Abstract
Arrhythmogenic right ventricular cardiomyopathy (ARVC) is an inherited heart muscle disease characterized by fibrofatty replacement of the myocardium and ventricular arrhythmias, associated with mutations in the desmosomal genes. Only a missense mutation in the DES gene coding for desmin, the intermediate filament protein expressed by cardiac and skeletal muscle cells, has been recently associated with ARVC. We screened 91 ARVC index cases (53 negative for mutations in desmosomal genes and an additional 38 carrying desmosomal gene mutations) for DES mutations. Two rare missense variants were identified. The heterozygous p.K241E substitution was detected in 1 patient affected with a severe form of ARVC who also carried the p.T816RfsX10 mutation in plakophilin-2 gene. This DES substitution, showing an allele frequency of <0.01 in the control population, is predicted to cause an intolerant amino acid change in a highly conserved protein domain. Thus, it can be considered a rare variant with a possible modifier effect on the phenotypic expression of the concomitant mutation. The previously known p.A213V substitution was identified in 1 patient with ARVC who was negative for mutations in the desmosomal genes. Because a greater prevalence of p.A213V has been reported in patients with heart dilation than in control subjects, the hypothesis that this rare variant could have an unfavorable effect on cardiac remodeling cannot be ruled out. In conclusion, our data help to establish that, in the absence of skeletal muscle involvement suggestive of a desminopathy, the probability of DES mutations in ARVC is very low. These findings have important implications in the mutation screening strategy for patients with ARVC.
Arrhythmogenic right ventricular cardiomyopathy (ARVC; MIM 107970) is an inherited cardiac disease characterized by progressive fibrofatty myocardial replacement, primarily of the right ventricle.1–3 Clinical manifestations occur most often between the second and fourth decade of life and are characterized by ventricular arrhythmias, heart failure, and sudden death.1,2 ARVC is the second most common cause of unexpected sudden death among young people and athletes4 and affects men more frequently than women, with a 3:1 ratio.5 The disease is familial in about half the cases and typically shows an autosomal dominant inheritance, with reduced penetrance and variable expression.2 Mutations in the genes encoding several components of the cardiac desmosome6–10 are associated with ARVC in about 50% of probands.3 The presence of multiple mutations in desmosomal genes have been reported in a significant proportion of cases.11,12 Nondesmosomal ARVC genes have also been identified,3 providing evidence that ARVC pathogenesis extends beyond the desmosomes. Currently, only a novel mutation in the DES gene encoding for desmin, the intermediate filament protein expressed by cardiac cells, has been identified in a proband showing a typical form of ARVC.13 We report the mutation screening of the DES gene in a cohort of Italian patients with ARVC to establish the prevalence of DES mutations.
Methods
We included 91 unrelated patients of Italian descent with ARVC. The clinical evaluation consisted of a detailed personal and family history, physical examination, 12-lead electrocardiogram (ECG), 2-dimensional echocardiogram, signal-averaged ECG, and stress test ECG, performed according to previously reported methods.14 In all probands, a clinical diagnosis of ARVC was made using the revised 2010 Task Force criteria.15 None of these patients showed skeletal muscle disease. Available family members of the probands carrying DES variants were also clinically evaluated. All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki. The ethics committee review board of the University of Padua (Padua, Italy) approved the study protocol. All the participants provided written informed consent before inclusion in the study. Of the 91 patients, 53 were ARVC index cases and were negative for mutations in the desmosomal genes, and 38 patients carried desmosomal gene mutations (11 in the plakophilin-2 [PKP2] gene, 13 in the desmoplakin [DSP] gene, 7 in the desmoglein-2 [DSG2] gene, 2 in the desmocollin-2 [DSC2] gene, and 5 with multiple mutations).
