Background: Naturally occurring G26R mutation of apoA-I is associated with hereditary amyloidosis.
Results: The G26R mutation destabilizes the N-terminal helix bundle in full-length protein and also enhances amyloid fibril formation by the N-terminal 1–83 fragment.
Conclusion: The G26R mutation has dual critical roles in apoA-I structure and fibril-forming propensity.
Significance: The findings provide new insight into amyloid fibril formation by apoA-I.
Keywords: Amyloid, Apolipoproteins, Fluorescence, Mutant, Protein Stability, Apolipoprotein A-I, Fibril
Abstract
A number of naturally occurring mutations of apolipoprotein (apo) A-I, the major protein of HDL, are known to be associated with hereditary amyloidosis and atherosclerosis. Here, we examined the effects of the G26R point mutation in apoA-I (apoA-IIowa) on the structure, stability, and aggregation propensity to form amyloid fibril of full-length apoA-I and the N-terminal fragment of apoA-I. Circular dichroism and fluorescence measurements demonstrated that the G26R mutation destabilizes the N-terminal helix bundle domain of full-length protein, leading to increased hydrophobic surface exposure, whereas it has no effect on the initial structure of the N-terminal 1–83 fragment, which is predominantly a random coil structure. Upon incubation for extended periods at neutral pH, the N-terminal 1–83 variants undergo a conformational change to β-sheet-rich structure with a great increase in thioflavin T fluorescence, whereas no structural change is observed in full-length proteins. Comparison of fibril-forming propensity among substituted mutants at Gly-26 position of 1–83 fragments demonstrated that the G26R mutation enhances the nucleation step of fibril formation, whereas G26K and G26E mutations have small or inhibiting effects on the formation of fibrils. These fibrils of the 1–83 variants have long and straight morphology as revealed by atomic force microscopy and exhibited significant toxicity with HEK293 cells. Our results indicate dual critical roles of the arginine residue at position 26 in apoA-IIowa: destabilization of the N-terminal helix bundle structure in full-length protein and enhancement of amyloid fibril formation by the N-terminal 1–83 fragment.
Introduction
Apolipoprotein A-I (apoA-I)2 is the major protein component of plasma high density lipoprotein (HDL). It functions as a critical mediator in reverse cholesterol transport, a process by which excess cholesterol in peripheral cells is transferred via HDL to the liver for catabolism (1, 2). The amino acid sequence of human apoA-I (243 residues) contains 11- and 22-residue repeats of amphipathic α-helices (3) that fold into two tertiary structure domains, comprising an N-terminal α-helix bundle spanning residues 1–187 and a separate less organized C-terminal region spanning the remainder of the molecule (4–6). The helical segments in the N-terminal domain are involved in the activation of lecithin-cholesterol acyltransferase to convert cholesterol to cholesterol ester (7, 8), whereas the C-terminal helix is involved in the strong lipid binding properties of this protein (5, 9, 10).
Naturally occurring mutations in human apoA-I are known to be associated with low plasma HDL levels and hereditary amyloidosis (11). The hereditary amyloidogenic mutations are clustered in the N terminus (residues 1–100) and in residues 154–178 of the protein (12, 13). In most cases, the N-terminal fragment of these mutated apoA-I is the predominant form of protein found in amyloid fibril deposits (14). Although the structural consequences of these mutations have not been defined yet, it is thought that an unstable N-terminal helix bundle conformation associated with amyloidogenic mutations promotes proteolytic cleavage of full-length protein, resulting in the release of the N-terminal fragment to form amyloid fibril deposits (13, 15). Consistent with this, the N-terminal 1–93 fragments of apoA-I variants have been shown to aggregate in vitro to form amyloid fibrils at acidic pH (16, 17).
Nonhereditary forms of wild-type (WT) apoA-I are also commonly observed to be deposited as amyloid fibrils in atherosclerotic plaques (18), suggesting that mutations in the protein are not essential for its conversion into amyloid. In fact, a recent study demonstrated that oxidative modification of apoA-I methionine residues induces amyloid formation by the native, intact protein (19). In addition, a peptide comprising residues 46–59 of apoA-I was shown to form amyloid fibrils with cross-β structure (20), suggesting that this region is one of the core amyloidogenic segments in the apoA-I sequence (15). However, the molecular mechanisms underlying the amyloidogenic propensity of apoA-I are still poorly understood.
In the present study, we examined the effects of the Iowa (G26R) mutation, the first and most common amyloidogenic mutation found in apoA-I (21–24), on the structure, stability, and aggregation propensity to form amyloid fibril for full-length apoA-I and the N-terminal fragment (amino acids 1–83) of apoA-I. As with other amyloidogenic variants, the N-terminal 1–83 fragment is the major constituent of the Iowa variant found in amyloid deposits (21). The results demonstrated that the N-terminal 1–83 fragment of apoA-I has a strong propensity to form amyloid fibrils at neutral pH and that the G26R mutation greatly enhances the amyloid fibril formation of this N-terminal fragment.
MATERIALS AND METHODS
Preparation of apoA-I Proteins
The cDNA encoding the N-terminal fragment 1–83 of apoA-I was obtained by PCR methods using full-length human apoA-I cDNA as a template. The mutations in human apoA-I to create G26R and/or single tryptophan variants (W@8, W@50, or W@72) were made using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The cDNA was ligated into a thioredoxin fusion expression vector pET32a(+) (Novagen, Madison, WI) and transformed into the Escherichia coli strain BL21 Star (DE3) (Invitrogen). The resulting thioredoxin-apoA-I fusion proteins were expressed and purified as described (5, 10). Cleavage of the thioredoxin fusion protein with thrombin leaves the target apoA-I with two extra amino acids, Gly-Ser, at the amino terminus. The apoA-I preparations were at least 95% pure as assessed by SDS-PAGE. In all experiments, apoA-I variants were freshly dialyzed from 6 m guanidine hydrochloride solution into the appropriate buffer before use.
Circular Dichroism (CD) Spectroscopy
Far-UV CD spectra were recorded from 185 to 260 nm at 25 °C using a Jasco J-600 spectropolarimeter. The apoA-I solutions of 50 μg/ml in 10 mm Tris buffer (pH 7.4) were subjected to CD measurements in a 2-mm quartz cuvette, and the results were corrected by subtracting the buffer base line. The α-helix content was derived from the molar ellipticity at 222 nm ([θ]222) using the equation: % of α-helix = ((−[θ]222 + 3,000)/(36,000 + 3,000)) × 100 (25).
Attenuated Total Reflection-Fourier Transform Infrared (ATR-FTIR) Spectroscopy
ATR-FTIR spectra were recorded on a Jasco FTIR spectrometer FT/IR-6200 equipped with an ATR PRO610P-S reflectance accessory. An aliquot of apoA-I samples (0.3 mg/ml) in 10 mm Tris buffer (pH 7.4) was spread on the germanium waveguide, and the excess water was removed under nitrogen flow. ATR-FTIR spectra were recorded over the wave number range of 1,000–3,500 cm−1 at a resolution of 2 cm−1 and 256 accumulations under continuous nitrogen purge. To evaluate secondary structure, the amide I area (1,600–1,700 cm−1) in the spectra was deconvoluted using a Spectra Manager Software (Jasco, Tokyo, Japan).
