Background: Regulation of G6PD expression by nutrients occurs by changes in accumulation of spliced mRNA without changes in transcriptional activity of the gene.
Results: Refeeding enhances SRSF3 binding to G6PD mRNA. Loss of SRSF3 inhibits G6PD expression.
Conclusion: SRSF3 is a target for nutritional regulation of splicing.
Significance: Regulation of RNA splicing is a novel target for nutrient action.
Keywords: Alternative Splicing, Chromatin Immunoprecipitation (ChIP), Fatty Acid, Insulin, RIP, RNA Splicing, SRSF3, Exonic Splicing Enhancer, Glucose-6-phosphate Dehydrogenase, Intron Retention
Abstract
Expression of G6PD is controlled by changes in the degree of splicing of the G6PD mRNA in response to nutrients in the diet. This regulation involves an exonic splicing enhancer (ESE) in exon 12 of the mRNA. Using the G6PD model, we demonstrate that nutrients and hormones control the activity of serine-arginine-rich (SR) proteins, a family of splicing co-activators, and thereby regulate the splicing of G6PD mRNA. In primary rat hepatocyte cultures, insulin increased the amount of phosphorylated SR proteins, and this effect was counteracted by arachidonic acid. The results of RNA affinity analysis with nuclear extracts from intact liver demonstrated that the SR splicing factor proteins SRSF3 and SRSF4 bound to the G6PD ESE. Consequently, siRNA-mediated depletion of SRSF3, but not SRSF4, in liver cells inhibited accumulation of both mRNA expressed from a minigene containing exon 12 and the endogenous G6PD mRNA. Consistent with the functional role of SRSF3 in regulating splicing, SRSF3 was observed to bind to the ESE in both intact cells and in animals using RNA immunoprecipitation analysis. Furthermore, refeeding significantly increased the binding of SRSF3 coincident with increased splicing and expression of G6PD. Together, these data establish that nutritional regulation of SRSF3 activity is involved in the differential splicing of the G6PD transcript in response to nutrients. Nutritional regulation of other SR proteins presents a regulatory mechanism that could cause widespread changes in mRNA splicing. Nutrients are therefore novel regulators of mRNA splicing.
Introduction
Nutritional status is a central regulator of cellular function. At a cellular level, the macronutrients, carbohydrate, amino acids, and fatty acids can modulate signal transduction pathways and gene expression. Nutrients can also affect circulating levels of hormones in animals. Although key enzymes in the pathways of intermediary metabolism are primary targets for regulation by nutritional status, our current understanding of the molecular details causing this regulation have emphasized transcriptional mechanisms or changes in the kinetic properties of key regulatory enzymes. We have gathered a body of data indicating that alternative RNA splicing is a significant, yet underappreciated mechanism by which nutrients regulate gene expression (1, 2).
Alternative splicing is regulated in a developmental and tissue-specific pattern and in response to extracellular stimuli such as hormones and growth factors (3, 4). cis-Acting elements in the RNA and trans-acting factors are key determinants of this process; mutations in these elements or changes in the amount or activity of trans-acting proteins alter the cellular pattern of mRNA and the protein isoforms that they encode. Greater than 90% of human transcripts appear to undergo alternative splicing, resulting in changes in the amount of a protein expressed, production of multiple proteins from a single gene, and the presence or absence of specific domains within a protein (5). Thus, the consequences of regulating this process have large implications for cellular fate and metabolism.
Serine-arginine-rich (SR)3 proteins are a family of trans-acting splicing factors that activate splicing and play key roles in the regulation of alternative splicing (6). The canonical SR protein family members (SRSFs 1–12) are RNA-binding proteins with long, unstructured repeats of alternating serines and arginines forming RS domains in their C terminus. The members of this family also contain one or two RNA recognition motifs in the N terminus (6). SR proteins play a critical role in the recognition of alternatively included exons, as well as constitutive exons that have weak splice sites (6). These proteins enhance the splicing process by binding to exonic splicing enhancers (ESEs) to recruit components of the spliceosome, specifically U1 small nuclear ribonucleoprotein, U2AF35, and U2 small nuclear ribonucleoprotein to intronic splice sites and/or by inhibiting the binding of negative regulators of splicing (6, 7). The serines in the RS domains can be phosphorylated (6). Phosphorylation enhances the ability of the SR protein to recruit components of the spliceosome to splice sites, regulates the intracellular and intranuclear localization of SR proteins, and enhances the binding of the proteins to RNA (8). SR proteins are known to be substrates of several kinases including SRPK, Clk/Sty, and Akt and as such are candidates for mediating the impact of extracellular signals upon the splicing process (9–11). The involvement of hormones and other extracellular signals in regulating splicing factor activity is poorly understood.
G6PD is an enzyme that is integral to the process of de novo lipogenesis in liver and adipose tissue by its generation of reducing equivalents in the form of NADPH + H+ (12). We previously showed that the nutritional status of the animal regulates expression of G6PD by changes in the efficiency by which the primary transcript is spliced and without changes in the transcriptional rate of the gene (13–15). Starvation or consumption of a diet containing polyunsaturated fatty acids reduces the efficiency of intron removal from the primary G6PD transcript (15), subsequently decreasing expression of the enzyme, and therefore, fewer reducing equivalents are available for lipogenesis. G6PD precursor mRNA containing retained introns adjacent to exon 12 accumulates in the nucleus when splicing is inhibited (15). Feeding a high carbohydrate, low fat diet to rodents after a short term fast induces efficient splicing of the mRNA, and mature mRNA accumulates in the absence of a change in the accumulation of the primary transcript. The increase in G6PD mRNA results in greater expression of the enzyme and an increase in lipogenic capacity as compared with fasted animals (13). These in vivo feeding experiments are recapitulated in primary rat hepatocytes in culture in which treatment with insulin induces the accumulation of spliced G6PD mRNA, and the polyunsaturated fatty acid, arachidonic acid, attenuates mRNA splicing (2, 15, 16). A splicing regulatory element in exon 12 of the G6PD transcript mediates the effect of nutrients upon the splicing of this precursor mRNA (2). The absence of transcriptional regulation of this gene by nutrients has made it a useful model for deciphering the molecular mechanisms involved in this post-transcriptional regulation.
We hypothesize that these splicing-related changes involve the differential binding of splicing regulatory proteins to the regulatory element in exon 12. In this report, we demonstrate that SR proteins are candidates for nutrient regulation of splicing. In this regard, SR protein amount and phosphorylation in rat hepatocytes is increased by insulin, and arachidonic acid inhibits this effect. Within the family of SR proteins, SRSF3 binds to the splicing regulatory element, and the binding of SRSF3 to the regulatory element in intact liver decreases during starvation and increases upon refeeding. Loss of SRSF3 decreases accumulation of spliced G6PD mRNA. These data are the first to demonstrate that nutritional status can regulate SR protein activity and introduces a new paradigm by which mRNA splicing can be regulated.
EXPERIMENTAL PROCEDURES
All of the animal experiments were conducted in conformity with the Public Health Service policy on Humane Care and Use of Laboratory Animals. Additionally the Institutional Animal Care and Use Committee of the Division of Laboratory Animal Resources at West Virginia University approved all experimental procedures.
Hepatocyte Preparation
Hepatocytes were isolated from male Sprague-Dawley rats (∼200 g) as previously described (15). Hepatocytes (3 × 106) were plated in collagen-coated 60-mm dishes containing Hi/Wo/BA medium (Waymouth MB752/liter plus 20 mm HEPES, pH 7.4, 0.5 mm serine, 0.5 mm alanine, 0.2% BSA) plus 5% newborn calf serum. After 4 h, the cells were washed with serum-free medium, and a MatrigelTM overlay was added (0.3 mg/ml; BD Pharmingen). After 24 h in culture, the medium was replaced with Hi/Wo/Ba alone or Hi/Wo/Ba containing 80 nm insulin or 80 nm insulin plus 175 μm arachidonic acid conjugated to BSA (15), and the medium was replenished every 12 h. The cells were harvested for RNA isolation or extract preparation after 24 h with these treatments. Nuclear extracts (from 6–12 plates/treatment) were prepared by the method of Dignam et al. (17). The proteins were separated by size using SDS-PAGE and subjected to Western analysis as described below.
Nuclear Extract Preparation from Whole Liver
Male Sprague-Dawley rats (∼200 g) were either fasted for 24 h or fasted for 24 h and then refed a high glucose diet (fat-free/high glucose USB® diet, #1810092 from Purina Mills containing 1% safflower oil as a source of essential fatty acid) for 16 h prior to euthanasia. Liver nuclei were isolated by the method of Schibler et al. (18), and the nuclear proteins were extracted using the Dignam protocol (17). All buffers contained protease and phosphatase inhibitors (1 mm Na3VO4, 1 mm PMSF, 50 mm NaF, 1 mm benzamidine, 0.5 μg/ml leupeptin, 2 μg/ml aprotinin, 10 mm β-glycerophosphate, and 0.1 mm Na2MoO4). A portion of the liver was used for measurement of G6PD mRNA abundance.
