Abstract
This review covers developments in the study of polymicrobial communities, biofilms and selected areas of host response relevant to dental plaque and related areas of oral biology. The emphasis is on recent studies in which proteomic methods, particularly those using mass spectrometry as a readout, have played a major role in the investigation. The last 5–10 years have seen a transition of such methods from the periphery of oral biology to the mainstream, as in other areas of biomedical science. For reasons of focus and space, the authors do not discuss biomarker studies relevant to improved diagnostics for oral health, as this literature is rather substantial in its own right and deserves a separate treatment. Here, global gene regulation studies of plaque-component organisms, biofilm formation, multispecies interactions and host–microbe interactions are discussed. Several aspects of proteomics methodology that are relevant to the studies of multispecies systems are commented upon.
Keywords: biofilm, dental plaque, Fusobacterium nucleatum, host response, microbial proteomics, multidimensional protein identification technology, Porphyromonas gingivalis, Prevotella intermedia, quantitative proteomics, Treponema denticola
Organisms comprising dental plaque were among the first bacteria to be observed and described by van Leeuwenhoek during the dawn of microbiology in the mid-17th century. It is now known that dental plaque is a complex and dynamic biofilm that develops on tooth surfaces through the sequential and ordered accumulation of several hundred species of bacteria [1,2]. In the early stages of development, plaque comprises predominantly oral streptococci and actinomyces, and usually exists in commensal harmony with the host. However, later population shifts lead to overrepresentation of acidophiles or of Gram-negative obligate anaerobes in subgingival plaque, which, respectively, contribute to the onset and progression of the most common oral diseases, caries and periodontal disease [3–5].
Spatial and temporal communities develop within plaque. These tend to be polymicrobial, with constituent species providing mutual nutritional and physiological support. Additional synergistic interactions can occur through the provision of attachment sites and interspecies signaling, all of which can serve to facilitate the retention of organisms and their adaptation to a multispecies community [3–5]. The interdependence of plaque constituents contributes to the inability to cultivate many oral bacterial species in the laboratory. Indeed, it has been estimated that over half of the bacteria in dental plaque have yet to be cultured [6]. Not all interbacterial interactions in plaque benefit the participant species. Conflict between organisms can occur through competition for space and nutrients, the production of bacteriocins or toxic metabolites and antagonistic signaling [3,5].
Specific plaque inhabitants have well-defined virulence properties. Streptococcus mutans ferments dietary carbohydrate to produce lactic acid, which eventually lowers the pH to levels at which demineralization of tooth enamel (caries) occurs [7,8]. Porphyromonas gingivalis, a Gram-negative black-pigmented anaerobe that is associated with several forms of severe periodontal disease, produces a variety of virulence factors, such as proteases, that contribute to gingival tissue destruction [9–11]. However, it is important to remember that these virulence factors are expressed in the context of a heterotypic microbial community. Furthermore, potential pathogens are often present in the absence of disease, indicating that host and other microbial factors make an essential contribution to the transition of plaque from a commensal to a pathogenic entity. Oral diseases can therefore be considered as community-based opportunistic infections. It has also been proposed that the translocation of oral bacteria to remote sites, and their colonization of tissues, can lead to systemic diseases such as coronary artery disease and preterm delivery of low-birth-weight infants [12]. Unraveling the basis of dental plaque development, therefore, will ultimately contribute both to oral health and to general overall health. Here, recent discoveries in biofilms, plaque and related areas of oral biology in which the tools and techniques of proteomics and protein biochemistry played a major role are selectively reviewed. It should be clear that these tools have now become so closely integrated with traditional genetics, various array technologies, next-generation sequencing and so on that proteomics is now essentially fully integrated into microbiological research.
Stress responses of oral pathogens
Global regulators
Free-living bacteria are exposed to ever-changing environmental conditions, such as nutritional deprivation, temperature shift, osmotic change, and exposure to antibiotics and DNA damage agents. To adapt to and survive these stresses, they have global response systems that adjust the rates of intracellular metabolic processes. These responses are controlled by global regulators, which include alternative sigma factors, such as RpoS and RpoH; small molecule effectors, such as (p)ppGpp; gene repressors, such as LexA; and inorganic molecules, such as polyphosphate [13,14]. The response pathways extensively overlap and are induced to various extents by the same environmental stresses [15,16]. All of these global stress responses include functions that can increase genetic variability. In particular, upregulation and activation of error-prone DNA polymerases, downregulation of error-correcting enzymes and movement of mobile genetic elements are common features of several stress responses [1,13]. Using these mechanisms, bacterial genetic changes are induced under a variety of stressful conditions, including the biofilm state [17].
