Background: RapA proteins promote bacterial aggregation and are composed of Ra/CHDL domains similar to the extracellular domains of eukaryotic cadherins.
Results: RapA2 shows a calcium-dependent cadherin-like β-sheet conformation and specifically recognizes acidic exopolysaccharides.
Conclusion: RapA2 is an extracellular calcium binding lectin.
Significance: RapA2 biological role sheds light on the biochemical function of Ra/CHDL domains found in numerous predicted extracellular or surface-exposed proteins.
Keywords: Bacteria, Calcium, Carbohydrate Binding Protein, Lectin, Polysaccharide, Rhizobium, Cadherin-like Domain (CHDL)
Abstract
In silico analyses have revealed a conserved protein domain (CHDL) widely present in bacteria that has significant structural similarity to eukaryotic cadherins. A CHDL domain was shown to be present in RapA, a protein that is involved in autoaggregation of Rhizobium cells, biofilm formation, and adhesion to plant roots as shown by us and others. Structural similarity to cadherins suggested calcium-dependent oligomerization of CHDL domains as a mechanistic basis for RapA action. Here we show by circular dichroism spectroscopy, light scattering, isothermal titration calorimetry, and other methods that RapA2 from Rhizobium leguminosarum indeed exhibits a cadherin-like β-sheet conformation and that its proper folding and stability are dependent on the binding of one calcium ion per protein molecule. By further in silico analysis we also reveal that RapA2 consists of two CHDL domains and expand the range of CHDL-containing proteins in bacteria and archaea. However, light scattering assays at various concentrations of added calcium revealed that RapA2 formed neither homo-oligomers nor hetero-oligomers with RapB (a distinct CHDL protein), indicating that RapA2 does not mediate cellular interactions through a cadherin-like mechanism. Instead, we demonstrate that RapA2 interacts specifically with the acidic exopolysaccharides (EPSs) produced by R. leguminosarum in a calcium-dependent manner, sustaining a role of these proteins in the development of the biofilm matrix made of EPS. Because EPS binding by RapA2 can only be attributed to its two CHDL domains, we propose that RapA2 is a calcium-dependent lectin and that CHDL domains in various bacterial and archaeal proteins confer carbohydrate binding activity to these proteins.
Introduction
Rhizobia are soil bacteria that establish symbiotic interactions with leguminous plants, inducing nitrogen-fixing root nodules under conditions of nitrogen starvation. Rhizobia are able to colonize several and diverse niches such as the bulk soil, the rhizosphere, and the vicinity of plant tissues. It was proposed that the ability of rhizobia to form different types of biofilms on a variety of surfaces might contribute to their ability to colonize (1). For example, the biofilm formed by Rhizobium leguminosarum under laboratory conditions on abiotic surfaces may reflect a survival mechanism on soil particles (1, 2). The acidic exopolysaccharide (EPS)2 and a functional type I secretion system PrsDE were found to be important for the development of a mature biofilm (3, 4).
PrsDE is responsible for the secretion of a family of proteins with affinity for the rhizobial cell surface named as “Rap” for rhizobium adhering proteins (3, 5, 6). Rap proteins contain one or several so-called “Ra” (Rhizobium-adhering) domains, which consist of 100–120 amino acids with a high degree of similarity displaying at least 20 conserved residues. Rap proteins include the RapA1 and RapA2 paralogues that were proposed as agglutinins and others of unknown function such as RapB, RapC, and RL2702 (herein RapD) (5, 6). Furthermore, the EPS-glycanases PlyA, PlyB, and PlyC, secreted by the PrsDE system, also contain a Ra domain in their C terminus and can thus also be considered as members of the Rap family (5, 7).
The smallest members of the Rap family (∼25 kDa) are RapA1 and RapA2, composed solely by two Ra domains. RapA1 is an extracellular calcium-binding protein with affinity for the cellular surface of R. leguminosarum biovars and Rhizobium etli. Remarkably, RapA1 binds to the cell surface at one pole. Exogenously added RapA1 caused aggregation of R. leguminosarum bv. trifolii cells, which was inhibited by incubation with a crude EPS preparation of the same strain (6). Therefore, it was argued that the surface receptor of RapA1 may be some structure of the acidic EPS. RapA1 overproduction increased the attachment of R. leguminosarum R200 cells to clover roots (8), suggesting its involvement in colonization or biofilm formation.
Interestingly, the C-terminal Ra domain of RapA1 has been identified as a cadherin-like domain (CHDL) that is also found in many other proteins from taxonomically diverse bacteria (9). Cadherins are a family of eukaryotic calcium-binding proteins that contain characteristic repeat sequences (cadherin domains) in their extracellular regions and are responsible for the initiation and maintenance of cell-cell contacts through a calcium-dependent mechanism (10). The bacterial CHDL domains (∼110 amino acids) were predicted to have an immunoglobulin β-sandwich-fold with Greek-key topology, similar to the characteristic fold of eukaryotic cadherins (9). The CHDL domains were often found in association with other protein domains encoding enzymatic activities, substrate binding, or other functions (9, 11). It has been proposed that CHDL domains may confer adhesion functions via protein-protein interactions (12, 13) and/or carbohydrate binding ability (9, 11). The predicted structural similarity to cadherins (9, 11) and the documented calcium binding and cellular autoaggregation properties of RapA1 (6) might lead to a straightforward hypothesis that RapA proteins mediate cellular agglutination by a cadherin-like mechanism. In this scenario, RapA2 with its two tandem Ra/CHDL domains, would mediate homophilic protein-protein interactions between Ra/CHDL domains on the surface of two neighboring cells (trans interactions).
Here we further expand the set of bacterial proteins harboring a CHDL domain, emphasizing the need to understand the function of this broadly conserved protein domain. To gain insight into the function of the bacterial CHDL domain, we have structurally characterized RapA2 from R. leguminosarum bv. viciae strain 3841 (14, 15). We show that RapA2 is structurally similar to cadherins and propose that all homologous Ra domains are bacterial CHDL domains. However, biophysical evidence strongly suggests that RapA2 does not mediate cell-cell contacts via calcium-dependent protein-protein interactions as is the case for cadherins. Instead, we demonstrate that RapA2 directly binds to EPS/capsular polysaccharides (CPS) and that calcium modulates both RapA2 folding and its carbohydrate binding ability. Our findings suggest that other bacterial CHDL domains would confer carbohydrate binding properties to their cognate proteins.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
R. leguminosarum bv. viciae strains 3841 (16, 17) and A412 (A34 prsD1::Tn5) were used in this study (18). R. leguminosarum bv. trifolii strain R200 is a spontaneous mutant of strain 2046 (6). R. leguminosarum was grown at 28 °C in TY (19) or in Y minimal medium (20) supplemented with 0.2% w/v mannitol and antibiotics streptomycin (400 μg/ml) or kanamycin (30 μg/ml). Sinorhizobium meliloti strain Rm 1021 (21) was grown on Yeast Mannitol medium (YM) (22) agar plates or in glutamate-mannitol-salts (GMS) medium (23) supplemented with biotin, thiamine, trace elements, and streptomycin (500 μg/ml). Escherichia coli BL21(DE3) was grown at 37 °C in Luria-Bertani (LB) medium supplemented with ampicillin (200 μg/ml).
Cloning and Construction of Recombinant Strains
The open reading frames of RapA2 (pRL100541) and RapB (RL3911) were PCR-amplified with sense and antisense primers containing sites for NdeI and XhoI restriction endonucleases, respectively using genomic DNA of R. leguminosarum bv. viciae strain 3841. The amplified fragments were cloned into the pET22b vector resulting in constructs expressing C-terminal histidine-tagged RapA2 and RapB. Primer sequences were as follows: 5′-GAGGAGAGTCATATGGCTTCGCC-3′ and 5′-GCTGATCACTCGAGAGCGTCGGTGC-3′ for RapA2 and 5′-GAAAATCACACATATGGTAGAAATG-3′ and 5′-CCCTTCGCTCGAGGATGAAATG-3′ for RapB. The constructs were verified by sequencing of the complete open reading frame.