Genomic DNA was isolated from peripheral blood lymphocytes using standard procedures. Primers for polymerase chain reaction amplification of the DES gene were designed using PRIMER3 software (available at: http://frodo.wi.mit.edu/; Supplemental Table 1). Polymerase chain reaction products were analyzed using denaturing high-performance liquid chromatography and/or direct sequencing. Denaturing high-performance liquid chromatography analysis was performed using the WAVE Nucleic Acid Fragment Analysis System 3500HT equipped with the DNASep HT cartridge (Transgenomic, Omaha, Nebraska). Temperatures for sample analysis were selected using WAVE Navigator software (Transgenomic; Supplemental Table 1). Polymerase chain reaction fragments showing an aberrant, temperature-modulated denaturing high-performance liquid chromatography heteroduplex profile were sequenced using the BIG DYE dideoxy-terminator chemistry (PerkinElmer, Waltham, Massachusetts) on an ABI 3730XL DNA sequencer (PE Applied Biosystems, Foster City, California). Chromas, version 1.5, software (Technelysium, South Brisbane, Australia) and Lasergene package computer programs (DNASTAR, Madison, Wisconsin) were used to edit, assemble, and translate the sequence. Numbering of the DES nucleotides starts at ATG and refers to GenBank accession number NM_001927 for the cDNA; the translation sequence was compared to the reference sequence NP_001918. A control group of 300 healthy and unrelated subjects (600 alleles) from the Italian population was used to exclude that novel variants could be common DNA polymorphisms. All the controls were matched to the probands by ancestry. The amplification refractory mutation system was used to screen the control population for the presence of nucleotide changes identified in the DES gene (c.638C>T, c.721A>G, and c.736-11A>G). The primer sequence, amplicon size, and amplification annealing temperatures are reported in Supplemental Table 2. Mutation screening was performed for all available family members of index cases in which a DES variant was detected.
Results
A systematic mutation screening in the DES coding regions was performed in 53 ARVC unrelated index cases negative for mutations in the desmosomal genes. Because the occurrence of multiple mutations in ARVC has been reported,11,12 the DNA of another 38 consecutive and unrelated patients with desmosomal gene mutations (11 in the PKP2 gene, 13 in the DSP gene, 7 in the DSG2 gene, 2 in the DSC2 gene, and 5 carrying multiple mutations) was investigated for DES mutations.
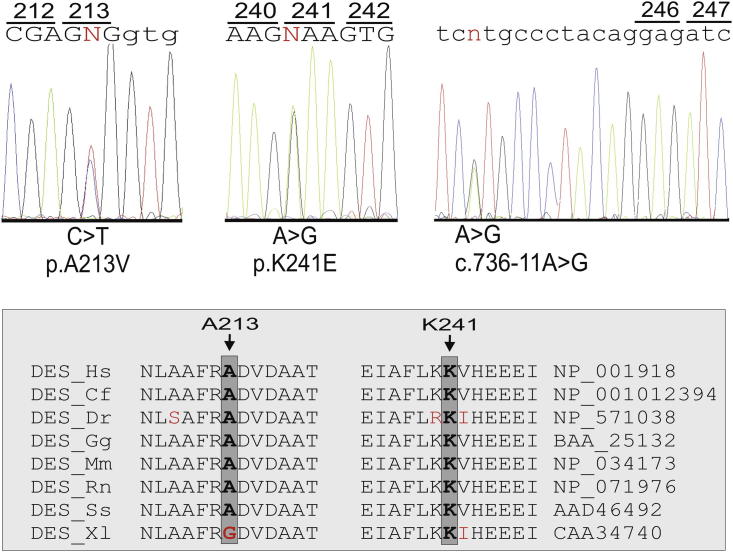
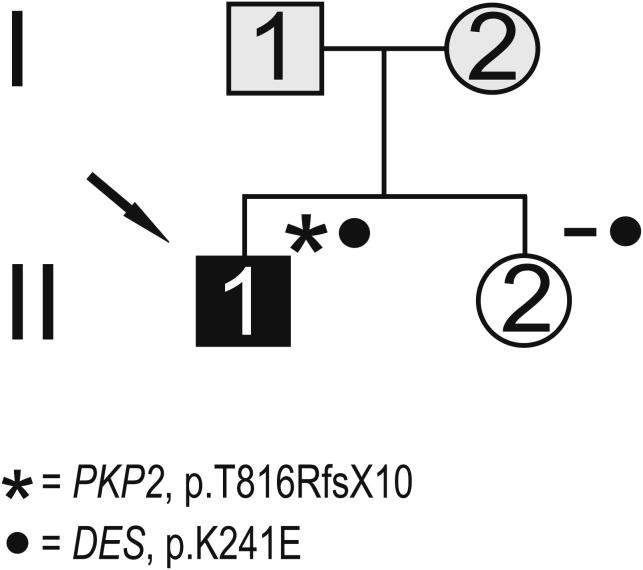