Toxin–antitoxin module organization & stress-response regulation
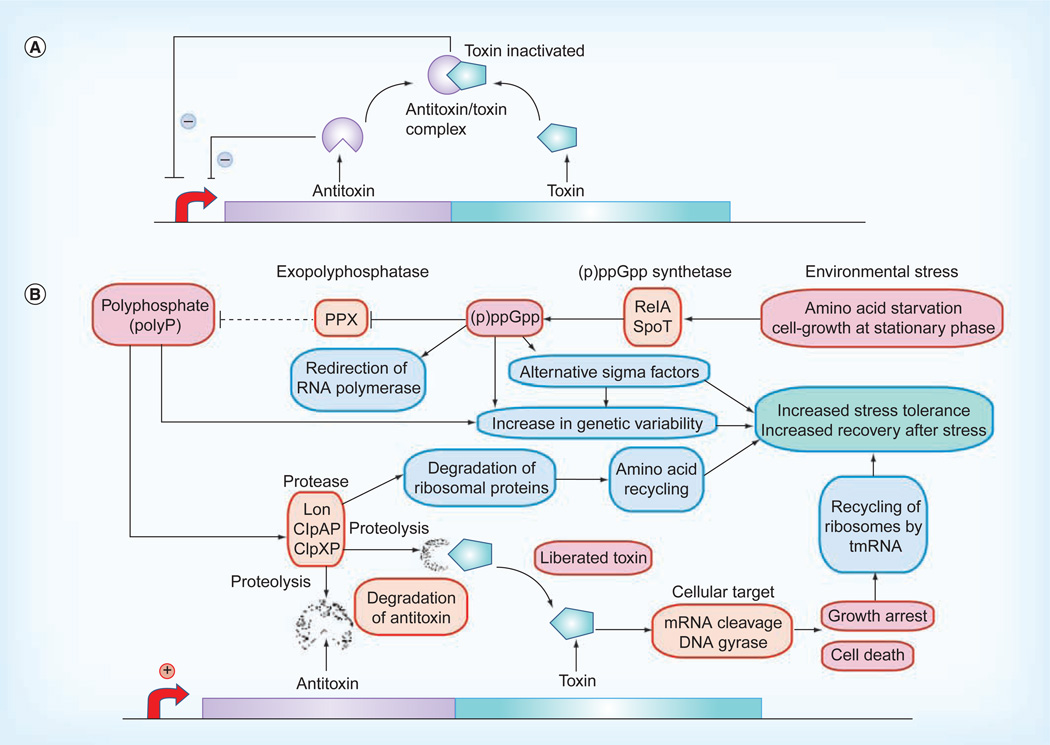
Toxin–antitoxin (TA) systems are widely distributed, two-component systems implicated in programmed cell death and/or growth inhibition, and influence biofilm formation and persistence [18,19]. TA modules encode a stable ‘toxin’ protein, whose activity results in either growth arrest or cell death, and an unstable ‘antitoxin’ that counteracts toxin activity (Figure 1). Antitoxins are either antisense RNAs that suppress toxin expression (type I TA system) or labile proteins that bind and inactivate their cognate toxin (type II TA system). The protein antitoxins of type II TA systems also act as transcriptional repressors that autoregulate TA expression. Toxins typically target translation through mRNA cleavage and DNA replication through DNA gyrase inhibition [19]. Environmental stresses such as amino acid starvation and cell growth at stationary phase activate chromosomal TA modules through antitoxin degradation by Lon (DNA-binding ATP-dependent protease La), the ClpP (caseinolytic protease) complex ClpXP or ClpAP, or unknown protease (Figure 1).
Figure 1. Toxin–antitoxin module stress-response regulator.
(A) Toxin–antitoxin (TA) module genetic organization. TA systems are typically two-gene operons with the antitoxin encoded upstream of the toxin gene. Antitoxins bind their cognate toxins and inhibit toxin activity. Antitoxins and TA complexes repress TA module transcription. Proteomic methods have played an important role in elucidating TA module structure and function [86,87]. (B) The stress-response regulator hypothesis of chromosomal TA function. Environmental stresses such as amino acid starvation activate chromosomal TA modules through sequential events starting with (p)ppGpp biosynthesis. ppGpp inhibits exopolyphosphatase and thereby leads to polyP accumulation; polyP in turn activates Lon to degrade idle ribosomal proteins. Antitoxins are degraded either by Lon, ClpAP or ClpXP proteases, and activated toxins inhibit cell growth and/or lead to cell death. tmRNA: Transfer-messenger RNA.
Interestingly, our proteomics study of a mixed species biofilm revealed that P. gingivalis suppressed the expression of SpoT (PGN1757; a GTP pyrophosphokinase), which is involved in the production of (p)ppGpp (Figure 1), when in communication with Streptococcus gordonii and Fusobacterium nucleatum. This phenomenon was consistent with the other results of biosynthetic pathway analyses and reflected the less-stressed state of P. gingivalis in the three-species mixed community relative to the monospecies culture [20].
The oral spirochete Treponema denticola is a Gram-negative, motile, asaccharolytic, anaerobic spirochete that has been shown to be strongly associated with chronic periodontitis. Recently, it was reported that genes encoding annotated homologs of TA systems (TDE1455, TDE1978 and TDE 1979), putative prophage and a family of putative transposases in T. denticola ATCC 35405 were upregulated when growing as a biofilm [21]. Intact bacteriophage particles were isolated from this strain, and their functional capability was confirmed, thus a higher potential for genetic mobility in the biofilm state was indicated.
Clp proteolytic complex
Clp proteolytic complexes, consisting of a proteolytic core ClpP flanked by Clp ATPases, are widely conserved in bacteria and play vital roles in adaptation under stress conditions [22]. In addition, Clp complexes are required for the optimal expression of virulence in a number of pathogens [23]. The Clp ATPases contain either one or two nucleotide-binding domain(s), and the length of the spacing between these domains, as well as the presence of specific signature sequences, forms the basis for the subfamily classification ClpA, ClpB, ClpC, ClpE, ClpL and ClpX. P. gingivalis possesses the ClpP proteolytic subunit along with the ATPases ClpB, ClpC and ClpX. In this organism, absence of the ClpXP complex diminished its tolerance to high temperature but elevated monospecies biofilm formation and enhanced heterotypic biofilm formation with S. gordonii [24].
A ClpP knockout mutant of S. mutans differentially regulates more than 100 genes, such as stress-related genes, genes encoding bacteriocin-related peptides and transcriptional regulators, genes in genomic islands and those associated with a putative CRISPR (clustered, regularly spaced short palindromic repeats) locus. Comparison of the proteomes between wild-type and ClpP-deficient strains revealed that 33 proteins were downregulated, while 21 proteins, including several transcriptional regulators, were upregulated in the ClpP-deficient strain [25].
pH stress
During the onset of periodontitis, the pH of the periodontal pocket increases with pocket depth and the severity of the inflammatory host response [26]. F. nucleatum is believed to play an important role in the microbial succession associated with the development of periodontal diseases. In comparison with other plaque inhabitants, F. nucleatum has the greatest ability to neutralize acidic environments [27]. In this organism, 22 soluble cytoplasmic proteins were differentially expressed in response to external pH as identified by 2D gel electrophoresis and mass spectrometry [28]. Eight differentially expressed proteins associated with increased energy (ATP) production via the 2-oxoglutarate and Embden–Meyerhof pathways appeared to be directed toward either cellular biosynthesis or the maintenance of internal homeostasis. Interestingly, several upregulated proteins under the low pH (pH 6.4) condition, such as NAD-specific glutamate dehydrogenase, glutamate formiminotransferase and oxygen-insensitive NADPH nitroreductase (dihydropteridine reductase), were associated with folate derivative biosynthesis and its closely related pathways.