Protein Production and Purification
Production of RapA2 and RapB by E. coli BL21(DE3) was induced by the addition of 0.5 mm isopropylthiogalactoside to cultures with an absorbance of 0.8 and incubation for 3 h. Harvested cells were resuspended in a buffer containing 20 mm Tris-HCl, 0.5 m NaCl, pH 8.0, with the addition of 1 mm phenylmethanesulfonyl fluoride and 10 mm imidazole. Cells were disrupted in a French pressure cell at 18,000 p.s.i. The cell extract was centrifuged at 100,000 × g for 1 h. The supernatant was applied to a Ni-NTA HiTrap Chelating column (GE Healthcare). Fusion proteins were eluted with an imidazole gradient (10–500 mm) in 20 mm Tris-HCl, 0.5 m NaCl, pH 8.0. Fractions containing RapA2 or RapB were concentrated by ultrafiltration and applied to Superdex-75 HR 10/30 column (Pharmacia Corp.) pre-equilibrated in 20 mm Tris-HCl and 150 mm NaCl, pH 8.0. Decalcified protein preparations were obtained by incubation with 10 mm EGTA on ice for 1 h and re-purification by size exclusion chromatography. Protein yield was ∼10 mg of RapA2 and 5 mg of RapB per liter of bacterial culture and purity greater than 98% for both proteins as judged by SDS-PAGE.
Sequence Analysis and Protein Structure Prediction
PSI-BLAST (24) searches were performed using inclusion thresholds E < 10−5 with a maximum of 20,000 target sequences against the NCBI non-redundant protein database. Multiple sequence alignments were performed using the MUSCLE program (25) at EMBL-EBI using default settings. The resulting alignments were colored, and the consensus was calculated using the Chroma program (26). Secondary structure prediction and protein fold recognition of RapA2 were performed using the 3D-Jury MetaServer in which results of various prediction servers are jointly evaluated (27). Four templates with the highest 3D-Jury scores (Protein Data Bank codes 1edh, 2a4e, 2a62, and 2o72 for mouse E-cadherin, mouse cadherin-11, mouse cadherin-8, and human E-cadherin, respectively) were chosen to predict the tertiary structure of RapA2 using the Deep View-SwissPdbViewer program version 4.0.4 and the automated mode of the Swiss Model workspace.
Spectroscopic Measurements
All measurements were done with protein samples dissolved in 20 mm Tris-HCl and 150 mm NaCl, pH 8.0, with the addition of CaCl2 at the indicated concentration. Far-UV circular dichroism (CD) measurements were carried out at 25 °C on a Jasco J-815 spectropolarimeter. The protein was diluted to 7 μm (0.18 mg/ml) and placed in a cell with a 1-mm path length. Spectra were acquired over the wavelength range of 196–260 nm. Thermally induced denaturation of RapA2 at varying calcium concentrations was followed by CD spectroscopy. Measurements were performed in a Peltier-thermostatted cell holder using a 1-mm path length cell. The spectra of RapA2 with or without added calcium were recorded as a function of temperature ranging from 15 to 95 °C with a 1 °C/min thermal gradient and a spectrum recorded every 5 °C. The reversibility of protein unfolding was examined by scanning the same sample and recording the CD spectra after rapid cooling to 15 °C. Spectra in the near-UV region (260–320) were recorded at 25 °C in a 10-mm path cell using a protein concentration of 75 μm and various concentrations of CaCl2. Each spectrum was the average of at least eight scans to reduce noise. Binding of 8-anilinonaphthalene-1-sulfonic acid (ANS) to RapA2 was measured with a JASCO FP-6500 spectrofluorimeter using 50 μm ANS, 75 μm RapA2, and 0.5 or 1 mm CaCl2. The excitation wavelength was set at 370 nm, and spectra were recorded from 400 to 600 nm at 25 °C.
Isothermal Titration Calorimetry
A VP-ITC calorimeter (Microcal Inc.) was used for calcium binding studies, and data were analyzed using the software Origin 7.0 supplied by Microcal. Ligand and protein solutions were prepared in 20 mm Tris-HCl and 150 mm NaCl, pH 8.0. Titration was carried out at 25 °C. RapA2 (400 μm) in the sample cell was titrated with 33 injections, 8 μl each of CaCl2 (10 mm). The injections were made at 180-s intervals. A control experiment was carried out to measure the ligand dilution heat that was subtracted from the calcium binding thermogram before data analysis.
Light Scattering
A Precision Detector PD2010 light-scattering instrument connected to an FPLC system and a LKB 2142 differential refractometer were used. RapA2 (300 μl, 1.5 mg/ml) was loaded on a Superdex 75 HR 10/30 column; size exclusion chromatography was performed at 0.4 ml/min in buffer composed of 20 mm Tris-HCl and 150 mm NaCl, pH 8.0, with CaCl2 added at 0.1 or 4 mm. The 90° light scattering, refractive index, and absorbance of the eluting material were recorded and analyzed with the Discovery32 software supplied by Precision Detectors. The 90° light scattering detector was calibrated using BSA as a standard.
For dynamic light scattering (DLS) studies, the protein was dissolved in 20 mm Tris-HCl and 150 mm NaCl pH 8.0 with the addition of the indicated concentration of CaCl2 (0–4 mm). DLS was measured with a Malvern Nanos S instrument model ZEN 1600. Diameter distribution was calculated from the average of 20 measurements.
Native Gel Electrophoresis
Native polyacrylamide gel electrophoresis was carried out according to Laemmli et al. (28) but without the addition of SDS or reducing agent to the standard solutions. When needed, 2 mm CaCl2 was added to the gel and to the running buffer. RapA2 and RapB (20 μg each) were preincubated alone or in a mixture in PBS buffer for 1 h at room temperature in the presence or absence of 2 mm CaCl2.
Preparation and Quantification of Extracellular Polysaccharides
CPS and EPS of R. leguminosarum strains were obtained from cells and culture supernatants, respectively, from cultures grown for 2 days in Y-mannitol minimal medium. To isolate CPS, the cell pellets were washed gently with 20 mm Tris-HCl, pH 7.0, and 1 mm MgSO4 followed by vigorous stirring in a washing buffer containing 0.5 m NaCl for 1 h at room temperature and centrifugation at 10,000 × g for 1 h. CPS in the supernatant was precipitated with 2 volumes of cold ethanol, recovered by centrifugation at 10,000 × g for 30 min, and subsequently washed with increasing concentrations of ethanol (70–90% v/v). The samples were air-dried, resuspended in sterile water, and stored at −20 °C. EPS was precipitated from culture supernatants by the addition of two volumes of cold ethanol followed by centrifugation. Washing and subsequent treatments were as described for CPS preparations. Succinoglycan was obtained from a 4-day culture of S. meliloti in GMS medium. Cells were removed by centrifugation, and succinoglycan was precipitated from culture supernatants by the addition of 3 volumes of ethanol. Quantification of polysaccharides was done by determination of hexoses by the sulfuric acid-anthrone method (29) and determination of hexuronic acids by the meta-hydroxybiphenyl method (30). Proteinase K treatment of acidic EPS and CPS was done according to Jefferson and Cerca (31). Briefly, samples of R. leguminosarum acidic EPS or CPS (1 mg/ml) were incubated with proteinase K (2 mg/ml) in a 2-ml volume. Proteinase was heat-inactivated at 80 °C for 30 min. Then treated polysaccharides were precipitated with ethanol, exhaustively washed, dried, and resuspended in sterile water.
Binding Assays
Purified RapA2 (40 μg) was mixed with polysaccharide suspensions containing 0.5 mg of EPS or CPS in 200 μl of PBS. Incubations were carried out at room temperature with gentle agitation for 20 min. Polysaccharides (0.25% in mixture reactions) were separated from supernatant by centrifugation at 100,000 × g for 1.5 h as described for xanthan gum (32). The efficacy of EPS sedimentation at 100,000 × g was confirmed by quantification of hexuronic acids in both fractions. Pellets were washed once with PBS, and the presence of protein in the pellet and supernatant fractions was assessed by SDS-PAGE. The binding of RapA2 to the different polysaccharides was characterized by a modified ELISA technique. Flat-bottomed Microlon 600 microplates (Greiner) were coated with 100 μl of the EPS suspension in coating buffer (50 mm sodium carbonate, pH 9.7) at 4 °C overnight. Binding of RapA2 (50 ng/ml in PBS, 0.05% Tween 20, and 0.5 mm CaCl2) was detected with a monoclonal anti-polyhistidine antibody followed by an alkaline phosphatase-conjugated secondary antibody. 4-Nitrophenyl phosphate was used as substrate in the phosphatase reaction. For binding inhibition assays RapA2 was preincubated for 30 min at room temperature with serial 10-fold dilutions of the putative inhibitory compound before the addition to the wells of the microplates. The initial concentration of the inhibiting polysaccharides was 100 μg/ml. The mono-/disaccharides were prepared as 200 mm stocks in PBS. The pH of the glucuronic acid (GlcA) stock was adjusted to 7.0. The effect of calcium on RapA2 binding to EPS was evaluated by preincubation of the protein with increasing concentrations of EGTA before the addition to the EPS-coated wells. The protective effect of MgCl2 on EPS was jointly evaluated in the same experiments. The data represent the mean values of a representative experiment of three independent assays done in quadruplicate.