Urea Denaturation Study
For monitoring chemical denaturation, lipid-free proteins at a concentration of 50 μg/ml were incubated overnight at 4 °C with urea at various concentrations. KD at a given denaturant concentration was calculated from the change in [θ]222 determined by CD in 10 mm Tris buffer (pH 7.4). The Gibbs free energy of denaturation in the absence of denaturant, ΔGD°, the midpoint of denaturation, D½, and m value, which reflects the cooperativity of denaturation in the transition region, were determined by the linear equation, ΔGD = ΔGD° − m[denaturant], where ΔGD = − RT ln KD (26, 27).
Fluorescence Measurements
Fluorescence measurements were carried out with a Hitachi F-4500 fluorescence spectrophotometer at 25 °C. To monitor the exposure of hydrophobic sites on the apoA-I variants, 8-anilino-1-naphthalenesulfonic acid (ANS) fluorescence spectra were collected from 400 to 600 nm at an excitation wavelength of 395 nm in the presence of 50 μg/ml protein and an excess of ANS (250 μm) (5). To access the local environment of apoA-I single Trp variants, Trp emission fluorescence was recorded from 300 to 420 nm using a 290-nm excitation wavelength to avoid tyrosine fluorescence. In quenching experiments of Trp fluorescence, Trp emission spectra of proteins were recorded at increasing concentrations of KI (0–0.56 m) using a 5 m stock solution containing 1 mm NaS2O3 to prevent the formation of iodine. After correction for dilution, the integrated fluorescence intensities were plotted according to the Stern-Volmer equation, F0/F = 1 + Ksv [KI], where F0 and F are the fluorescence intensities in the absence and presence of quencher, respectively, and Ksv is the Stern-Volmer constant. Quenching parameters were obtained by fitting to the modified Stern-Volmer equation, F0/(F0 − F) = 1/fa +1/faKsv[KI], where fa is the fraction of Trp residue accessible to the quencher.
Kinetics of protein aggregation and fibril formation were monitored using fluorescent dyes, thioflavin T (ThT), and ANS. apoA-I variants (0.3 mg/ml) in 10 mm Tris buffer (150 mm NaCl, 0.02% NaN3, pH 7.4) were incubated at 37 °C with agitation on an orbital rotator at 10 rpm in the presence of 10 μm ThT or 10 μm ANS. ThT fluorescence was recorded from 460 to 600 nm at an excitation wavelength of 445 nm. The time-dependent increase in ThT fluorescence intensity was fitted to a sigmoidal equation (28, 29)
 |
where F is the fluorescence intensity, F0 is the initial base line during the lag phase, and Fmax is the final base line after the growth phase has ended. k is the apparent rate constant for the growth of fibrils, and tm is the time to 50% of maximal fluorescence. The lag time is calculated as tm − 2/k.
Atomic Force Microscopy (AFM)
For analysis by AFM, 5 ml of each sample solution (0.6 mg/ml) in 10 mm Tris buffer was diluted with distilled water (45 ml), and 10 ml of the mixture was deposited on freshly cleaved mica (The Nilaco Corp., Tokyo, Japan). After washing the mica with distilled water (20 ml), samples were imaged under ambient conditions at room temperature using a NanoScope® IIIa tapping mode AFM (Veeco, Plainview, NY) and microcantilever OMCL-AC160TS-R3 (Olympus, Tokyo, Japan).
Cytotoxicity Assay
Cytotoxicity was measured using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (30). HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 units/ml penicillin, 100 μg/ml streptomycin at 37 °C in 5% CO2 atmosphere. Cells were plated at a density of 4 × 104 cells/well on poly-l-lysine-coated 24-well plates in DMEM containing 2% FBS. After a 24-h incubation, apoA-I variants in 10 mm phosphate-buffered saline were added and cells were further incubated with DMEM containing 2% FBS for 24 h. Cell viability was quantitatively determined by reduction of MTT. After the addition of MTT to each well (final concentration was 0.5 mg/ml) for 3 h, the medium was removed, and the resulting formazan crystals were solved in dimethyl sulfoxide. Absorbance values of formazan were measured at 570 nm with subtracted absorbance at 650 nm.
RESULTS
Physicochemical Characterization of apoA-I G26R Variants
We first investigated the effects of the G26R mutation on the structure and stability of full-length apoA-I and its N-terminal 1–83 fragment. CD spectra (Fig. 1A) show that full-length apoA-I forms α-helical structure, whereas the 1–83 fragment is predominantly in the random coil structure at neutral pH (16), and that the G26R mutation has a small effect on their overall secondary structure (17). However, the G26R mutation greatly reduces stability of full-length apoA-I against urea denaturation (Fig. 1B) such that the midpoint of denaturation is 1.8 m urea (versus 2.7 m urea for WT apoA-I) and the free energy of denaturation is 2.4 kcal/mol (versus 3.5 kcal/mol for WT). As shown in Fig. 1C, such a destabilization of full-length protein by the G26R mutation leads to exposure of more hydrophobic surfaces as reflected by the great increase in ANS fluorescence (17). In contrast, both the 1–83 and the 1–83/G26R fragments exhibited a little increase in ANS fluorescence, consistent with both fragments forming the random coil structure (Fig. 1A). These results indicate that the G26R mutation significantly destabilizes the helix bundle structure of full-length apoA-I, but that this mutation has negligible effect on the random coil structure of the 1–83 fragment.
FIGURE 1.
G26R mutation greatly affects structure and stability of full-length apoA-I. A, far-UV CD spectra of apoA-I WT (a), G26R (b), 1–83 (c), and 1–83/G26R (d). Protein concentration was 50 μg/ml. deg, degree. B, urea-induced denaturation curves of apoA-I WT (●) and G26R (○) monitored by CD. Protein concentration was 50 μg/ml. C, ANS fluorescence spectra in the presence of apoA-I variants. apoA-I WT (a), G26R (b), 1–83 (c), 1–83/G26R (d), and free ANS for comparison (e). Protein and ANS concentrations were 50 μg/ml and 250 μm, respectively.
Amyloid Fibril Formation of apoA-I N-terminal Fragment
The propensity to form amyloid fibrils of apoA-I G26R variants was assessed using the amyloidophilic dye, ThT (31, 32). On incubation for several days at 37 °C, pH 7.4 with agitation, there was no increase in ThT fluorescence for WT and the G26R variant in full-length apoA-I (Fig. 2A) (19, 24). In contrast, the N-terminal 1–83 fragments exhibited large increases in ThT fluorescence in a time-dependent manner, in which the G26R mutation greatly enhanced the amyloid fibril formation. Kinetic analysis of sigmoidal increases in ThT fluorescence indicated that the G26R mutation greatly reduces the lag time for fibril formation of the 1–83 fragments (22.5 and 11.4 h for 1–83 and 1–83/G26R, respectively). With increasing protein concentration, the development of ThT fluorescence was accelerated (Fig. 2B), consistent with the nucleation-elongation mechanism of aggregation that is common to most amyloid fibrils (33).
FIGURE 2.
Fibril formation was monitored by ThT fluorescence for apoA-I variants incubated at pH 7.4. A, ▵, apoA-I WT; ▾, G26R; □, 1–83; ●, 1–83/G26R. Protein concentration was 0.3 mg/ml. a. u., arbitrary units. Error bars indicate S.E. B, effect of protein concentration on ThT fluorescence of apoA-I 1–83/G26R. ▴, 0.1 mg/ml; ●, 0.3 mg/ml; ▿, 0.5 mg/ml. Error bars indicate S.E.