Mutation analysis resulted in the identification of several nucleotide substitutions (Table 1). A heterozygous nucleotide change, c.721A>G, resulting in the p.K241E substitution, was identified in 1 patient (Figure 1). It was not detected in 600 control chromosomes, in single nucleotide polymorphism database (dbSNP; available at: http://www.ncbi.nlm.nih.gov/projects/SNP/), or in the Exome Variant Server (available at: http://evs.gs.washington.edu/EVS/). Very recently, it was reported in the 1000 Genomes Project database (available at: http://www.1000genomes.org) with a minor allele frequency of <0.01 (1000GENOMES_2_220285054). This nonconservative amino acid substitution, which was considered intolerant by Condel (available at: http://bg.upf.edu/condel/analysis), SIFT (available at: http://sift.jcvi.org/) and PolyPhen-2 (available at: http://genetics.bwh.harvard.edu/pph2/) analysis, involves a highly conserved residue in the 1B α-helix subdomain (Figure 1). The index case carrying the p.K241E variant had no family history of juvenile sudden death, ARVC, or skeletal muscle disease. He was diagnosed at 25 years old after a sustained ventricular tachycardia. The basal ECG showed the presence of negative T waves in leads V1 to V5 in the absence of complete right bundle branch block (QRS ≥120 ms). The 2-dimensional echocardiogram showed severe dilation of the right ventricle (RV) with marked hypokinesia of the free wall, reduced RV ejection fraction, and mild dilation of the left ventricle. He was affected by a severe form of ARVC and received an implantable cardioverter defibrillator at 34 years old. He was found to carry a pathogenic frame shift mutation (p.T816RfsX10) in PKP2 gene. The patient's sister, carrying only the same DES variant, was fully asymptomatic clinically and did not show any cardiac abnormality on the echocardiogram, ECG, or magnetic resonance imaging scan (Figure 2).
Table 1.
Nucleotide substitutions identified in Desmin gene in patients affected with arrhythmogenic right ventricular cardiomyopathy (ARVC)
| Exon | Nucleotide Change | Amino Acid Change | dbSNP/1000Genomes ID | MAF |
|---|---|---|---|---|
| 1 | c.75A>G | p.P25P | rs1318299 | 0.089 |
| 1 | c.93T>C | p.S31S | rs2017800 | 0.100 |
| 1 | c.408C>T | p.L136L | rs111828114 | 0.021 |
| Intron 1 | c.579-38C>T | − | rs12991025 | 0.456 |
| 2 | c.638C>T | p.A213V | rs41272699 | 0.008; 0.012∗ |
| 3 | c.669T>C | p.I223I | rs75882680 | 0.022 |
| 3 | c.721A>G | p.K241E | 1000GENOMES_2_220285054 | <0.01; 0.000∗ |
| Intron 3 | c.735+20C>T | − | rs151226355 | 0.005; 0.013∗ |
| Intron 3 | c.736-35C>A | − | rs41272701 | 0.018 |
| Intron 3 | c.736-11A>G | − | novel (GenBank JX114779) | 0.000∗ |
| 4 | c.828C>T | p.D276D | rs1058261 | 0.347 |
| 5 | c.1014G>C | p.L338L | rs12920 | 0.347 |
| 6 | c.1104G>A | p.A368A | rs1058284 | 0.342 |
ID = identification number; MAF = minor allele frequency.
MAF estimated in the Italian control population for nucleotide changes not reported in SNP databases or showing MAF <0.01.
Figure 1.