RESULTS
RapA2 Is Predicted to Be Composed by Two Conserved Cadherin-like Domains
To update the number of proteins harboring a domain similar to Ra/CHDL, a PSI-BLAST search in the NCBI non-redundant protein database was performed using the second Ra domain of R. leguminosarum bv. viciae 3841 RapA2 (encoded by gene pRL100451) as the sequence query (residues 118–236). After three iterations, 405 sequences were retrieved with an E value <10−5 from a variety of taxonomic groups. The most populated were the Proteobacteria (260 proteins) and Cyanobacteria (66 proteins) groups, but also the Bacteroidetes, Planctomycetes, Actinobacteria, Firmicutes, Green sulfur bacteria, and Verrumicrobia were represented as well as the eukaryotes and archaea. Multiple sequence alignments with representative homologues from each group were constructed using the MUSCLE program (25) (supplemental Fig. S1). This analysis revealed the presence of several conserved residues likely important for structure and function. Furthermore, in some large proteins the presence of consecutive repetitions of Ra/CHDL was detected, as in a putative exported RTX (repeat in toxin) protein from Ralstonia eutropha (YP_841825, 2426 amino acids), which contains 5 repetitions of the domain.
Protein folding prediction of RapA2 from strain 3841 (1–236) was performed using the Metaserver (27) and the Deep View program (33). The template structures used for RapA2 modeling were two type I: the N-terminal domains 1 and 2 of mouse E-cadherin (PDB code 1edh) and the human E-cadherin (PDB code 2o72), and two type II cadherins: the mouse cadherin-8 (PDB code 2a62) and mouse cadherin-11 (PDB 2a4e), from which the higher 3D-Jury scores were obtained. According to this model, RapA2 may comprise two homologous Ra/CHDL domains (with 57% identity between them) composed mostly of β-sheets joined by a linker region (supplemental Fig. S2). Despite the limited sequence similarity between RapA2 and cadherin domains, the structural alignment suggests a clear resemblance of the overall topology of RapA2 and cadherin domains, as they share the length and relative position of several β strands and amino acid residues (Fig. 1).
FIGURE 1.
Structural alignment of RapA2 and eukaryotic cadherins. The amino acid sequence of RapA2 was aligned to template sequences according to the MetaServer predictions. The template structures and their PDB entries were: ectodomains 1 and 2 (EC1–2) of mouse E-cadherin (PDB code 1edh), human E-cadherin EC1–2 (PDB 2o72), mouse cadherin-8 EC1–3 (PDB code 2a62), and mouse cadherin-11 EC1–2 (PDB code 2a4e). The predicted RapA2 secondary structure is from SWISS-MODEL, and the template structures were from the Protein Data Bank. Secondary structure elements are shown as a gray background (light gray for β-strands, dark gray for α-helices). Conserved residues are in bold, asterisks denote identical residues, and circles indicate similar residues. The conserved calcium binding motifs in cadherins (DXNDN, DXD and LDRE) are underlined in the sequence of PDB code 1edh. The acidic amino acids of RapA2 mutated to alanine are shown in italics and indicated with arrows.
Calcium Triggers Conformational Change of RapA2
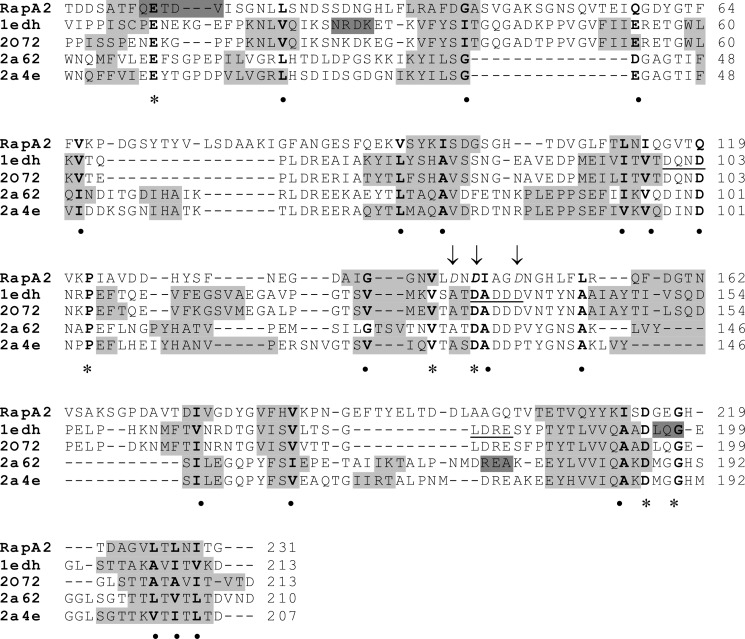
To experimentally verify the predicted structural similarity of RapA2 to cadherins, we studied its secondary structure in the presence and absence of calcium. Recombinant RapA2 was overexpressed as a soluble protein in E. coli and purified to homogeneity. The secondary structure of RapA2 was studied by CD spectroscopy. In the absence of calcium the far-UV CD spectrum of RapA2 showed a minimum near 210 nm (Fig. 2A). This spectrum is likely a mixture of unstructured regions with some elements of β-sheet structure. Interestingly, upon the addition of calcium in increasing concentrations (0–600 μm), a broad negative band centered at 216 nm appeared in the spectrum of RapA2, indicative of β-sheet conformation. Ellipticity values at 208 nm measured for each calcium concentration are shown in Fig. 2B. An apparent equilibrium dissociation constant (KD) of 40 μm was estimated.
FIGURE 2.
Effect of calcium on RapA2 secondary structure. A, shown are far-UV CD spectra at increasing calcium concentrations, ranging from 0 to 600 μm. B, shown is molar ellipticity at 208 nm measured at increasing calcium concentrations. MRW, mean residue weight.
Near-UV CD spectroscopy (250–350 nm) is dominated by aromatic residues. The spectra of RapA2 possessing Phe and Tyr but no Trp residues showed a broad positive signal (268–290 nm), and a decrease in ellipticity was seen upon the addition of calcium at 150, 300, and 400 μm (data not shown), pointing to a change in RapA2 tertiary structure.
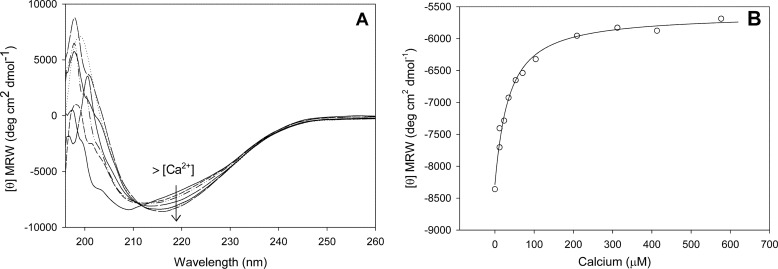
The presence of solvent-exposed hydrophobic clusters was examined by ANS binding (Fig. 3). Binding of ANS to RapA2 was higher in the absence of calcium, showing that calcium induces a conformational change that reduces the exposition of hydrophobic clusters.
FIGURE 3.
ANS binding to RapA2 at increasing calcium concentrations. Intensity of fluorescence emission of ANS when bound to RapA2 at 0, 0.5 or 1 mm calcium is shown. AU, arbitrary units.