The hallmark feature of amyloid proteins is the increase in β-strand conformation. To determine the secondary structural change in the 1–83 fragments, CD spectra were collected for the samples before and after incubation. As shown in Fig. 3A, a spectrum of the 1–83 variant after incubation displayed a single minimum at ∼216 nm, signifying the formation of β-sheet-rich structure. Similar conversion of CD spectra was also observed for the 1–83/G26R (Fig. 3B). In contrast to the case of structural transition of the N-terminal fragment of apoA-I at acidic pH (16, 17), the transient helical structure was not observed during incubation at neutral pH (supplemental Fig. S1).
FIGURE 3.
Far-UV CD (A and B) and ATR-FTIR (C and D) spectra of apoA-I 1–83 and 1–83/G26R variants before (dashed line) and after incubation for 5 days (solid line) at pH 7.4. Protein concentration was 50 μg/ml. deg, degree. a. u., arbitrary units.
The secondary structure of fibrils was also estimated by ATR-FTIR spectroscopy. FTIR spectra of the 1–83 and 1–83/G26R variants showed that the band at ∼1,630 cm−1 became evident after incubation, indicating the conversion to the β-sheet structures (Fig. 3, C and D). These results indicate that the N-terminal 1–83 fragment of apoA-I forms amyloid fibrils with β-strand conformation at pH 7.4 and that the G26R mutation greatly enhances its fibril-forming propensity.
Single Trp Variants of apoA-I 1–83/G26R
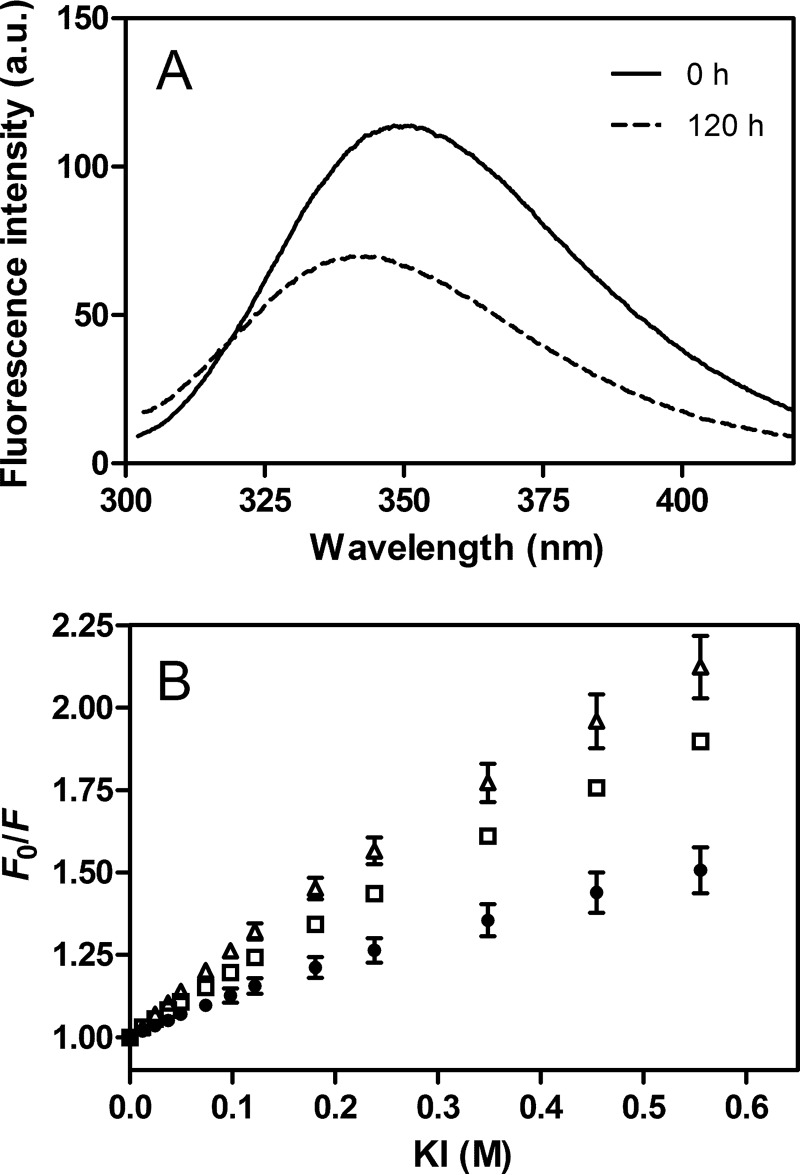
To probe the local conformation of the 1–83/G26R fragment in the fibril structure, we substituted intrinsic Trp residues (Trp-8, Trp-50, and Trp-72) to Phe to generate single Trp variants, W@8, W@50, and W@72, in the 1–83/G26R fragment. Such substituted mutations did not affect the kinetics of amyloid fibril formation as monitored by ThT fluorescence (data not shown). Trp fluorescence emission spectra of 1–83/G26R W@50 (Fig. 4A) demonstrated a significant blue-shift in wavelength of maximum fluorescence upon transition to the fibrils, indicating transfer of the Trp residue into a more hydrophobic environment. Reduction of fluorescence intensity is likely to be due to the intermolecular energy transfer among Trp residues in close proximity (34). Comparison of wavelength of maximum fluorescence among three single Trp mutants indicates that Trp-8 is located in a more solvent-exposed environment than Trp-50 and Trp-72 in the fibril structure (Table 1).
FIGURE 4.
Trp fluorescence of amyloid fibrils of apoA-I 1–83/G26R single Trp variants. A, Trp fluorescence spectra of apoA-I 1–83/G26R W@50, before (dashed line) and after incubation for 5 days (solid line). The excitation wavelength was 290 nm. a. u., arbitrary units. B, Stern-Volmer plot for KI quenching of amyloid fibrils of apoA-I 1–83/G26R W@8 (▵), W@50 (●), and W@72 (□). Error bars indicate S.E.
TABLE 1.
Fluorescence parameters for amyloid fibrils of apoA-I 1–83/G26R single Trp variants
| apoA-I variants | WMFa | KI quenching parameters |
|
|---|---|---|---|
| fa | KSV | ||
| nm | m−1 | ||
| 1–83/G26R W@8 | 345 | 0.89 ± 0.19 | 3.0 ± 1.1 |
| 1–83/G26R W@50 | 343 | 0.60 ± 0.13 | 1.9 ± 0.4 |
| 1–83/G26R W@72 | 343 | 0.91 ± 0.14 | 2.1 ± 0.5 |
a WMF, wavelength of maximum fluorescence. Estimated error is within ±1 nm.
We also compared Trp quenching behaviors of three single Trp mutants of 1–83/G26R to the aqueous quencher KI (Fig. 4B). As summarized in Table 1, the fraction of Trp residue accessible to the quencher of 1–83/G26R W@50 is much smaller than other variants, indicating that Trp-50 is the least exposed to the solvent in the fibril structure. These results suggest that Trp-50 is located in the core region of the 1–83/G26R fibrils, consistent with the prediction that residues 15–20 and 50–57 are the most aggregation-prone regions in the apoA-I N-terminal fragment (17).