Various species of Streptococcus, one of the most abundant genera in the mouth, are associated with oral health, as well as with dental caries. Cariogenic streptococci, such as S. mutans and Streptococcus sobrinus, depend on a biofilm lifestyle for survival and persistence in the oral cavity and have developed sophisticated mechanisms to cope with environmental stress [29]. Stress tolerance mechanisms, especially acid tolerance mechanisms utilized by S. mutans, have been extensively investigated [30]. Interspecies microarray and proteomic comparisons between S. mutans and S. sobrinus were used to identify novel pathways that participate in the S. sobrinus-specific acid stress response. The results revealed that metabolic alterations that enhance energy generation and upregulation of the malolactic fermentation enzyme activity constitute important acid resistance properties in S. sobrinus [31].
Oxidative stress
Using a 2D gel electrophoresis and mass spectrometry approach, 78 proteins that displayed differential intracellular concentration in response to either O2- or H2O2-induced oxidative stress were identified in F. nucleatum [32]. Three major protein systems were altered in response to oxidative stress: proteins of the alkyl hydroperoxide reductase/thioredoxin reductase system were elevated, presumably to reduce the amount of reactive oxygen species in the cell; glycolytic enzymes were elevated or modified by oxidation (i.e., d-glyceraldehyde, 3-phosphate dehydrogenase and fructose 6-phosphate aldolase), with an accompanying decrease in ATP production; and levels of molecular chaperones and related proteins (i.e., ClpB, DnaK, HtpG and HrcA) were increased, presumably to counter the harmful effects of reactive oxygen species.
Iron starvation
Because iron is an essential nutrient for most bacteria, pathogens have developed specific iron acquisition systems to counteract iron deprivation in the host. Many aerobic and facultative anaerobic bacteria produce siderophores, low-molecular-mass iron chelators that remove iron complexed to host iron-carrying proteins and deliver it to bacterial cells.
Unlike aerobic or facultative anaerobic bacteria, P. gingivalis does not produce siderophores to chelate environmental iron. Instead, P. gingivalis stores heme on its surface in the form of micro-oxo bis-heme, which has inherent catalase activity that helps to protect the cell from oxidative stress [33]. The initiation and progression of periodontal disease are associated with bleeding at the site of disease, thereby providing an elevated level of hemoglobin. The Arg- and Lys-gingipain proteases of P. gingivalis are involved in proteolysis of hemoglobin from which the defensive dimeric heme pigment is formed. Environmental heme availability has been reported to affect the virulence of P. gingivalis, although the exact effects are still unclear. A combination of large-scale quantitative proteomics and DNA microarray analysis was used to identify differences in P. gingivalis W50 protein and transcript abundances between heme-excess and heme-limited continuous culture conditions [34]. This approach identified 160 genes and 70 proteins including aspartate and glutamate catabolic enzymes, which were differentially regulated by heme availability.
Fumarate respiration is the most widespread type of anaerobic respiration. Conversion of aspartate to succinate via fumarate respiration, which is catalyzed by a heterotrimeric succinate–quinone oxidoreductase complex consisting of two cytoplasmic enzymes FrdA (PG1615) and FrdB (PG1614), and a transmembrane protein FrdC (PG1616), is important for energy and efficient growth [35]. Under heme limitation, FrdA and FrdB showed a three- to four-fold decrease in abundance, whereas all enzymes on the pathway of glutamate catabolism significantly increased. These results demonstrated a shift from an energy-efficient pathway of aspartate catabolism to a less efficient process of glutamate catabolism.
Heme limitation also resulted in an increase in abundance of PG1374, a protein that plays a role in epithelial cell invasion. The greater abundance of a number of transcripts/proteins linked to invasion of host cells, the oxidative stress response, iron/heme transport and virulence of the bacterium indicated that there was a broad response of P. gingivalis to heme availability.
Prevotella intermedia plays important roles in the initiation and development of periodontitis by stimulating host cells to release proinflammatory cytokines and proteinases such as matrix metalloproteinases [36]. Although P. intermedia is also found at healthy sites, the profile of degradative enzymes produced by the organism varies depending on the site, suggesting that this organism alters its enzyme profile to become more virulent under certain conditions. Furthermore, P. intermedia has been shown to be resistant to many antibiotics including penicillins, cephalosphorins and tetracyclines, thus the elimination of this organism may also be critical owing to the fact that it can serve as a reservoir of antibiotic resistance [37,38]. As hemin is an indispensable nutrient and hemin acquisition mechanisms have been shown to be induced in iron-depleted conditions in P. intermedia, proteomic approaches were applied to detect expression changes that were affected by iron [39]. Forty protein spots were analyzed by 2D fluorescence difference gel electrophoresis in association with mass spectrometry, and 19 proteins were identified. Two homologs of hemin uptake receptor protein, PIN0009 and PINA0611, were upregulated 38-times more in iron-depleted conditions than control. PIN0009 is encoded by a gene that is the first of a seven-gene genomic locus of a hemin acquisition system, which is known as the hmu locus in P. gingivalis. PIN0009 is present on a megaplasmid, suggesting that the megaplasmid-encoded hmu locus was acquired later and retained due to the importance of hemin acquisition. The second protein, PINA0611, is a homolog of numerous TonB-dependent outer membrane receptors. Additionally, a hemoglobin-binding protein and a thioredoxin-like protein were identified as iron- regulated proteins.
Biofilms
Biofilm formation is a complex process that requires the coordinate expression and simultaneous regulation of many genes for reversible and irreversible attachment, microcolony formation, formation of a stable 3D structure and dispersion. Biofilm-associated cells grow significantly more slowly than planktonic cells [40]. To illustrate which proteins changed in abundance between the planktonic and biofilm growth states, the cell envelope fraction of P. gingivalis W50 cultivated in a chemostat was analyzed [41]. Proteins were labeled with H2 16O or H2 18O then identified and quantified by gel-enhanced LC-MALDI TOF/TOF MS. Within the identified 81 cell envelope proteins, 24 increased and 18 decreased in abundance in the biofilm. The heme-binding lipoprotein HmuY, GAPDH and some of the cell-surface-located C-terminal domain (CTD) family proteins, including RgpA, HagA, CPG70 and PG99, increased in the biofilm cells. Concurrently, the heme-binding protein IhtB, TonB-dependent receptor P92, Kgp, lipoprotein RagB, endopeptidase PepO, a glutamine cyclotransferase-related protein, fumarate reductases FrdAB, and ribosomal proteins L10, L13 and L25 significantly decreased. The same research group analyzed P. gingivalis ATCC33277 outer membrane proteins using a proteomics approach and reported 24 identified proteins that were categorized into a CTD protein family. In that study, they demonstrated that the CTD was involved in a coordinated process of export and attachment to the cell surface [42]. Tannerella forsythia is a Gram-negative, anaerobic, fusiform bacterium that is found together with P. gingivalis and T. denticola in human subgingival plaque. These bacterial species are classified into the ‘red complex’, and members of this consortium are strongly implicated as periodontal pathogens [43]. With the use of 2D-PAGE, SDS-PAGE and LC-MALDI-TOF/TOF MS, 221 proteins of T. forsythia outer membrane preparations were identified, of which 197 were predicted to be located in the cell envelope [44]. Thirteen proteins were found to share sequence similarity over approximately 60 amino acid residues at their extreme C-terminal ends, which are similar to the CTD of a family proteins in P. gingivalis and certain other members of the phylum Bacteroidetes, and thus were designated as CTD family proteins in T. forsythia. They include surface-layer protein A (TfsA), surface-layer protein B (TfsB), BspA and a possible internalin-related protein (TF1032), while the remainder are annotated as hypothetical proteins. By using replicated 2D gels stained with a carbohydrate-specific fluorescent dye, it was revealed that the CTD family proteins exhibited higher molecular weight than expected because of glycosylation.