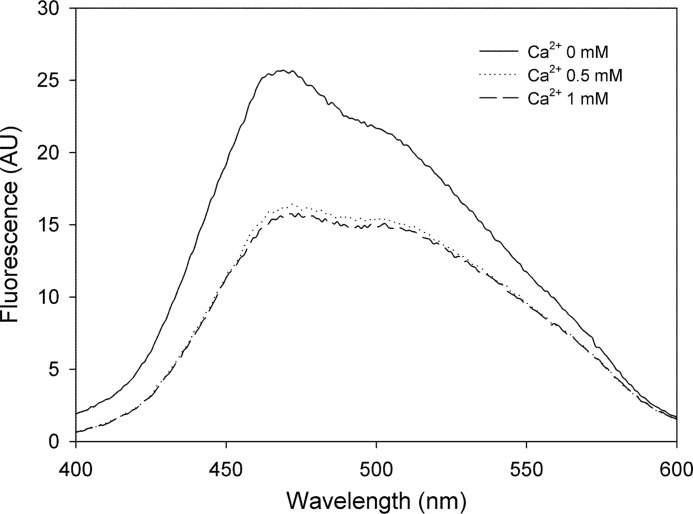
To further investigate the role of calcium on RapA2 folding, the thermal stability of the protein in the presence or absence of calcium was analyzed. The apo (RapA2)- and holo-form (RapA2-Ca2+) were submitted to increasing temperatures, and their loss of secondary structure were followed by the measured ellipticity at 209 nm (Fig. 4A). RapA2 incubated without added calcium was marginally stable, showing a non-cooperative unfolding transition with a midpoint (Tm) near 38 °C. On the contrary, calcium-bound RapA2 was far more stable, displaying a highly cooperative unfolding transition with a Tm of 50 °C. This result shows that the apo-form of RapA2 is sensitive to subtle changes in temperature, whereas in the presence of calcium it adopts a compact stable conformation. Thermal stability curves denote a calcium-dependent cooperative folding for RapA2, suggesting a one-step process.
FIGURE 4.
Analysis of the effect of calcium on RapA2 thermal stability. A, far-UV CD spectra of RapA2 were recorded at temperatures ranging from 0 to 95 °C. The ellipticity at 209 nm of RapA2 in the absence (apo-form, line) or in the presence of 0.5 mm CaCl2 (holo-form, dashed line) is depicted at different temperatures. B, the reversibility of thermal-induced denaturation of the apo- and holo-forms of RapA2 was determined. Far-UV CD spectra were recorded at 15 °C and 95 °C, and after cooling to 15 °C the samples submitted to 95 °C.
Thermal denaturation of the apo and holo states of RapA2 followed by cooling showed that both forms recover some of their original conformation (Fig. 4B). In the presence of calcium, RapA2 almost recovered its native structure, showing that unfolding is a reversible process in an adequate environment.
Binding of other divalent cations to RapA2 was also explored. The addition of increasing zinc concentration (supplemental Fig. S3A) did not modify the position of the CD spectrum, but an increase in ellipticity was detected at 20 μm ZnSO4, probably due to protein aggregation, as judged by the observation of sample turbidity. On the other hand, adding magnesium had no effect on the secondary structure of RapA2 at any of the concentrations tested (10 μm–2 mm MgCl2) (supplemental Fig. S3B).
Altogether these results show that in the absence of calcium RapA2 displays a conformation similar to a molten globule state, whereas folding of the protein into a compact structure depends specifically on the presence of calcium ions.
Calcium Binding Analysis by Isothermal Titration Calorimetry (ITC)
It has been proposed that in eukaryotic cadherins three calcium ions bind close to the linker between cadherin domains (binding sites underlined in Fig. 1). A single binding site model was fitted to the data obtained from calcium titration of RapA2 by CD (Fig. 2B). However, it should be noted that these data are also compatible with a stoichiometry of two bound calcium per protein molecule, with both binding sites displaying similar affinity. To obtain the stoichiometry (n) of the binding reaction and an accurate determination of binding constants, we used ITC. A one-set of site model could be accurately fitted to the calcium binding isotherm of RapA2 (Fig. 5). RapA2 binds one calcium ion with a dissociation constant of 177 μm. The binding reaction proceeds with a negative enthalpy change (ΔH) and negative entropy (ΔS) (Table 1).
FIGURE 5.
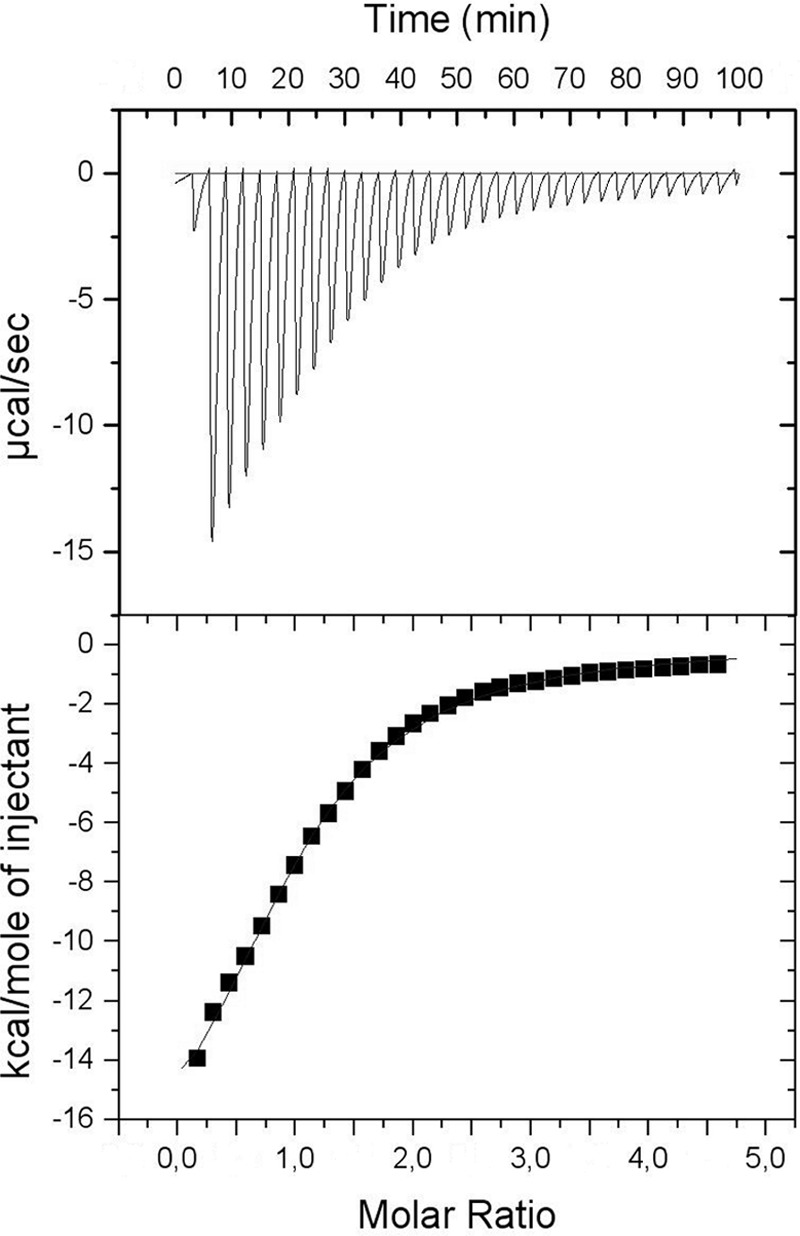
Isotherm of calcium binding to RapA2 measured by ITC. Protein and ligand were prepared in 20 mm Tris-HCl, 150 mm NaCl, pH 8.0. RapA2 (400 μm) in the sample cell was titrated with CaCl2 (10 mm). The upper panel shows the observed heats for each injection of CaCl2 after base-line correction. The lower panel depicts the binding enthalpies versus the calcium/protein molar ratio. The data (■) best fitted to the one set of site binding model. Best-fit parameters are listed in Table 1.
TABLE 1.
Thermodynamic parameters of calcium binding to RapA2
| Model | n | KA | KD | ΔH | ΔS |
|---|---|---|---|---|---|
| m−1 | μm | kcal·mol−1 | cal·mol−1·K−1 | ||
| One set of site | 0.99 ± 0.01 | (5.6 ± 0.2) × 10−3 | 177 | −20.7 ± 0.5 | −52.3 |
From the calcium binding sites of cadherin domains, only the central site is partially conserved in the second Ra domain of RapA2 (Fig. 1) and other bacterial CHDL domains (supplemental Fig. S1). To explore the role of the site shared with cadherins in the binding of calcium, the Asp residues located in the 145–149 cluster and a neighboring Asp (Asp-143) were mutated to Ala (pointed in Fig. 1). The spectra of these proteins in the absence of added calcium were similar to that of the parent protein, but in all cases (including a triple mutant) calcium addition caused a change toward a more structured β-sheet conformation (not shown). These observations suggest that in the Ra/CHDL domains other sites/residues are implicated in calcium binding.
