyzes the first step of the biosynthesis of flavonoids, condenses a phenylpropanoid-CoA starter substrate (e.g. p-coumaroyl-CoA (1k)) with three malonyl-CoA extender molecules, and then cyclizes the tetraketide intermediate by Claisen acylation to give a chalcone 2 (Fig. 1A). Several type III PKSs from microorganisms and plants utilize long chain fatty acyl-CoA esters as the starter substrate, although they differ in the cyclization mechanism used. Azotobacter vinelandii ArsC and Mycobacterium tuberculosis PKS18 produce triketide and tetraketide alkyl-2-pyrones (2, 3) (Fig. 1B). Conversely, A. vinelandii ArsB and Sorghum bicolor alkylresorcinol synthases (SbARSs) produce tetraketide alkylresorcinols (3, 4), whereas Neurospora crassa 2′-oxoalkylresorcylic acid synthase (NcORAS) and Oryza sativa alkylresorcylic acid synthase (OsARAS) produce pentaketide and tetraketide alkylresorcylic acids, respectively (5–7) (Fig. 1B).
FIGURE 1.
Reactions catalyzed by type III PKSs. A, CHS condenses a p-coumaroyl-CoA with three molecules of malonyl-CoA and cyclizes the tetraketide intermediate to produce a chalcone. B, several type III PKSs iteratively condense a long chain fatty acyl-CoA ester with a number of acetate units derived from malonyl-CoA and cyclize the linear polyketide intermediate to produce polyketides with distinct ring structures. Both ORS and ARAS consume four molecules of malonyl-CoA. ORS catalyzes decarboxylative aldol cyclization, whereas ORAS catalyzes aldol cyclization. In contrast, ArsC condenses a long chain acyl-CoA ester with two or three molecules of malonyl-CoA to produce triketide or tetraketide 2-pyrone. ORS also produces triketide and tetraketide pyrones depending on the CoA ester substrate used. ARS and ARSA produce tetraketide alkylresorcinols and alkylresorcylic acids, respectively.
After extensive studies in the past few decades, we now know much about the catalytic mechanisms, gene regulation, and biological functions of type III PKSs (1, 8). Although type III PKSs are found in bacteria (9) and fungi (5, 10), they are more widely distributed in green land plants (Embryophyta). Each plant has a set of taxon-specific type III PKSs, which produce metabolites that are involved in UV protection (flavonoids), antimicrobial defense (stilbenes, bibenzyls, and alkylresorcinols), flower pigmentation (anthocyanins), spore/pollen protection (hydroxyalkyl pyrones), pollen tube growth (flavonols), and legume nodulation (isoflavonoids). The overall significance and scope of their roles suggest that type III PKSs have successfully co-evolved with land plants. This led us to investigate what major contributions ancestral type III PKSs might have made during the early evolution of land plants, especially during the colonization of land by ancestral plants. Our approach to gain insight into this question was to study the enzymatic properties of a modern-day plant type III PKS thought to closely resemble ancestral plant type III PKSs.
Bryophytes, comprising liverworts, mosses, and hornworts, are the simplest and earliest diverging lineages of land plants and offer unique windows into the early evolution of land plants. The model moss Physcomitrella patens is currently the only bryophyte whose genome has been sequenced (11), and its genome contains at least 17 putative type III PKS genes (12). Among them, PpORS (formerly PpCHS11) was shown to be basal to all plant type III PKS genes in phylogenetic trees. Therefore, it was proposed to encode an extant enzyme that might closely resemble the most recent common ancestor (MRCA) of plant type III PKSs (13). In this study, we cloned PpORS and characterized the enzymatic properties of the recombinant PpORS to demonstrate that PpORS is a 2′-oxoalkylresorcinol synthase with substrate preference for very long chain fatty acyl-CoA esters. We then identified putative active site residues by performing structure modeling and mutagenesis studies. We also investigated the expression patterns of PpORS and Phypa126819, a P. patens PKS gene closely related to PpORS, by expressed sequence tag (EST) abundance and microarray analyses, and carried out phytochemical analysis in an attempt to learn about in planta function of PpORS. These studies should help us to understand the roles that type III PKSs may have played during early evolution of land plants.
EXPERIMENTAL PROCEDURES
Materials
Expression plasmids for ArsB and ArsC were provided by Dr. Nobutaka Funa (University of Tokyo). p-Coumaroyl-CoA and cinnamoyl-CoA were purchased from TransMIT (Giessen, Germany). Eicosanoyl-CoA (1c, C20-CoA), docosanoyl-CoA (1b, C22-CoA), and tetracosanoyl-CoA (1a, C24-CoA) were from Avanti Polar Lipids (Alabaster, AL). Other acyl-CoA esters and Fast Blue B salt (ZnCl2) were from Sigma-Aldrich. [2-14C]Malonyl-CoA (55.2 mCi/mmol) was purchased from NEN/PerkinElmer Life Sciences. Wheat bran was obtained from Old Fashion Food (Regina, SK, Canada). Syntheses of 5-pentadecylresorcinol and 4-hydroxy-6-tridecyl-2-pyrone were reported previously (14, 15).
Cloning of PpORS
P. patens (Hedw.) Bruch and Schimp subspecies patens, strain Gransden2004, was cultivated on sterile peat pellets (Jiffy-7; Jiffy Products International AS, Kristansand, Norway) for 1–1.5 months at 25 °C under continuous light. Upper halves of gametophores without gametangia were collected with scissors. A full-length cDNA library was prepared by the oligo-capping method (16), and the cDNAs were cloned into the DraIII sites of pME18S-FL3 vector (AB009864). The full-length cDNA database in PHYSCObase was searched using the sequence of Contig1663 (17) as a query, and five corresponding clones (pph23j16, ppsp2a16, ppsp2k15, ppsp2n21, and ppsp12p08) were obtained. The coding region of PpORS was amplified by PCR from the ppsp2a16 clone using the primers shown in supplemental Table S1. The PCR products produced under standard PCR conditions were digested with restriction enzymes and subcloned into pET32a and pET28a expression vectors (Novagen) to give pET32-PpORS and pET28-PpORS, respectively.
Heterologous Production and Purification of Recombinant Proteins
Protein production and purification by Ni2+-chelation chromatography were performed as described previously (18) except that purification buffer was 20 mm potassium phosphate (KPi, pH 7.6) containing 200 mm NaCl. The enzyme solution was buffer-exchanged to 0.1 m KPi buffer, pH 7.6, using a 10DG column (Bio-Rad) for functional assays.
Enzyme Assay, Kinetic Analysis, and Product Determination
The standard assay mixture (100 μl) contained purified enzyme (10–20 μg), 0.1 mm starter-CoA (e.g. C24-CoA), and 0.1 mm [2-14C]malonyl-CoA (11 mCi/mmol) in 0.1 m KPi buffer, pH 7.6. Reactions to measure substrate preference and kinetic parameters were performed in 0.1 m KPi buffer, pH 7.6, containing 10% glycerol and 0.1% Triton X-100. After incubation at 30 °C for 20–40 min, the reaction was stopped by acidification (7.5 μl of 1 n HCl), and the reaction products were extracted with ethyl acetate (200 μl). The radioactive products were separated and quantified by thin layer chromatography (TLC) and phosphorimaging as described previously (15). Protein concentration was measured by an adapted Lowry method (Bio-Rad) with BSA as standard. The specific enzyme activity was expressed in pmol of the product produced s−1 mg−1 (picokatals mg−1).
