Abstract
We report the characterization of a highly germline competent C57BL/6N mouse embryonic stem cell line, JM8. To simplify breeding schemes, the dominant Agouti coat color gene was restored in JM8 cells by targeted repair of the C57BL/6 nonagouti mutation. These cells provide a robust foundation for large-scale mouse knockout programs that aim to provide a public resource of targeted mutations in the C57BL/6 genetic background.
C57BL/6 is one of the best characterized inbred strains of mice and is the reference strain for the mouse genome sequence1. However, genetic manipulation of the mouse genome is carried out predominantly in embryonic stem cells derived from the 129 strain of mice. A reliance on 129 embryonic stem cells is not ideal, particularly for genetic studies of immunology, neurobiology and physiology. Recently, large-scale knockout programmes have been established to mutate all protein-coding genes in mouse using a combination of gene trapping and gene targeting strategies2. In planning these resources, C57BL/6 embryonic stem cells were chosen for the production of mutant alleles as this genetic background is better suited for the large-scale phenotyping efforts that will follow.
Several laboratories have reported the establishment of C57BL/6 embryonic stem cell lines from the C57BL/6J and C57BL/6N substrains of mice3-10, however, the use of these cell lines for high throughput genetic engineering in mice has met with limited success due to low germline transmission rates10. The value of embryonic stem cell resources critically depends on achieving high germline transmission rates among individual clones. Embryonic stem cells derived from the 129 strain of mice have proven to be robust. For example, 80% of clones from the BayGenomics and Sanger Institute gene trap resources were able to colonize the germline of mice following blastocyst injection (W.C.S. and K.L., unpublished). Compared to 129 strains, C57BL/6 embryonic stem cells are less able to maintain a normal karyotype and, consequently, germline transmission of manipulated clones is compromised8.
As a foundation for the international mouse knockout program2, we sought to establish both feeder-dependent and feeder-free C57BL/6 embryonic stem cells with reliable and robust germline colonization. We established several male cell lines from C57BL/6J and C57BL/6N blastocysts on fibroblast feeder cells using standard methods11. C57BL/6N embryonic stem cells showed better growth properties and morphology compared to C57BL/6J embryonic stem cells (data not shown). Furthermore, our attempts to culture C57BL/6J embryonic stem cells on gelatin in the presence of LIF were unsuccessful, whereas C57BL/6N embryonic stem cells readily adapted to feeder-free culture conditions.
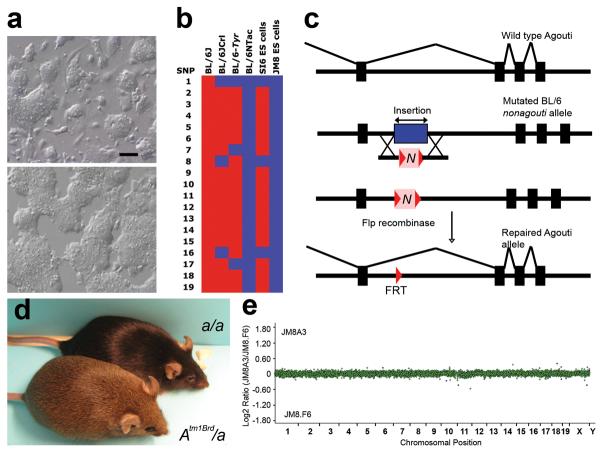
A parental male embryonic stem cell line derived from C57BL/6N blastocysts, JM8, possessed a normal male (XY) karyotype (data not shown) and exhibited a normal undifferentiated morphology when cultured on feeder cells and on gelatin-treated plates (Fig. 1a). Injections of early-passage JM8 cells into albino C57BL/6-Tyrc-Brd blastocysts produced chimeras with high coat color chimerism, a sex distortion in favour of males and a high proportion of chimeras with 100% contribution to both somatic tissues and the germline (Supplementary Data 1 online). To confirm the genetic purity of the JM8 cell line, we genotyped a panel of 19 SNP markers which distinguish the N and J substrains of C57BL/6 mice (Fig. 1b and Supplementary Table 1 online; J.L.M. and D.R.B., unpublished). This analysis confirmed their C57BL/6N origin. Although the J and N substrains are very closely related — only 102 of 139,561 SNPs genotyped in these lines are discordant — several phenotypic differences have been noted12,13.