RNA Affinity Assay
RNA oligonucleotides corresponding to nucleotides 43–72 and 79–93 of exon 12 of G6PD were purchased from IDT. The RNA oligonucleotides were attached to adipic acid dihydrazide-agarose beads (Sigma) as previously described (19). Each RNA-bead complex was mixed with binding buffer containing rat liver nuclear extract (250 μg of protein), 20 mm HEPES, pH 7.4, 9% glycerol, 70 mm KCl, 0.2 mm EDTA, pH 8.0, 1 μg/ml tRNA, 2.5 mm ATP, 2 mm MgCl2, and 0.2 mm DTT. The binding reactions were incubated at 30 °C for 30 min, after which the bead mixtures were placed into columns. Unbound proteins were washed from the RNA-bead complexes with 20 mm HEPES, pH 7.4, 5% glycerol, 0.2 mm EDTA pH 8.0, and 0.1 mm DTT. The bound proteins were eluted by gravity flow in wash buffer plus 250 mm KCl. The appropriate concentration of KCl for elution was determined empirically. The eluate was desalted and concentrated with Amicon® Ultra centrifugal filter devices (3,000 molecular weight cutoff; Millipore). The concentrated proteins were separated by size using SDS-PAGE, and bound SR proteins were detected by Western analysis.
Western Analysis
Following PAGE, the proteins were transferred to PVDF membranes (Bio-Rad) and probed with the antibodies indicated in the figure legends. Phosphorylated SR proteins were detected using the supernatant from hybridoma cells (ATCC) expressing the monoclonal antibody mAb104 or a commercially prepared monoclonal antibody 1H4 (Invitrogen). These two antibodies detect SRSFs 1–6. Total SR protein abundance was measured using the monoclonal antibody 16H3 (Invitrogen; referred to as Pan-SR) that detects SRSF4, SRSF6, SRSF5, and SRSF3, regardless of phosphorylation state. The amount of SRSF3 was also measured using a specific anti-SRSF3 antibody (7B4; Invitrogen). Lamin A/C (Cell Signaling) was used as a loading control. Secondary antibodies were conjugated to horseradish peroxidase, and the signals were detected using ECL Plus (GE Healthcare), followed by visualization on film and a Typhoon 9410 Imager (GE Healthcare). Signals were quantified with ImageJ (National Institutes of Health) or ImageQuant TL (Molecular Dynamics), respectively.
RNA Isolation and Measurement
Total RNA was isolated using TRI Reagent® (Molecular Research Center) and digested with DNase I (Turbo DNA-free; Invitrogen) according to the manufacturer's protocol. G6PD and cyclophilin B mRNAs were quantified by real time RT-PCR (ICYCLER; Bio-Rad) using the QuantiTect SYBR Green kit (Qiagen) and the primers and probes listed in supplemental Table S1. The amount of each mRNA was calculated using a relative standard curve.
Chromatin Immunoprecipitation
Male C57BL/6 mice (6 weeks of age) were fed the high carbohydrate diet as described above for 1 week. The mice were starved for 18 h (starved group) or were starved for 18 h and refed the high carbohydrate diet for 12 h (refed group) prior to euthanasia. The cross-linking reaction and ChIP assay are modifications of existing protocols (20, 21). Immediately after euthanasia, the livers were removed and immersed in four volumes of phosphate-buffered saline containing 1.25% formaldehyde, 1 mm Na3VO4, 1 mm PMSF, 50 mm NaF, 1 mm benzamidine, 0.5 μg/ml leupeptin, 2 μg/ml aprotinin, 10 mm β-glycerophosphate, 30 mm p-nitrophenyl phosphate, and 0.1 mm Na2MoO4, minced, and mixed by rotation for 12 min at room temperature. The cross-linking reaction was stopped with the addition of glycine to a final concentration of 125 mm. The samples were homogenized in a Dounce homogenizer (eight strokes, loose pestle), incubated 15 min on ice, and then centrifuged 5 min at 1,500 × g at 4 °C. The pellets were resuspended in three volumes of cell lysis buffer (5 mm HEPES, pH 8.0, 85 mm KCl, 0.5% Nonidet P-40, 1 mm Na3VO4, 1 mm PMSF, 50 mm NaF, 1 mm benzamidine, 0.5 μg/ml leupeptin, 2 μg/ml aprotinin, 10 mm β-glycerophosphate, 30 mm p-nitrophenyl phosphate, and 0.1 mm Na2MoO4), homogenized in a Dounce homogenizer (15 strokes, loose pestle), incubated on ice for 15 min, and centrifuged at 3500 × g for 5 min at 4 °C. The pellets were resuspended in 1 volume of nuclear lysis buffer (1% SDS, 50 mm Tris-HCl, pH 8.1, 10 mm EDTA, and protease and phosphatase inhibitors as listed above), homogenized with a Dounce homogenizer (five strokes, loose pestle), aliquoted into 1-ml volumes, and incubated on ice for 10 min. The samples were sonicated with continuous pulse in four 15-s bursts. The probe was chilled between each pulse. The samples were then cleared by centrifugation at 19,000 × g for 10 min. Supernatants were diluted 1:5 with dilution buffer (0.01% SDS, 1.1% Triton X-100, 16.7 mm MOPS, pH 7.3, 1.2 mm EDTA, 167 mm NaCl, 1 mm Na3VO4, 1 mm PMSF, 50 mm NaF, 1 mm benzamidine, 0.5 μg/ml leupeptin, 2 μg/ml aprotinin, 10 mm β-glycerophosphate, 30 mm p-nitrophenyl phosphate, and 0.1 mm Na2MoO4) and stored at −80 °C until use.
For immunoprecipitation, protein A/G-agarose beads (Santa Cruz) were washed five times in dilution buffer and blocked in dilution buffer containing 5 mg/ml BSA and 250 μg/ml sonicated salmon sperm DNA. An aliquot from each sample was removed prior to immunoprecipitation to serve as the “input.” The remainder of each sample was precleared with blocked agarose beads for 2 h with rotation and centrifuged. The supernatants were transferred into fresh tubes, to which the following antibodies were added (7.5–15 μg): phosphorylated SR (1H4; Invitrogen), heterogeneous nuclear ribonucleoprotein (hnRNP) M (Invitrogen), or RNA polymerase II (Covance). The “no antibody” control was treated identically except it received no primary antibody. The samples were immunoprecipitated overnight with rotation at 4 °C, after which the samples were rotated with blocked beads for 2 h and washed once with each of the following buffers: low salt buffer (20 mm MOPS, pH 7.3, 150 mm NaCl, 2 mm EDTA, 0.1% SDS, 1% Triton X-100), high salt buffer (20 mm MOPS, pH 7.3, 500 mm NaCl, 2 mm EDTA, 0.1% SDS, 1% Triton X-100), LiCl buffer (10 mm MOPS, pH 7.3, 1 mm EDTA, 0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate), and then washed twice with buffer containing 10 mm MOPS, pH 7.3 and 1 mm EDTA. All of the wash buffers contained protease and phosphatase inhibitors as listed above. The immunoprecipitated proteins were eluted from the agarose beads with 0.1 m NaHCO3 and 1% SDS. Immunoprecipitated and input samples were reverse-cross-linked with 200 mm NaCl at 65 °C for 6 h and digested with proteinase K (180 μg/ml) for 1 h at 45 °C, and the DNA was purified. Immunoprecipitated DNA was measured using real time PCR (QuantiTect probe PCR reagents; Qiagen), and the primers and dual labeled probes (IDT) are listed in supplemental Table S1. The amount of DNA that was immunoprecipitated relative to the amount present in total input chromatin was determined with the following formulas: ΔCt = Ct(input) − Ct(IP), % total = 2ΔCt × 1.7, where 1.7% is the percentage that the input chromatin represents of the total chromatin (22). This value was then normalized to the signal found in the no antibody control.
RNA EMSA
The RNA EMSA protocol is a modification of existing methods (23, 24). Briefly, 100 fmol of a 5′-hexachlorofluorescein (HEXTM)-labeled RNA probe (IDT) corresponding to the G6PD regulatory element (supplemental Table S1) was mixed with 0.27 μg of purified recombinant GST-conjugated-SRSF3 (Abnova) in 1× binding buffer (10 mm Tris, pH 7.5, 1 mm MgCl2, 100 mm KCl, 0.1 mm DTT, 5% glycerol) (25) plus/minus unlabeled competitor oligonucleotides (0, 1, 5, and 10 pmol; supplemental Table S1; IDT) in a total reaction volume of 20 μl. The reactions were incubated for 30 min at room temperature and then loaded onto a prerunning 5% native polyacrylamide gel. The gel was imaged directly on a Typhoon 9410 Imager, and the signals were quantified using ImageQuant TL software.
siRNA Transfection
HepG2 (human hepatoma) cells stably expressing an RNA splicing reporter (pβ-gal ex12-ex13) or HeLa cells were used for knockdown of SR proteins. One day after plating, HepG2 cells (5 × 105) or HeLa cells (2 × 105) were transfected with 75 nm siRNA pools using TransIT-siQUEST® transfection reagent (Mirus) in minimum essential medium containing 10% FBS. SiGENOME SMARTpools (Dharmacon) consisting of four siRNA duplexes (supplemental Table S1) were used for each target mRNA. SiGENOME nontargeting siRNA pool #1 was used as the negative control. Total RNA and whole cell protein extracts were collected after 48 h. Cell lysates were prepared using radioimmune precipitation assay buffer plus phosphatase and protease inhibitors (Thermo Scientific). HeLa cells were first transfected with the siRNA and then 24 later transiently transfected with the splicing reporter (pβ-gal ex12-ex13) using TransIT®-LT1 transfection reagent (Mirus) in complete minimum essential medium. Total RNA and whole cell protein extracts were collected after an additional 24 h.