Calcium Has No Effect on the Oligomerization State of RapA2
The requirement of calcium for proper folding of RapA2 is in agreement with its predicted cadherin-like structure. Next we explored whether the previously documented autoaggregative properties of RapA proteins also depend on a cadherin-like adhesion mechanism. Cadherins mediate cell-cell contact through rigidification followed by dimerization of their extracellular domains, and these processes are promoted and maintained by bound calcium ions (34, 35).
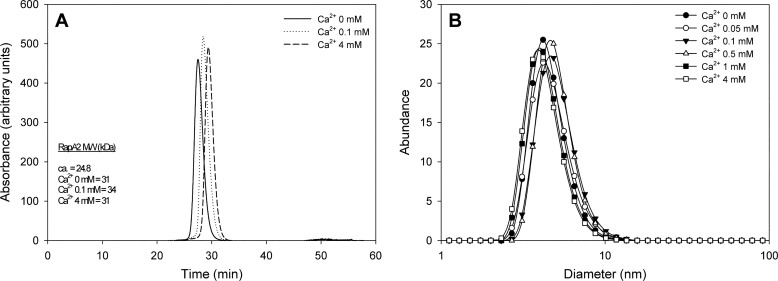
To determine the oligomerization state of RapA2 at different calcium concentrations we first employed size exclusion chromatography and static light scattering (Fig. 6A). Purified RapA2 (with or without added calcium) was passed through a gel filtration column, a light scattering detector, and a refractive index detector. The oligomeric state of RapA2 was revealed by the calculated native molecular weight. We observed that, in contrast to cadherins, calcium does not induce RapA2 oligomerization. Moreover, the increase of the size exclusion chromatography retention time in the presence of calcium suggests that the ion induces a more compact structure of the protein. The protein samples were further analyzed by DLS to rule out the possible loss of protein oligomers within the chromatographic column used in size exclusion chromatography-static light scattering. DLS experiments showed the presence of only one species of RapA2 in solution, with a diameter distribution centered at 4.19 nm regardless of the calcium concentration employed (Fig. 6B).
FIGURE 6.
Hydrodynamic properties and diameter distribution of RapA2 at different CaCl2 concentrations. A, shown is molecular mass of RapA2 at various CaCl2 concentrations obtained by size exclusion chromatography and static light scattering. The theoretical and experimentally determined Mr are indicated. B, diameter distribution of RapA2 was measured by DLS at CaCl2 concentrations ranging from 0 to 4 mm.
These results show that even though calcium is needed for RapA2 folding and stability, it has no effect on the formation of higher order oligomers. Thus, the previously reported cell-agglutinating property of RapA proteins (6) is not likely mediated by RapA oligomerization.
The possibility that RapA2 interacts with other members of the Rap family through Ra/CHDL domains was also investigated. RapB (RL3911, 30 kDa), which bears a Ra domain in its N terminus, was expressed and purified to homogeneity. RapA2 and RapB (20 μg each) were incubated alone or in a mixture without calcium or supplemented with 2 mm CaCl2 followed by native gel electrophoresis (supplemental Fig. S4). We did not detect protein complex formation in any of the conditions tested, suggesting that in vitro, RapA2 and RapB are not able to establish protein-protein interactions.
RapA2 Specifically Recognizes Acidic Exopolysaccharides with Identical or Similar Structures
Because a cadherin-like protein-protein interaction mechanism to explain the role of Rap proteins in adhesion and aggregation was ruled out, the binding of RapA2 to carbohydrate structures produced by rhizobia was studied, as suggested by a previous report (6).
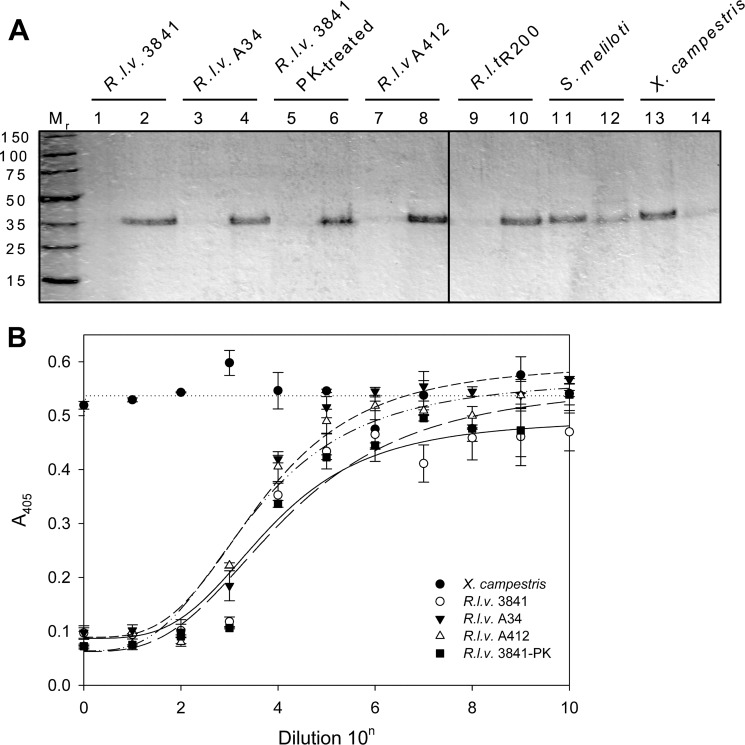
It was shown that the acidic EPS from R. leguminosarum strains is a polymer of octasaccharide repeating units containing glucose (Glc), GlcA, and galactose (Gal) (5:2:1) (36, 37). Purified RapA2 was mixed with the EPS obtained from several closely related wild type strains, such as R. leguminosarum bv. viciae (strains 3841 and A34) and R. leguminosarum bv. trifolii R200. We also assayed the EPS produced by the protein secretion mutant prsD (A412) and a proteinase K-treated EPS from strain 3841. Two other exopolysaccharides were also included in the assay; they were the succinoglycan (polymer of Glc and Gal (7:1)) obtained from S. meliloti Rm1021 and a purified commercial preparation of xanthan gum (composed of Glc, mannose (Man), and GlcA (2:2:1)) produced by Xanthomonas campestris pv. campestris. After 30 min of incubation with RapA2, the polymers were separated by ultracentrifugation and washed, and the presence of RapA2 bound to polysaccharide (in the pellet) or free in solution (in the supernatant) was assessed by SDS-PAGE (Fig. 7A). Most of the RapA2 protein was found to be associated with the EPS fraction obtained from the R. leguminosarum wild type strains 3841, A34, and R200. As we expected, xanthan gum and succinoglycan were not able to retain RapA2. Interestingly, EPS from the prsD mutant strain A412 was able to bind RapA2 despite the lack of secreted Rap and other proteins. Furthermore, EPS from strain 3841 treated with proteinase K was also capable to bind RapA2, showing that there are no other extracellular proteins involved in RapA2-EPS interaction.
FIGURE 7.
Interaction of RapA2 with bacterial soluble EPS. A, binding of RapA2 to different soluble polysaccharides was examined. The protein was added to the EPS preparation, and after sedimentation of the EPS by centrifugation at 100,000 × g, the pellet (even numbers) and supernatant fractions (odd numbers) were assayed for the presence of proteins by SDS-PAGE. B, shown is inhibition of the binding of RapA2 to EPS of R. leguminosarum bv. viciae strain 3841 by different soluble EPS preparations. Inhibitory EPS was serially diluted by a factor of 10, starting with a concentration of 100 μg ml−1. R.l.v., R. leguminosarum bv. viciae; R.l.t., R. leguminosarum bv. trifolii; PK, pretreatment with proteinase K.