Steady-state kinetic parameters of PpORS for C10-CoA and C24-CoA were determined in the presence of 0.1 mm malonyl-CoA and 9 μm PpORS. The concentration of starter substrate varied from 5 to 80 μm, and the reaction time was 10 min. Km and Vmax were calculated by fitting the data to the Michaelis-Menten equation using a nonlinear curve-fitting program (GraphPad Prism v.5.03).
Large scale reactions for product determination were performed with hexanoyl-CoA (C6-CoA), decanoyl-CoA (C10-CoA), or C24-CoA as the starter substrate under the standard assay conditions except that the concentration of malonyl-CoA was 0.2 mm and the reaction was run for 2 h. After standard work-up procedures, the reaction products were dissolved in methanol for MS analysis. Mass spectra were recorded using a Finnigan-Matt TSQ-700 mass spectrometer equipped with electrospray ionization and a Harvard syringe pump. Solutions were electrosprayed at 4.5 kV, with a capillary temperature of 75 °C. Flow rate was varied between 1 and 10 μl/min, and tube lens voltage was varied between −40 V and −80 V, depending on the compound. 2-Pyrone products of wild-type and mutant PpORS were compared with those produced by ArsC (2) or P. patens CHS (PpCHS) (17) by co-spotting on aluminum-backed silica 60 TLC sheets (EMD). Enzyme products were detected by staining with Fast Blue B salt (0.1% in H2O) (19).
Structure Modeling and Site-directed Mutagenesis
The structure of PpORS was first modeled with I-TASSER, which utilizes an ab initio multiple-threading approach (20). The quality of the model was further improved by a 10,000-step minimization in NAMD with AMBER ff99SBildn force fields, explicit solvation in TIP3P water, and Particle Mesh Ewald (21, 22).
The PpORS mutants (Q218T, V277G, A286F, Q218T/V277G, Q218T/A286F, V277G/A286F, Q218T/V277G/A286F) were generated from the pET28-PpORS plasmid using the QuikChange Site-directed Mutagenesis kit (Stratagene) and mutagenic primers shown in supplemental Table S1. The recombinant mutant proteins were produced in Escherichia coli BL21(DE3) or Tuner(DE3) cells. Gene expression in the Tuner(DE3) cells was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Protein purification was carried out in the same manner as the wild-type PpORS except that the KPi buffer solutions contained 10% glycerol and 0.1% Triton X-100.
Characterization of PpORS Paralogs, EST Abundance, and Expression Analysis
Two putative genes homologous to PpORS were identified by tblastn search against the JGI Physcomitrella_patens.1_1 database with PpORS as the query sequence. Genomic sequences of these putative genes were manually translated into amino acid sequences based upon exon-intron architecture and homology to other type III PKSs. EST abundance of the two putative PpORS paralogs, Phypa126819 and Phypa72618, was then examined by blastn searches against the NCBI EST database, and the EST profile of PpORS was obtained by examining corresponding ESTs (Ppa.5302) in individual NCBI P. patens UniGene libraries. The expression patterns of PpORS and Phypa126819 were determined with whole genome microarrays (CombiMatrix, Mukileto, WA) based on all gene models v1.2 (11). RNA samples were obtained from protonema from liquid cultures, juvenile gametophores grown on solid medium (23), and freshly isolated protoplasts (24). The microarray experiments were done in biological triplicates. Data analysis with the Expressionist software (Genedata, Basel, Switzerland) was performed as described previously (25).
Phytochemical Analysis
Plants were grown on solid medium with (protonemata) and without (gametophores) ammonium tartrate as described previously (12). Dried and ground tissue (protonema, gametophore, or wheat bran, 0.5 g each) was extracted with 10 ml of acetone for 3 h with a wrist shaker, and the extract was filtered and vacuum-dried. The residue was dissolved in 6 ml of methanol. A portion (0.5 ml) was made basic (pH ∼10) by the addition of 0.15 ml of 0.1 m KOH and incubated at 40 °C for 4 h. The resulting hydrolyzed solution was acidified to pH ∼2 with 10 μl of 6 n HCl, and partitioned with hexanes (0.4 ml). The organic layer was vacuum-dried, and the residue was dissolved in 30 μl of methanol. Extracts were analyzed before and after alkali treatment by silica TLC (toluene/acetone/acetic acid 75/25/1, v/v/v), and stained with Fast Blue B salt (0.1% in H2O).
Phylogenetic Analysis
Phylogenetic analysis with the Bayesian inference method was performed using the MrBayes program (v. 3.2-cvs) (26), as described previously (13) with some modifications. The search was initialized at a user-defined tree, which was generated from the amino acid sequences by the default slow/accurate option in ClustalW. The Markov Chain Monte Carlo analysis was run for one million generations with four chains, and trees were sampled after every 100 generations. After all trees sampled during the first 250,000 generations were discarded, a consensus tree was constructed based on the remaining trees and displayed using MEGA4 (27).
RESULTS
Cloning and Heterologous Production of PpORS
The full-length coding region of PpORS was obtained from the moss gametophore cDNA library. Attempts to clone the gene from protonema of the moss were unsuccessful, suggesting that PpORS is not expressed during the protonema stage (see below). As discussed earlier (12), the third ATG codon among the four in-frame candidate start codons was assumed to be the translation initiation site of PpORS and used to produce the recombinant PpORS. The enzyme was produced both as a thioredoxin (Trx)-His6-tagged protein (Trx-PpORS, 61 kDa) and as a His6-tagged protein (PpORS, 44 kDa). The Trx tag increased the stability of the recombinant enzyme and had little effect on the product profile. Thus, Trx-PpORS was used for the large scale reactions. The deduced amino acid sequence of PpORS (ABU87504) contains the conserved catalytic residues, Cys185, His323, and Asn356 (28) and the G385FGPG loop (29). The sequence identity of PpORS to other type III PKSs was generally low, and it was 20% to NcORAS, 34% to SbARS1 (4), and 36% to PpCHS and Medicago sativa CHS (MsCHS). Sequence alignments of PpORS with other type III PKSs are shown in supplemental Fig. S1.
In Vitro Analysis of PpORS Activity
We first tested the substrate preference of PpORS. PpORS produced a single major product (3j–3e) when the chain length of the starter fatty acyl-CoA substrate was C6 to C16 (Fig. 2). PpORS also produced the same type of compounds (3d–3a) from C18- to C24-CoA substrates, as evidenced by the progressively increasing RF value of the product with increasing chain length of the starter substrate (Fig. 2). From C18- to C24-CoA substrates, PpORS produced two additional types of compounds, one with lower RF values (4d–4a) and another with higher RF values (5d–5a). Compounds 5c–5a were the major products from C20- to C24-CoA substrates (Fig. 2). Based on ESI-MS (negative mode) and TLC data, the products, 3j–3a, were determined to be triketide alkylpyrones (Fig. 1B). ESI-MS analysis of 3h obtained from C10-CoA yielded a molecular ion peak [M−H]− at m/z 237. Minor peaks were also observed at 273 [M+Cl]− and 475 [M+M−H]−, providing further confirmation of the mass. Compound 3h co-migrated on TLC with the reaction product of ArsC from the same starter substrate (C10-CoA), and both products were stained yellow-orange with Fast Blue B salt, similarly to 4-hydroxy-6-methyl-2-pyrone (λmax = 469 nm in methanol) (supplemental Fig. S2, A and B). ArsC was shown to produce triketide alkylpyrones from the starter C6- to C12-CoA substrates and produces both triketide and tetraketide alkylpyrones from C14- to C22-CoAs (2). Therefore, we concluded that 3h is 4-hydroxy-6-nonyl-2-pyrone (Fig. 1B).
FIGURE 2.