DES variants identified in patients with ARVC. (Top) Sequence electropherograms showing p.A213V and p.K241E amino acid substitutions and intronic nucleotide change c.736-11A>G detected in DES gene. (Bottom) ClustalW (European Molecular Biology Laboratory–European Bioinformatics Institute, Cambridgeshire, United Kingdom) alignment showing evolutionary conservation of highly conserved 1B desmin subdomain among species. Organism names and Genbank Accession numbers are reported on the sides (Cf = Canis familiaris; Dr = Danio rerio; Gg = Gallus gallus; Hs = Homo sapiens; Mm = Mus musculus; Rn = Rattus norvegicus; Ss = Sus scrofa; Xl = Xenopus laevis).
Figure 2.
Pedigree of patient with ARVC carrying DES p.K241E rare variant. Index patient indicated by arrow. Black, white, and gray symbols represent clinically affected subjects, unaffected subjects, and subjects with unknown disease status, respectively. Presence (asterisk) or absence (−) of gene variations shown.
The known c.638C>T nucleotide substitution, resulting in a p.A213V amino acid change, was identified in 1 patient who was negative for mutations in the desmosomal ARVC genes (Figure 1). This variant currently appears in single nucleotide polymorphism database (rs41272699, minor allele frequency 0.8%). In the present study, it was detected in the healthy controls at an allele frequency of 1.2%. The index patient carrying the p.A213V variant was diagnosed at 47 years old after a presyncopal episode. The basal ECG showed the presence of negative T waves in leads V1 to V3 in the absence of complete right bundle branch block (QRS ≥120 ms). Echocardiography showed mild RV dilation with kinetic abnormalities in the RV free wall. Magnetic resonance imaging confirmed the presence of mild dilation of the right ventricle, together with kinetic alterations of the RV free wall, and showed adipose tissue infiltration along the RV free wall. No family history of juvenile sudden death, ARVC, or skeletal muscle disease was reported. No family members were available for clinical and genetic testing.
A heterozygous nucleotide change close to the acceptor splice site of exon 4 (c.736-11A>G) was identified in 1 patient in whom no mutation was detected in the desmosomal ARVC genes (Figure 1). He was examined at 46 years old because of effort dyspnea. The basal ECG showed the presence of negative T waves in leads V1 to V3 in the absence of complete right bundle branch block and an epsilon wave in the right precordial leads. The 2-dimensional echocardiogram revealed the presence of severe RV dilation, depression of the RV fractional area change, apical and subtricuspid akinesia, and apical sacculations. The detected unclassified variant, which was absent in 600 control chromosomes and in the above-mentioned databases, was identified in 2 of 5 affected and 3 unaffected members of the family. No skeletal muscle and cardiac biopsy samples were available to perform additional studies to check for perturbed splicing of the DES mRNA transcript. On the basis of the absence of co-segregation with the disease phenotype and the identification in unaffected family members, this intronic variant was unlikely the causal mutation in this family.