RNA Immunoprecipitation (RIP)
The RIP protocol is a modification of existing methods (26, 27). For HepG2 cells, 5 × 106 cells were cross-linked in serum-free DMEM containing 1% formaldehyde for 10 min with agitation. The reaction was stopped by the addition of glycine to a final concentration of 125 mm and incubated for 5 min. The cells were washed twice with ice-cold 1× PBS and collected in ice-cold RIPA++ buffer (1× radioimmune precipitation assay buffer, 1× HaltTM protease and phosphatase inhibitor mixture, and 2 units/μl SUPERase·InTM RNase inhibitor; Invitrogen), followed by sonication in a Diagenode BioRuptor (two 15-min cycles, medium intensity, 30-s on/off pulses). The lysates were cleared by centrifugation, and the total protein content of the lysates was determined using the Bradford method (Thermo Scientific). Lysate containing 500 μg of protein was suspended in IP buffer (1× PBS, 2× HaltTM protease and phosphatase inhibitor mixture, and 2 units/μl SUPERase·InTM RNase inhibitor) and precleared with protein G Dynabeads at 4 °C for 2 h with rotation. A portion of the lysate was saved for the input sample.
In a separate tube, protein G Dynabeads (Invitrogen) were washed twice with 1× PBS + 0.02% Tween 20 (PBS/T). Antibodies (10–20 μg; SRSF3, Invitrogen; RNA polymerase II, Covance; and normal mouse IgG, Millipore) were added to the beads and incubated at room temperature for 1 h with rotation. The antibody-bound beads were washed three times with PBS/T and then mixed with the precleared lysates. The bead/lysate mixtures were incubated overnight at 4 °C with rotation and then washed six times with PBS/T. RNA bound to the immunoprecipitated proteins was released by digestion with proteinase K. The RNA was extracted with 1 ml of TriReagent, and DNA was removed by DNase I digestion. The G6PD splicing regulatory element was detected in the immunoprecipitated RNA by real time RT-PCR using the primers indicated in supplemental Table S1. Amplification in the absence of RT was used as a control for chromatin contamination.
In experiments with HepG2 cells, the cells were transiently transfected as described above with FLAG®-tagged SRSF3 (gift of Dr. Sandri-Goldin) (28). The cells were cross-linked 48 h after transfection, and SRSF3 was immunoprecipitated with anti-FLAG® antibody (Sigma-Aldrich).
For detection of SRSF3 binding to the regulatory element in whole liver, 8–10-week-old male C57BL/6J (The Jackson Laboratory) were fasted for 18 h or were fasted for 18 h and then refed a low fat, high carbohydrate diet (Basal Mix TD.00235 supplemented with 1% safflower oil; Harlan Laboratories) for 12 h. The livers were removed, minced with a razor blade, and cross-linked in four volumes of 1× PBS containing 1% formaldehyde for 10 min at room temperature and with mild agitation. The reaction was stopped with glycine as previously described. The samples were centrifuged for 4 min at 300 × g at 4 °C to pellet the cross-linked tissue. The pellets were resuspended in two volumes of ice-cold RIPA++ buffer and homogenized using a Dounce homogenizer (15 strokes, loose pestle), followed by sonication. All of the subsequent steps were as described for HepG2 cells.
Statistical Analyses
For Figs. 1 and 2, one-way analysis of variance was performed, and if the overall p value was significant (p < 0.05), Dunnett's multiple comparison test was performed to make pairwise comparisons to the insulin treatment (medium alone versus insulin and insulin versus insulin + arachidonic acid; GraphPad Prism, version 4.0). All other statistical comparisons were made using Student's t test.
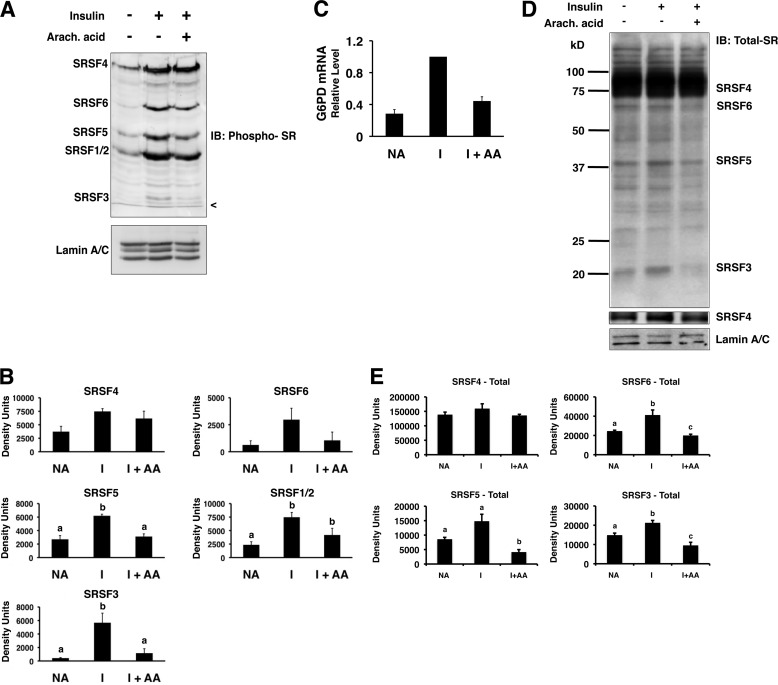
FIGURE 1.
Insulin and arachidonic acid regulate the amount of phosphorylated SR proteins in the nuclei of primary rat hepatocytes. Primary rat hepatocytes were incubated in high glucose (27.5 mm) medium alone (NA), insulin (I; 80 mm), or insulin plus arachidonic (Arach.) acid (175 μm). After 24 h, nuclear extract was prepared and analyzed by Western blotting. A, a representative immunoblot (IB) using an antibody against phosphorylated SR proteins (mAB104) and an antibody against lamin A/C is shown. The identities of the SR proteins are listed on the left side of the gel. The < symbol indicates the dye front of the gel. B, quantitation of the immunoblot data from n = 4 independent hepatocyte isolations. The amounts of the phosphorylated SR proteins in each treatment were measured by densitometry. C, total RNA was isolated from the hepatocytes after 24 h with the indicated treatments, and the amount of G6PD mRNA was measured by real time RT-PCR. The value for the amount of G6PD mRNA with insulin treatment (1.5 ± 0.3; n = 3) was set at 1, and the values for treatments with medium alone and for those with insulin plus arachidonic (Arach.) acid (I+AA) are expressed relative to the insulin treatment. D, representative immunoblots using antibodies against total SR proteins (16H3; to detect SRSF4, SRSF5, SRSF6, and SRSF3) or lamin A/C are shown. A second panel shows SRSF4 at a lighter exposure. The identities of the detected proteins are listed on the right side of the gels. E, quantitation of the immunoblot data from three independent experiments. The amounts of the SR proteins in each treatment were measured by densitometry. Columns with different letters are significantly different (p < 0.05).
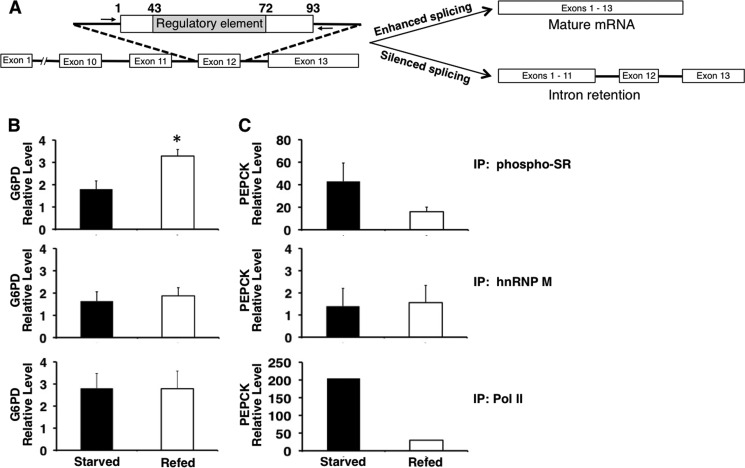
FIGURE 2.
Refeeding increases the binding of phosphorylated SR proteins to the regulatory element of G6PD exon 12 in vivo. ChIP was performed in livers of mice that were starved for 16 h or starved and then refed a high carbohydrate diet for 12 h. A, scheme of the G6PD RNA containing the regulatory element and observed changes in splicing. The arrows denote the location of the primers. The forward primer ends 6 nt upstream of the 3′ splice site of intron 11, and the reverse primer starts 3 nt downstream of the 5′ splice site in intron 12. B, immunoprecipitation (IP) used antibodies that detect: phosphorylated SR protein (phospho-SR), hnRNP M, and RNA polymerase II. The phospho-SR antibody (1H4) detects SRSF1, SRSF2, SRSF3, SRSF4, SRSF5, and SRSF6. The immunoprecipitated DNA was measured using real time PCR and primers adjacent to exon 12 (A). The amount of DNA detected is expressed relative to the input chromatin as described under “Experimental Procedures.” The asterisk indicates p < 0.05, n = 3 mice. C, detection of PEPCK in the immunoprecipitated samples. PEPCK DNA was measured using real time PCR and primers to exon 2 (n = 2–3 mice).