The binding of RapA2 to EPS was further analyzed by means of a binding inhibition assay using a modified ELISA technique (38). Microtiter wells coated with EPS of R. leguminosarum bv. viciae 3841 were incubated with RapA2 or with RapA2 preincubated with serial dilutions of a putative inhibitory compound in the presence of 0.5 mm CaCl2. Bound RapA2 was immunodetected using anti-His tag antibodies. EPS from R. leguminosarum strains 3841, A34, and A412 (prsD) or proteinase K-treated EPS produced similar concentration-dependent inhibition of the binding of RapA2 to the immobilized EPS from strain 3841 (Fig. 7B). These results confirm that RapA2 binding is not mediated by other proteins but by a direct recognition of some structure of the EPS. To note, xanthan gum from X. campestris (Fig. 7B) or succinoglycan obtained from S. meliloti (not shown) were not able to inhibit the binding of RapA2 to immobilized EPS, confirming the narrow specificity of RapA2 for exopolysaccharides of R. leguminosarum strains.
It has been shown that EPS and CPS of some strains of R. leguminosarum are composed of the same octasaccharide repeating unit (see above) (36, 37). Some reports accounted for differences in the degree of non-carbohydrate substitutions (39). CPS obtained from strains 3841, A34, or A412 also showed an inhibitory effect similar to that of the EPSs (not shown), indicating that a similar epitope is present on both polysaccharide species.
Next we tested whether the three sugars of the repeating unit (Glc, Gal, GlcA) and others (Man, galacturonic acid, maltose, and lactose) are individually able to inhibit the binding of RapA2 to the EPS at concentrations ranging from 0 to 100 mm (Fig. 8A). Glc or Gal had no effect on the binding of RapA2 to the EPS. Interestingly, GlcA was able to inhibit RapA2 binding to the EPS at concentrations higher than 10 mm, indicating that GlcA residues might be part of the structure recognized by RapA2 on the EPS/CPS. To note, galacturonic acid, which is also an acidic monosaccharide, had no effect on binding. In addition, neither Man or the disaccharides affected the binding to the EPS.
FIGURE 8.
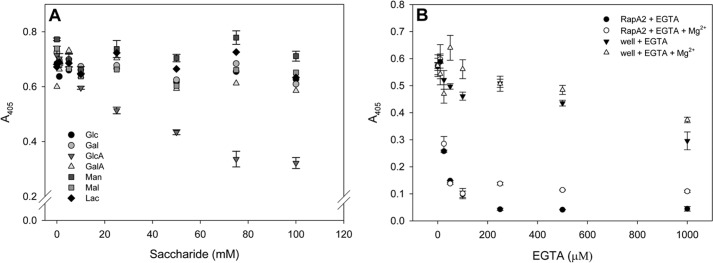
Inhibitory properties of monosaccharides and EGTA on RapA2-EPS interaction. A, inhibition curves of RapA2 binding to immobilized EPS were obtained by preincubation of the protein with increasing concentrations of the indicated sugars ranging from 0 to 100 mm. B, the effect of EGTA on RapA2 binding to the EPS was determined by preincubation of RapA2 with various concentrations of the chelating agent (0–1 mm). As a control, EGTA was also added to the EPS-coated wells before RapA2 addition. In some cases MgCl2 was added at 500 μm to evaluate its effect on the immobilized EPS in the wells.
The effect of calcium on RapA2 binding to immobilized EPS was assessed by preincubation of the protein with various concentrations of EGTA (Fig. 8B). Because EPS binding to the wells can be disrupted by the chelator, immobilized EPS was also incubated with the same concentrations of EGTA before the addition of RapA2. In some cases 500 μm MgCl2 was added to protect the structure of the EPS bound to the wells. The presence of the chelating agent had a negative effect on the stability of bound polysaccharide only at concentrations higher than 500 μm. Interestingly, 25 μm EGTA decreased the binding activity of RapA2 by 50%. At a higher concentration of EGTA, the binding activity was completely abolished. A modest protective effect of MgCl2 was observed, probably due to stabilization of EPS in the wells, as RapA2 was shown not to bind Mg2+ ions by CD spectroscopy. These observations show the strict requirement of calcium for the binding of RapA2 to the EPS and support a regulatory role of calcium on RapA2 activity in vivo.
DISCUSSION
Here we have structurally and functionally analyzed the RapA2 protein, which consists of two homologous, so-called Ra domains and was originally shown to be involved in autoaggregation and adhesion of R. leguminosarum cells (6, 8). Due to the expansion of the sequence databases, it is now clear that the Ra domains are conserved far beyond rhizobia (9). In the present study we show that Ra domains are present not only in phylogenetically very distant bacteria but also in archaea and some eukaryotes (supplemental Fig. S1), underscoring the importance to understand the biological function of a domain present in all kingdoms of life. Interestingly, the C-terminal Ra domain of RapA2 has bioinformatically been shown to possess structural features similar to eukaryotic cadherins and belong to a so-called CHDL domain family (9). Taken that involvement in adhesion and calcium binding have been documented not only for RapA but also for several other proteins in the Rap family, it is rather straightforward to assume that the CHDL domains might function as adhesion modules via a cadherin-like mechanism. Calcium-dependent oligomerization of the CHDL domains would thus be a crucial prerequisite for this function. However, data obtained in this work very clearly show that the RapA2 protein forms neither homo-oligomers (Fig. 6) nor hetero-oligomers with other Rap proteins (supplemental Fig. S4), excluding the possibility of a cadherin-like mechanism for RapA2. This illustrates the need for caution and experimental evidence in assigning a function to structurally similar proteins.
However, RapA-coated beads induced bacterial agglutination and a crude preparation of EPS (that might have contained extracellular proteins) inhibited this effect (6). Here we have rigorously verified the binding of RapA2 to the EPS and shown that it is highly specific. EPS samples that lack proteins secreted by PrsDE or completely devoid of proteins retained RapA2, showing that binding to EPS is not mediated by other proteins. Furthermore, no binding of RapA2 to the succinoglycan from S. meliloti (which also contains Glc and Gal residues) was observed nor to xanthan from X. campestris despite bearing GlcA residues that confer negative charge to the polymer. It seems that RapA proteins recognize a particular combination of sugars and glycosidic linkages. Interestingly, binding of RapA2 to immobilized EPS was partially blocked in the presence of glucuronic acid. A concentration-dependent decrease in binding reaching a 60% inhibition at 75 mm GlcA was observed. It is not surprising that monosaccharide-protein interactions show equilibrium dissociation constants (KD) in the mm range. However, it has been observed that in the biological context, avidity is increased 100–10.000-fold due to the multivalency of the interaction (40). Because the RapA2 protein contains no other elements than the two homologous Ra/CHDL domains, it suggests that each domain constitutes a carbohydrate binding module. This notion is further strengthened by the fact that the PlyA and PlyB glycanases, which are together with the Rap proteins secreted by the PrsDE system (18), also contain a Ra/CHDL domain in their C-terminal end (6, 18). PlyA and PlyB cleave EPS and carboxymethylcellulose only when they are in contact with the cell surface (7, 41). The Ra/CHDL domain in glycanases could be involved in carbohydrate recognition or in positioning the enzyme for optimal cleavage of EPS. The modular organization of Ply glycanases would resemble that of bacterial glycoside hydrolases, which are active on complex polysaccharides (42, 43). These glycosidases exhibit a fibronectin-type III (FN3) module with a β-sheet structure fused to its N terminus. Although the exact function of this FN3 module was not established, it was suggested that it could help in polysaccharide recognition and degradation. Moreover, if Ra domains of all Rap proteins recognize the same structure on the EPS/CPS, RapA binding would inevitably affect EPS characteristics; i.e. favoring or impeding the action of glycanases that determine EPS molecule length. Furthermore, it is tempting to propose that the polar location on the cell surface of RapA proteins is related to the place where EPS/CPS chains emerged immediately after they are synthesized (44). We hypothesize that RapA proteins may contribute to the assembly of the EPS component of the biofilm matrix and that its activity may be coordinated with glycanase secretion and activity on the bacterial surface.