Discussion
Desmin is encoded by a single copy gene located on chromosome 2q35. In the skeletal and cardiac muscle cells, it forms a flexible 3-dimensional scaffold around the myofibrillar Z-discs, connecting them to the plasma membrane, nuclear lamina, mitochondria, and desmosomes and aligning the myofibrils.16 Therefore, desmin plays key structural and signaling roles in myocytes and is critical for cytoskeletal organization and maintaining the cardiomyocyte structure.17
Currently, >50 different DES mutations have been identified worldwide. Mutations in the DES gene result in formation of inclusion bodies, weakening of the desmin cytoskeleton, disruption of subcellular organelle organization, and, eventually, myofibril degradation. These muscle disorders are referred to as desmin-related myopathies or desminopathies. They often present in young childhood, with patients experiencing increasing muscle weakness, and are associated with a wide spectrum of clinical phenotypes, even within the same family. These range from scapuloperoneal, limb girdle, and distal phenotypes with variable cardiac or respiratory involvement to pure cardiomyopathies.18
To the best of our knowledge, only 1 DES mutation (p.N116S) was identified in a German patient with ARVC.13 The amino acid substitution leads to aggresome formation in cardiac and skeletal muscle without signs of an overt clinical myopathy.13,19 However, the 17-year-old mutation carrier showed concomitant subclinical skeletal muscle alterations. All other reported ARVC-related DES mutations underlie a clinically heterogeneous phenotype, frequently associated with muscle alterations (Table 2). The p.S13F mutation was identified in 39 family members from 8 Dutch families.20,21 This mutation was frequently associated with a myopathy and with a wide spectrum of cardiomyopathies. ARVC was found in only 2 cases. The p.N342D mutation has been described previously in patients affected with desmin-related myopathies22; van Spaendonck-Zwarts et al21 and Otten et al23 were the first to demonstrate the association of this mutation with RV cardiomyopathy. Among 21 carriers of the p.N342D substitution from 3 different families showing a variable neuromuscular phenotype with cardiac involvement, ARVC was diagnosed in only 1. More recently, the p.P419S mutation has been identified by exome sequencing in a large Swedish family showing myofibrillar myopathy and ARVC, previously linked to chromosome 10q22.3 markers (ARVC7 locus, MIM 609160).24 No additional DES mutations were found in a set of 50 Dutch patients with an ARVC-like disorder23 and in a large series of 143 unrelated North American probands diagnosed with ARVC.12
Table 2.
Desmin mutations associated with arrhythmogenic right ventricular cardiomyopathy (ARVC)
| Mutation | Index Cases (n) | Population | Phenotype | Investigators |
|---|---|---|---|---|
| c.38C>T, p.S13F | 8 | Dutch | Desminopathy (ARVC in 2 relatives) | van Tintelen et al20; van Spaendonck-Zwarts et al21 |
| c.347A>G, p.N116S | 1 of 22 | German | ARVC | Klauke et al13 |
| c.1024A>G, p.N342D | 3 | Dutch | Desminopathy (plus ARVC in 1 relative) | van Spaendonck-Zwarts et al21; Otten et al23 |
| c.1255C>T, p.P419S | 1 | Swedish | Myofibrillar myopathy plus ARVC | Hedberg et al24 |
In the present study, we focused on mutation screening of the DES gene, a recent player in the growing field of ARVC-associated genes, in a large and well-studied cohort of Italian patients with a typical form of ARVC. Two rare missense variants, both with a possible modifier effect, were identified in 2 of 91 index cases (2.2%). These findings help to establish that, in the absence of skeletal muscle involvement suggestive of a desminopathy, the probability of DES mutations in ARVC is very low. The p.K241E substitution was found in 1 patient who carried a 2-base pair deletion (c.2447_2448delCC) in exon 12 of PKP2 gene. This frame shift mutation (p.T816RfsX10), identified in another ARVC proband,12 is expected to cause an important change in the protein structure, introducing a premature stop codon in the ARM-domain 8 of PKP2. The p.K241E substitution, recently reported in the 1000 Genomes Project database with a very low minor allele frequency, involves a highly conserved residue and is predicted to cause an intolerant amino acid change. On the basis of these data, this DES substitution can be considered a rare variant with a possible modifier effect on the phenotypic expression of the concomitant PKP2 mutation. The previously known DES p.A213V amino acid change was identified in 1 patient with ARVC who was negative for mutations in the desmosomal genes. In the published data, this substitution has been associated with 3 completely different clinical phenotypes: restrictive cardiomyopathy, dilated cardiomyopathy and isolated distal myopathy.25–27 However, the identification of this substitution in healthy controls has raised the hypothesis that the p.A213V shift represents a rare polymorphism rather than a true disease-causing mutation. Recently, a high prevalence of p.A213V substitution was reported in patients with heart dilation of various origins.28 Although p.A213V desmin did not show a clear in vitro functional effect in transfected HeLa cells, Kostareva et al28 speculated that its altered biophysical properties can predispose to cardiac vulnerability in the presence of unfavorable factors. Therefore, overrepresentation of p.A213V in patients with cardiac dilation supports the hypothesis that this rare variant might have an unfavorable effect on cardiac remodelling under stressful conditions. Likewise, the p.A213V substitution detected in a patient with ARVC in the present study could play a predisposing role in heart remodeling in the presence of an additional, as yet unidentified, disease-causing mutation.