Effect of Gly-26 Substituted Mutations on Amyloid Fibril Formation of apoA-I 1–83 Fragment
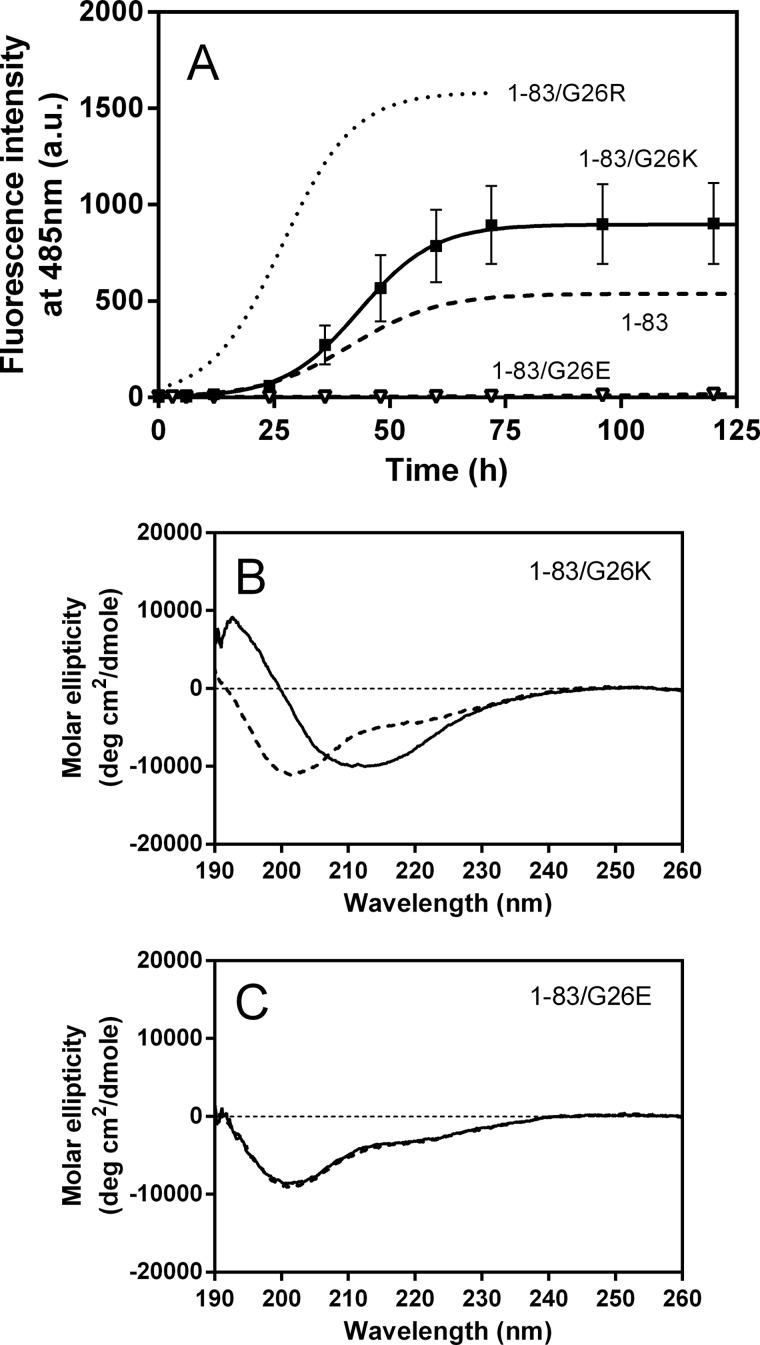
To explore the role of arginine residue at position 26 on the fibril-forming propensity of the apoA-I 1–83 fragment, we generated two additional substituted variants of the 1–83 fragment, 1–83/G26K and 1–83/G26E; glycine 26 is substituted with a positively and negatively charged amino acid, respectively. Fig. 5A shows the increases in ThT fluorescence of these 1–83 variants over the time course. The G26K mutation enhanced the fibril formation of the 1–83 fragment, but the effect was much smaller than the G26R. In contrast, no significant increase in ThT fluorescence was observed in the G26E mutant over time. Consistent with this trend in development of ThT fluorescence, the CD spectra reflected conversion from random coil to β-sheet in 1–83/G26K (Fig. 5B), whereas there was no structural change in 1–83/G26E (Fig. 5C).
FIGURE 5.
Changes in ThT fluorescence and CD spectra of apoA-I 1–83 variants over incubation time at pH 7.4. A, ThT fluorescence of 1–83 (dashed line), 1–83/G26R (dotted line), 1–83/G26K (■), and 1–83/G26E (▿). Protein and ThT concentrations were 0.3 mg/ml and 10 μm, respectively. a. u., arbitrary units. Error bars indicate S.E. B and C, far-UV CD spectra of 1–83/G26K (B) and 1–83/G26E (C) before (dashed line) and after incubation for 5 days (solid line). Protein concentration was 50 μg/ml. deg, degree.
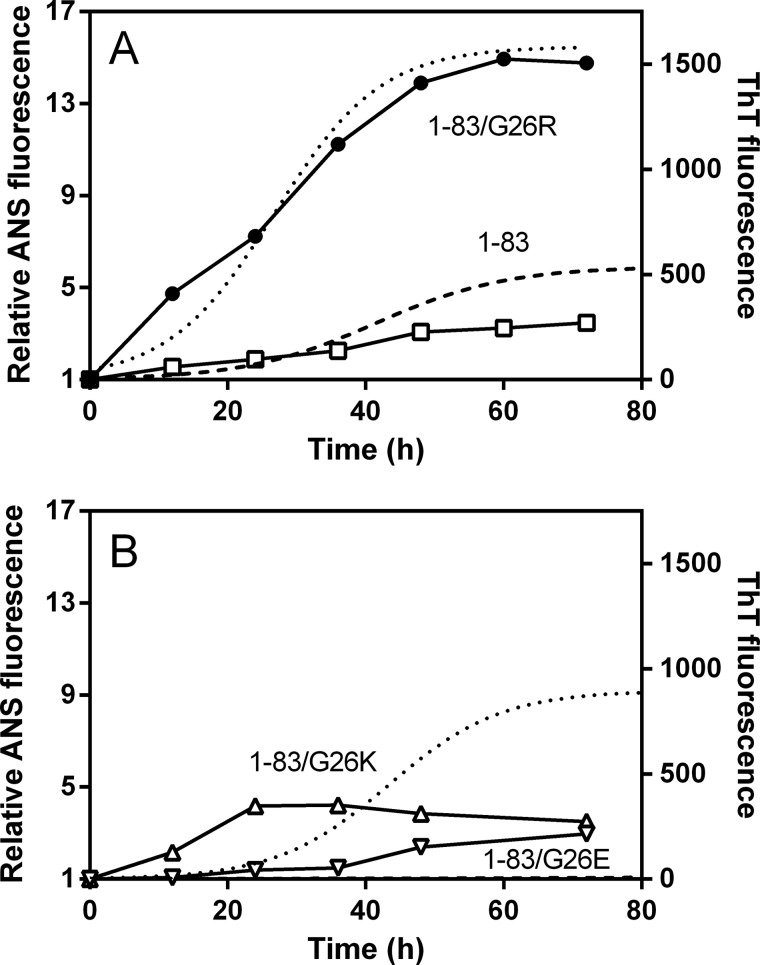
We also performed ANS binding studies to the apoA-I 1–83 variants to compare the aggregated state over incubation time. ANS has been used to detect amyloid fibrils because it binds to the fibrillar or prefibrillar states of proteins (35). Similarly to the 1–83/G26R protein (Fig. 1C), all 1–83 variants exhibited a small increase in ANS fluorescence in freshly prepared sample. On incubation, ANS fluorescence increased with similar kinetics to the increase in ThT fluorescence for the 1–83 and 1–83/G26R variants (Fig. 6A). In contrast, the development of ANS fluorescence was observed at lag phase of ThT fluorescence for 1–83/G26K, and despite the lack of ThT signal, a significant increase in ANS fluorescence was detected for 1–83/G26E (Fig. 6B). Such discrepancies in increases in ThT and ANS fluorescence suggest that the 1–83/G26K and 1–83/G26E variants predominantly form prefibril oligomers before the fibril formation (36).