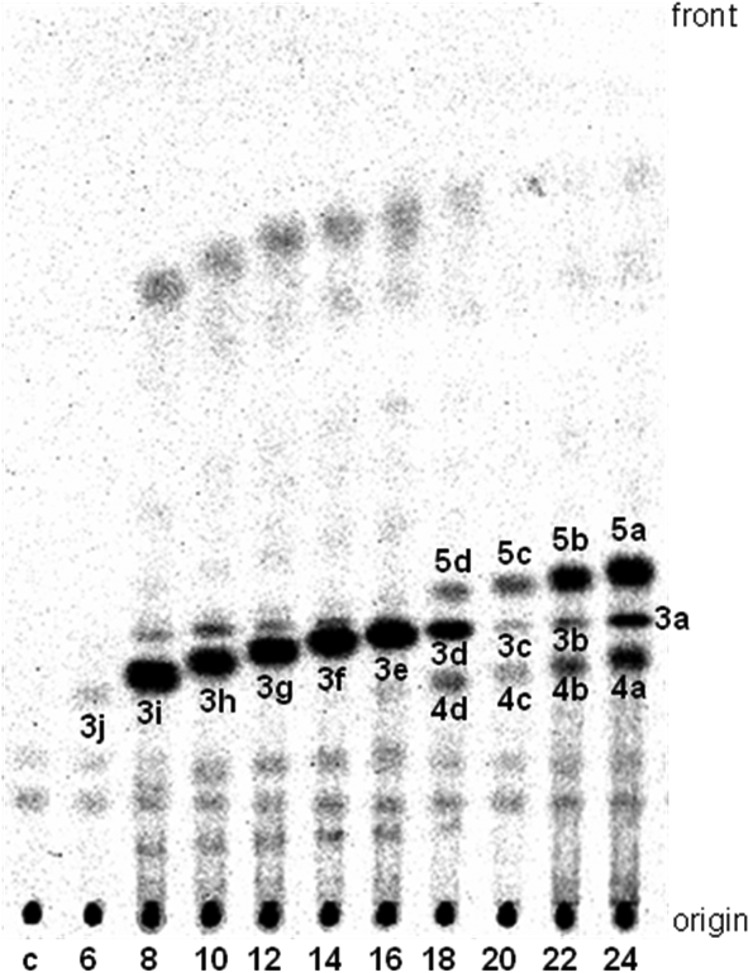
Radio silica thin layer chromatogram of reaction products produced by PpORS from fatty acyl-CoA starter substrates of varying chain lengths and [2-14C]malonyl-CoA. The number below each lane indicates the chain length of the fatty acyl-CoA substrate; e.g. 6 for C6-CoA and 8 for C8-CoA, etc. The control sample (lane c) received [2-14C]malonyl-CoA only. Specific activity of PpORS when determined with C24-CoA was 0.32 ± 0.12 picokatals/mg (±S.D., n = 3). PpORS also produced additional minor products from C8- to C16-CoA substrates. The yields of these minor products were 3–7% of those of the major products (3j–3e) and remained uncharacterized.
Compound 4a as well as 3a showed migration and staining patterns on TLC identical to those of the ArsC products from C24-CoA (supplemental Fig. S2C). Negative mode ESI-MS of the PpORS products from C24-CoA yielded two major ion peaks at m/z 433 and 475. The same molecular ion peaks were also obtained from an ESI-MS analysis of the ArsC products from C24-CoA. Based on these data, we concluded that 3a and 4a are 4-hydroxy-6-tricosyl-2-pyrone ([M−H]− at m/z 433) and 4-hydroxy-6-(2-oxo-pentacosyl)-2-pyrone ([M−H]− at m/z 475), respectively (Fig. 1B).
Compound 5a was stained violet with Fast Blue B salt, similarly to olivetol (λmax = 500 nm in methanol) (supplemental Fig. S2, A and C), suggesting that 5a contains a resorcinol ring. However, compounds 5a–5c exhibited lower RF values compared with the tetraketide alkylresorcinols produced by ArsB from the same substrates. After reduction with NaBH4, 5a and 4a were converted to polar compounds with lower RF values, whereas 3a remained intact, suggesting that 5a contains an oxo group as does 4a (data not shown). Positive mode ESI-MS of TLC-purified 5a yielded a molecular ion peak at m/z 950 corresponding to a protonated dimer [M+M+H]+ of 5-(2′-oxo)pentacosylresorcinol (supplemental Fig. S3). Based on these results, we concluded that 5a–5d are pentaketide 2′-oxoalkylresorcinols.
Other CoA esters were also examined as starter substrates for PpORS. PpORS produced a single triketide alkylpyrone product from palmitoleoyl-CoA (C16:1-CoA) or oleoyl-CoA (C18:1-CoA) with specific activity comparable with that for C16-CoA (data not shown). On the other hand, no enzyme activity was observed when the starter substrate was p-coumaroyl-, cinnamoyl-, benzoyl-, acetyl-, butyryl-, malonyl-, or arachidonoyl (C20:4)-CoA.
Enzymatic Properties and Kinetics of PpORS
We next tested the effects of different reaction conditions on the PpORS activity and product profile. The optimal activity for production of the pentaketide 2′-oxoalkylresorcinol (5a) from C24-CoA was observed at pH 7.5. The ratio of 5a to 3a decreased progressively as the pH increased, and the ratio was 2.3 at pH 6.5 and 0.67 at pH 8.0 (Fig. 3A). Similarly, the ratio of 5a to 3a varied depending on the concentration of the extender substrate (malonyl-CoA). More 5a was produced at higher concentrations of malonyl-CoA; however, overall activity decreased at 200 μm malonyl-CoA, possibly due to substrate inhibition (Fig. 3B). Incubation time had no effect on the product profile, and the production of 5a, 3a, and 4a from C24-CoA increased steadily as incubation time increased up to 1 h (data not shown).
FIGURE 3.
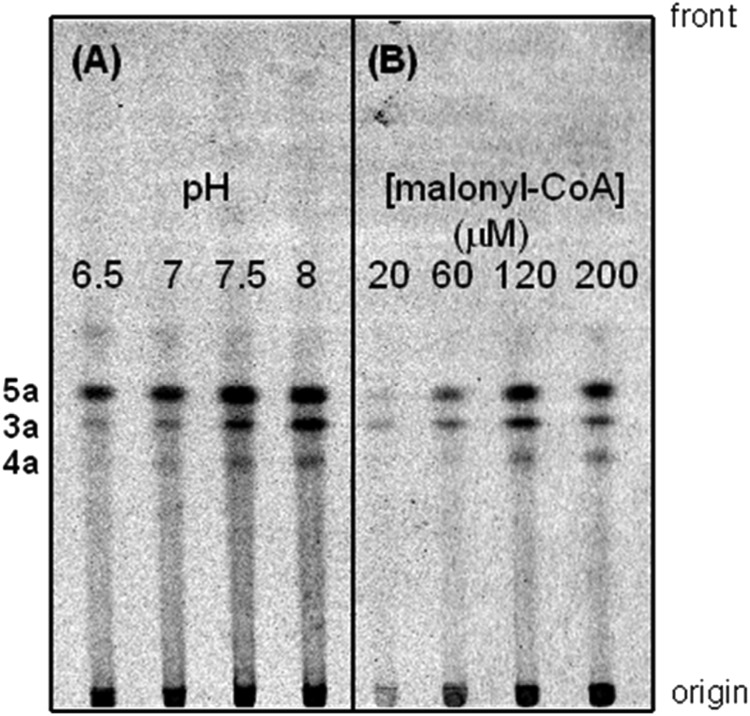
Effects of pH and malonyl-CoA concentration on PpORS activity. A, the enzyme reaction was carried out in the presence of 0.1 mm C24-CoA and 0.1 mm [2-14C]malonyl-CoA in 0.1 m KPi buffer at different pH values. B, the enzyme reaction was performed in 0.1 m KPi buffer at pH 7.6. The concentration of [2-14C]malonyl-CoA varied, whereas the concentration of C24-CoA was constant at 0.1 mm.