Future studies should be planned to screen for novel genes other than desmosomal genes, especially in the Next Generation Sequencing era, to better define the etiopathogenesis of ARVC. The maturation of the recently developed Next Generation Sequencing technologies provides unprecedented sequencing capacity at dramatically lower cost and will be particularly useful for mutation screening in patients affected with this highly genetic heterogeneous disease. However, the use of Next Generation Sequencing to identify gene mutations could face huge challenges, in particular, in small families and sporadic cases, to discern the disease-causing variants from those that by chance alone are carried by the affected persons.
Acknowledgment
We are grateful to all the family members who kindly participated in the present study and to Mrs. Paola Marcon for her invaluable help in collecting family data.
Footnotes
This study was supported by grant GGP09293 from the Italian Telethon Foundation (Rome, Italy); grant 13317 from the Association Française contre les Myopathies (Évry, France); Fondazione Cassa di Risparmio, Padova e Rovigo (Rovigo, Italy).
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of our report.
See page 404 for disclosure information.
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.amjcard.2012.10.017
Disclosures
The authors have no conflicts of interest to disclose.
Supplementary data
References
- 1.Thiene G., Nava A., Corrado D., Rossi L., Pennelli N. Right ventricular cardiomyopathy and sudden death in young people. N Engl J Med. 1988;318:129–133. doi: 10.1056/NEJM198801213180301. [DOI] [PubMed] [Google Scholar]
- 2.Nava A., Thiene G., Canciani B., Scognamiglio R., Daliento L., Buja G.F., Martini B., Stritoni P., Fasoli G. Familial occurrence of right ventricular dysplasia: a study involving nine families. J Am Coll Cardiol. 1988;12:1222–1228. doi: 10.1016/0735-1097(88)92603-4. [DOI] [PubMed] [Google Scholar]
- 3.Basso C., Bauce B., Corrado D., Thiene G. Pathophysiology of arrhythmogenic cardiomyopathy. Nat Rev Cardiol. 2011;9:223–233. doi: 10.1038/nrcardio.2011.173. [DOI] [PubMed] [Google Scholar]
- 4.Corrado D., Basso C., Schiavon M., Thiene G. Screening for hypertrophic cardiomyopathy in young athletes. N Engl J Med. 1998;339:364–369. doi: 10.1056/NEJM199808063390602. [DOI] [PubMed] [Google Scholar]
- 5.Bauce B., Frigo G., Marcus F.I., Basso C., Rampazzo A., Maddalena F., Corrado D., Winnicki M., Daliento L., Rigato I., Steriotis A., Mazzotti E., Thiene G., Nava A. Comparison of clinical features of arrhythmogenic right ventricular cardiomyopathy in men versus women. Am J Cardiol. 2008;102:1252–1257. doi: 10.1016/j.amjcard.2008.06.054. [DOI] [PubMed] [Google Scholar]
- 6.McKoy G., Protonotarios N., Crosby A., Tsatsopoulou A., Anastasakis A., Coonar A., Norman M., Baboonian C., Jeffery S., McKenna W.J. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease) Lancet. 2000;355:2119–2124. doi: 10.1016/S0140-6736(00)02379-5. [DOI] [PubMed] [Google Scholar]