Figure 1.
Properties and origin of Agouti JM8 C57BL/6N embryonic stem cells. (a) JM8 parental embryonic stem cells cultured on (top) SNL feeder cells16 in medium containing 15% serum and (bottom) gelatin-coated plates in medium supplemented with 10% serum and LIF. Scale bar: 100 μm (b) Genotype of embryonic stem cells and mice using a panel of 19 SNPs that differentiate between the N and J substrains of C57BL/6 (see Supplementary Table). JM8 embryonic stem cells were homozygous for all N alleles (shown in blue), confirming their origin from the C57BL/6N substrain. (c) Schematic of targeting strategy to restore Agouti function in JM8 cells. Exons (black boxes), retrotransposon (blue box), neo-TK selection cassette (N). For full details see Methods online. (d) F1 offspring from a male chimera, generated by injection of JM8A3 cells into a C57BL/6-Tyrc-Brd blastocyst, crossed to a C57BL/6-Tyrc-Brd female. Both mice shown are embryonic stem cell-derived and the mouse carrying the corrected allele (Atm1Brd, bottom) has an agouti coat. (e) CGH plot showing no copy number variants in JM8A3 cells compared to JM8.F6 cells. Relative copy number is plotted as the log ratio of hybridization signals of probes from JM8A3 DNA compared to JM8.F6. The JM8.F6 line has no copy number variants relative to C57BL/6NTac mice (see Supplementary Fig. 1).
Genetic variability within a population of embryonic stem cells is the critical determinant of their performance and this can only be assessed by the analysis of large numbers of subclones. We therefore isolated a set of clonal sublines either on feeders or on gelatin and tested these for their germline transmission potential (Supplementary Data 1). In this set of experiments, most clones exhibited very high rates of chimera formation, producing large numbers of chimeras in which a high percentage or even all of the cells were derived from the injected embryonic stem cells. More importantly, 80% of the chimeras were male, indicating that sex-conversion of female host blastocysts had occurred in the majority of cases. Upon breeding, we observed an exceptionally high rate of clonal germline transmission, calculated as the fraction of total clones injected that produce at least one male chimera with embryonic stem cell contribution to the germline. From injections of approximately 40 blastocysts per subline, 76% (16 of 21) of the sublines gave rise to at least one germline male chimera. Moreover, culturing cells under feeder-free conditions does not appear to compromise the pluripotency of JM8 embryonic stem cells.
Two sublines in particular, JM8.F6 (feeder-dependent) and JM8.N4 (feeder-free) produced favourable results and were tested for their ability to support high-throughput gene targeting. Targeted JM8 embryonic stem cell clones generated for the large-scale knockout programme (W.C.S. et al., manuscript in preparation) were expanded and microinjected as above. From injections of 320 targeted clones the clonal germline transmission rate is at least 65% (Table 1 and Supplementary Data 2 online). This should be regarded as a minimum estimate of germline potential since the re-injection of clones is expected, in some cases, to recover failures. Targeted clones derived from the feeder-free JM8.N4 cell line appear to be particularly proficient, producing a 70% clonal rate of germline transmission.
Table 1.
Germline transmission of JM8 targeted clones.
| Microinjections | Male chimeras | Testcrosses | ||||||
|---|---|---|---|---|---|---|---|---|
| Cell line | Clones injected |
Clones at birth |
% of clones injected |
Clones at weaning |
% of clones injected |
Clones set up |
Clones with GLTa |
% clones injected with GLTb |
| JM8 | 61 | 61 | 100% | 52 | 85% | 52 | 38 | 62% |
| JM8.F6 | 108 | 104 | 96% | 87 | 81% | 85 | 67 | 62% |
| JM8.N4 | 151 | 151 | 100% | 133 | 88% | 126 | 104 | 69% |
| JM8A3 | 11 | 11 | 100% | 10 | 91% | 10 | 9 | 82% |
|
| ||||||||
| Total | 331 | 327 | 99% | 282 | 85% | 273 | 218 | 67% |
GLT, germline transmission.