Recently, iTRAQ technology was applied to identify and quantify alterations in global protein expression of T. forsythia during growth in a biofilm [45]. In total, 348 proteins were identified and quantified with the expression of 44 proteins being significantly altered between biofilm and planktonic cells. Upregulated proteins in biofilms were putative transport proteins and S-layer proteins. The butyrate production pathway was markedly downregulated in biofilms. Given these data, the possible alterations in the nature of host interactions were indicated.
One of the challenges of researching T. denticola, another member of the ‘red complex’ consortium, is the difficulty in cultivating them. Veith et al. developed methods for the cultivation of this bacterium in continuous culture, which has the advantages of allowing the generation time and environmental factors to be rigorously controlled [46]. They prepared membrane-enriched and cytoplasm-enriched samples and identified 219 proteins, including numerous virulence factors, lipoproteins, ABC transporter proteins and enzymes involved in the metabolism of nine different amino acids. A 2D western blot analysis using sera from mice immunized with formalin-killed T. denticola cells suggested that Msp, PrcA, OppA, OppA10, MglB, TmpC and several flagellar filament proteins are antigenic.
Aggregatibacter (Actinobacillus) actinomycetemcomitans is a Gram-negative, capnophilic bacterium associated with localized aggressive periodontitis and refractory forms of chronic periodontitis. The transcription and translation of genes/proteins responsible for biofilm determinants such as fimbria, lipopolysaccharide (LPS) and EPS production were determined in A. actinomycetemcomitans strains D7 and HK1651 under various stress conditions. The results indicated that anaerobic conditions, nutrient stress and iron limitation each upregulate known biofilm determinants of A. actinomycetemcomitans to contribute to biofilm formation [47]. A 2D electrophoresis-based proteomic analysis of A. actinomycetemcomitans NCTC9710 was performed to investigate the influence of microenvironments, including biofilm/planktonic mode of growth, aerobic/anaerobic atmospheres, iron depletion and presence of serum or blood. Differential expression of a number of the secreted surface-associated proteins was observed under different growth conditions, and these included the glycolytic enzyme triose phosphate isomerase, thiol peroxidase, [Cu,Zn]-superoxide dismutase and YfiD protein [48].
Multispecies interactions
Dental plaques are polymicrobial in nature, and certain diseases of the oral cavity are believed to be the result of changes to communities rather than only due to the influence of individual organisms [49,50]. Within these communities, interbacterial signaling and communication can occur, by which one organism influences gene and protein expression in another. In order to address questions of interactions among oral microbes in a tractable system, the response of P. gingivalis in a simplified microbial community consisting of P. gingivalis, S. gordonii and F. nucleatum has been examined [20]. Whole-cell proteomics was used to determine changes in protein levels between P. gingivalis alone or coincubated with S. gordonii or S. gordonii and F. nucleatum. P. gingivalis in contact with S. gordonii showed limited changes that may reflect fine tuning of the proteome to allow attachment and accretion. Similar low level changes occur in the transcriptome, but these are targeted to pathways that control expression of adhesins and signaling molecules [51,52]. This is in keeping with models of dental plaque where S. gordonii is an early colonizer, binding to tooth surfaces, while P. gingivalis is a late colonizer generally interacting with the developing biofilm. By broadening the scope of the model to include F. nucleatum, an organism that provides physiological support to P. gingivalis [53,54], we begin to see more significant changes in protein levels in a number of biological pathways.
Forty-four proteins annotated as involved in the cell envelope, such as outer membrane constituents, showed significant changes in the three species community [20]. Four proteins showed increased abundance compared with P. gingivalis alone but most had reduced abundance, including MreB (PGN0234), a bacterial actin homolog that influences cell shape.
Several results indicated possible physiological support from the microbial community. DNA repair proteins showed a general decrease, implying reduced DNA damage when all three organisms were present. P. gingivalis also showed a significant increase in protein synthesis [20]. Most ribosomal proteins and translation initiation and elongation factors demonstrated increased abundance. These studies were not conducted in culture media so the results likely indicate increased translation, consistent with physiological support from the community, rather than rapid growth.
Physiological support may come in terms of nutrient transfer. Proteins for several vitamin synthesis pathways, thiamine (vitamin B1), pyridoxal phosphate (vitamin B6) and cobalamin (vitamin B12), showed reduced abundance in the three-species community [20]. However, neither of the two proteins in P. gingivalis known to employ thiamine as a cofactor showed decreased levels, implying that the demand for thiamine was unchanged.