- 7.Rampazzo A., Nava A., Malacrida S., Beffagna G., Bauce B., Rossi V., Zimbello R., Simionati B., Basso C., Thiene G., Towbin J.A., Danieli G.A. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2002;71:1200–1206. doi: 10.1086/344208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerull B., Heuser A., Wichter T., Paul M., Basson C.T., McDermott D.A., Lerman B.B., Markowitz S.M., Ellinor P.T., MacRae C.A., Peters S., Grossmann K.S., Michely B., Sasse-Klaassen S., Birchmeier W., Dietz R., Breithardt G., Schulze-Bahr E., Thierfelder L. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet. 2004;36:1162–1164. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- 9.Pilichou K., Nava A., Basso C., Beffagna G., Bauce B., Lorenzon A., Frigo G., Vettori A., Valente M., Towbin J., Thiene G., Danieli G.A., Rampazzo A. Mutations in desmoglein-2 gene are associated with arrhythmogenic right ventricular cardiomyopathy. Circulation. 2006;113:1171–1179. doi: 10.1161/CIRCULATIONAHA.105.583674. [DOI] [PubMed] [Google Scholar]
- 10.Syrris P., Ward D., Evans A., Asimaki A., Gandjbakhch E., Sen-Chowdhry S., McKenna W.J. Arrhythmogenic right ventricular dysplasia/cardiomyopathy associated with mutations in the desmosomal gene desmocollin-2. Am J Hum Genet. 2006;79:978–984. doi: 10.1086/509122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauce B., Nava A., Beffagna G., Basso C., Lorenzon A., Smaniotto G., De Bortoli M., Rigato I., Mazzotti E., Steriotis A., Marra M.P., Towbin J.A., Thiene G., Danieli G.A., Rampazzo A. Multiple mutations in desmosomal proteins encoding genes in arrhythmogenic right ventricular cardiomyopathy/dysplasia. Heart Rhythm. 2010;7:22–29. doi: 10.1016/j.hrthm.2009.09.070. [DOI] [PubMed] [Google Scholar]
- 12.Xu T., Yang Z., Vatta M., Rampazzo A., Beffagna G., Pilichou K., Scherer S.E., Saffitz J., Kravitz J., Zareba W., Danieli G.A., Lorenzon A., Nava A., Bauce B., Thiene G., Basso C., Calkins H., Gear K., Marcus F., Towbin J.A. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2010;55:587–597. doi: 10.1016/j.jacc.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klauke B., Kossmann S., Gaertner A., Brand K., Stork I., Brodehl A., Dieding M., Walhorn V., Anselmetti D., Gerdes D., Bohms B., Schulz U., Zu Knyphausen E., Vorgerd M., Gummert J., Milting H. De novo desmin-mutation N116S is associated with arrhythmogenic right ventricular cardiomyopathy. Hum Mol Genet. 2010;19:4595–4607. doi: 10.1093/hmg/ddq387. [DOI] [PubMed] [Google Scholar]
- 14.Nava A., Bauce B., Basso C., Muriago M., Rampazzo A., Villanova C., Daliento L., Buja G., Corrado D., Danieli G.A., Thiene G. Clinical profile and long term follow-up of 37 families with arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2000;36:2226–2233. doi: 10.1016/s0735-1097(00)00997-9. [DOI] [PubMed] [Google Scholar]
- 15.Marcus F.I., McKenna W.J., Sherrill D., Basso C., Bauce B., Bluemke D.A., Calkins H., Corrado D., Cox M.G., Daubert J.P., Fontaine G., Gear K., Hauer R., Nava A., Picard M.H., Protonotarios N., Saffitz J.E., Sanborn D.M., Steinberg J.S., Tandri H., Thiene G., Towbin J.A., Tsatsopoulou A., Wichter T., Zareba W. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. Eur Heart J. 2010;31:806–814. doi: 10.1093/eurheartj/ehq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazarides E. Intermediate filaments as mechanical integrators of cellular space. Nature. 1980;238:249–256. doi: 10.1038/283249a0. [DOI] [PubMed] [Google Scholar]
- 17.McLendon P.M., Robbins J. Desmin-related cardiomyopathy: an unfolding story. Am J Physiol Heart Circ Physiol. 2011;301:H1220–H1228. doi: 10.1152/ajpheart.00601.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin J., Bulst S., Thirion C., Schmidt F., Bötzel K., Krause S., Pertl C., Kretzschmar H., Walter M.C., Giese A., Lochmüller H. Divergent molecular effects of desmin mutations on protein assembly in myofibrillar myopathy. J Neuropathol Exp Neurol. 2010;69:415–424. doi: 10.1097/NEN.0b013e3181d71305. [DOI] [PubMed] [Google Scholar]