Fraction of total clones injected that produce at least one male chimera with GLT
Copy number changes are common in cultured embryonic stem cells and, if transmitted, are likely to contribute to phenotypic variation14. Early passage JM8.F6 and JM8.N4 cells were expanded and examined by comparative genome hybridization (CGH) using a tiling path BAC array (0.2 Mb resolution; Supplementary Fig. 1 online). Both sublines exhibited a normal XY karyotype. For the JM8.F6 subline, CGH analysis revealed no copy number variants in a comparison with genomic DNA from C57BL/6NTac mice. Interestingly, the JM8.N4 subline harboured a copy number gain of a 1.7 Mb region (79,225,351-80,900,647) of chromosome 10. However, the hybridization signal suggested that this variant was present only in a subset of the cells.
Blastocysts of various strains can be used in combination with C57BL/6 embryonic stem cells (Fig. 2). We routinely used albino C57BL/6-Tyrc-Brd blastocysts as hosts — these mice have been maintained as a closed colony for 20 years and selected to be highly fecund for blastocyst production. The injection of C57BL/6N embryonic stem cells into C57BL/6-Tyrc-Brd blastocysts was a particularly favorable strain combination for germline transmission. However, in test crosses to detect embryonic stem cell contribution to the germline we were obliged to cross the chimeras to albino C57BL/6-Tyrc-Brd animals, producing hybrid F1 animals from the C57BL/6-Tyrc-Brd and C57BL/6N substrains. To obtain pure inbred C57BL/6N G1 mice, a separate cross with C57BL/6N mice is required after identifying chimeras with germline colonization. Although this can be avoided with strains like C57BL/6J-AW-J, C57BL/6J-Tyrc-2J or BALB/c (Figure 2), they are specialist strains with limited availability and poor fecundity, thus their use should be avoided.
Figure 2.
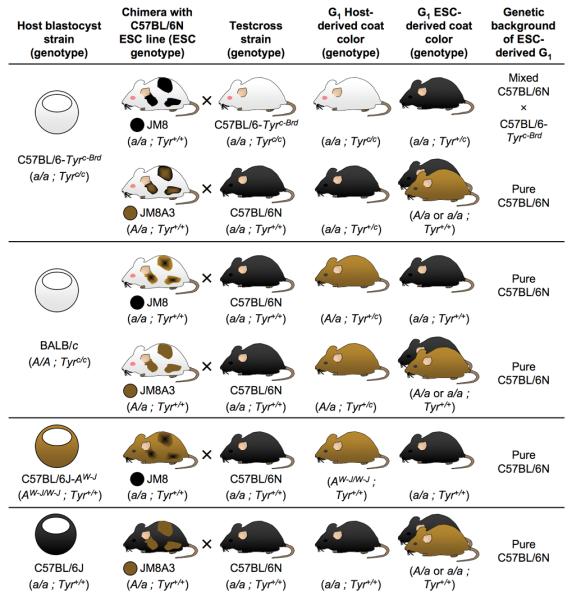
Coat color of chimeric mice and their offspring from injections of JM8 and JM8A3 cells into blastocysts from several common mouse strains. Possible test crosses to detect germline contribution are indicated, along with the expected coat color(s) and genetic composition of embryonic stem cell-derived (ESC-derived) and host-derived G1 offspring. For a full explanation of chimera coat colors see Supplementary Note online.
Currently, most transgenic facilities inject 129-derived embryonic stem cells into C57BL/6 host blastocysts. To capitalize on the widespread experience with, and availability of, the C57BL/6 host blastocyst, we repaired the Agouti locus in JM8.F6 embryonic stem cells by gene targeting. Restoring Agouti function to C57BL/6N ES cells allows visualization of embryonic stem cell derived mice by coat color and permits the recovery of pure inbred mice from test crosses with C57BL6/N mice.