RESULTS
Previous work in our laboratory identified three members of the hnRNP family (K, L, and A2/B1) whose binding to a splicing regulatory element in the G6PD mRNA increased during starvation and occurred coincident with a decrease in exon 12 splicing (19). In contrast to these inhibitory proteins, enhanced splicing during refeeding could involve SR proteins that bind within exons and are splicing enhances. Because SR proteins undergo regulatory phosphorylation, we hypothesized that this family of proteins may also be targets for regulation by nutritional status. We used three approaches to test whether SR proteins can be intermediates in the nutritional regulation of splicing: 1) measurement of changes in SR protein activity, 2) changes in binding of SR proteins to an ESE in response to changes in nutritional status, and 3) loss-of-function via siRNA-mediated depletion of candidate SR proteins.
Phosphorylation of SR Proteins Changes with Insulin and Arachidonic Acid Treatment
As a first approach to addressing a physiological role for SR proteins in nutrient-regulated splicing, we asked whether the hormones and nutrients involved in regulating gene expression could alter SR protein amount or phosphorylation. Primary rat hepatocytes are the most insulin-responsive liver cell model and regulate their metabolism in response to fatty acids in a manner similar to intact liver (16, 29). Treatment of primary rat hepatocytes with insulin and a high glucose medium reflects the humoral milieu of the refed state, and these treatments significantly increased the amount of phosphorylated SRSF1, SRSF2, and SRSF3 as detected with the monoclonal antibody, mAb104, that detects a phosphorylated epitope in these proteins (Fig. 1, A and B). The increase in phosphorylated SRSF4 and SRSF6 upon treatment with insulin was not significantly different from untreated hepatocytes, possibly because of variation in the response. In contrast, treatment of hepatocytes with the nonesterified fatty acid, arachidonic acid, significantly decreased the amounts of phosphorylated SRSF3 and SRSF5 by 50–80%. Equal amounts of protein were used for the analysis as indicated by the control protein, lamin A/C (Fig. 1A). These changes in the amounts of phosphorylated SR proteins by insulin and fatty acids were coincident with changes in the accumulation of G6PD mRNA (Fig. 1C). Previous work in our laboratory has demonstrated that changes in G6PD mRNA by insulin and arachidonic acid are solely caused by changes in splicing of the primary transcript and mediated through a splicing regulatory element in exon 12 of the mRNA (2, 16). Thus, changes in the amount of phosphorylated SR proteins may play a role in regulated splicing.
The detected changes in SR protein phosphorylation could reflect either a change in the phosphate content of the proteins or a change in the total amount of the protein. To measure the amounts of total SR proteins, we used an antibody (16H3) that detects the SRSF proteins 3, 4, 5, and 6 regardless of phosphorylation state. Incubation of hepatocytes with insulin resulted in a 1.2-fold increase in the abundance of SRSF3 and SRSF6, a 2-fold increase in SRSF5, and little or no change in the amount of SRSF4 (Fig. 1, D and E). The addition of arachidonic acid to the hepatocytes decreased the abundance of SRSF3, SRSF5, and SRSF6 by 50% or more but had little or no effect on the abundance of SRSF4. Thus, the changes in phosphorylated SR protein amounts are caused in part by changes in the abundance of these proteins within the nucleus.
Refeeding Increases the Binding of SR Proteins to the Splicing Regulatory Element
As a second approach to test whether SR proteins are involved in nutrient-regulated splicing, we measured SR protein binding to an ESE in the livers of mice that were starved or refed. Binding was measured using ChIP analysis, which can detect RNA-protein interactions because splicing occurs co-transcriptionally, resulting in close proximity between splicing factors bound to the newly transcribed RNA and chromatin (5, 30). When subjected to cross-linking agents, the RNA-bound splicing factors become covalently attached to the chromatin adjacent to the nascent RNA transcript (31). Exon 12 of G6PD mRNA contains a regulatory region that has ESE activity and is involved in mediating the effects of insulin and arachidonic acid on G6PD mRNA splicing (2). As previously demonstrated and diagrammed in Fig. 2A, silenced splicing of the G6PD transcript results in intron retention surrounding exon 12 (15).
Starvation and refeeding cause the greatest changes in G6PD mRNA accumulation (7-fold or more) (13). Immunoprecipitation with a monoclonal antibody that recognizes phosphorylated RS domains allowed detection of binding by most members of the SR protein family. Refeeding significantly increased the binding of phosphorylated SR proteins to the splicing regulatory element by 1.8-fold above the fasting level (Fig. 2B). To control for potential cross-reactivity between this antibody and the phosphorylated C-terminal domain of RNA polymerase II (32, 33), we performed a second immunoprecipitation with an antibody against polymerase II. Dietary treatment did not alter the occupancy of polymerase II across this region of exon 12, indicating that an increase in SR protein binding was occurring (Fig. 2B). The lack of change in polymerase II occupancy on the G6PD chromatin is consistent with the absence of transcriptional regulation of this gene (14). hnRNP M immunoprecipitation was performed as a negative control because previous experiments have determined that this RNA-binding protein does not bind to the splicing regulatory element in vitro (2). As expected, binding of hnRNP M to the regulatory element was low, and this binding was not altered by the nutritional status of the animal (Fig. 2B). Refeeding did not cause a generalized increase in splicing factor binding to newly transcribed mRNA because SR protein binding to phosphoenolpyruvate carboxykinase (PEPCK) mRNA was not increased by refeeding (Fig. 2C). These data are the first to demonstrate regulation of SR protein binding to an RNA element in the whole animal.
SRSF3 and 4 Bind to the G6PD ESE
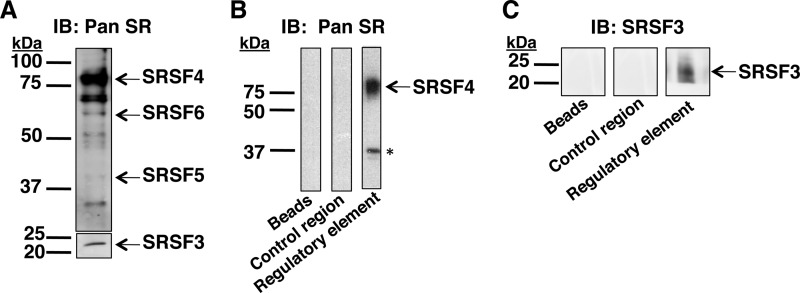
The ChIP assay cannot discriminate between SR proteins bound to RNA polymerase II and SR proteins bound to the RNA. Thus, an RNA affinity assay was used to verify that SR proteins bind to the RNA and also to determine which SR proteins were bound. RNA oligonucleotides corresponding to the splicing regulatory element (nucleotides 43–72 of G6PD exon 12; Fig. 2A) were attached to agarose beads and used to affinity purify RNA-binding proteins from rat liver nuclear extracts. An RNA oligonucleotide corresponding to a region of exon 12 outside of the regulatory element (control region; nucleotides 79–93) was used to detect nonspecific binding of proteins to RNA, whereas the beads alone were included to detect background binding (Fig. 3). Liver nuclear extracts contained SRSF3, SRSF4, SRSF5, and SRSF6 (Fig. 3A). The additional bands in the upper panel of Fig. 3A are not unexpected, because the 16H3 antibody will detect other proteins with SR repeats (33). SRSF3 was detected in a second immunoblot using more nuclear protein, because it is more difficult to detect with this antibody, presumably because of its lower abundance in liver compared with other SR proteins. Following pulldown with the splicing regulatory element, a band the size of SRSF4 was detected (Fig. 3B). The band near 37 kDa is not SRSF1 or 2, which migrate at this size, because the 16H3 antibody does not detect these SR proteins. To verify the identities of the bound proteins, the blots were reprobed with a different antibody (1H4), which detects phosphorylated forms of the SR proteins including SRSF1 and SRSF2. SRSF4 binding was also detected with this antibody, but the band near 37 kDa was not detected (data not shown). In addition, SRSF3 binding to the regulatory element was also observed (Fig. 3C). An SRSF3-specific antibody was used for detection to better visualize this protein. Neither SR protein bound to the control RNA oligonucleotide nor the beads alone. Thus, SRSF3 and SRSF4 are candidate SR proteins involved in regulating splicing of the G6PD mRNA.
FIGURE 3.