The pyrimidine biosynthesis pathway also appeared to be downregulated in the three-species community [20]. However, the proteins responsible for incorporating finished ribonucleotides into RNA were unchanged or increased, implying an alternative source of nucleotide. Decreases in DNA repair and replication have also been observed between planktonic P. gingivalis cells and those in biofilms [55], likely due to decreased growth rate in the biofilm. However, as the three species community was studied in nonculture media, growth rate differences are an unlikely explanation. In addition, uracil permease (PGN1223) showed increased abundance in the three species community, and P.gingivalis is known to take up nucleosides and nucleobases that may be important nutrients [56].There has been speculation that lactate and formate may be important sources of nutrient transfer to P. gingivalis from the Streptococcus species common in the oral cavity. S. gordonii is known to be able to support Veillonella alcalescens in vitro through nutrient transfer of lactate [57], and P. gingivalis has been shown to take up lactate and formate [58]. Analysis of P. gingivalis grown under anaerobic and microaerobic conditions indicated that the lactate permease transcript (PGN1128) and lactate consumption from the growth medium were reduced under microaerophilic conditions compared with anaerobic growth. The authors theorized that under anaerobic conditions, lactate may be an important source of nutrient transfer from Streptococcus to P. gingivalis. However, in the anaerobic three species community, P. gingivalis showed reduced levels of lactate permease, and it may also be reduced in the P. gingivalis plus S. gordonii coincubations. While nutrient transfer appears to be occurring in the coincubations, lactate does not seem to be a significant part of the transfer [20]. The formate–tetrahydrofolate ligase (PGN1111) and a formate/nitrate transporter (PGN0314) transcript as well as formate utilization from the media [58] were increased under microaerophilic conditions. The three species community also showed an increase in formate–tetrahyrofolate ligase (PGN1111) though the transporter was not detected [20], making formate a likely candidate for nutrient transfer.
Technical considerations
The work described in the study by Kuboniwa et al. [20] and subsequent follow-up papers currently in preparation are limited by several factors. Unlike metagenomic or metaproteomic studies, where the prevailing philosophy is ‘any information about the system under investigation is better than no information’, more traditional analytical concerns have greater relevance to a tractable model community consisting of several organisms with fully sequenced and annotated genomes. As microbial community complexity increases, a metaproteomics study of a system consisting of hundreds or thousands of different organisms will tend toward being dominated by a few proteotypic peptides that are particularly amenable to the analytical approach being employed. Typically, such a peptide will extract and enrich well, lack major issues with fixed regions of secondary structure, respond well to a range of HPLC conditions and have a higher than average molar response factor for the electrospray process that is most often used to ionize the peptide prior to mass/charge analysis in the mass spectrometer. It has been widely observed, regardless of sample type or origin, that the more complex the sample, the greater the dominance of such proteotypic peptides. Proteins or classes of proteins that are the putative sources of such highly responsive proteolytic fragments will therefore tend to be overrepresented in the dataset. The obvious problem with such a scenario is that the relationship of the experimental observations to what is really occurring in nature is tenuous at best as the system being sampled increases in complexity. At the metaproteomic level, results are generally qualitative only. With a simpler system, such as described in [20], the quality of the calculated relative abundance ratios will be compromised and limited by increasing complexity as well, but at least it is often possible to calculate a number that can be interpreted in a straightforward manner. Thus, multiple conditions of interest can be compared quantitatively as well as qualitatively. Methods exist today by which one can statistically demonstrate the effects on the dataset of system complexity. One approach to model community studies works as follows: first, verify that the numbers of organisms of each species in the model are present in roughly equal quantities. This may not be the best assumption in terms of the biology, but the calculations employed make this assumption unless an explicit correction factor is employed. Second, determine that proteome qualitative coverage is roughly equal for all organisms, that is, sampling statistics are the same for each proteome. The greater the deviation from this assumption of equal coverage, the less reliable the numbers from subsequent abundance ratio calculations. Differences in coverage can be corrected to a limited degree by adjusting the numbers of unique peptides required for a protein assignment for each organism. Ideally, this minimum nonredundant spectral count value, typically n = 2–5 in most instances, will be constant for all proteomes extracted from the model community. Third, determine a set of qualitative false discovery rates (FDRs) as a function of the number of unique peptides required for positive identification. Select a value for n that provides the best compromise in terms of coverage at the individual protein level, coverage at the proteome level and the desired level of statistical confidence in the data. Fourth, normalize the data such that the total spectral counts, number of heavy–light pairs and so on are equal for each proteome being compared in terms of relative abundance ratios. Normalization factors will generally be quite modest if the experiments were done correctly and the level of complexity is tractable. With the system described in [20], normalization factors are generally less than 20% of the total observed signal for any given proteome. Fifth, proceed to calculating relative abundance ratios for the biological states of interest using methods appropriate to metabolic isotopic labeling, post-harvest isotopic labeling, native abundance peak areas or spectral counting, as one chooses. The formalisms employed for quantitative significance testing will differ depending on the approach. Computational details and references for two of these methods, native abundance peak area (summed signal intensity) and spectral counting can be found in [20]. Quantitative FDRs will necessarily become greater in magnitude (tending toward less significance) on average, as the complexity of the system increases, and will of necessity be less favorable than those typically employed in single-organism studies. The danger of generating an unchanging ‘flat’ set of uninformative quantitative FDR values is significant with such data, and it is especially important to work with log-transformed abundance values that are, if not normally distributed, at least unimodal and somewhat symmetric about a measure of central tendency. This will seldom be the case when working with the untransformed data. Once one has established a reasonable target qualitative FDR for the system, abundance ratio calculations can proceed more or less as per single organism studies, with the caveat that observed p and q values will generally not be as easy to interpret in terms of an obvious cutoff value as with less complex analyses. Biological intuition plays at least an equal role relative to purely statistical guidelines in terms of setting such cutoff values. We tend to look at cutoff values as dynamic, rather than static, and with modern computational methods, it is straightforward to observe the finished dataset under a range of cutoffs, much as one would observe a time-lapse video. Such a presentation of the data has proven to be a much more useful guide to biological insight than a simple static assignment of a single q or p cutoff. Our standard practice is to include a user-adjustable cutoff value as a part of every database we construct. The human readable form of the fully processed dataset normally consists of a relational database such as MySQL or FileMaker.
Metaproteomic analysis of oral microbiota
To examine the feasibility of metaproteomic approaches for obtaining useful information from commensal microbial communities sampled against a high background of host proteins, salivary microbiota collected from oral squamous cell carcinoma patients and healthy persons were analyzed for taxonomy and metabolic activities [59]. First, human salivary proteins were examined by a 3D peptide separation method that identified over 2000 host origin proteins [60], then a metaproteomic analysis was performed on the salivary microbiota, whose peptide dataset was coexistent with that of the host proteins. To identify microbial species and generate a phylogenetic tree, the Uniprot/Swiss-Prot protein sequence database [101], the NR database [102], the metagenomic software MEGAN [61] and the 16S-rRNA-based Human Oral Microbiome Database [103] were used. The Clusters of Orthologous Groups prokaryotic database [62,104] and the MetaCyc [105] were used for functional analysis of the peptide dataset. Sixty-three percent of the peptides were reported to be identifiable and distributed among five bacterial phyla (61%), archea (0.5%) and viruses (0.8%), whereas the rest were identifiable only at the level of multicellular organisms or bacteria. Twenty-nine percent were assignable at the genus level, and most belonged to Streptococcus (17%). Eleven percent of all peptides could be assigned to species, including periodontal pathogens such as P. gingivalis and T. denticola. The pathway analysis indicated that peptides were linked to translation (37%), followed by glycolysis (19%), amino acid metabolism (8%) and energy production (8%), thus salivary microbes appear to be actively engaged in protein synthesis.