FIGURE 6.
Changes in ANS fluorescence of apoA-I 1–83 variants over incubation time at pH 7.4. A, ANS fluorescence of 1–83 (□) and 1–83/G26R (●). ThT fluorescence of 1–83 (dashed line) and 1–83/G26R (dotted line) is also shown for comparison. B, ANS fluorescence of 1–83/G26K (▵) and 1–83/G26E (▿). ThT fluorescence of 1–83/G26K (dotted line) and 1–83/G26E (dashed line) is also shown for comparison. Protein and ANS concentrations were 0.3 mg/ml and 10 μm, respectively.
The morphology of amyloid fibrils formed by the 1–83 fragments was estimated by AFM (Fig. 7). AFM images of the 1–83, 1–83/G26R, and 1–83/G26K variants incubated for 5 days at 37 °C demonstrated the formation of long and straight fibrils with heights of ranging from 5 to 10 nm. The coexistence of large aggregates is likely to be due to the strong hydrophobic interactions resulting in the formation of amorphous aggregates as well as amyloid fibrils (37). AFM image of 1–83/G26E reveals that there is no fibril formation with this variant.
FIGURE 7.
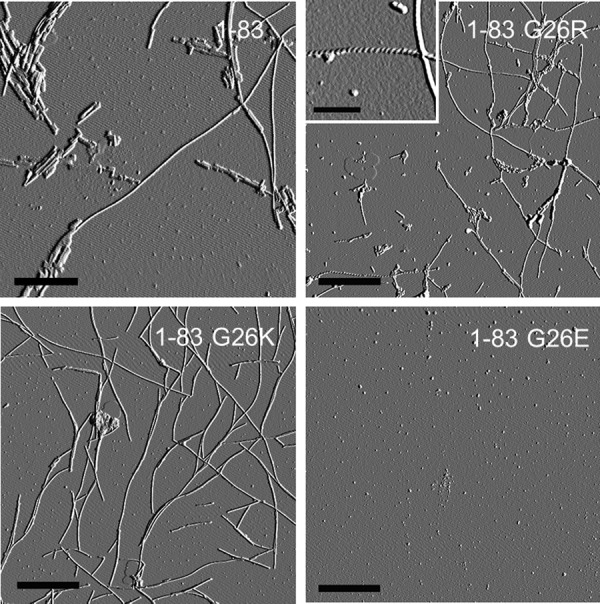
AFM images of apoA-I 1–83 variants after 5 days of incubation at pH 7.4. Scale bars represent 1 μm except for the inset in 1–83/G26R (0.2 μm) showing the twisted morphology.
Cytotoxicity of apoA-I Amyloid Fibrils
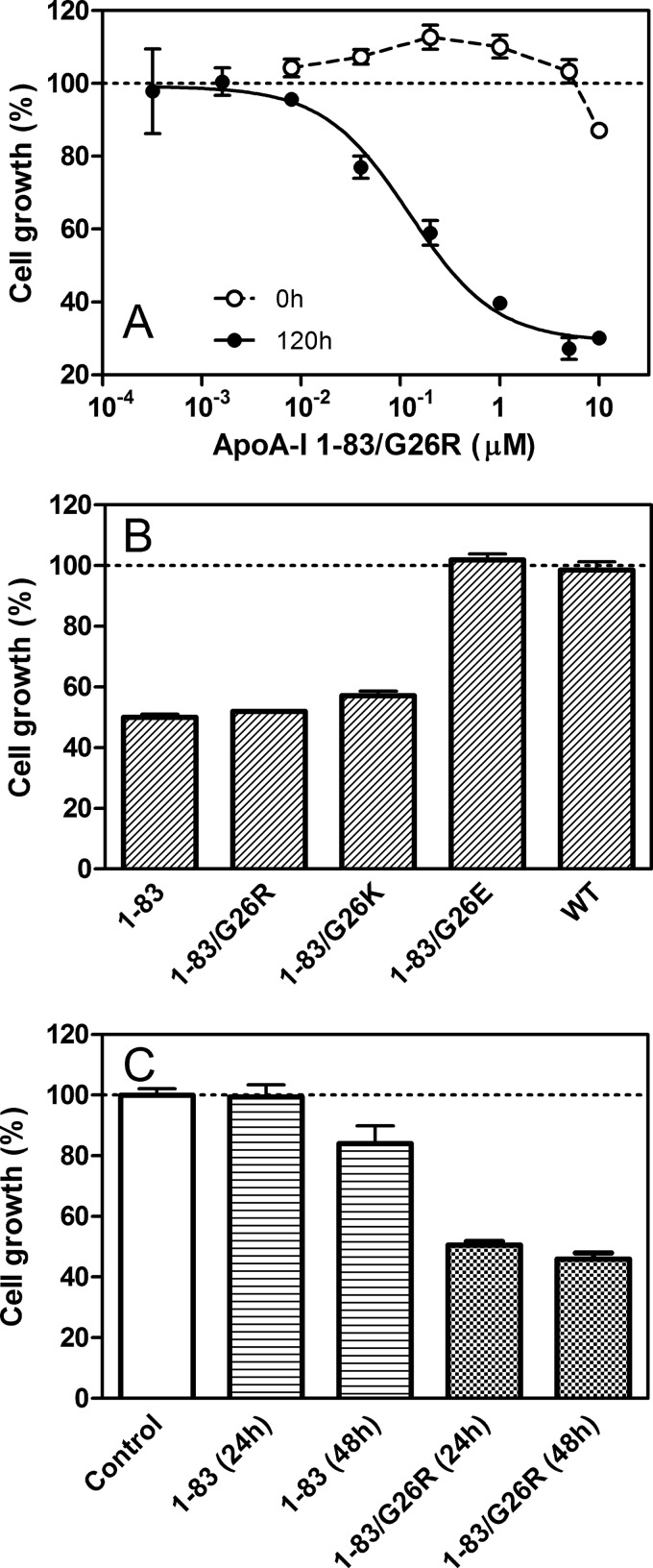
The cytotoxicity of amyloid fibrils of apoA-I 1–83 variants was examined by using the MTT assay. As shown in Fig. 8A, the 1–83/G26R fibrils prepared by incubation for 120 h induced high cell toxicity with HEK293 cells in a dose-dependent manner (effective concentration giving 50% cytotoxicity (EC50) is 0.12 μm), whereas a freshly prepared sample had no toxicity. The fibrils of the 1–83 and 1–83/G26K variants exhibited similar toxicity to 1–83/G26R, whereas the 1–83/G26E variant as well as WT apoA-I did not exhibit any toxicity (Fig. 8B), indicating that the formation of the fibril structure is critical to the cytotoxicity of the 1–83 fragments. We also tested the cytotoxicity of the prefibrillar state of apoA-I 1–83 and 1–83/G26R variants after incubation for 24 or 48 h. As shown in Fig. 8C, apoA-I 1–83 had no or a small effect on the cell growth up to a 48-h incubation, whereas the 1–83/G26R variant exhibited strong toxicity even at a 24-h incubation. These results suggest that the G26R mutation promotes the development of the intermediate aggregates of the 1–83 fragments responsible for the strong cytotoxicity (38, 39).
FIGURE 8.