SRSF3 and SRSF4 bind to the splicing regulatory element. An RNA affinity assay was performed using liver nuclear extracts (250 μg of protein) and RNA oligonucleotide-bead complexes representing the regulatory element (nucleotides 43–72 of exon 12 of G6PD), a control region (nucleotides 79–93 of exon 12 of G6PD), or with the beads alone as a control for nonspecific binding to the beads. A, the nuclear extracts used in the affinity assay were analyzed by Western analysis with an antibody (16H3, Pan SR) that detects most SR proteins independent of phosphorylation state. Because of its lower abundance in liver nuclear extracts, SRSF3 was detected in a second blot using more nuclear protein. B and C, proteins eluted from the bead/oligonucleotide complexes were detected by Western analysis using the Pan SR antibody (B) or an antibody against SRSF3 (C). The protein marker (kDa) is indicated to the left of each set of gels. The asterisk indicates a nonspecific band that was not detected when the blot was reprobed with the phosphorylated SR protein-specific antibody (1H4; not shown). Each RNA affinity assay is a representative of three experiments performed using two independent nuclear extract preparations; all experiments showed the same results. IB, immunoblot.
Purified SRSF3 Binds to the G6PD Regulatory Element
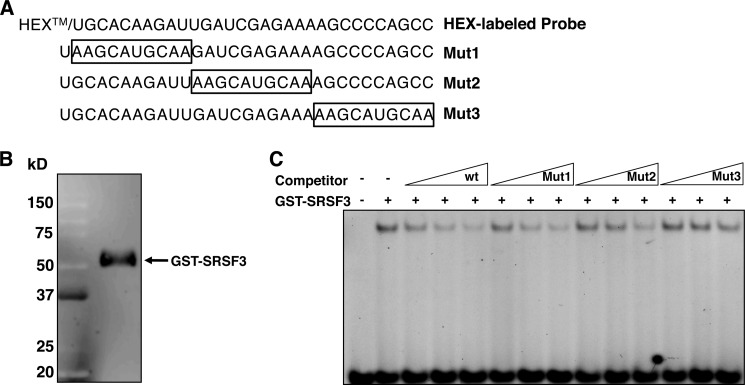
RNA EMSA was used to confirm the binding of SRSF3 to the regulatory element. Purified SRSF3 conjugated to GST was incubated with an RNA oligonucleotide representing the 30-nt regulatory element in exon 12 (Fig. 4, A and B). This complex resulted in a single shifted band (Fig. 4C). The shifted band was competed to a maximum of 10% by increasing concentrations of the unlabeled wild type RNA (Fig. 4C). We next tested the ability of the three mutated sequences to compete for SRSF3 binding (Fig. 4A). These mutations represent the same sequence changes previously used by us that abrogated the inhibition of splicing by arachidonic acid using splicing reporter constructs in functional assays in rat hepatocytes (2). Mutation of the first 10 nt of this sequence resulted in an RNA that competed for SRSF3 binding as effectively as the wild type sequence. Mutation of the second 10 nt attenuated the competition to only 33% of the wild type. Mutation of the final 10 nt resulted in little to no competition for SRSF3 binding (70% at the highest concentration of competitor). Thus, SRSF3 can bind to the splicing regulatory element and appears to bind to the latter half of this element.
FIGURE 4.
Purified SRSF3 binds to the regulatory element in exon 12. RNA EMSA was performed using a fluorescently labeled probe representing the regulatory element in exon 12. A, the sequence of the regulatory element and the mutant competitors (Mut1, Mut2, and Mut3). The box denotes the mutated sequence. B, purified recombinant GST-tagged SRSF3 (270 ng) visualized by Western analysis using an SRSF3 specific antibody. The GST tag shifts the apparent molecular mass to 55 kDa. C, EMSA. Each reaction contained 270 ng of SRSF3 and 100 fmol of probe. Competitions used 10×, 50×, and 100× molar excess of competitor RNA. wt refers to the unlabeled probe.
siRNA Depletion of SRSF3 Decreases G6PD mRNA Accumulation
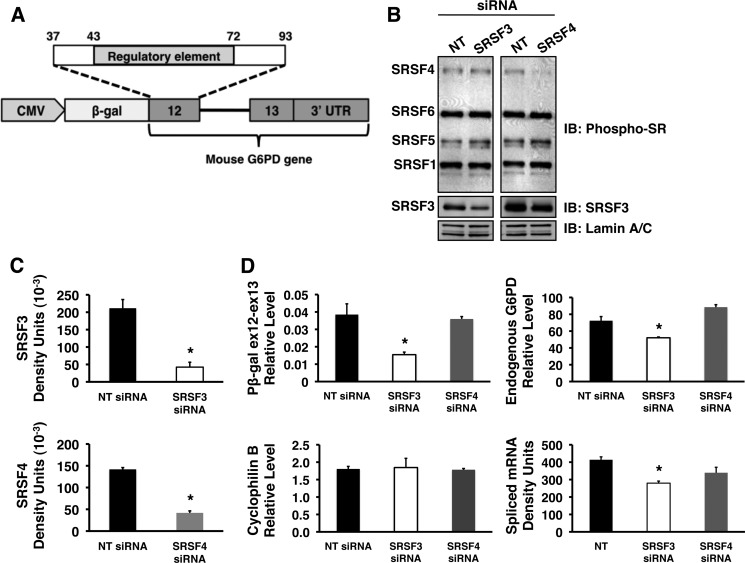
To test the functional significance of SR proteins in regulating splicing of G6PD mRNA in cells, we used a loss of function approach. siRNA-mediated knockdowns of SRSF3 and SRSF4 were performed in a clonal isolate of HepG2 cells stably expressing a G6PD splicing reporter (Fig. 5A). HepG2 cells were used as a liver cell model because they can be transfected with a higher efficiency than primary rat hepatocytes. The splicing reporter, pβ-gal ex12-ex13 (Fig. 5A), contains exon 12, intron 12, exon 13, and the 3′-UTR of G6PD ligated to the β-galactosidase gene. This construct retains splicing regulation by insulin and fatty acid (2) and permits evaluation of SR protein activity involving the exon 12 ESE without interference from regulatory elements in other exons of the G6PD precursor mRNA. Knockdowns of SRSF3 and SRSF4 were 80 and 70%, respectively, across three separate experiments (Fig. 5, B and C). In each case, knockdown of the specific SR protein did not result in a significant difference in the expression of other members of the SR protein family or the control proteins lamin A/C (Fig. 5B and data not shown). Knockdown to greater than 80% could be achieved but resulted in decreased expression of the other SR proteins. The 80% decrease in SRSF3 amount was accompanied by a significant inhibition (60%) of the expression of the G6PD reporter RNA in the absence of an effect on cyclophilin B mRNA, an endogenous control (Fig. 5D, left graphs). The decrease in G6PD reporter expression represented a decrease in the amount of spliced RNA for this reporter (Fig. 5D, bottom right graph). In addition, a significant decrease was also observed in the expression of the endogenous G6PD mRNA (Fig. 5D, top right graph). In contrast, depletion of SRSF4 did not inhibit expression of the splicing reporter or the endogenous G6PD gene (Fig. 5D).
FIGURE 5.
siRNA-mediated depletion of SRSF3 reduces the splicing of a G6PD reporter and the endogenous G6PD mRNA. HepG2 cells stably expressing a splicing reporter were transiently transfected with siRNA pools directed against SRSF3, SRSF4, or with a nontargeting control siRNA (NT) for 48 h, after which the cells were harvested for isolation of total RNA and preparation of whole cell protein lysate. A, schematic representation of the splicing reporter, pβ-gal ex12-ex13. The construct contains mouse genomic DNA encompassing exon 12 (nucleotides 37–93), intron 12, and exon 13 of the G6PD gene ligated to β-galactosidase and under the transcriptional control of a CMV promoter. B, representative Western blot of whole cell lysate probed with antibodies against phosphorylated SR proteins (top panels), SRSF3 (middle panels), and lamin A/C (bottom panels). C, quantitation of SRSF3 and SRSF4 knockdown for three separate siRNA experiments in HepG2 cells. The asterisk indicates a significant difference (p < 0.05). D, total RNA isolated from HepG2 cells following knockdown was used to measure the amounts of RNA for the splicing reporter (pβ-gal ex12-ex13), the endogenous G6PD transcript, cyclophilin B, and the amount of spliced reporter RNA (spliced RNA). The asterisk indicates a significant difference (p < 0.05), n = 3 independent knockdown experiments. IB, immunoblot.