The steady-state kinetic parameters of PpORS for two representative substrates, C10-CoA and C24-CoA were estimated according to the Michaelis-Menten kinetics model. The Km and kcat/Km values for C24-CoA were 26 (±2.1, n = 3) μm and 0.70 m−1 s−1, and those for C10-CoA were 63 (±5.9) μm and 1.1 m−1 s−1, respectively (supplemental Fig. S4).
Structure Modeling and Site-directed Mutagenesis
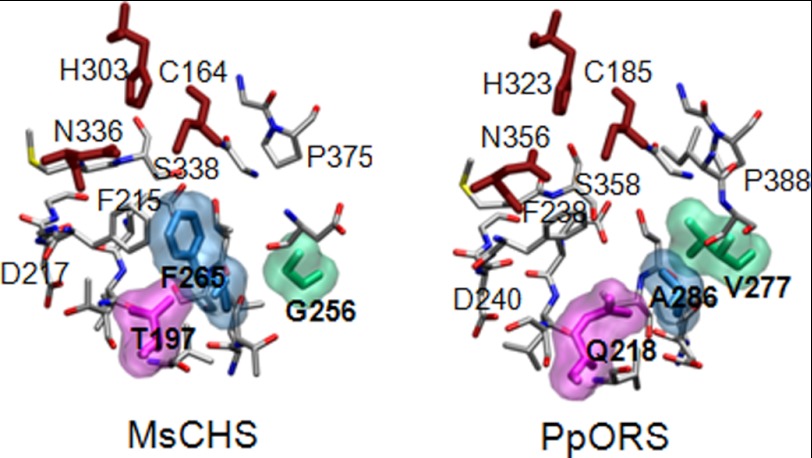
The modeled PpORS structure closely resembles the x-ray structures of MsCHS and other type III PKSs. Active site residues known to be critical for enzyme function are found at similar positions in the PpORS model compared with known type III PKS structures. Thus, the Cys185-His323-Asn356 catalytic triad, Phe238, Ser358, the G385FGPG loop and others are almost superimposable on the corresponding active site residues of MsCHS (Fig. 4). On the other hand, Thr197, Phe215, and G256 of MsCHS are uniquely substituted by Gln218, Val277, and Ala286, respectively, in PpORS (supplemental Fig. S1). Because these unique substitutions might play roles in the differential enzyme activity of PpORS, these three residues were mutated individually and in combination to the corresponding MsCHS residues by site-directed mutagenesis.
FIGURE 4.
Comparison of the active sites of MsCHS (Protein Data Bank ID code 1bi5) and the PpORS model. The Cys-His-Asn catalytic triads are shown in brick red. The PpORS specific active site residues, Gln218, Val277, and Ala286, are shown as space-filling model with the corresponding Thr197, Gly211, and Gln212 residues in MsCHS. According to the MolProbity parameters (43), the modeled PpORS structure was found to be in the 69th percentile of overall quality (MolProbity score 2.12) compared with all known crystal structures. The model achieved the following scorings: 1.75% for Ramachandran outliers, 8.06% for poor rotamers, and 1.47 for Clashscore.
Among the seven single and multiple mutants studied, only the A286F mutant and the V277G/A286F double mutant exhibited activity, and the rest were inactive with all starter substrates examined. Compared with the wild-type enzyme (Fig. 2), it was evident that both mutants lost the ability to produce pentaketide oxoresorcinols and instead produced triketide alkylpyrones (3a–3j) from C6- to C24-CoA starter substrates (Fig. 5). Compound 3j produced by the A286F mutant co-migrated on TLC with 4-hydroxy-6-pentyl-2-pyrone ([M−H]− at m/z 181) produced by PpCHS from C6-CoA. Both mutants exhibited similar substrate preference in that C8- and C16-CoA esters were most preferred. However, the V277G/A286F double mutant was better than the A286F mutant at accepting C22- and C24-CoA esters as the starter substrate. Specific activity for the formation of 3a was 0.019 and 0.072 picokatals mg−1 for the A286F and V277G/A286F mutants, respectively. Tetraketide alkylpyrones (4a–4c) were also produced by both mutants at lower levels. Like the wild-type enzyme, neither mutant accepted p-coumaroyl-, cinnamoyl-, C2-, and C4-CoA esters as starter substrate.
FIGURE 5.
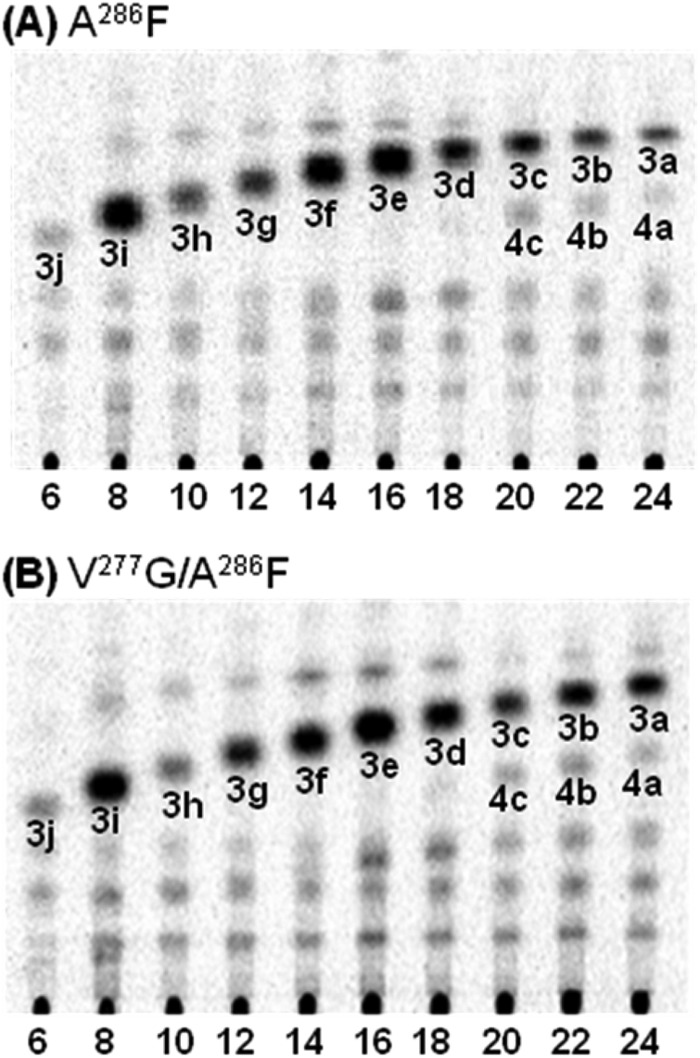
Radio silica thin layer chromatograms of reaction products produced by the A286F (A) and the V277G/A286F (B) mutants of PpORS from fatty acyl-CoA starter substrates of varying chain lengths and [2-14C]malonyl-CoA. The number below each lane indicates the chain length of the fatty acyl-CoA substrate. Only lower portions of the chromatograms are shown for brevity.
Expression Profile of PpORS and Phypa126819
Based on homology to PpORS, two gene models, Phypa72618 and Phypa126819, were identified as PpORS paralogs in the Physcomitrella genome. Deduced amino acid sequences of the two gene models were 46–48% identical to the PpORS sequence, assuming three putative introns at conserved positions. Proposed gene structures and sequence alignments of the deduced amino acids of the two gene models are provided in supplemental Fig. S5. The open reading frame of Phypa72618 contained two in-frame stop codons toward its 3′-end, and no corresponding EST was found in the NCBI EST database, suggesting that it is a pseudogene. On the other hand, the presence of a single Phypa72618 EST clone (BY962703) provided support for our deduced sequence of its gene product and also suggested that Phypa126819 may be a functional gene.