In another metaproteomic study, reverse-phase nanoliterscale liquid chromatography/tandem mass spectrometry was applied to identify proteins of bacterial origin present in endodontic infections [63]. Proteins, mainly of cell wall or membrane from Enterococcus faecalis, Enterococcus faecium, P. gingivalis, F. nucleatum and T. denticola, were identified from all the samples tested and included adhesins, autolysins, proteases, virulence factors and antibiotic resistance proteins.
Host–microbe interactions
Proteomic approaches have been used to decipher the molecular dialog between oral bacteria and host cells both in tissue culture models and in vivo. In the subgingival compartment, gingival epithelial cells (GECs) are among the first host cells encountered by oral bacteria. Oral organisms such as P. gingivalis can respond on a global scale to secreted GEC components. In addition, P. gingivalis can internalize and survive within GECs with concomitant adaptation of the expressed proteome of the organism [64]. Multidimensional protein identification technology analysis has found differential expression of 60 P. gingivalis proteins in response to secreted GEC components: 35 upregulated and 25 downregulated [65]. Several proteins involved in hemin uptake showed reduced expression levels. In addition, similar to P. gingivalis under oxidative stress, there were increases in the levels of Clp stress response system proteins (ClpC, CplP and ClpX), as well as several heat shock proteins. The overall conclusion is that the extracellular epithelial cell environment is hemin sufficient but places more stress upon P. gingivalis. The broadly based changes to the P. gingivalis proteome in response to the GEC secretome also allow P. gingivalis to prepare for contact and invasion, and the transition to the intracellular lifestyle. ClpC and ClpXP are necessary for entry of P. gingivalis into host epithelial cells [24]. Another protein upregulated by secreted GEC components is SerB, a haloacid dehydrogenase family phosphoserine phosphatase enzyme. SerB impacts host cell signal transduction pathways that control the cytoskeletal architecture, and treatment of epithelial cells with SerB induces actin microfilament and tubulin microtubule rearrangements [66,67]. Furthermore, SerB is required for intracellular persistence as a mutant lacking SerB is compromised in intracellular survival [67]. An additional role for SerB concerns involvement in the stealth-like properties of P. gingivalis [9]. P. gingivalis is capable of both inhibiting secretion of IL-8 from GECs and antagonizing IL-8 production stimulated by other organisms [68–71]. SerB activity is required for this innate immune suppression, known as localized chemokine paralysis [66,68].
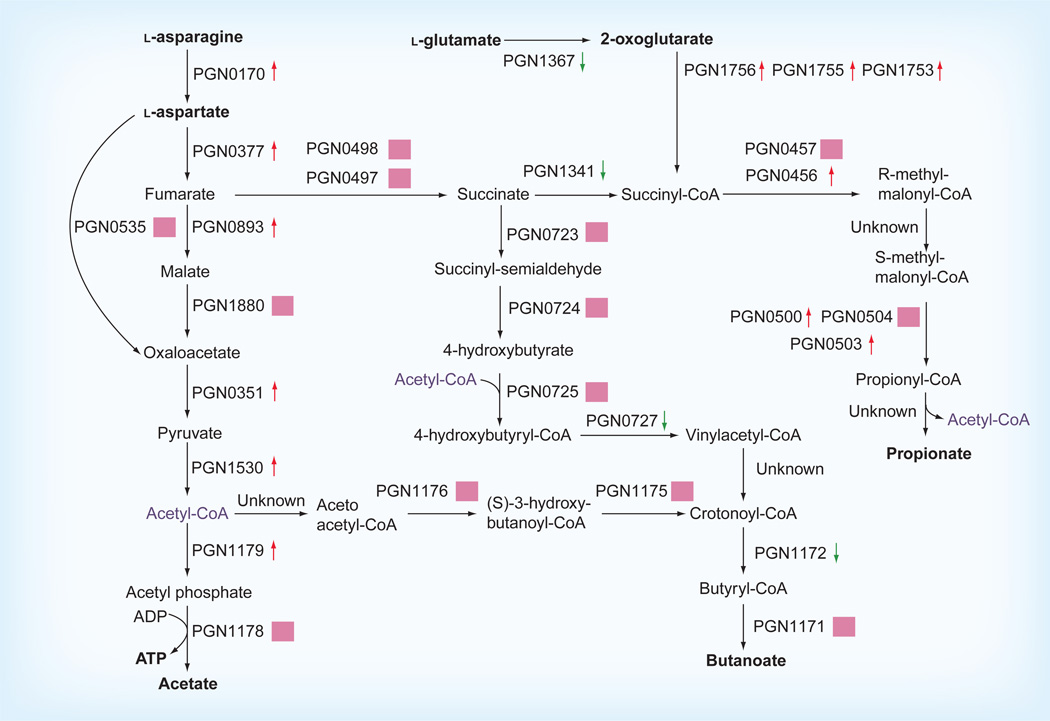
The proteome of internalized P. gingivalis has also been examined using multidimensional protein identification technology [72,73]. Similar to cells exposed to GEC-conditioned media, the internalized P. gingivalis showed decreases in hemin uptake proteins as well as increases in stress-related proteins including heat shock proteins, alkyl hydroperoxide reductases and two Clp proteases, ClpB and ClpC. Internalized cells also significantly reduced the expression of FimA, a subunit protein of long fimbriae, presumably as fimbriae are superfluous after internalization. Proteases were also downregulated in internalized P. gingivalis, indicating that the organism may control expression of compounds that are toxic to the host cells in order to prolong survival. Internalized P. gingivalis showed a general increase in the translational machinery. Most translation initiation, elongation and termination proteins as well as tRNA synthetases and ribosomal proteins had increased abundance in internalized P. gingivalis. The transcriptional machinery also showed increased abundance. Combined with the increase in energy metabolism, these results indicate that the interior of GECs is an energy-rich environment for P. gingivalis. There were also extensive changes in expression levels of proteins involved in diverse metabolic pathways (Figure 2) [72]. For example, the main energy-producing pathway from asparagine/aspartate to acetate and ATP had increased levels of protein components. Increased abundance was also seen for proteins in the pathway for production of the cytotoxin propionate, while the butyrate pathway had reduced or unchanged protein levels. The overall trend is to an increase in energy production in internalized cells and a shift in the production of cytotoxic metabolic by-products from butyrate to propionate. Given that butyrate is a more potent apoptosis-inducing agent than propionate, this is indicative of a shift to a physiology that supports long-term survivability of host cells. Indeed, GECs infected with P. gingivalis are resistant to chemically induced apoptosis and display an accelerated progression through the cell cycle [74,75]. A proteomic analysis of host cells using an antibody array with more than 600 cell cycle-related targets revealed that infection of GECs with P. gingivalis induces broadly based changes in the level and phosphorylation status of proteins that exert multilevel control on the eukaryotic cell cycle. Pathways that were impacted by P. gingivalis included those involving cyclins, p53 and PI3K [74].