- 19.Brodehl A., Hedde P.N., Dieding M., Fatima A., Walhorn V., Gayda S., Šarić T., Klauke B., Gummert J., Anselmetti D., Heilemann M., Nienhaus G.U., Milting H. Dual color photoactivation localization microscopy of cardiomyopathy-associated desmin mutants. J Biol Chem. 2012;287:16047–16057. doi: 10.1074/jbc.M111.313841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Tintelen J.P., Van Gelder I.C., Asimaki A., Suurmeijer A.J., Wiesfeld A.C., Jongbloed J.D., van den Wijngaard A., Kuks J.B., van Spaendonck-Zwarts K.Y., Notermans N., Boven L., van den Heuvel F., Veenstra-Knol H.E., Saffitz J.E., Hofstra R.M., van den Berg M.P. Severe cardiac phenotype with right ventricular predominance in a large cohort of patients with a single missense mutation in the DES gene. Heart Rhythm. 2009;6:1574–1583. doi: 10.1016/j.hrthm.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 21.van Spaendonck-Zwarts K.Y., van der Kooi A.J., van den Berg M.P., Ippel E.F., Boven L.G., Yee W.C., van den Wijngaard A., Brusse E., Hoogendijk J.E., Doevendans P.A., de Visser M., Jongbloed J.D., van Tintelen J.P. Recurrent and founder mutations in the Netherlands: the cardiac phenotype of DES founder mutations p.S13F and p.N342D. Neth Heart J. 2012;20:219–228. doi: 10.1007/s12471-011-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalakas M.C., Park K.Y., Semino-Mora C., Lee H.S., Sivakumar K., Goldfarb L.G. Desmin myopathy, a skeletal myopathy with cardiomyopathy caused by mutations in the desmin gene. N Engl J Med. 2000;342:770–780. doi: 10.1056/NEJM200003163421104. [DOI] [PubMed] [Google Scholar]
- 23.Otten E., Asimaki A., Maass A., van Langen I.M., van der Wal A., de Jonge N., van den Berg M.P., Saffitz J.E., Wilde A.A., Jongbloed J.D., van Tintelen J.P. Desmin mutations as a cause of right ventricular heart failure affect the intercalated disks. Heart Rhythm. 2010;7:1058–1064. doi: 10.1016/j.hrthm.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 24.Hedberg C., Melberg A., Kuhl A., Jenne D., Oldfors A. Autosomal dominant myofibrillar myopathy with arrhythmogenic right ventricular cardiomyopathy 7 is caused by a DES mutation. Eur J Hum Genet. 2012;20:984–985. doi: 10.1038/ejhg.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowles N.E., Jimenez S., Vatta M., Chrisco M., Szmuszkovicz J., Capetanaki Y. Familial restrictive cardiomyopathy caused by a missense mutation in the desmin gene. Pediatric Res. 2002;51(Suppl 2) [Google Scholar]
- 26.Goudeau B., Rodrigues-Lima F., Fischer D., Casteras-Simon M., Sambuughin N., de Visser M., Laforet P., Ferrer X., Chapon F., Sjöberg G., Kostareva A., Sejersen T., Dalakas M.C., Goldfarb L.G., Vicart P. Variable pathogenic potentials of mutations located in the desmin alpha-helical domain. Hum Mutat. 2006;27:906–913. doi: 10.1002/humu.20351. [DOI] [PubMed] [Google Scholar]
- 27.Kostareva A., Gudkova A., Sjoberg G., Kiselev I., Moiseeva O., Karelkina E., Goldfarb L., Schlyakhto E., Sejersen T. Desmin mutations in a St. Petersburg cohort of cardiomyopathies. Acta Myol. 2006;25:109–115. [PubMed] [Google Scholar]
- 28.Kostareva A., Sjoberg G., Gudkova A., Smolina N., Semernin E., Shlyakhto E., Sejersen T. Desmin A213V substitution represents a rare polymorphism but not a mutation and is more prevalent in patients with heart dilation of various origins. Acta Myol. 2011;30:42–45. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.