To investigate the expression pattern of PpORS, we first analyzed EST abundance within the EST libraries prepared from different Physcomitrella tissues. As compiled in the NCBI UniGene database (Fig. 6A), PpORS ESTs were found in Physcomitrella EST libraries prepared both from gametophytes and sporophytes. EST counts were particularly high in the libraries of ppls (upper half part of gametophores) and ppaa (gametangia, shoot tip with antheridia and archegonia) but were lower in ppgs (green sporophytes) and ppsp (sporophytes with surrounding archegonia) libraries. In a sharp contrast, no EST was found in any of the libraries prepared from protonema and regenerated protoplasts. The results obtained from our microarray analysis agreed with the expression pattern inferred from the EST counts. Thus, PpORS was expressed in gametophores at mid-level compared with all transcripts, but its expression was not detected in protonema and freshly prepared protoplasts (Fig. 6, B and C). Phypa126819 was not expressed above the detection limit in all three tested tissue types in the microarray analysis.
FIGURE 6.
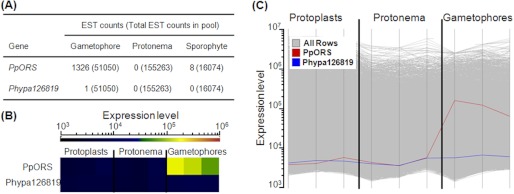
EST profile and microarray analysis of the expression of PpORS and its paralog Phypa123819 in different Physcomitrella tissues. A, EST profile of PpORS is from the NCBI UniGene transcriptome database, whereas the only EST corresponding to Phypa123819 (BY962703) was found by blastn search against the NCBI EST database. B and C, tile display (B) and profile display (C) of the expression patterns of PpORS and Phypa123819 are shown. In the profile display the expression patterns of all other transcripts are also shown as gray lines in the background. Gene expression was assayed in three different tissue types (protoplasts, protonema, and gametophores) in three biological replicates. In B, the color of the tiles indicates the expression level of each gene in a given tissue type. As the color approaches red, the gene is expressed at a higher level. The same data are presented graphically in C. PpORS was expressed in gametophores only, whereas Phypa123819 was not expressed in all tissues tested. Values below 104 are in the background and thus were considered as not expressed.
Phytochemical Analysis
To examine whether PpORS products exist in planta either in monomeric or in esterified forms, we attempted to detect putative PpORS products from the moss gametophore before and after alkaline treatment. Wheat bran extracts, a positive control, yielded a major band on TLC, which stained violet with Fast Blue B salt. The extracts were determined to contain 5-nonadecylresorcinol ([M−H]− at m/z 375) and 5-heneicosayresorcinol ([M−H]− at m/z 403), in agreement with the literature (30) (supplemental Fig. S6). However, we failed to detect any resorcinol derivatives from the extracts of the moss gametophores as well as protonemata. No band that responded to the dye in the characteristic manner of resorcinol derivatives was detected. These results led us to conclude that the moss gametophore tissues do not contain (oxo)alkylresorcinols, either as monomeric or esterified forms. Alternatively, the amounts of (oxo)alkyresorcinols present are below the detection limit of this study (∼200 ng/g of tissue estimated based on the sensitivity of the dye staining).
Phylogenetic Analysis
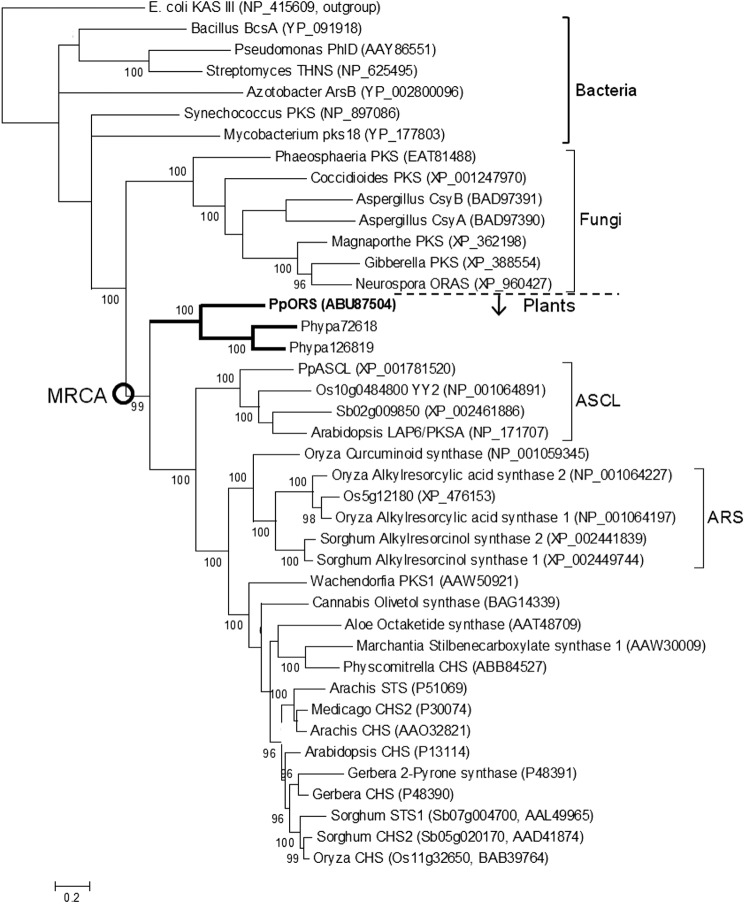
Expanding previous phylogenetic analyses of type III PKSs (4, 13, 31), a phylogenetic tree was constructed with PpORS, the two P. patens paralogs, and other long chain acyl-CoA-utilizing type III PKSs including the gramineous ARSs (SbARS1, SbARS2, OsARAS1, and OsARAS2) (Fig. 7). The tree shows the expected progressive evolution from bacterial to fungal to plant enzymes. PpORS and its two moss paralogs form their own clade at the base of the plant clade. Thus, they are direct descendants of the MRCA of the plant type III enzyme family. The rest of the plant enzymes, in turn, form a sister clade to the PpORS clade. They themselves are divided into two sister clades, one made of anther-specific chalcone synthase-like enzymes (ASCLs) (15) and the other made of non-ASCLs. The gramineous ARSs belong to one of the two sister clades of the non-ASCL clade, reflecting their close evolutionary relationship among themselves. More importantly, PpORS and its moss paralogs are clearly separated from the gramineous ARSs, indicating that the gramineous ARSs are not direct descendents of the PpORS lineage and have evolved independently from the PpORS lineage.
FIGURE 7.
Bayesian-inferred phylogram of type III PKSs. Numbers above branches are posterior probabilities, and branch lengths are proportional to expected numbers of amino acid substitutions per site. Only those nodes with posterior probability of >95 are shown. Convergence was reached with average S.D. of split frequencies of 0.0080 and a potential scale reduction factor of 1.001. The tree was built with E. coli β-ketoacyl-(acyl-carrier protein) synthase III as outgroup. The MRCA of the plant enzymes is marked with a circle.