The nonagouti mutation in C57BL/6 strains is due to the presence of an 11.8 kilobase pair (kbp) retrotransposon in the first intron of the Agouti gene which abolishes transcription of Agouti mRNA15. We designed a targeting strategy to delete the retrotransposon from the locus and restore Agouti gene function (Fig. 1c and Supplementary Fig. 2 online). The retrotransposon was replaced with a selectable marker, which was subsequently removed by Flp recombinase. The resulting Agouti allele contains a net deletion of 607 bp of intron sequence relative to the 129 strain Agouti allele and the retention of a single Flp recombinase target (FRT) site.
We injected three independent clones into albino C57BL/6-Tyrc-Brd blastocysts. All three produced chimeras with high percentage agouti coat color contribution and germline colonization, with one clone, JM8A3, showing particularly high frequencies (Table 1 and Supplementary Data 2). Since these cells are heterozygous for the corrected Agouti allele, testcrosses with C57BL/6N mice yield embryonic stem cell-derived offspring with either an agouti or a black coat (Fig. 1d). We analyzed gross copy number variation of JM8A3 DNA by array CGH against the JM8.F6 line and found no differences at the 0.2 Mb resolution of the assay (Fig. 1e). To assess the suitability of JM8A3 cells for high throughput gene targeting, we performed targeting experiments in the JM8A3 subline and measured the clonal transmission rate of targeted clones as above. We obtained a clonal germline transmission rate of 80% from the injection of eleven targeted clones (Table 1 and Supplementary Data 2).
In summary, we have derived robust, highly stable, germline competent embryonic stem cells from the C57BL/6 inbred genetic background that are suitable for high-throughput genetic manipulation. These cells are easily propagated using standard embryonic stem culture conditions, in the presence and absence of feeder cells, obviating the need for expensive specialty medium6. The JM8.F6 subline was further modified to introduce a dominant agouti coat color marker with no adverse effects on genome stability or germline transmission potential. Agouti JM8 cells take advantage of the widespread use of C57BL/6J mice for blastocyst injection by enabling visual assessment of coat color contribution and germline transmission. Moreover, C57BL/6N mice may be used in test crosses to identify chimeras with germline colonization and to produce pure inbred C57BL/6N G1 mice (Table 2). This saves one generation of breeding and facilitates the use of this cell line for both small and large-scale mouse genetics programs. JM8, JM8.F6, JM8.N4 and JM8A3 embryonic stem cells are available upon request from the KOMP (www.komp.org) and EUCOMM (www.eummcr.org) repositories.
METHODS
Mouse strains and embryonic stem cell derivation
C57BL/6J (Jackson Laboratory), C57BL/6JCrl (Charles River Laboratories) and C57BL/6NTac (Taconic) were obtained from commercial breeders. The C57BL/6-Tyrc-Brd mice were maintained as a closed colony by A.B17. Embryonic stem cell lines were established from C57BL/6J and C57BL/6N blastocysts on feeder cells using standard methods11.
Media and reagents
For embryonic stem cell culture, 500 ml Knockout DMEM (Invitrogen) was supplemented with 5 ml 100× l-glutamine (Invitrogen), 5 ml 100× β-mercaptoethanol (0.36 ml β-mercaptoethanol in 500 ml PBS, filter sterilised), 100 U/ml LIF (Millipore) and 10% to 15% foetal bovine serum (Invitrogen, Lot 40F1150K). Trypsin solution was prepared by adding 20 ml 2.5% trypsin solution (Invitrogen) and 5 ml chicken serum (Invitrogen) to 500 ml filter-sterilised PBS containing 0.1 g EDTA (Sigma) and 0.5 g d-glucose (Sigma).