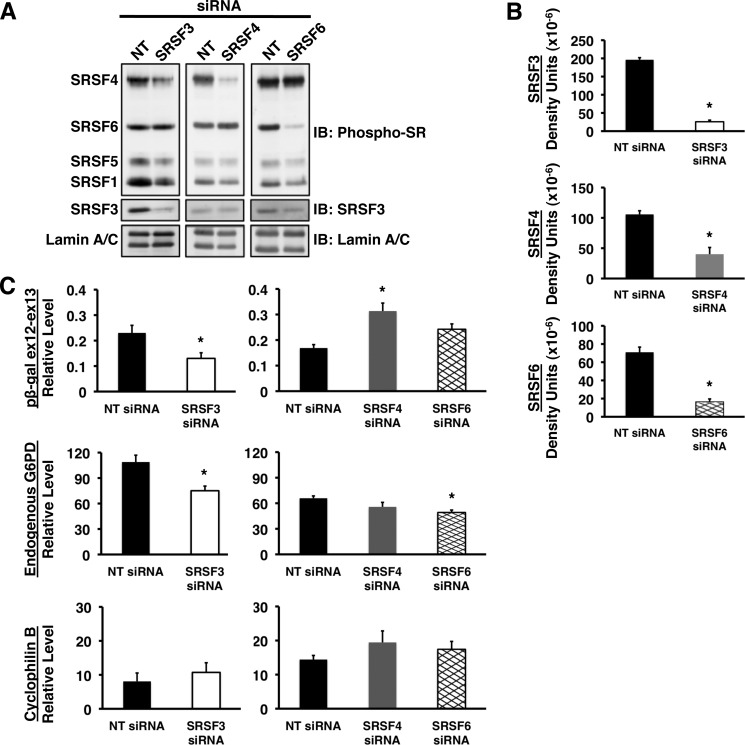
To test for cell type-specific effects of knockdown, we employed a similar strategy in HeLa cells transiently expressing the reporter construct (Fig. 5A). Depletion of SRSF3 by 80% or more (Fig. 6, A and B) resulted in significant decreases in accumulation of RNA from both the transiently transfected splicing reporter and the endogenous G6PD gene (Fig. 6C). Accumulation of the reporter mRNA was inhibited by 44%, and accumulation of mRNA for the endogenous G6PD gene was inhibited by 30% in comparison with cells transfected with nontargeting siRNA. These decreases are physiologically significant because the total elimination of G6PD expression causes embryonic lethality (34). Similar to the results with HepG2 cells, knockdown of SRSF4 by 90% or more (Fig. 6, A and B) did not inhibit accumulation of either the reporter or the endogenous G6PD mRNAs and, in fact, increased expression 1.9-fold compared with nontargeting siRNA (Fig. 6C). As a negative control, siRNA against SRSF6 was used because SRSF6 does not bind to the exon 12 regulatory element (Fig. 3B). siRNA treatment depleted SRSF6 by 94% compared with the nontargeting siRNA, but this decrease did not inhibit expression of the splicing reporter RNA (Fig. 6). A small decrease in endogenous G6PD expression was observed in these cells upon SRSF6 knockdown, which may reflect a potential interaction with other exons within the G6PD transcript (Fig. 6C). As in the HepG2 cell, knockdown of each SR protein did not significantly impact the expression of the other SR proteins (Fig. 6A and data not shown). Together, these data demonstrate that SRSF3 is involved in splicing of G6PD RNA across multiple cells types.
FIGURE 6.
siRNA-mediated depletion of SRSF3 reduces splicing of G6PD in HeLa cells. HeLa cells were transiently transfected with the splicing reporter (Fig. 5A) and siRNA against SRSF3, SRSF4, SRSF5, and the nontargeting control. A, whole cell lysates were prepared from HeLa cells 24 h after transfection. Western analysis was performed with antibodies against multiple SR proteins (mAB104), SRSF3, and lamin A/C. A representative blot is shown. B, quantitation of SR protein knockdown for five (SRSF3) and four (SRSF4 and SRSF6) separate experiments. C, the expression of the transiently transfected G6PD reporter (pβ-gal ex12-ex13), endogenous G6PD gene, and cyclophillin B gene in each of these experiments was measured by real time RT-PCR. The asterisk indicates a significant difference (p < 0.05). IB, immunoblot.
Refeeding Enhances Binding of SRSF3 to the Splicing Regulatory Element in Vivo
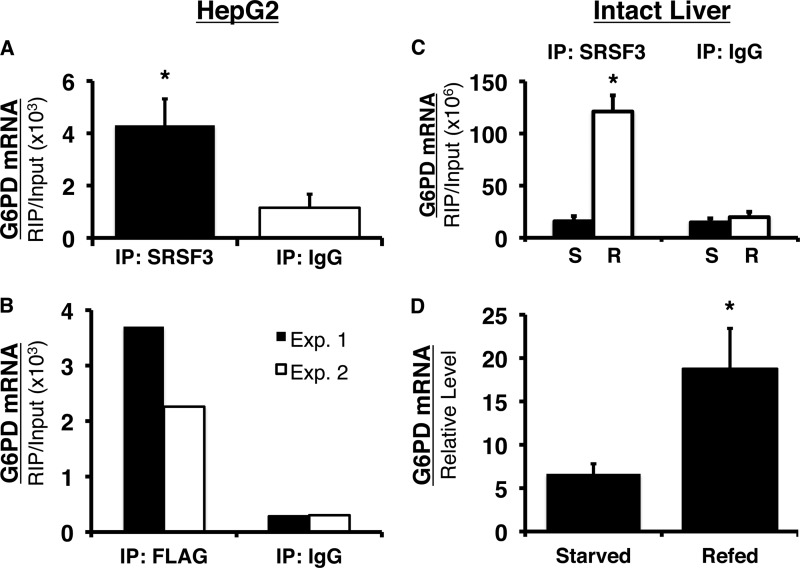
To verify that SRSF3 binds to the splicing regulatory element in intact cells, we used RIP and primers within exon 12 that amplify the region containing the regulatory element. In HepG2 cells, a 6-fold enrichment of SRSF3 binding was observed as compared with immunoprecipitation using the isotype control, IgG (Fig. 7A). To verify that the detected binding was not due to cross-reactivity of the antibody with other SR proteins, HepG2 cells were transiently transfected with SRSF3 containing a FLAG tag (Fig. 7B). When RIP was performed with an anti-FLAG antibody, SRSF3 binding to the splicing regulatory element increased 13- and 7.5-fold compared with immunoprecipitation with IgG in experiments 1 and 2, respectively (Fig. 7B).
FIGURE 7.
SRSF3 specifically binds to the splicing regulatory element in vivo and refeeding enhances the binding of SRSF3 to the splicing regulatory element in mouse liver. A, HepG2 cells were cross-linked with formaldehyde, and SRSF3 was immunoprecipitated from the lysates. G6PD RNA bound to SRSF3 was detected using primers, which amplify the region between nucleotides 11 and 85 in exon 12 (the regulatory element spans nucleotides 43–72). Immunoprecipitation (IP) with IgG was used as control for nonspecific binding to antibody. The amount of RNA detected is expressed relative to the input RNA in each sample. The data are the means ± S.E. of four separate immunoprecipitations. B, HepG2 cells were transiently transfected with FLAG-tagged SRSF3. The cells were chemically cross-linked 48 h after transfection, and SRSF3 was immunoprecipitated with anti-FLAG antibody. The results of two separate experiments are shown. C, mice were either starved for 18 h or starved for 18 h and refed a high carbohydrate diet for 12 h. RIP was performed on the chemically cross-linked livers using an antibody against SRSF3 or IgG. G6PD RNA immunoprecipitated with SRSF3 or IgG was detected with real time RT-PCR and primers to the region of exon 12 (nucleotides 18–89) that contains the splicing regulatory element (nucleotides 43–72). The amount of detected RNA is expressed relative to the input RNA in each sample. The data are the means ± S.E. of three mice/group. D, total RNA was isolated from a portion of each liver used in the RIP analysis. G6PD mRNA was measured using real time RT-PCR as previously described. Each bar is the mean ± S.E. of three mice. The asterisk indicates a significant difference (p < 0.05).
The ability of nutritional status to regulate binding of SRSF3 to the G6PD regulatory element was measured using RIP in the livers of mice that were starved or starved and then refed followed by formaldehyde cross-linking (Fig. 7C). SRSF3 was present in 6-fold greater abundance on the regulatory element in the refed livers as compared with IgG; furthermore, SRSF3 binding was enhanced by 6-fold or more in refed livers as compared with its binding to the regulatory element in the livers of fasted mice. This dietary paradigm causes a 7-fold or more increase in G6PD mRNA abundance (13) and a similar fold increase in the splicing of exon 12 (15). To the best of our knowledge, these are the first data to demonstrate that nutritional status regulates the presence of an SR protein on a splicing regulatory element and that this binding alters the splicing of the transcript.
DISCUSSION
Understanding how nutrients and hormones affect the splicing process introduces a new paradigm for regulating cellular function. Because the majority of genes undergo alternative splicing, this process is central in determining the protein composition of cells. G6PD was the first gene identified in which nutritional status regulates its expression by alternative splicing, specifically, intron retention (1, 2). In this report, we demonstrate that SR proteins are targets for the actions of insulin and the polyunsaturated fatty acid arachidonic acid, resulting in changes in their amount and phosphorylation state and enhanced presence at an ESE within the G6PD gene during active transcription. Of the SR proteins, SRSF3 is playing a specific role in the regulation of G6PD mRNA splicing. A specific role of this protein in nutrient-regulated splicing is supported by the specific binding of SRSF3 to the splicing regulatory element in the G6PD mRNA, the decrease in G6PD RNA splicing upon SRSF3 depletion, and the regulated binding of SRSF3 to the G6PD regulatory element in the livers of starved and refed mice.
SRSF3 functions in both alternative splicing and mRNA transport to the cytosol (35, 36). Our data suggest that SRSF3 is regulating splicing of G6PD mRNA rather than transport because the loss of SRSF3 decreased the accumulation of spliced RNA (Figs. 5 and 6). The binding of SR proteins to exon 12 as detected using ChIP (Fig. 2) requires that the proteins be present during active transcription of the RNA and possibly associated with the C-terminal domain of RNA polymerase II. Although the ChIP data does not specifically identify which SR proteins are present, the in vitro binding data (Figs. 3 and 4) indicate that only SRSF3 and SRSF4 bind to exon 12.