Figure 2. Response of energy and cytotoxin metabolism in Porphyromonas gingivalis to internalization.
Proteins catalyzing each step from the preferred metabolites l-glutamate, l-aspartate and l-asparagine to ATP and cytotoxin production are shown by their P. gingivalis PGN designation. Red up arrows indicate statistically significant increases in protein levels 18 h after internalization into human gingival epithelial cells. Green down arrows indicate decreased levels and pink squares no statistically significant change. Acetyl-CoA appears as a substrate and product at multiple points and is shown in purple. The metabolic precursors l-asparagine, l-aspartate, l-glutamate and 2-oxoglutarate as well as ATP and the cytotoxic end products acetate, butanoate and propionate are shown in bold. The map shows a shift toward l-asparagine/l-aspartate metabolism and acetate and propionate production and a shift away from production of the more toxic butanoate after internalization. Data taken from [72].
While epithelial cells provide barrier and signaling functions in the gingival crevice, oral organisms will also encounter the phagocytic cells, polymorphonuclear granulocytes and monocytes. Using 2D electrophoresis and mass spectrometry, Saba et al. found that monocytic cells (THP-1) responded to P. gingivalis LPS through upregulation of deoxyribonuclease, actin, carbonic anhydrase 2, α-enolase, adenylyl cyclase-associated protein (CAP1), protein disulfide isomerase, glucose-regulated protein (grp78) and 70-kDa heat shock protein (HSP70) [76]. Stimulation with live P. gingivalis caused upregulation of CAP1, tubulin β-2 chain, ATP synthase β chain, tubulin α-6 chain, protein disulfide isomerase, vimentin, 60-kDa heat shock protein and nucleolin; and downregulation of carbonic anhydrase II, β-actin and HSP70. The differential outcome to challenge with P. gingivalis or its LPS indicates preferential signal pathway activation in host immune responses to infection. ‘Multiplex technology’, basically a reagent kit for a targeted set of proteins, was used in conjunction with other techniques to show that human blood monocytes, differentiated dendritic cells and macrophages respond to P. gingivalis LPS through the production of proinflammatory cytokines [77]. Moreover, the long-lived myeloid inflammatory cells, particularly dendritic cells, responded rapidly to P. gingivalis LPS by secreting chemokines, cytokines and proteases capable of driving T-helper polarization. In the absence of activation of immunosuppressive pathways, these results were interpreted as evidence for a role for myeloid cell–LPS interactions both in host defense and in tissue damage [77].
Proteomic analysis of P. gingivalis in vivo has been performed using mouse subcutaneous chambers [78]. 2D electrophoresis and mass spectrometry of P. gingivalis recovered from chambers showed downregulation of four proteins and upregulation of 14 proteins, including the DNA-binding response regulator RprY and a TPR domain protein. The relevance of the TPR domain protein was corroborated in mouse abscess experiments as a mutant defective in production of this protein was less virulent than the parental strain.
The gingival compartment is bathed in gingival crevicular fluid (GCF), a serum exudate. A label-free mass spectrometric analysis of GCF identified 154 proteins of human and microbial origin [79]. The proportion of bacterial, viral and yeast protein was increased in GCF from diseased sites, whereas host defense-related proteins, such as cystatin-B and defensins, were only identified in healthy sites. The sensitive nature of the analysis allowed the identification of novel GCF proteins such as l-plastin, which was detected only in disease, and annexin-1, which was elevated in healthy sites.
In addition to oral diseases, a role for periodontal bacteria in severe systemic conditions, such as coronary artery disease, is emerging [80]. The role of the TLR2 pathway in atherosclerosis associated with a high-fat diet and P. gingivalis infection in the ApoE+/− mouse model has been investigated by proteomics [81]. 2D electrophoresis and mass spectrometry of aortic samples from ApoE+/−–TLR2+/+ mice fed the high-fat diet and infected with P. gingivalis identified six proteins upregulated greater than two-fold: Vesl-2 protein, Sod-2 protein, fumarate hydratase, myosin light chain polypeptide 3, aconitase and gelsolin.
Expert commentary
Recent applications of proteomics to the oral microbiome, host response to oral pathogens and plaque specifically have paralleled other areas of biology in terms of the increasingly ambitious nature of such studies. The difficulty and expense involved in performing proteomics at the same level of coverage and quantitative reliability as what can often be more easily achieved at the transcriptome level, continues to be an issue for all but a few specialist labs. As a consequence, one can expect to see more emphasis on targeted studies and particularly posttranslational modifications (PTMs), both areas that are in most instances not as demanding of instrument time as whole-cell or community proteogenomic studies, and where mass spectrometer technology has generally been more successful. PTM studies often tend to require more time for sample preparation and selective enrichment, however. The recognition of the importance of PTMs to gene regulation in general is an even more important driving force in the direction of more targeted studies. Clinical studies at any level of complexity in particular have been problematic, so much so, that a panel discussion at a recent meeting expressed [82] the collective view that proteomics has so far largely failed to improve the speed and accuracy of medical diagnosis, and that reproducibility issues were a large part of the problem. Here, a careful distinction needs to be made between proteomics that contributes substantially to the development of new diagnostics through biomarker discovery and validation, and the direct application of mass spectrometry-based proteomics in the clinical setting itself. In the fields of oral biology and periodontology, the tools of proteomics are being used for research, not clinical practice. For example, research directed toward establishing a reliable set of protein biomarkers predictive of the early stages of periodontal disease, that could in time lead to an inexpensive, chip-based diagnostic tool, is one ongoing area of investigation. Nonetheless, despite problems on the clinical side, the contributions to basic dental science discussed in the present review are substantial. A particularly important contribution of proteomics to plaque-related science is the dataset published by Madan et al. that provides novel information regarding the potential mechanisms linking atherogenesis to infection, inflammation and the immune response [81]. The comprehensive and quantitatively reproducible measurement of host response at the cellular level remains a major challenge owing to the shear scale of the problem and our incomplete knowledge of human genomics. Despite major technical challenges, the use of proteomic methods for the simultaneous analysis of both microbial gene expression and the host response to microbial challenge is expected to be a major area of emphasis in future years.