We have also shown that calcium is needed for optimal binding of RapA2 to EPS. In bacteria, dedicated transporters and channels maintain cytosolic free calcium in concentrations ranging from 0.1 to 2 μm (45). This is 1 or 2 orders of magnitude lower than the concentration in the external milieu, which is typically in the millimolar range. Here we have presented evidence that at low calcium concentrations, as those prevailing in the bacterial cytosol, RapA2 displays a molten globule-like conformation, with a non-cooperative thermal unfolding behavior. This is likely to facilitate its secretion by the PrsDE system, as it appears that unfolded or partially folded substrates are required for secretion by type I systems (46, 47). Acquisition of RapA2 native β-sheet conformation is expected to occur in the more favorable extracellular environment, where calcium concentrations outweigh the measured KD of 177 μm. Similar observations have been made for other type I secretion system substrates such as the RTX (repeat in toxin) domain of the CyaA adenylate cyclase toxin of Bordetella pertussis (48) and for a non-RTX-bearing protein such as the HasA hemophore from Serratia marcescens (49). As the transition to a folded conformation is frequently coupled to ligand binding, it is reasonable to assume that RapA2 secretion and folding are coordinated with calcium and EPS binding on the cell surface.
Microcalorimetric titration of RapA2 with calcium returned a negative enthalpy change (ΔH) that denotes the exothermic character of the binding reaction and negative entropy. These data help to explain the conformational change of RapA2 to a more stable conformation, observed upon calcium binding by CD and fluorescence spectroscopy. In cadherins, the binding of three calcium ions to the linker between two cadherin domains induces the change to a β-sheet conformation. Here, we show that unlike cadherins, the stoichiometry of RapA2 is of one bound calcium ion per protein molecule. It seems that binding of only one calcium ion triggers RapA2 conformational change to a compact β-structure.
Of the three specific motifs of cadherins that participate in calcium binding (DXD, LDRE, and DXNDN), only the DXD motif was found to be present in the prokaryotic CHDL domains as a DXDXD motif (9). Using the updated NCBI-nr proteins database and multiple alignments of 65 Ra/CHDL representative sequences, we confirmed these observations. To note, in the RapA proteins, the DXDXD motif is replaced by DXXXD, but an additional Asp residue was found in the vicinity of this motif (Fig. 1). Mutational analysis of these residues and of other acidic residues conserved in CHDL domains (not shown) did not allow us to identify the calcium binding site. As roles for calcium in prokaryotic organisms are just recently emerging, only few types of calcium-binding proteins are known so far in bacteria. These correspond to EF-hand proteins (50), proteins with β-propeller structure (51), and βγ-crystallins (52, 53). RapA2 does not display any of the conserved domains of these proteins; therefore, it is an interesting possibility that RapA proteins and others harboring Ra/CHDL domains may constitute a new class of calcium-binding proteins. X-ray or NMR structure information will be necessary to identify the calcium binding site in RapA2 and CHDL-containing proteins.
The Ra/CHDL domain was found in a considerable number of predicted extracellular proteins from bacteria that have a mutualistic or pathogenic relationship with humans. Mechanisms involving carbohydrate-containing structures have been implicated in the development of biofilms in human tissues (54, 55). Furthermore, bacterial sugar-bearing polymers were shown to play a role in immunomodulation and suppression of inflammatory diseases (56). Therefore, CHDL-containing proteins could be involved in the modulation of the structure of the biofilm matrix, which in turn may influence host colonization or immunomodulation of host response.
In conclusion, the role of RapA2 has been redefined by means of biophysical and biochemical approaches showing that RapA proteins are lectins that display extraordinary structural flexibility and depend on calcium binding for folding and function. Furthermore, the elucidation of RapA2 biological role aids in the definition and the understanding of the biochemical function of Ra/CHDL domains, which are part of many predicted extracellular or surface-exposed proteins.
Acknowledgments
We gratefully acknowledge Marta Bravo and Susana Raffo for DNA sequencing and technical assistance and Dr. Gastón Paris for help with static light scattering experiments. We specially thank Lic. Silvina Salinas for her valuable help with ITC studies.
This work was supported by a grant from Agencia Nacional de Promoción Científica y Tecnológica (FONCYT, PICT 20334; to A. Z.) a grant (PIP 545) from Consejo Nacional de Investigaciones Científicas y Tecnológicas (to A. Z.), a grant from Agencia Nacional de Promoción Científica y Tecnológica (PICT 2010-0376) to J. J. C., and a grant from the Swedish Research Council to N. A.

This article contains supplemental Figs. S1–S4.
- EPS
- exopolysaccharide
- Ra
- rhizobium-adhering
- CPS
- capsular polysaccharide
- ANS
- 8-anilinonaphthalene-1-sulfonic acid
- DLS
- dynamic light scattering
- GlcA
- glucuronic acid
- ITC
- isothermal titration calorimetry
- CHDL
- cadhering-like domain.
REFERENCES
- 1. Downie J. A. (2010) The roles of extracellular proteins, polysaccharides, and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 34, 150–170 [DOI] [PubMed] [Google Scholar]
- 2. Rinaudi L. V., Giordano W. (2010) An integrated view of biofilm formation in rhizobia. FEMS Microbiol. Lett. 304, 1–11 [DOI] [PubMed] [Google Scholar]
- 3. Russo D. M., Williams A., Edwards A., Posadas D. M., Finnie C., Dankert M., Downie J. A., Zorreguieta A. (2006) Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J. Bacteriol. 188, 4474–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams A., Wilkinson A., Krehenbrink M., Russo D. M., Zorreguieta A., Downie J. A. (2008) Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J. Bacteriol. 190, 4706–4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krehenbrink M., Downie J. A. (2008) Identification of protein secretion systems and novel secreted proteins in Rhizobium leguminosarum bv. viciae. BMC Genomics 9, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ausmees N., Jacobsson K., Lindberg M. (2001) A unipolarly located, cell surface-associated agglutinin, RapA, belongs to a family of Rhizobium-adhering proteins (Rap) in Rhizobium leguminosarum bv. trifolii. Microbiology 147, 549–559 [DOI] [PubMed] [Google Scholar]
- 7. Zorreguieta A., Finnie C., Downie J. A. (2000) Extracellular glycanases of Rhizobium leguminosarum are activated on the cell surface by an exopolysaccharide-related component. J. Bacteriol. 182, 1304–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mongiardini E. J., Ausmees N., Pérez-Giménez J., Julia Althabegoiti M., Ignacio Quelas J., López-García S. L., Lodeiro A. R. (2008) The rhizobial adhesion protein RapA1 is involved in adsorption of rhizobia to plant roots but not in nodulation. FEMS Microbiol. Ecol. 65, 279–288 [DOI] [PubMed] [Google Scholar]
- 9. Cao L., Yan X., Borysenko C. W., Blair H. C., Wu C., Yu L. (2005) CHDL. A cadherin-like domain in Proteobacteria and Cyanobacteria. FEMS Microbiol. Lett. 251, 203–209 [DOI] [PubMed] [Google Scholar]
- 10. Nollet F., Kools P., van Roy F. (2000) Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J. Mol. Biol. 299, 551–572 [DOI] [PubMed] [Google Scholar]
- 11. Fraiberg M., Borovok I., Bayer E. A., Weiner R. M., Lamed R. (2011) Cadherin domains in the polysaccharide-degrading marine bacterium Saccharophagus degradans 2-40 are carbohydrate binding modules. J. Bacteriol. 193, 283–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fraiberg M., Borovok I., Weiner R. M., Lamed R. (2010) Discovery and characterization of cadherin domains in Saccharophagus degradans 2-40. J. Bacteriol. 192, 1066–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dickens N. J., Beatson S., Ponting C. P. (2002) Cadherin-like domains in α-dystroglycan, α/ϵ-sarcoglycan, and yeast and bacterial proteins. Curr. Biol. 12, R197–R199 [DOI] [PubMed] [Google Scholar]