SRSF3 clearly binds to the regulatory element both in vitro and in intact cells and tissues; however, this element does not contain a canonical SRSF3-binding site. SRSF3-binding sites that have been identified using SELEX or CLIP-seq are CU- and CA-rich sequences (37, 38). Putative SRSF3-binding sites conforming to these known consensus sequences occur just 5′ of the regulatory element and overlapping the 3′ end of the element, respectively. Results using RNA EMSA support the binding of SRSF3 to the 3′ end of the regulatory element (Fig. 4). Although the EMSA data confirm that SRSF3 can bind the regulatory element, in vivo, SRSF3 may be interacting with another RNA-binding protein. The chemical cross-linking used in the RIP analysis permits both protein-RNA and protein-protein cross-links. UV cross-linking could discriminate between these possibilities but cannot be applied to intact tissue. Nonetheless, sequences are present within G6PD exon 12 and/or the regulatory element that support SRSF3 recruitment.
Most remarkable is the large change in SRSF3 binding with refeeding. This is the first demonstration of regulated binding of SR proteins to mRNA in response to nutritional status. Although both SRSF3 and SRSF4 bound to the splicing regulatory element in vitro, only the binding of SRSF3 was relevant for increased splicing of G6PD mRNA. In this regard, knockdown of SRSF4 failed to decrease expression of G6PD mRNA (Figs. 5 and 6). This does not completely rule out involvement of SRSF4, because its loss could have been compensated for by the closely related SRSF6 (39); however, knockdown of SRSF6 did not decrease G6PD RNA accumulation. The involvement of SRSF3 in the nutritional regulation of splicing is further supported by the regulation of SRSF3 phosphorylation and amount in response to insulin and arachidonic acid. Upon phosphorylation, SR proteins move from the cytoplasm to the nucleus and from sites of concentration within the nucleus, called speckles, to nascent RNA at transcription sites (8). In addition, phosphorylation enhances binding to RNA regulatory elements and protein-protein interactions (40, 41). SR proteins bound to the G6PD regulatory element during transcription were phosphorylated based on their detection with a phosphorylation-specific antibody in ChIP analysis (Fig. 2). The increase in binding of SR proteins during refeeding is in contrast with our earlier data demonstrating an opposite pattern of binding by proteins that silence splicing. In this regard, members of the hnRNP family of splicing silencing proteins, K, L, and A2/B1, bind to the G6PD regulatory element in the starved state, and this binding is decreased by refeeding (19). Competition between SRSF3 and splicing silencers has been demonstrated in regulation of the alternative splicing of the insulin receptor (36). A similar competition for binding may be part of the regulatory mechanism controlling G6PD splicing in response to nutrients and nutritional status.
External stimuli and hormones can exert an effect upon splicing by increasing or decreasing the expression of splicing factors and/or by changing their activity (9, 10, 42, 43). A considerable body of literature supports a broad role of insulin in alternative splicing. The results from splicing-sensitive microarrays performed with insect cells indicated that greater than 150 genes undergo alternative splicing in response to insulin treatment (44). In mammalian cells, insulin regulates alternative splicing of PKCβII (45, 46). The effect of insulin upon alternative splicing of PKCβII is coincident with an increase in the phosphorylation of SRSF5 (11). In hepatocytes in culture, insulin increases G6PD mRNA and increases SR protein phosphorylation, including phosphorylation of SRSF3 (Fig. 1). The coincident increase in G6PD splicing and SRSF3 binding with refeeding likely involves a regulatory role for insulin because insulin is required for the diet-mediated increases in G6PD expression (1).
Previous evidence from our lab demonstrates that the increase in splicing of G6PD that we observe with insulin treatment is due to an increase in its signaling through the PI3K pathway, whereas arachidonic acid treatment attenuates insulin signaling by inhibiting PI3K activity (47). Arachidonic acid decreases the amount of phosphorylated SR proteins and causes a coincident decrease in the accumulation of G6PD mRNA (Fig. 1). The presence of a consensus sequence for Akt phosphorylation in the RS domain of SRSF3 suggests that this protein can be a target of insulin action directly. Insulin action could also be indirect via stimulation of the SR protein kinase Clk/Sty, which is known to phosphorylated upon insulin treatment (10). Consistent with this concept, the phosphorylation-specific antibody used in these studies detects phosphorylations by multiple kinases including Clk/Sty. Identifying the relevant kinases involved in SR protein phosphorylation in intact liver and primary rat hepatocyte cultures is the subject of ongoing investigations in the laboratory. The decrease in phosphorylation of SR proteins with arachidonic acid treatment may be the result of reduced insulin signal transduction, or alternatively, may be due to an increase in phosphatase activity. The bioactive lipid, ceramide, reduces phosphorylation of SR proteins via activation of protein phosphatase 1 (48), and arachidonic acid has been shown to induce ceramide production from sphingomyelin in HL-60 cells (49).
The finding that insulin, arachidonic acid, and nutritional status regulate the phosphorylation and/or amount of the SR proteins has consequences beyond the regulation of G6PD expression. SR proteins enhance the splicing of multiple genes; thus, nutritional status could impact the splicing of many transcripts in addition to G6PD. In this regard, splicing of the insulin receptor mRNA is also regulated by SRSF3, and the resulting isoform switch may be involved in liver development and/or liver regeneration (36). Increased SRSF3 abundance in tumors and cancer cells and identification of multiple SRSF3 targets in differentiating neural cells highlight a much broader role of this protein in organisms (50).
In summary, these data indicate that nutrients and nutritional status induce changes in the activity of SR proteins, which impacts the splicing of a metabolic gene. Nutrient-regulated splicing provides another layer of control over gene expression beyond the well described effects upon transcription (29). We hypothesize that the RNA transcripts of other metabolic genes are regulated by nutrients at the step of splicing. Fully understanding the process by which our metabolic organs regulate splicing in response to nutrients and hormones will provide a new paradigm for nutrient regulation of cellular function.
Acknowledgments
We thank Dr. Roz Sandri-Goldin (University of California, Irvine) for the gift of the plasmid expressing the FLAG-tagged SRSF3. We thank Peter Stoilov for advice and critically reading the manuscript. We also thank Matthew Myers for assistance with conducting the experiments in Fig. 4.
This work was supported, in whole or in part, by National Institutes of Health Grants DK46897 (to L. M. S.) and T32 HL090610 (to A. B. K.). This work was also supported by a grant from the West Virginia Graduate Student Fellowships in Science, Technology, Engineering and Math Program (to C. M. W.).

This article contains supplemental Table S1.
- SR
- serine-arginine-rich
- SRSF
- SR splicing factor
- G6PD
- glucose-6-phosphate dehydrogenase
- ESE
- exonic splicing enhancer
- RS
- arginine-serine-rich
- RIP
- RNA immunoprecipitation
- PEPCK
- phosphoenolpyruvate carboxykinase
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- nt
- nucleotide.