DISCUSSION
Pentaketide 2′-oxoalkylresorcinols were previously detected among the reaction products of NcORAS and long chain fatty acyl CoA substrates (5). However, based on a careful time course study, the authors concluded that NcORAS instead produced pentaketide 2′-oxoalkylresorcylic acids and that the detected 2′-oxoalkylresorcinols were indirect products formed by nonenzymatic (or enzymatic (6)) decarboxylation of the resorcylic acids. We also examined the possibility that PpORS may produce 2′-oxoalkylresorcylic acids. We monitored the PpORS reaction with C24-CoA and [2-14C]malonyl-CoA at different time intervals up to 1 h and observed a steady increase of the formation of 5a and no evidence for the formation of a 2′-oxoalkylresorcylic acid. The same products were produced at different reaction pH values and substrate concentrations. Furthermore, we have demonstrated that 6-tridecyl-β-resorcylic acid, an alkylresorcylic acid, is stable up to several hours in 0.1 m KPi buffer (pH 7.8) (14). Based on these results, we conclude that PpORS is a pentaketide 2′-oxoalkylresorcinol synthase. Thus, PpORS condenses a very long chain fatty acyl-CoA with four molecules of malonyl-CoA and cyclizes the pentaketide intermediate to produce 2′-oxoalkylresorcinol, through an aldol reaction accompanied by decarboxylation (Fig. 1B).
PpORS produces different major products from starter substrates of different chain lengths; triketide alkylpyrones from the C6 to C16 substrates and pentaketide oxoresorcinols from the C20 to C24 substrates. The shift is not abrupt, and from C18-CoA, the enzyme produced significant amounts of a triketide alkylpyrone (3d) and a pentaketide oxoalkylresorcinol (5d) along with a tetraketide alkylpyrone (4d). This “substrate-directed product specificity” is not uncommon for type III PKS catalyzed reactions. Notably, NcORAS produces triketide alkylpyrones from C4 to C8 starter substrates, tri- and tetraketide alkylpyrones and tetraketide alkylresorcinols from C10 to C14 substrates, and tetra- and pentaketide alkylresorcylic acids from C16 to C20 substrates (5). Also, OsARASs produce alkylresorcylic acids and do not produce tetraketide alkylpyrones when the starter substrate is longer than C14-CoA (7). This raises a question as to the chemical nature of the in planta products of these enzymes. It has been postulated that enzymes that produce more than one product are advantageous in secondary metabolism because they generate chemical diversity at low cost (32). PpORS may well produce in planta alkylpyrones and 2′-oxoalkylresorcinols from fatty acyl-CoA esters of varying chain lengths for similar or different functions. In that case, substrate availability will determine the type of products made by the enzyme in planta. On the other hand, triketide and tetraketide pyrones are produced by most type III PKSs when nonphysiological substrates are given (1). Even with physiological substrates, most type III PKSs produce pyrones as in vitro derailment products. For example, CHS produces bisnoryangonin (a triketide pyrone) and coumaroyltriacetic acid lactone (a tetraketide pyrone) in addition to a chalcone (33). Furthermore, type III PKS mutants often produce pyrones when their active sites are compromised. The two PpORS mutants (A286F and V277G/A286F) in which putative active site residues were mutated failed to produce 2′-oxoalkylresorcinols, but still produced alkylpyrones (Fig. 5). These findings suggest that alkylpyrones produced by PpORS in vitro might be derailment products due to nonphysiological substrates or suboptimal reaction conditions. In that case the in planta products might be very long chain 2′-oxoalkylresorcinols.
Long chain alkylresorcinols have been found in higher plants including gramineous cereals. They are particularly abundant in the bran layer of cereal grains and are thought to exert antifungal activity (19). Long chain (C19-C25) 2′-oxoalkylresorcinols were found as minor components in wheat and rye grains and etiolated rice seedlings (34, 35). All plant (2′-oxo)alkylresorcinols identified to date are extractable monomers. To the best of our knowledge, no extractable alkylresorcinols have been detected in mosses, and the absence of alkylresorcinols in Sphagnum mosses is well documented (36). In A. vinelandii, monomeric alkylresorcinols and alkylpyrones produced by ArsB and ArsC from C20- and C22-CoA esters are the major lipid components of the protective cyst coat (2). PpORS is unique in that it produces exclusively 2′-oxoalkylresorcinols but does not produce alkylresorcinols. In addition to presenting an interesting mechanistic problem for future study, it might also bear significant implications for in planta function of PpORS because it implies important roles for the oxo group in the products. PpORS is expressed in gametophores, and its expression was not largely affected either by light/dark cycle (12) or by UV-B exposure (31). Moreover, PpORS is not expressed in moss protonemata and protoplasts (Fig. 6). Taken together with our failure to detect monomeric or esterified resorcinol derivatives from the moss gametophore extracts, these data suggest that 2′-oxoalkylresorcinols produced by PpORS might be constituents of gametophore-specific materials, such as a cuticle (37) or lignin-like materials (38). The plant cuticle is a waxy covering that protects plant from desiccation. In these materials, chemical components could be bound, at least partly, through alkaline-resistant linkages such as ether bonds. In this context, it is worthwhile to note that the ASCL-produced tetraketide 2′-oxoalkylpyrones have been proposed to be reduced by tetraketide α-pyrone reductases before being incorporated into sporopollenin, a biopolymer found in the pollen and spore walls (39). The resultant hydroxyl group of the hydroxyalkylpyrones might then form ether or ester linkages in the sporopollenin polymer. The oxo group in 2′-oxoalkylresorcinols might also be reduced in a similar manner in planta.
Thr197, Gly256, and Phe265 (numbering of MsCHS) that are highly conserved in CHS and many other type III PKSs are uniquely replaced with Gln218, Val277, and Ala286 in PpORS, respectively. All three residues are situated at the opposite side of the active site cavity from the nucleophilic Cys residue to which the growing polyketide chain is attached during catalysis (Fig. 4). Numerous studies have shown that Thr197 and Gly256 play critical roles in determining both substrate preference and the extent of condensation reactions by controlling the size and shape of active site cavity (reviewed in Ref. 1). For example, substitutions of Thr197, Gly256, and Ser338 of MsCHS with the corresponding residues found in 2-pyrone synthase were sufficient to convert the T197L/G256L/S338I triple mutant of MsCHS to a functional 2-pyrone synthase (40). However, only a few mutational studies have been done on Phe265, which sits at the entrance to the active site. Whereas the F233A mutant of a bacterial type III PKS, RppA, was devoid of enzymatic activity (41), the F265V mutant of MsCHS exhibited similar substrate selectivity as the wild-type enzyme (42). We could not properly access the functional role of the Gln218 residue of PpORS because the CHS-like substitution of the Gln218 to a Thr apparently disturbed the folding processes and made the mutant protein insoluble when produced in E. coli BL21(DE3) cells. Replacement of Val277 with a Gly had similar detrimental effects on protein structure, suggesting that Gln218 and Val277 are critical for proper folding and structural integrity of PpORS. Substitution of Ala286 with a bulky Phe had dual effects on enzyme activity. First, the A286F mutant completely lost the ability to form pentaketide 2′-oxoalkylresorcinols. Second, the ability of the mutant to accept the starter substrate gradually decreased as the chain length grew longer than C16. Both effects, although the former was more drastic, could be explained by the constriction of the active site cavity by the bulky Phe residue. The V277G/A286F double mutant exhibited a slightly higher activity than the A286F mutant in accepting C22- and C24-CoA substrates. Introducing a smaller Gly residue in place of a Val could have expanded the active site cavity to better accommodate very long chain substrates, but it failed to allow the mutant to form the pentaketide product.