- 14. Young J. P., Crossman L. C., Johnston A. W., Thomson N. R., Ghazoui Z. F., Hull K. H., Wexler M., Curson A. R., Todd J. D., Poole P. S., Mauchline T. H., East A. K., Quail M. A., Churcher C., Arrowsmith C., Cherevach I., Chillingworth T., Clarke K., Cronin A., Davis P., Fraser A., Hance Z., Hauser H., Jagels K., Moule S., Mungall K., Norbertczak H., Rabbinowitsch E., Sanders M., Simmonds M., Whitehead S., Parkhill J. (2006) The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7, R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Butcher B. G., Lin Y.-P., Helmann J. D. (2007) The yydFGHIJ operon of Bacillus subtilis encodes a peptide that induces the LiaRS two-component system. J. Bacteriol. 189, 8616–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnston A. W., Beringer J. E. (1975) Identification of the Rhizobium strains in pea root nodules using genetic markers. J. Gen. Microbiol. 87, 343–350 [DOI] [PubMed] [Google Scholar]
- 17. Downie J. A., Ma Q. S., Knight C. D., Hombrecher G., Johnston A. W. (1983) Cloning of the symbiotic region of Rhizobium leguminosarum. The nodulation genes are between the nitrogenase genes and a nifA-like gene. EMBO J. 2, 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Finnie C., Hartley N. M., Findlay K. C., Downie J. A. (1997) The Rhizobium leguminosarum prsDE genes are required for secretion of several proteins, some of which influence nodulation, symbiotic nitrogen fixation, and exopolysaccharide modification. Mol. Microbiol. 25, 135–146 [DOI] [PubMed] [Google Scholar]
- 19. Beringer J. E. (1974) R factor transfer in Rhizobium leguminosarum. J. Gen Microbiol. 84, 188–198 [DOI] [PubMed] [Google Scholar]
- 20. Sherwood M. T. (1970) Improved synthetic medium for the growth of Rhizobium. J. Appl Bacteriol. 33, 708–713 [DOI] [PubMed] [Google Scholar]
- 21. Leigh J. A., Signer E. R., Walker G. C. (1985) Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. U.S.A. 82, 6231–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miller K. J., Gore R. S., Johnson R., Benesi A. J., Reinhold V. N. (1990) Cell-associated oligosaccharides of Bradyrhizobium spp. J. Bacteriol. 172, 136–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zevenhuizen L. P. T. M., Van Neerven A. R. W. (1983) (1,2)-β-d Glucan and acidic oligosaccharides produced by Rhizobium meliloti. Carbohydr. Res. 118, 127–134 [Google Scholar]
- 24. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST. A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edgar R. C. (2004) MUSCLE. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goodstadt L., Ponting C. P. (2001) CHROMA. Consensus-based colouring of multiple alignments for publication. Bioinformatics 17, 845–846 [DOI] [PubMed] [Google Scholar]
- 27. Ginalski K., Elofsson A., Fischer D., Rychlewski L. (2003) 3D-Jury. A simple approach to improve protein structure predictions. Bioinformatics 19, 1015–1018 [DOI] [PubMed] [Google Scholar]
- 28. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 29. Loewus M. W., Briggs D. R. (1952) The number of catalytically active sites present on the chymotrypsin molecule. J. Biol. Chem. 199, 857–864 [PubMed] [Google Scholar]
- 30. Filisetti-Cozzi T. M., Carpita N. C. (1991) Measurement of uronic acids without interference from neutral sugars. Anal. Biochem. 197, 157–162 [DOI] [PubMed] [Google Scholar]
- 31. Jefferson K. K., Cerca N. (2006) Methods in Molecular Biology. Cell-Cell Interactions: Methods and Protocols, pp. 119–126, Humana Press Inc., Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 32. Sandford P. A., Pittsley J. E., Knutson C. A., Watson P. R., Cadmus M. C., Janes A. (1977) Variation in Xanthomonas campestris NRRL B-1459. Characterization of xanthan products of differing pyruvic acid content: American Chemical Society Symposium Series No. 45, pp. 192–210 (Sandford P. A., Laskin A., eds) American Chemical Society, Washington, D. C [Google Scholar]
- 33. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer. An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 34. Nagar B., Overduin M., Ikura M., Rini J. M. (1996) Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature 380, 360–364 [DOI] [PubMed] [Google Scholar]
- 35. Pertz O., Bozic D., Koch A. W., Fauser C., Brancaccio A., Engel J. (1999) A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 18, 1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robertsen B. K., Aman P., Darvill A. G., McNeil M., Albersheim P. (1981) Host-symbiont interactions. V. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 67, 389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laus M. C., Logman T. J., Van Brussel A. A., Carlson R. W., Azadi P., Gao M. Y., Kijne J. W. (2004) Involvement of exo5 in production of surface polysaccharides in Rhizobium leguminosarum and its role in nodulation of Vicia sativa subsp. nigra. J. Bacteriol. 186, 6617–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen C. P., Song S. C., Gilboa-Garber N., Chang K. S., Wu A. M. (1998) Studies on the binding site of the galactose-specific agglutinin PA-IL from Pseudomonas aeruginosa. Glycobiology 8, 7–16 [DOI] [PubMed] [Google Scholar]
- 39. McNeil M., Darvill J., Darvill A. G., Albersheim P., van Veen R., Hooykaas P., Schilperoort R., Dell A. (1986) The discernible, structural features of the acidic polysaccharides secreted by different Rhizobium species are the same. Carbohydr. Res. 146, 307–326 [Google Scholar]
- 40. Zeng X., Andrade C. A., Oliveira M. D., Sun X. L. (2012) Carbohydrate-protein interactions and their biosensing applications. Anal. Bioanal. Chem. 402, 3161–3176 [DOI] [PubMed] [Google Scholar]
- 41. Finnie C., Zorreguieta A., Hartley N. M., Downie J. A. (1998) Characterization of Rhizobium leguminosarum exopolysaccharide glycanases that are secreted via a type I exporter and have a novel heptapeptide repeat motif. J. Bacteriol. 180, 1691–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abbott D. W., Boraston A. B. (2007) The structural basis for exopolygalacturonase activity in a family 28 glycoside hydrolase. J. Mol. Biol. 368, 1215–1222 [DOI] [PubMed] [Google Scholar]
- 43. Kataeva I. A., Seidel R. D., 3rd, Shah A., West L. T., Li X. L., Ljungdahl L. G. (2002) The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 68, 4292–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dazzo F. B., Truchet G. L., Sherwood J. E., Hrabak E. M., Gardiol A. E. (1982) Alteration of the trifoliin A-binding capsule of Rhizobium trifolii 0403 by enzymes released from clover roots. Appl. Environ. Microbiol. 44, 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Watkins N. J., Knight M. R., Trewavas A. J., Campbell A. K. (1995) Free calcium transients in chemotactic and non-chemotactic strains of Escherichia coli determined by using recombinant aequorin. Biochem. J. 306, 865–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Delepelaire P. (2004) Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 1694, 149–161 [DOI] [PubMed] [Google Scholar]
- 47. Holland I. B., Schmitt L., Young J. (2005) Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (review). Mol. Membr. Biol. 22, 29–39 [DOI] [PubMed] [Google Scholar]
- 48. Chenal A., Guijarro J. I., Raynal B., Delepierre M., Ladant D. (2009) RTX calcium binding motifs are intrinsically disordered in the absence of calcium. Implication for protein secretion. J. Biol. Chem. 284, 1781–1789 [DOI] [PubMed] [Google Scholar]
- 49. Wolff N., Sapriel G., Bodenreider C., Chaffotte A., Delepelaire P. (2003) Antifolding activity of the SecB chaperone is essential for secretion of HasA, a quickly folding ABC pathway substrate. J. Biol. Chem. 278, 38247–38253 [DOI] [PubMed] [Google Scholar]
- 50. Michiels J., Xi C., Verhaert J., Vanderleyden J. (2002) The functions of Ca2+ in bacteria. A role for EF-hand proteins? Trends Microbiol. 10, 87–93 [DOI] [PubMed] [Google Scholar]
- 51. Orans J., Johnson M. D., Coggan K. A., Sperlazza J. R., Heiniger R. W., Wolfgang M. C., Redinbo M. R. (2010) Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility. Proc. Natl. Acad. Sci. U.S.A. 107, 1065–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aravind P., Mishra A., Suman S. K., Jobby M. K., Sankaranarayanan R., Sharma Y. (2009) The βγ-crystallin superfamily contains a universal motif for binding calcium. Biochemistry 48, 12180–12190 [DOI] [PubMed] [Google Scholar]
- 53. Aravind P., Suman S. K., Mishra A., Sharma Y., Sankaranarayanan R. (2009) Three-dimensional domain swapping in nitrollin, a single-domain βγ-crystallin from Nitrosospira multiformis, controls protein conformation and stability but not dimerization. J. Mol. Biol. 385, 163–177 [DOI] [PubMed] [Google Scholar]
- 54. Vu B., Chen M., Crawford R. J., Ivanova E. P. (2009) Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14, 2535–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ghafoor A., Hay I. D., Rehm B. H. (2011) Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 77, 5238–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Begun J., Gaiani J. M., Rohde H., Mack D., Calderwood S. B., Ausubel F. M., Sifri C. D. (2007) Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 3, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]