REFERENCES
- 1. Salati L. M., Szeszel-Fedorowicz W., Tao H., Gibson M. A., Amir-Ahmady B., Stabile L. P., Hodge D. L. (2004) Nutritional regulation of mRNA processing. J. Nutr. 134, 2437S–2443S [DOI] [PubMed] [Google Scholar]
- 2. Szeszel-Fedorowicz W., Talukdar I., Griffith B. N., Walsh C. M., Salati L. M. (2006) An exonic splicing silencer is involved in the regulated splicing of glucose-6-phosphate dehydrogenase mRNA. J. Biol. Chem. 281, 34146–34158 [DOI] [PubMed] [Google Scholar]
- 3. Kalsotra A., Cooper T. A. (2011) Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 12, 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stamm S. (2008) Regulation of alternative splicing by reversible protein phosphorylation. J. Biol. Chem. 283, 1223–1227 [DOI] [PubMed] [Google Scholar]
- 5. Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 [DOI] [PubMed] [Google Scholar]
- 6. Long J. C., Caceres J. F. (2009) The SR protein family of splicing factors. Master regulators of gene expression. Biochem. J. 417, 15–27 [DOI] [PubMed] [Google Scholar]
- 7. Zhu J., Mayeda A., Krainer A. R. (2001) Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8, 1351–1361 [DOI] [PubMed] [Google Scholar]
- 8. Lin S., Fu X. D. (2007) SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 623, 107–122 [DOI] [PubMed] [Google Scholar]
- 9. Blaustein M., Pelisch F., Tanos T., Muñoz M. J., Wengier D., Quadrana L., Sanford J. R., Muschietti J. P., Kornblihtt A. R., Cáceres J. F., Coso O. A., Srebrow A. (2005) Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat. Struct. Mol. Biol. 12, 1037–1044 [DOI] [PubMed] [Google Scholar]
- 10. Jiang K., Patel N. A., Watson J. E., Apostolatos H., Kleiman E., Hanson O., Hagiwara M., Cooper D. R. (2009) Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCbetaII messenger ribonucleic acid. Endocrinology 150, 2087–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel N. A., Kaneko S., Apostolatos H. S., Bae S. S., Watson J. E., Davidowitz K., Chappell D. S., Birnbaum M. J., Cheng J. Q., Cooper D. R. (2005) Molecular and genetic studies imply Akt-mediated signaling promotes protein kinase CβII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. J. Biol. Chem. 280, 14302–14309 [DOI] [PubMed] [Google Scholar]
- 12. Rognstad R. (1979) Rate-limiting steps in metabolic pathways. J. Biol. Chem. 254, 1875–1878 [PubMed] [Google Scholar]
- 13. Amir-Ahmady B., Salati L. M. (2001) Regulation of the processing of glucose-6-phosphate dehydrogenase mRNA by nutritional status. J. Biol. Chem. 276, 10514–10523 [DOI] [PubMed] [Google Scholar]
- 14. Stabile L. P., Hodge D. L., Klautky S. A., Salati L. M. (1996) Posttranscriptional regulation of glucose-6-phosphate dehydrogenase by dietary polyunsaturated fat. Arch Biochem. Biophys 332, 269–279 [DOI] [PubMed] [Google Scholar]
- 15. Tao H., Szeszel-Fedorowicz W., Amir-Ahmady B., Gibson M. A., Stabile L. P., Salati L. M. (2002) Inhibition of the splicing of glucose-6-phosphate dehydrogenase precursor mRNA by polyunsaturated fatty acids. J. Biol. Chem. 277, 31270–31278 [DOI] [PubMed] [Google Scholar]
- 16. Stabile L. P., Klautky S. A., Minor S. M., Salati L. M. (1998) Polyunsaturated fatty acids inhibit the expression of the glucose-6-phosphate dehydrogenase gene in primary rat hepatocytes by a nuclear posttranscriptional mechanism. J. Lipid Res. 39, 1951–1963 [PubMed] [Google Scholar]
- 17. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schibler U., Hagenbüchle O., Wellauer P. K., Pittet A. C. (1983) Two promoters of different strengths control the transcription of the mouse alpha-amylase gene Amy-1a in the parotid gland and the liver. Cell 33, 501–508 [DOI] [PubMed] [Google Scholar]
- 19. Griffith B. N., Walsh C. M., Szeszel-Fedorowicz W., Timperman A. T., Salati L. M. (2006) Identification of hnRNPs K, L and A2/B1 as candidate proteins involved in the nutritional regulation of mRNA splicing. Biochim. Biophys. Acta 1759, 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Friedman J. R., Larris B., Le P. P., Peiris T. H., Arsenlis A., Schug J., Tobias J. W., Kaestner K. H., Greenbaum L. E. (2004) Orthogonal analysis of C/EBPβ targets in vivo during liver proliferation. Proc. Natl. Acad. Sci. U.S.A. 101, 12986–12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yin L., Wang Y., Dridi S., Vinson C., Hillgartner F. B. (2005) Role of CCAAT/enhancer-binding protein, histone acetylation, and coactivator recruitment in the regulation of malic enzyme transcription by thyroid hormone. Mol. Cell. Endocrinol. 245, 43–52 [DOI] [PubMed] [Google Scholar]
- 22. Frank S. R., Schroeder M., Fernandez P., Taubert S., Amati B. (2001) Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15, 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hellman L. M., Fried M. G. (2007) Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2, 1849–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Motta-Mena L. B., Smith S. A., Mallory M. J., Jackson J., Wang J., Lynch K. W. (2011) A disease-associated polymorphism alters splicing of the human CD45 phosphatase gene by disrupting combinatorial repression by heterogeneous nuclear ribonucleoproteins (hnRNPs). J. Biol. Chem. 286, 20043–20053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Setzer D. R. (1999) Measuring equilibrium and kinetic constants using gel retardation assays. Methods Mol. Biol. 118, 115–128 [DOI] [PubMed] [Google Scholar]
- 26. Selth L. A., Gilbert C., Svejstrup J. Q. (2009) RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harb. Protoc. 2009, pdb.prot5234 [DOI] [PubMed] [Google Scholar]
- 27. Ule J., Jensen K., Mele A., Darnell R. B. (2005) CLIP. A method for identifying protein-RNA interaction sites in living cells. Methods 37, 376–386 [DOI] [PubMed] [Google Scholar]
- 28. Sciabica K. S., Dai Q. J., Sandri-Goldin R. M. (2003) ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 22, 1608–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hillgartner F. B., Salati L. M., Goodridge A. G. (1995) Physiological and molecular mechanisms involved in nutritional regulation of fatty acid synthesis. Physiol. Rev. 75, 47–76 [DOI] [PubMed] [Google Scholar]
- 30. Kornblihtt A. R. (2007) Coupling transcription and alternative splicing. Adv. Exp. Med. Biol. 623, 175–189 [DOI] [PubMed] [Google Scholar]
- 31. Listerman I., Sapra A. K., Neugebauer K. M. (2006) Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 13, 815–822 [DOI] [PubMed] [Google Scholar]
- 32. Doyle O., Corden J. L., Murphy C., Gall J. G. (2002) The distribution of RNA polymerase II largest subunit (RPB1) in the Xenopus germinal vesicle. J. Struct. Biol. 140, 154–166 [DOI] [PubMed] [Google Scholar]
- 33. Neugebauer K. M., Stolk J. A., Roth M. B. (1995) A conserved epitope on a subset of SR proteins defines a larger family of pre-mRNA splicing factors. J. Cell Biol. 129, 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Longo L., Vanegas O. C., Patel M., Rosti V., Li H., Waka J., Merghoub T., Pandolfi P. P., Notaro R., Manova K., Luzzatto L. (2002) Maternally transmitted severe glucose 6-phosphate dehydrogenase deficiency is an embryonic lethal. EMBO J. 21, 4229–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang Y., Steitz J. A. (2001) Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7, 899–905 [DOI] [PubMed] [Google Scholar]
- 36. Sen S., Talukdar I., Webster N. J. (2009) SRp20 and CUG-BP1 modulate insulin receptor exon 11 alternative splicing. Mol. Cell. Biol. 29, 871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Änkö M. L., Müller-McNicoll M., Brandl H., Curk T., Gorup C., Henry I., Ule J., Neugebauer K. M. (2012) The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 13, R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schaal T. D., Maniatis T. (1999) Selection and characterization of pre-mRNA splicing enhancers. Identification of novel SR protein-specific enhancer sequences. Mol. Cell. Biol. 19, 1705–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Longman D., Johnstone I. L., Cáceres J. F. (2000) Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19, 1625–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao S. H., Manley J. L. (1997) Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 11, 334–344 [DOI] [PubMed] [Google Scholar]
- 41. Yeakley J. M., Tronchère H., Olesen J., Dyck J. A., Wang H. Y., Fu X. D. (1999) Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J. Cell Biol. 145, 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Massiello A., Roesser J. R., Chalfant C. E. (2006) SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 20, 1680–1682 [DOI] [PubMed] [Google Scholar]
- 43. Sumanasekera C., Watt D. S., Stamm S. (2008) Substances that can change alternative splice-site selection. Biochem. Soc. Trans. 36, 483–490 [DOI] [PubMed] [Google Scholar]
- 44. Hartmann B., Castelo R., Blanchette M., Boue S., Rio D. C., Valcárcel J. (2009) Global analysis of alternative splicing regulation by insulin and wingless signaling in Drosophila cells. Genome Biol. 10, R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Apostolatos A., Song S., Acosta S., Peart M., Watson J. E., Bickford P., Cooper D. R., Patel N. A. (2012) Insulin promotes neuronal survival via the alternatively spliced protein kinase CδII isoform. J. Biol. Chem. 287, 9299–9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chalfant C. E., Watson J. E., Bisnauth L. D., Kang J. B., Patel N., Obeid L. M., Eichler D. C., Cooper D. R. (1998) Insulin regulates protein kinase CβII expression through enhanced exon inclusion in L6 skeletal muscle cells. A novel mechanism of insulin- and insulin-like growth factor-I-induced 5′ splice site selection. J. Biol. Chem. 273, 910–916 [DOI] [PubMed] [Google Scholar]
- 47. Talukdar I., Szeszel-Fedorowicz W., Salati L. M. (2005) Arachidonic acid inhibits the insulin induction of glucose-6-phosphate dehydrogenase via p38 MAP kinase. J. Biol. Chem. 280, 40660–40667 [DOI] [PubMed] [Google Scholar]
- 48. Jenkins G. M., Cowart L. A., Signorelli P., Pettus B. J., Chalfant C. E., Hannun Y. A. (2002) Acute activation of de novo sphingolipid biosynthesis upon heat shock causes an accumulation of ceramide and subsequent dephosphorylation of SR proteins. J. Biol. Chem. 277, 42572–42578 [DOI] [PubMed] [Google Scholar]
- 49. Jayadev S., Linardic C. M., Hannun Y. A. (1994) Identification of arachidonic acid as a mediator of sphingomyelin hydrolysis in response to tumor necrosis factor α. J. Biol. Chem. 269, 5757–5763 [PubMed] [Google Scholar]
- 50. Ankö M. L., Morales L., Henry I., Beyer A., Neugebauer K. M. (2010) Global analysis reveals SRp20- and SRp75-specific mRNPs in cycling and neural cells. Nat. Struct. Mol. Biol. 17, 962–970 [DOI] [PubMed] [Google Scholar]