Cytotoxicity of apoA-I 1–83 variants. A, effect of apoA-I 1–83/G26R concentration on the cell growth. apoA-I 1–83/G26R was added to the medium of HEK293 cells before (○) or after incubation for 5 days (●). Error bars indicate S.E. B, 1–83 variants or WT apoA-I after incubation for 5 days was added to the medium of HEK293 cells at a final concentration of 0.2 μm. Error bars indicate S.E. C, apoA-I 1–83 or 1–83/G26R after incubation for 24 or 48 h was added to the medium of HEK293 cells at a final concentration of 0.2 μm. Error bars indicate S.E.
DISCUSSION
It is known that apoA-I amyloid fibrils extracted from patients with hereditary apoA-I amyloidosis consist of the N-terminal 9–11-kDa fragments; the fibrillar deposits in the heart contain mainly the 1–93 fragment, whereas those in other organs contain fragments ranging from approximately residues 1–80 to 1–100 (12, 13). In the Iowa (G26R) mutation, the major protein constituent of the fibrils isolated from spleen and liver was found to be residues 1–83 of apoA-I (21). In the present study, we demonstrated that this N-terminal 1–83 fragment of apoA-I has a strong propensity to form amyloid fibrils at neutral pH and that the G26R mutation greatly enhances its fibril formation. We also found that the amyloid fibrils of the 1–83 fragments exhibit strong toxicity to HEK293 cells as with amyloid β fibrils (40). To our knowledge, the finding that the 1–83 fragment of apoA-I forms amyloid fibrils at physiological neutral pH is quite novel because the previous observations indicated that the fibril formation of apoA-I in vitro requires the acidic condition (16, 41, 42), methionine oxidation (19), or amyloidogenic mutations (23, 43).
Many plasma apolipoproteins including apoA-I display a high susceptibility to form or associate with amyloid fibrils both in vitro and in vivo (44, 45), perhaps due to their partially folded, flexible conformation in the lipid-free state (46). All known amyloidogenic mutations in human apoA-I are clustered in residues 26–107 and 154–178, both of which are located within the largely helical bundle structure comprised of residues 1–120 and 144–184 (15). Thus, it is expected that mutations in these positions destabilize the bundle structure of apoA-I, thereby promoting its proteolysis.
The results in the previous (24) and present studies demonstrated that the G26R mutation indeed destabilizes the helix bundle structure of full-length apoA-I (Fig. 1). Because the G26R substitution places the strong basic arginine residue into the nonpolar face of the amphipathic helix, it would prevent the optimal packing of the helical segments, resulting in the destabilization and increased solvent exposure of the N-terminal helix bundle (supplemental Fig. S2). In fact, our recent hydrogen-deuterium exchange and mass spectroscopy measurements of apoA-IIowa demonstrated that the G26R mutation induces widespread structural reorganization of residues in the central region of the apoA-I molecule despite a small change in the net helical contents (47). Similarly to the G26R mutation, the fibrillogenic L178H mutation, which is also located within the N-terminal helix bundle, was shown to exhibit a decreased protein stability and an altered lipid binding profile (43). In addition, oxidation of the methionine residues of apoA-I causes the destabilization of the tertiary and quaternary structure of the protein, leading to the formation of amyloid fibrils (19). Thus, it is likely that the destabilization and partial unfolding of the protein structure is a common feature for the fibril formation of full-length apoA-I.
The fluorescence results using single Trp mutants of the apoA-I 1–83/G26R fibrils (Fig. 3B and Table 1) indicate that the residues around Trp-50 are located in the core region of the fibrils formed by the N-terminal 1–83 fragment. The crystal structure of a C-terminal truncated apoA-I (PDB entry 3R2P) identified a segment, residues 44–55, in an extended conformation with the β-strand-like geometry (6). This implies the possibility that segment 44–55 provides a template for the cross-β-sheet conformation in amyloid fibrils (15). Consistent with this, a peptide comprising residues 46–59 of apoA-I was shown to aggregate to form amyloid-like fibrils (20), suggesting the exposure of this relatively unstructured segment being involved in the initiation and propagation of the fibril formation of apoA-I. In addition, the electron paramagnetic resonance study demonstrated that residues 41–56 are converted to β-sheet structure in apoA-IIowa, whereas residues 27–56 are largely α-helical in WT apoA-I (23). Thus, the predominantly random coil structure of the N-terminal fragment is expected to expose the region of residues 44–55 to facilitate protein aggregation.
Previous studies of apoA-I 1–93 fragment, however, demonstrated that the protein aggregation occurred only at acidic pH, not at neutral pH (16, 41), and that the G26R mutation had no effect on its aggregation propensity (17). Because the 1–93 fragment contains additional residues 84–93 (QEMSKDLEEV), which are rich in negatively charged amino acids at neutral pH, it is possible that this region may alter intermolecular or intramolecular electrostatic interactions of the N-terminal fragments, resulting in the inhibition of protein aggregation. In this regard, the fact that the G26E mutation in the 1–83 fragment strongly inhibits its fibril-forming propensity suggests that the optimal electrostatic interactions are important for the fibril formation of the1–83 fragment.
The finding that the G26K mutation exhibits less pronounced effect on the increase in ThT fluorescence when compared with the G26R mutation (Fig. 5A) is striking. Similar increasing behaviors of ANS in the 1–83/G26R and 1–83/G26K variants over 24 h (Fig. 6, A and B) indicate that the generation of prefibril oligomers is similar in both variants. Rather, the formation of the fibril structure appears to be much faster in the 1–83/G26R variant than 1–83/G26K. Although the replacement of arginine with lysine does not alter the net charge at neutral pH, this conservative substitution is known to affect many protein-protein interactions (48, 49). Thus, the differences in the hydrogen-bonding capacity (50) and relative hydrophobicity (51) in arginine and lysine residues may affect the intermolecular interactions of the 1–83 fragments, thereby altering the kinetics of fibril formation. Interestingly, molecular dynamic simulations suggested that arginine interacts with the aromatic residue of proteins due to cation-π electron interaction, leading to alteration of the protein stability in the partially unfolded state (52). It should be noted that the effects of the G26K mutation on the protein stability of full-length apoA-I appear to be similar to the G26R mutation (supplemental Fig. S3).
As a general mechanism of amyloid fibril formation, it is proposed that a subtle balance between the hydrophobic and electrostatic interactions determines the amyloidogenicity of the proteins (37). In lysozyme, a well known amyloidogenic protein, the extent of charge repulsion was supposed to determine the assembly pathway and morphology of fibrillar aggregates (53). Indeed, some of the amyloidogenic mutations in lysozyme, W64R and D67H, involve the introduction of additional positively charged residues into the clustered region of the protein (54). In this regard, it is interesting that many amyloidogenic mutations in apoA-I including the G26R mutation involve the additional positive charge or the loss of a hydrophobic residue within the N-terminal helix bundle (15).
In conclusion, this is the first study demonstrating that the N-terminal 1–83 fragment of apoA-I has a strong propensity to form amyloid fibrils at physiological neutral pH. Our results also indicate that the G26R point mutation in apoA-IIowa has dual critical roles in the protein structure and stability: destabilization of the N-terminal helix bundle in full-length protein and enhancement of amyloid fibril formation by the N-terminal 1–83 fragment. Thus, it is likely that many other naturally occurring mutations of apoA-I associated with hereditary amyloidosis would cause not only the destabilization of the protein structure but also an increase in susceptibility to form amyloid fibrils by enhancing the intermolecular aggregation.