Embryonic stem cell culture
JM8 embryonic stem cells were grown either on a feeder layer of SNL7 fibroblasts16 (neomycin and/or puromycin resistant) or on gelatinized tissue culture plates in embryonic stem cell medium containing 15% or 10% serum, respectively. For routine passage of stem cells grown on feeders, confluent cultures were washed twice with pre-warmed PBS and trypsinized for 15 minutes at 37°C. Ten volumes of pre-warmed medium was added, the cells were dispersed by passing gently through a pipette and transferred at a dilution of 1:3 into new plates containing feeders (plated for at least one week prior to use). Passage of cells on gelatinized plates was carried out in a similar manner except the cells were trypsinized for 7 min and passed at a dilution of 1:5 into freshly gelatinized plates (treated for 1 min with PBS containing 0.1% gelatin (Sigma)). The medium was replaced the next day and the cells typically reached confluence two days after passaging. For blastocyst injections, embryonic stem cells were grown for two days to confluence, trypsinized as described above, pelleted for 3 min at 1000 × g) and resuspended in HEPES-buffered embryonic stem cell media (pH 7.6) containing 15% serum.
SNP and CGH analysis
Genomic DNA was prepared by standard methods from early passage embryonic stem cells grown in the absence of feeders or mouse tail samples. SNP analysis was performed with a panel of 768 SNPs using the Illumina GoldenGate platform. 19 SNPs are polymorphic between N and J substrains (Supplementary Table). CGH analysis was performed as described14.
Targeting the non-agouti locus
Primers used for initial cloning of the Agouti breakpoint and genotyping the Atm1Brd allele were (5′ to 3′) GCTCCCCGCGGTGCTTCCAGATGTGGAAAGAAGTTC (includes SacII site) and CGCCGGTACCGATCTGGCACTGCCTTTAAGAGTA (includes KpnI site). The targeting vector endpoints are 2:154830701 and 2:154868845 (NCBI m37 coordinates).
Construction of the targeting vector (ATV-20) was as follows: A fragment of the wild-type agouti locus spanning the insertion site was amplified by PCR (using the primers above) from a 129S7 strain bacterial artificial chromosome (BAC, bMQ-37M9). A positive-negative selection cassette containing FRT-flanked neomycin phosphotransferase II (neo; confers G418/kanamycin drug resistance) and herpes simplex virus thymidine kinase (TK; sensitizes to the drug 1-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl)-5-iodouracil, FIAU) genes was cloned into BglII and EcoRI restriction sites which closely flank the insertion site. Use of these restriction sites results in a small deletion of a 607 bp BglII-EcoRI fragment in the final allele relative to wild-type agouti. We electroporated the resulting construct into recombination-competent E. coli18 carrying a C57BL/6J BAC (RP23-192A6) that contains the agouti locus (i.e. nonagouti allele). This resulted in the replacement of the nonagouti insertion in the BAC by the selection cassette. A fragment of this BAC containing the selection cassette was retrieved by gap repair recombineering to complete construction of the targeting vector. The 5′ and 3′ homology arms are nine and 13 kbp long respectively (Supplementary Fig. 2a). Vector DNA is isogenic with C57BL/6J except immediately around the selection cassette but targeting is efficient in a C57BL/6N background — for the targeting in JM8.F6 cells, seven out of fifty screened G418-resistant colonies were correctly targeted (14%). Clones were screened by Southern blot for the initial targeting (Supplementary Fig. 2b). A correctly targeted clone was expanded and electroporated with a pFLPe expression plasmid19 to remove the selection cassette and leave a single FRT site. Clones that had lost the selection cassette were selected in 200 nM FIAU and screened by PCR using the primers above (Supplementary Fig. 2c). The resulting Atm1Brd allele (MGI:3842513) confers an all-over agouti coat, unlike a spontaneously reversion isolated in a C57BL/6J colony, which has a white belly20 (Supplementary Fig. 2d).