PKS18, NcORAS, SbARS, and OsARAS also accept long chain fatty acyl-CoA esters as starter substrates. X-ray crystallography and mutagenesis studies of PKS18 indicated that Thr144, Cys205, Ala209, Ile220, His221, and Cys275 residues are situated along the long acyl binding tunnel and crucial in accepting the long chain acyl moiety of starter substrates (3). The corresponding residues in PpORS are Thr154, Ser215, and Ala219, Thr234, Leu235, and Ala286, respectively (supplemental Fig. S1). For NcORAS, Cys120, Thr121, Ser186, Met189, Phe210, and Ser340 were proposed to be involved in shaping the long acyl binding tunnel (6). The corresponding residues in PpORS are Ser153, Thr154, Ser215, Gln218, Phe238, and Ser358, respectively. On the other hand, for the gramineous ARSs, Tyr140 (numbering of SbARS1), Ala145, Ala205 (Cys in OsARAS) and Met265 were proposed to be important in determining their preference for long chain acyl-CoA substrates (4), and the corresponding residues in PpORS are Ser153, Thr158, Gln218, and Val277 (supplemental Fig. S1). Although caution should be taken in the absence of x-ray structures, PpORS appears to be more similar to NcORAS than to the gramineous ARSs with regard to the construction of the acyl binding site. This is unexpected because the overall sequence of PpORS is more similar to SbARS (34% identity) than to NcORAS (20% identity).
The apparently different mechanisms for controlling substrate preference in PpORS and the gramineous ARSs can be understood when the evolutionary relationships of these enzymes are considered. In phylogenetic trees constructed in this (Fig. 7) and other studies (13, 31), PpORS belongs to the basal clade along with its two moss paralogs, whereas the gramineous enzymes appear to have evolved later, independently from the PpORS lineage. In other words, PpORS and the gramineous enzymes appear not to be orthologs despite their similar activity. We searched available plant genome sequences for putative PpORS orthologs that belong to the PpORS clade, but were unable to find any. This suggests massive gene loss of PpORS orthologs in tracheophyte lineages. We have previously proposed an evolutionary scenario for the plant type III PKS family, in which ancestral acyl-CoA utilizing type III PKS genes have undergone repeated gene duplication-loss and functional diversification throughout evolution (13). Taken together, PpORS and the gramineous ARSs appear to represent an interesting example of enzyme evolution in which the same functions (the common use of long chain acyl-CoA starter substrates and the same type of cyclization) have evolved more than once within an enzyme family by adopting different structural strategy (different architecture of the binding sites).
In this study, PpORS was characterized to be a very long chain 2′-oxoalkylresorcinol synthase. A unique set of putative active site residues (Gln218, Val277, and Ala286) were identified through structure modeling and sequence alignments. Replacement of the Ala286 with a bulky Phe affected both the cyclization mode and the substrate preference. Expression profiling and phytochemical studies suggested that in planta products of PpORS might not exist in monomeric form and could be components of a polymeric structure that is specific to nonprotonemal tissues, such as the cuticle. More genetic and phytochemical studies are warranted to understand how the type III PKS family and land plants successfully co-evolved. This study represents the first step of such efforts.
Acknowledgments
We thank Matthew Endsin and Fatima Abbas for their contribution in the preparation of pET28-PpORS, and Li Li for the purification of Trx-PpORS. We also thank Dr. Nobutaka Funa (University of Tokyo) for expression plasmids of ArsB and ArsC. CGS-M postgraduate scholarship from NSERC to C. C. Colpitts is also acknowledged.
This work was supported by the Natural Sciences and Engineering Research Council, Canada, Discovery Grant (to D.-Y. S.).

This article contains supplemental Table S1 and Figs. S1–S6.
- PKS
- polyketide synthase
- ARAS
- alkylresorcylic acid synthase
- ARS
- alkylresorcinol synthase
- ASCL
- anther-specific chalcone synthase-like enzyme
- CHS
- chalcone synthase
- ESI
- electrospray ionizaton
- EST
- expressed sequence tag
- MRCA
- most recent common ancestor
- ORAS
- 2′-oxoalkylresorcylic acid synthase
- ORS
- 2′-oxoalkylresorcinol synthase
- SbARS
- S. bicolor ARS
- Trx
- thioredoxin.
REFERENCES
- 1. Abe I., Morita H. (2010) Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat. Prod. Rep. 27, 809–838 [DOI] [PubMed] [Google Scholar]
- 2. Funa N., Ozawa H., Hirata A., Horinouchi S. (2006) Phenolic lipid synthesis by type III polyketide synthases is essential for cyst formation in Azotobacter vinelandii. Proc. Natl. Acad. Sci. U.S.A. 103, 6356–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sankaranarayanan R., Saxena P., Marathe U. B., Gokhale R. S., Shanmugam V. M., Rukmini R. (2004) A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat. Struct. Mol. Biol. 11, 894–900 [DOI] [PubMed] [Google Scholar]
- 4. Cook D., Rimando A. M., Clemente T. E., Schröder J., Dayan F. E., Nanayakkara N. P., Pan Z., Noonan B. P., Fishbein M., Abe I., Duke S. O., Baerson S. R. (2010) Alkylresorcinol synthases expressed in Sorghum bicolor root hairs play an essential role in the biosynthesis of the allelopathic benzoquinone sorgoleone. Plant Cell 22, 867–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Funa N., Awakawa T., Horinouchi S. (2007) Pentaketide resorcylic acid synthesis by type III polyketide synthase from Neurospora crassa. J. Biol. Chem. 282, 14476–14481 [DOI] [PubMed] [Google Scholar]
- 6. Rubin-Pitel S. B., Zhang H., Vu T., Brunzelle J. S., Zhao H., Nair S. K. (2008) Distinct structural elements dictate the specificity of the type III pentaketide synthase from Neurospora crassa. Chem. Biol. 15, 1079–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matsuzawa M., Katsuyama Y., Funa N., Horinouchi S. (2010) Alkylresorcylic acid synthesis by type III polyketide synthases from rice Oryza sativa. Phytochemistry 71, 1059–1067 [DOI] [PubMed] [Google Scholar]
- 8. Dao T. T., Linthorst H. J., Verpoorte R. (2011) Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10, 397–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gross F., Luniak N., Perlova O., Gaitatzis N., Jenke-Kodama H., Gerth K., Gottschalk D., Dittmann E., Müller R. (2006) Bacterial type III polyketide synthases: phylogenetic analysis and potential for the production of novel secondary metabolites by heterologous expression in pseudomonads. Arch. Microbiol. 185, 28–38 [DOI] [PubMed] [Google Scholar]
- 10. Seshime Y., Juvvadi P. R., Kitamoto K., Ebizuka Y., Fujii I. (2010) Identification of csypyrone B1 as the novel product of Aspergillus oryzae type III polyketide synthase CsyB. Bioorg. Med. Chem. 18, 4542–4546 [DOI] [PubMed] [Google Scholar]