Acknowledgments
We thank Dr. Jiro Kasahara (Institute of Health Biosciences, The University of Tokushima) for help with cytotoxicity assays. We also thank Dr. Masafumi Tanaka (Kobe Pharmaceutical University) for advice on ATR-FTIR measurements. We are indebted to Dr. Naomi Sakashita (Institute of Health Biosciences, The University of Tokushima) and Dr. Galyna Gorbenko (Kharkov National University) for valuable advice.
This work was supported, in whole or in part, by National Institutes of Health Grant HL22633 (to M. C. P.). This work was also supported by Grant-in-Aid for Scientific Research 22590046 (to H. S.) from the Japan Society for the Promotion of Science.

This article contains supplemental Figs. S1–S3.
- apoA-I
- apolipoprotein A-I
- AFM
- atomic force microscopy
- ANS
- 8-anilino-1-naphthalenesulfonic acid
- ATR
- attenuated total reflection
- FTIR
- Fourier transform infrared spectroscopy
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ThT
- thioflavin T.
REFERENCES
- 1. Tall A. R. (2008) Cholesterol efflux pathways and other potential mechanisms involved in the athero-protective effect of high density lipoproteins. J. Intern. Med. 263, 256–273 [DOI] [PubMed] [Google Scholar]
- 2. Rader D. J., Alexander E. T., Weibel G. L., Billheimer J., Rothblat G. H. (2009) The role of reverse cholesterol transport in animals and humans and relationship to atherosclerosis. J. Lipid Res. 50, (suppl.) S189–S194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. (1992) The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33, 141–166 [PubMed] [Google Scholar]
- 4. Davidson W. S., Hazlett T., Mantulin W. W., Jonas A. (1996) The role of apolipoprotein AI domains in lipid binding. Proc. Natl. Acad. Sci. U.S.A. 93, 13605–13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saito H., Dhanasekaran P., Nguyen D., Holvoet P., Lund-Katz S., Phillips M. C. (2003) Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 278, 23227–23232 [DOI] [PubMed] [Google Scholar]
- 6. Mei X., Atkinson D. (2011) Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 286, 38570–38582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sorci-Thomas M., Kearns M. W., Lee J. P. (1993) Apolipoprotein A-I domains involved in lecithin-cholesterol acyltransferase activation. Structure:function relationships. J. Biol. Chem. 268, 21403–21409 [PubMed] [Google Scholar]
- 8. Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M., 3rd, Smith J. D., Gogonea V., Hazen S. L. (2007) The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 14, 861–868 [DOI] [PubMed] [Google Scholar]
- 9. Fang Y., Gursky O., Atkinson D. (2003) Lipid-binding studies of human apolipoprotein A-I and its terminally truncated mutants. Biochemistry 42, 13260–13268 [DOI] [PubMed] [Google Scholar]
- 10. Tanaka M., Koyama M., Dhanasekaran P., Nguyen D., Nickel M., Lund-Katz S., Saito H., Phillips M. C. (2008) Influence of tertiary structure domain properties on the functionality of apolipoprotein A-I. Biochemistry 47, 2172–2180 [DOI] [PubMed] [Google Scholar]
- 11. Sorci-Thomas M. G., Thomas M. J. (2002) The effects of altered apolipoprotein A-I structure on plasma HDL concentration. Trends Cardiovasc. Med. 12, 121–128 [DOI] [PubMed] [Google Scholar]
- 12. Rowczenio D., Dogan A., Theis J. D., Vrana J. A., Lachmann H. J., Wechalekar A. D., Gilbertson J. A., Hunt T., Gibbs S. D., Sattianayagam P. T., Pinney J. H., Hawkins P. N., Gillmore J. D. (2011) Amyloidogenicity and clinical phenotype associated with five novel mutations in apolipoprotein A-I. Am. J. Pathol. 179, 1978–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Obici L., Franceschini G., Calabresi L., Giorgetti S., Stoppini M., Merlini G., Bellotti V. (2006) Structure, function and amyloidogenic propensity of apolipoprotein A-I. Amyloid 13, 191–205 [DOI] [PubMed] [Google Scholar]
- 14. Genschel J., Haas R., Pröpsting M. J., Schmidt H. H. (1998) Apolipoprotein A-I induced amyloidosis. FEBS Lett. 430, 145–149 [DOI] [PubMed] [Google Scholar]
- 15. Gursky O., Mei X., Atkinson D. (2012) The crystal structure of the C-terminal truncated apolipoprotein A-I sheds new light on amyloid formation by the N-terminal fragment. Biochemistry 51, 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andreola A., Bellotti V., Giorgetti S., Mangione P., Obici L., Stoppini M., Torres J., Monzani E., Merlini G., Sunde M. (2003) Conformational switching and fibrillogenesis in the amyloidogenic fragment of apolipoprotein A-I. J. Biol. Chem. 278, 2444–2451 [DOI] [PubMed] [Google Scholar]
- 17. Raimondi S., Guglielmi F., Giorgetti S., Di Gaetano S., Arciello A., Monti D. M., Relini A., Nichino D., Doglia S. M., Natalello A., Pucci P., Mangione P., Obici L., Merlini G., Stoppini M., Robustelli P., Tartaglia G. G., Vendruscolo M., Dobson C. M., Piccoli R., Bellotti V. (2011) Effects of the known pathogenic mutations on the aggregation pathway of the amyloidogenic peptide of apolipoprotein A-I. J. Mol. Biol. 407, 465–476 [DOI] [PubMed] [Google Scholar]
- 18. Röcken C., Tautenhahn J., Bühling F., Sachwitz D., Vöckler S., Goette A., Bürger T. (2006) Prevalence and pathology of amyloid in atherosclerotic arteries. Arterioscler. Thromb. Vasc. Biol. 26, 676–677 [DOI] [PubMed] [Google Scholar]
- 19. Wong Y. Q., Binger K. J., Howlett G. J., Griffin M. D. (2010) Methionine oxidation induces amyloid fibril formation by full-length apolipoprotein A-I. Proc. Natl. Acad. Sci. U.S.A. 107, 1977–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wong Y. Q., Binger K. J., Howlett G. J., Griffin M. D. (2012) Identification of an amyloid fibril forming peptide comprising residues 46–59 of apolipoprotein A-I. FEBS Lett. 586, 1754–1758 [DOI] [PubMed] [Google Scholar]
- 21. Nichols W. C., Dwulet F. E., Liepnieks J., Benson M. D. (1988) Variant apolipoprotein AI as a major constituent of a human hereditary amyloid. Biochem. Biophys. Res. Commun. 156, 762–768 [DOI] [PubMed] [Google Scholar]
- 22. Rader D. J., Gregg R. E., Meng M. S., Schaefer J. R., Zech L. A., Benson M. D., Brewer H. B., Jr. (1992) In vivo metabolism of a mutant apolipoprotein, apoA-IIowa, associated with hypoalphalipoproteinemia and hereditary systemic amyloidosis. J. Lipid Res. 33, 755–763 [PubMed] [Google Scholar]
- 23. Lagerstedt J. O., Cavigiolio G., Roberts L. M., Hong H. S., Jin L. W., Fitzgerald P. G., Oda M. N., Voss J. C. (2007) Mapping the structural transition in an amyloidogenic apolipoprotein A-I. Biochemistry 46, 9693–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramella N. A., Schinella G. R., Ferreira S. T., Prieto E. D., Vela M. E., Ríos J. L., Tricerri M. A., Rimoldi O. J. (2012) Human apolipoprotein A-I natural variants: molecular mechanisms underlying amyloidogenic propensity. PLoS One 7, e43755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sparks D. L., Lund-Katz S., Phillips M. C. (1992) The charge and structural stability of apolipoprotein A-I in discoidal and spherical recombinant high density lipoprotein particles. J. Biol. Chem. 267, 25839–25847 [PubMed] [Google Scholar]