Blastocyst injection
Blastocysts (3.5 days post-coitum) were harvested from natural matings of C57BL/6-Tyrc-Brd albino mice and expanded for 1–2 hours in embryonic stem cell medium containing 20% fetal calf serum. Up to 25 cells (fewer less expanded blastocysts) were microinjected into the blastocyl cavity and five injected blastocysts transferred to the uteri of pseudopregnant F1 female mice (five blastocysts per uterine horn). To test for germline transmission, male chimeras were bred to C57BL/6-Tyrc-Brd albino female mice. For JM8, JM8.F6 and JM8.N4 the presence of black pups in litters from these crosses indicates germline transmission. In the case of JM8A cells (Agouti heterozygous), a mixture of black and agouti pups are obtained in the case of germline transmission. All animal studies were carried out at the Wellcome Trust Sanger Institute under the UK Home Office licenses 80/2020 and 80/2076.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Qin, F. Law, L. Delaney, J. Meneses, T. Creek, H. Tharagonnet and A. Beasley for help with embryonic stem cell culture; N. Adams, J. White, R. Ramirez-Solis and the Wellcome Trust Sanger Institute microinjection team for blastocyst injections and mouse breeding; N. Conte and M. Storer for help with CGH. This work was funded by the Wellcome Trust Sanger Institute (WT077187), grants from the National Institutes of Health (KOMP, U01-HG004080 to W.C.S., K.L. and A.B; U01-42430 to D.R.B) and a grant from the Sixth Framework Programme of the EU (EUCOMM, to W.C.S. and A.B.).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Mouse Genome Sequencing Consortium Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 2.International Mouse Knockout Consortium Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 3.Kontgen F, Suss G, Stewart C, Steinmetz M, Bluethmann H. Int Immunol. 1993;5 doi: 10.1093/intimm/5.8.957. [DOI] [PubMed] [Google Scholar]
- 4.Auerbach W, et al. Biotechniques. 2000;29:1024–1032. doi: 10.2144/00295st04. [DOI] [PubMed] [Google Scholar]
- 5.Schuster-Gossler K, et al. Biotechniques. 2001;31:1022–1026. doi: 10.2144/01315st01. [DOI] [PubMed] [Google Scholar]
- 6.Cheng J, Dutra A, Takesono A, Garrett-Beal L, Schwartzberg P. Genesis. 2004;39:100. doi: 10.1002/gene.20031. [DOI] [PubMed] [Google Scholar]
- 7.Shimizukawa R, et al. Genesis. 2005;42:47. doi: 10.1002/gene.20122. [DOI] [PubMed] [Google Scholar]
- 8.Hughes E, et al. Mammalian Genome. 2007;18:549. doi: 10.1007/s00335-007-9054-0. [DOI] [PubMed] [Google Scholar]
- 9.Keskintepe L, Norris K, Pacholczyk G, Dederscheck S, Eroglu A. Transgenic Research. 2007;16:751. doi: 10.1007/s11248-007-9125-8. [DOI] [PubMed] [Google Scholar]
- 10.Hansen G, et al. Genome Research. 2008;18 doi: 10.1101/gr.078352.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagy A, Gertesenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. [Google Scholar]
- 12.Stiedl O, et al. Behav. Brain Res. 1999;104:1. doi: 10.1016/s0166-4328(99)00047-9. [DOI] [PubMed] [Google Scholar]
- 13.Khisti R, Wolstenholme J, Shelton K, Miles M. Alcohol. 2006;40:119. doi: 10.1016/j.alcohol.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang Q, Conte N, Skarnes W, Bradley A. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17453. doi: 10.1073/pnas.0805638105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bultman S, et al. Genes Dev. 1994;8:481–490. doi: 10.1101/gad.8.4.481. [DOI] [PubMed] [Google Scholar]
- 16.McMahon A, Bradley A. Cell. 1990;62:1073. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- 17.Liu P, Zhang H, McLellan A, Vogel H, Bradley A. Genetics. 1998;150:1155–1168. doi: 10.1093/genetics/150.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Court DL, et al. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- 19.Farley F, Soriano P, Steffen L, Dymecki S. Genesis. 2000;28:106. [PubMed] [Google Scholar]
- 20.Dickie M. J. Hered. 1969;60:20–25. doi: 10.1093/oxfordjournals.jhered.a107920. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.