- 11. Rensing S. A., Lang D., Zimmer A. D., Terry A., Salamov A., Shapiro H., Nishiyama T., Perroud P. F., Lindquist E. A., Kamisugi Y., Tanahashi T., Sakakibara K., Fujita T., Oishi K., Shin-I. T., Kuroki Y., Toyoda A., Suzuki Y., Hashimoto S., Yamaguchi K., Sugano S., Kohara Y., Fujiyama A., Anterola A., Aoki S., Ashton N., Barbazuk W. B., Barker E., Bennetzen J. L., Blankenship R., Cho S. H., Dutcher S. K., Estelle M., Fawcett J. A., Gundlach H., Hanada K., Heyl A., Hicks K. A., Hughes J., Lohr M., Mayer K., Melkozernov A., Murata T., Nelson D. R., Pils B., Prigge M., Reiss B., Renner T., Rombauts S., Rushton P. J., Sanderfoot A., Schween G., Shiu S. H., Stueber K., Theodoulou F. L., Tu H., Van de Peer Y., Verrier P. J., Waters E., Wood A., Yang L., Cove D., Cuming A. C., Hasebe M., Lucas S., Mishler B. D., Reski R., Grigoriev I. V., Quatrano R. S., Boore J. L. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 [DOI] [PubMed] [Google Scholar]
- 12. Koduri P. K., Gordon G. S., Barker E. I., Colpitts C. C., Ashton N. W., Suh D.-Y. (2010) Genome-wide analysis of the chalcone synthase superfamily genes of Physcomitrella patens. Plant Mol. Biol. 72, 247–263 [DOI] [PubMed] [Google Scholar]
- 13. Jiang C., Kim S. Y., Suh D.-Y. (2008) Divergent evolution of the thiolase superfamily and chalcone synthase family. Mol. Phylogenet. Evol. 49, 691–701 [DOI] [PubMed] [Google Scholar]
- 14. Posehn S. E., Kim S. Y., Wee A. G., Suh D.-Y. (2012) Mapping the mechanism of the resorcinol ring formation catalyzed by ArsB, a type III polyketide synthase from Azotobacter vinelandii. ChemBioChem 13, 2212–2217 [DOI] [PubMed] [Google Scholar]
- 15. Colpitts C. C., Kim S. S., Posehn S. E., Jepson C., Kim S. Y., Wiedemann G., Reski R., Wee A. G., Douglas C. J., Suh D.-Y. (2011) PpASCL, a moss ortholog of anther-specific chalcone synthase-like enzymes, is a hydroxyalkylpyrone synthase involved in an evolutionarily conserved sporopollenin biosynthesis pathway. New Phytol. 192, 855–868 [DOI] [PubMed] [Google Scholar]
- 16. Maruyama K., Sugano S. (1994) Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138, 171–174 [DOI] [PubMed] [Google Scholar]
- 17. Jiang C., Schommer C. K., Kim S. Y., Suh D.-Y. (2006) Cloning and characterization of chalcone synthase from the moss, Physcomitrella patens. Phytochemistry 67, 2531–2540 [DOI] [PubMed] [Google Scholar]
- 18. Yamazaki Y., Suh D.-Y., Sitthithaworn W., Ishiguro K., Kobayashi Y., Shibuya M., Ebizuka Y., Sankawa U. (2001) Diverse chalcone synthase superfamily enzymes from the most primitive vascular plant, Psilotum nudum. Planta 214, 75–84 [DOI] [PubMed] [Google Scholar]
- 19. Kozubek A., Tyman J. P. H. (1995) Cereal grain resorcinolic lipids: mono and dienoic homologues are present in rye grains. Chem. Phys. Lipids 78, 29–35 [Google Scholar]
- 20. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781-1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J. L., Dror R. O., Shaw D. E. (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reski R. (1998) Development, genetics and molecular biology of mosses. Bot. Acta 111, 1–15 [Google Scholar]
- 24. Rother S., Hadeler B., Orsini J. M., Abel W. O., Reski R. (1994) Fate of a mutant macrochloroplast in somatic hybrids. J. Plant Physiol. 143, 72-77 [Google Scholar]
- 25. Richardt S., Timmerhaus G., Lang D., Qudeimat E., Corrêa L. G., Reski R., Rensing S. A., Frank W. (2010) Microarray analysis of the moss Physcomitrella patens reveals conserved transcriptional regulation of salt stress and abscisic acid signalling. Plant Mol. Biol. 72, 27–45 [DOI] [PubMed] [Google Scholar]
- 26. Ronquist F., Huelsenbeck J. P. (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- 27. Tamura K., Dudley J., Nei M., Kumar S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 28. Ferrer J.-L., Jez J. M., Bowman M. E., Dixon R. A., Noel J. P. (1999) Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 6, 775–784 [DOI] [PubMed] [Google Scholar]
- 29. Suh D.-Y., Fukuma K., Kagami J., Yamazaki Y., Shibuya M., Ebizuka Y., Sankawa U. (2000) Identification of amino acid residues important in the cyclization reactions of chalcone and stilbene synthases. Biochem. J. 350, 229–235 [PMC free article] [PubMed] [Google Scholar]
- 30. Ross A. B., Shepherd M. J., Schüpphaus M., Sinclair V., Alfaro B., Kamal-Eldin A., Aman P. (2003) Alkylresorcinols in cereals and cereal products. J. Agric. Food Chem. 51, 4111–4118 [DOI] [PubMed] [Google Scholar]
- 31. Wolf L., Rizzini L., Stracke R., Ulm R., Rensing S. A. (2010) The molecular and physiological responses of Physcomitrella patens to ultraviolet-B radiation. Plant Physiol. 153, 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Firn R. D., Jones C. G. (2000) The evolution of secondary metabolism: a unifying model. Mol. Microbiol. 37, 989–994 [DOI] [PubMed] [Google Scholar]
- 33. Yamaguchi T., Kurosaki F., Suh D.-Y., Sankawa U., Nishioka M., Akiyama T., Shibuya M., Ebizuka Y. (1999) Cross-reaction of chalcone synthase and stilbene synthase overexpressed in Escherichia coli. FEBS Lett. 460, 457–461 [DOI] [PubMed] [Google Scholar]
- 34. Seitz L. M. (1992) Identification of 5-(2-oxoalkyl)resorcinols and 5-(2-oxoalkenyl)resorcinols in wheat and rye grains. J. Agric. Food Chem. 40, 1541–1546 [Google Scholar]
- 35. Suzuki Y., Esumi Y., Saito T., Kishimoto Y., Morita T., Koshino H., Uzawa J., Kono Y., Yamaguchi I. (1998) Identification of 5-n-(2′-oxo)alkylresorcinols from etiolated rice seedlings. Phytochemistry 47, 1247–1252 [Google Scholar]
- 36. Avsejs L. A., Nott C. J., Xie S., Maddy D., Chambers F. M., Evershed R. P. (2002) 5-n-Alkylresorcinols as biomarkers of sedges in an ombrotrophic peat section. Org. Geochem. 33, 861–867 [Google Scholar]
- 37. Wyatt H. D., Ashton N. W., Dahms T. E. (2008) Cell wall architecture of Physcomitrella patens is revealed by atomic force microscopy. Botany 86, 385–397 [Google Scholar]
- 38. Ligrone R., Carafa A., Duckett J. G., Renzaglia K. S., Ruel K. (2008) Immunocytochemical detection of lignin-related epitopes in cell walls in bryophytes and the charalean alga Nitella. Pl. Syst. Evol. 270, 257–272 [Google Scholar]
- 39. Grienenberger E., Kim S. S., Lallemand B., Geoffroy P., Heintz D., de Azevedo Souza Cde A., Heitz T., Douglas C. J., Legrand M. (2010) Analysis of tetraketide α-pyrone reductase function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22, 4067–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jez J. M., Austin M. B., Ferrer J.-L., Bowman M. E., Schröder J., Noel J. P. (2000) Structural control of polyketide formation in plant-specific polyketide synthases. Chem. Biol. 7, 919–930 [DOI] [PubMed] [Google Scholar]
- 41. Funa N., Ohnishi Y., Ebizuka Y., Horinouchi S. (2002) Alteration of reaction and substrate specificity of a bacterial type III polyketide synthase by site-directed mutagenesis. Biochem. J. 367, 781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jez J. M., Bowman M. E., Noel J. P. (2002) Expanding the biosynthetic repertoire of plant type III polyketide synthases by altering starter molecule specificity. Proc. Natl. Acad. Sci. U.S.A. 99, 5319–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375-W383 [DOI] [PMC free article] [PubMed] [Google Scholar]