- 26. Tanaka M., Dhanasekaran P., Nguyen D., Ohta S., Lund-Katz S., Phillips M. C., Saito H. (2006) Contributions of the N- and C-terminal helical segments to the lipid-free structure and lipid interaction of apolipoprotein A-I. Biochemistry 45, 10351–10358 [DOI] [PubMed] [Google Scholar]
- 27. Kono M., Tanaka T., Tanaka M., Vedhachalam C., Chetty P. S., Nguyen D., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. (2010) Disruption of the C-terminal helix by single amino acid deletion is directly responsible for impaired cholesterol efflux ability of apolipoprotein A-I Nichinan. J. Lipid Res. 51, 809–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nielsen L., Khurana R., Coats A., Frokjaer S., Brange J., Vyas S., Uversky V. N., Fink A. L. (2001) Effect of environmental factors on the kinetics of insulin fibril formation: elucidation of the molecular mechanism. Biochemistry 40, 6036–6046 [DOI] [PubMed] [Google Scholar]
- 29. Zhou Z., Fan J. B., Zhu H. L., Shewmaker F., Yan X., Chen X., Chen J., Xiao G. F., Guo L., Liang Y. (2009) Crowded cell-like environment accelerates the nucleation step of amyloidogenic protein misfolding. J. Biol. Chem. 284, 30148–30158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tada H., Shiho O., Kuroshima K., Koyama M., Tsukamoto K. (1986) An improved colorimetric assay for interleukin 2. J. Immunol. Methods 93, 157–165 [DOI] [PubMed] [Google Scholar]
- 31. Naiki H., Higuchi K., Hosokawa M., Takeda T. (1989) Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal. Biochem. 177, 244–249 [DOI] [PubMed] [Google Scholar]
- 32. Biancalana M., Koide S. (2010) Molecular mechanism of thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta 1804, 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crespo R., Rocha F. A., Damas A. M., Martins P. M. (2012) A generic crystallization-like model that describes the kinetics of amyloid fibril formation. J. Biol. Chem. 287, 30585–30594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davidson W. S., Arnvig-McGuire K., Kennedy A., Kosman J., Hazlett T. L., Jonas A. (1999) Structural organization of the N-terminal domain of apolipoprotein A-I: studies of tryptophan mutants. Biochemistry 38, 14387–14395 [DOI] [PubMed] [Google Scholar]
- 35. Lindgren M., Hammarström P. (2010) Amyloid oligomers: spectroscopic characterization of amyloidogenic protein states. FEBS J. 277, 1380–1388 [DOI] [PubMed] [Google Scholar]
- 36. Lindgren M., Sörgjerd K., Hammarström P. (2005) Detection and characterization of aggregates, prefibrillar amyloidogenic oligomers, and protofibrils using fluorescence spectroscopy. Biophys. J. 88, 4200–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yanagi K., Ashizaki M., Yagi H., Sakurai K., Lee Y. H., Goto Y. (2011) Hexafluoroisopropanol induces amyloid fibrils of islet amyloid polypeptide by enhancing both hydrophobic and electrostatic interactions. J. Biol. Chem. 286, 23959–23966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003) Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 39. Fändrich M. (2012) Oligomeric intermediates in amyloid formation: structure determination and mechanisms of toxicity. J. Mol. Biol. 421, 427–440 [DOI] [PubMed] [Google Scholar]
- 40. Bieschke J., Russ J., Friedrich R. P., Ehrnhoefer D. E., Wobst H., Neugebauer K., Wanker E. E. (2010) EGCG remodels mature α-synuclein and amyloid-β fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 107, 7710–7715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Di Gaetano S., Guglielmi F., Arciello A., Mangione P., Monti M., Pagnozzi D., Raimondi S., Giorgetti S., Orrù S., Canale C., Pucci P., Dobson C. M., Bellotti V., Piccoli R. (2006) Recombinant amyloidogenic domain of ApoA-I: analysis of its fibrillogenic potential. Biochem. Biophys. Res. Commun. 351, 223–228 [DOI] [PubMed] [Google Scholar]
- 42. Ramella N. A., Rimoldi O. J., Prieto E. D., Schinella G. R., Sanchez S. A., Jaureguiberry M. S., Vela M. E., Ferreira S. T., Tricerri M. A. (2011) Human apolipoprotein A-I-derived amyloid: its association with atherosclerosis. PLoS One 6, e22532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petrlova J., Duong T., Cochran M. C., Axelsson A., Mörgelin M., Roberts L. M., Lagerstedt J. O. (2012) The fibrillogenic L178H variant of apolipoprotein A-I forms helical fibrils. J. Lipid Res. 53, 390–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hatters D. M., Howlett G. J. (2002) The structural basis for amyloid formation by plasma apolipoproteins: a review. Eur. Biophys. J. 31, 2–8 [DOI] [PubMed] [Google Scholar]
- 45. Teoh C. L., Griffin M. D., Howlett G. J. (2011) Apolipoproteins and amyloid fibril formation in atherosclerosis. Protein Cell 2, 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Uversky V. N., Fink A. L. (2004) Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim. Biophys. Acta 1698, 131–153 [DOI] [PubMed] [Google Scholar]
- 47. Chetty P. S., Ohshiro M., Saito H., Dhanasekaran P., Lund-Katz S., Mayne L., Englander W., Phillips M. C. (2012) Effects of the iowa and milano mutations on apolipoprotein A-I structure and dynamics determined by hydrogen exchange and mass spectrometry. Biochemistry 51, 8993–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zaiou M., Arnold K. S., Newhouse Y. M., Innerarity T. L., Weisgraber K. H., Segall M. L., Phillips M. C., Lund-Katz S. (2000) Apolipoprotein E;-low density lipoprotein receptor interaction. Influences of basic residue and amphipathic α-helix organization in the ligand. J. Lipid Res. 41, 1087–1095 [PubMed] [Google Scholar]
- 49. Vacca R. A., Giannattasio S., Graber R., Sandmeier E., Marra E., Christen P. (1997) Active-site Arg → Lys substitutions alter reaction and substrate specificity of aspartate aminotransferase. J. Biol. Chem. 272, 21932–21937 [DOI] [PubMed] [Google Scholar]
- 50. Hristova K., Wimley W. C. (2011) A look at arginine in membranes. J. Membr. Biol. 239, 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. White S. H., Wimley W. C. (1998) Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta 1376, 339–352 [DOI] [PubMed] [Google Scholar]
- 52. Shukla D., Trout B. L. (2010) Interaction of arginine with proteins and the mechanism by which it inhibits aggregation. J. Phys. Chem. B 114, 13426–13438 [DOI] [PubMed] [Google Scholar]
- 53. Hill S. E., Miti T., Richmond T., Muschol M. (2011) Spatial extent of charge repulsion regulates assembly pathways for lysozyme amyloid fibrils. PLoS One 6, e18171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Frare E., Polverino De Laureto P., Zurdo J., Dobson C. M., Fontana A. (2004) A highly amyloidogenic region of hen lysozyme. J. Mol. Biol. 340, 1153–1165 [DOI] [PubMed] [Google Scholar]