Five-year view
The human oral cavity is a complex ecosystem comprising hundreds of species of microorganisms along with host cells and fluids. The reductionist approach of dissecting this oral microbiome complexity into smaller, experimentally tractable units has provided significant insights. However, over the next few years, the trend is likely to continue towards the use of metaproteomic, metagenomic and metabolomic methods in order to provide greater insight into whole microbial ecosystem development and function. It will also be important to address the host side and examine differential expression in host cells such as epithelial cells, fibroblasts and phagocytic cells upon microbial community challenge. Ultimately, the goal will be to analyze in vivo samples, such as plaque, from healthy and diseased sites and biopsies of gingival tissues. Such an increase in experimental complexity will require concomitant increases in instrumental throughput and subsequent computation. Current constraints limiting the direct measurement of protein expression in microbial communities include, but are not limited to, those that relate to overall analytical throughput, dynamic range and proteome coverage. Future work in several contributing fields is expected to lead to incremental improvements in all three areas. Even a model community consisting of only three organisms represents a major challenge for a single mass spectrometer in terms of generating enough data in a reasonable amount of time to reliably measure quantitative trends [20], in a fashion that is both truly reproducible and comprehensive. As with the situation in metagenomic studies [83], the direct measurement of proteomes in dental plaque and other natural communities, that consist of hundreds of microbial species, is fraught with issues surrounding the interpretation of such data, and the inadequacy of ‘hit or miss’ sampling [84] in the face of such complexity. Instrumental throughput, for the papers cited in this review using mass spectrometry as the readout, was limited to roughly a single full scan (MS1) and ten collision-induced dissociation spectra (MS2) per 3-s data-dependent scan event. More recent instruments have increased the number of collision-induced dissociations that can be acquired per unit time by at least a factor of five. Even with such increases, with present separations technology, high proteome coverage in a model community would be limited to five or six organisms with inferred proteomes on the order of 2000 protein-encoding open reading frames. An important caveat is that the organisms need to be sufficiently dissimilar at the level of protein primary structure such that an adequate number of unique proteolytic fragments can be isolated for quantitation, that is, fragments that can only be mapped to a single open reading frame in a single organism. Otherwise, quantitation must be performed using some other broader phylogenetic category, typically based on homology across closely related organisms in the present context. For genetically similar organisms and for communities in which a high level of horizontal gene transfer is the norm, isolating an adequate number of unique peptides can be problematic, and proteome coverage, as conventionally defined in terms of individual proteins associated with a single organism, suffers. The combination of mass spectrometer hardware with inherently greater dynamic range and faster sampling [85], coupled with more emphasis on frequency-based quantitation (i.e., spectral counting), is likely to lead to major improvements in coverage at all levels, both qualitative and quantitative, and larger volumes of data. As a consequence, the computational approaches used with such data will continue to converge with those commonly being used with next-generation and ‘next-next generation’ approaches to transcriptomics and genome sequencing. As noted earlier, proteomics is now a mainstream technique, and it will become increasingly common to incorporate experimental proteomics data into genome browsers such that one may view genome sequence, message, protein, associated small molecule metabolomics data and all associated annotations from links accessible under a single graphical user interface. The use of such a unified computational ‘front end’ that allows the user to interact meaningfully with very large databases containing different kinds of data is expected to become the norm for this kind of research. This in itself is simply one reflection of the increasingly holistic nature of the science.
Key issues.
Lack of reproducibility still remains a problem with many proteomics investigations, especially in areas involving eukaryotic cell lines and tissues, for example, host response studies in the present context.
The interpretation of data from microbial community proteomic studies is heavily dependent on a thorough knowledge of sampling, coverage and sequence similarity issues.
‘Hit or miss’ sampling in large-scale polymicrobial proteomic studies can drastically reduce quantitative reproducibility. Power studies need to be done ahead of time to establish what level of quantitative change in protein expression can in fact be detected under a given set of conditions.
Subtle choices made with respect to database selection, construction and search parameters that would be less critical for single organism studies can be crucial for the success and reproducibility of more complex studies involving many microbes, such as heterotypic biofilms.
Studies that seek to acquire a comprehensive readout of protein expression by both microbe and host during microbial challenge will require higher throughput and dynamic range than what is presently feasible in the absence of multiple, dedicated mass spectrometers.
Acknowledgements
The authors thank David AC Beck for discussion and criticism and Fred Taub for computer support and database programming.
Funding was provided by the Ministry of Education, Culture, Sports, Science and Technology of Japan through grants-in-aid for Scientific Research (B), Scientific Research on Innovative Areas and Scientific Research for Challenging Exploratory Research, and by NIH NIDCR through grants DE014372 and DE11111.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:.
• of interest.
•• of considerable interest.
- 1.Kuboniwa M, Lamont RJ. Subgingival biofilm formation. Periodontol. 2000. 2010;52(1):38–52. doi: 10.1111/j.1600-0757.2009.00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2(13):1599–1607. doi: 10.1016/s1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- 3.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13(12):589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 4. Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol. 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. •• Provides an extensive overview of the development of plaque as a dynamic, multispecies biofilm.
- 5. Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 2007;71(4):653–670. doi: 10.1128/MMBR.00024-07. •• Mentions the concept of ‘system thinking’ and ‘holism’ in microbiology and describes interspecies interactions and community-based assays in detail.
- 6.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 7.Lemos JA, Burne RA. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology. 2008;154(Pt 11):3247–3255. doi: 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell RR. Changing concepts in caries microbiology. Am. J. Dent. 2009;22(5):304–310. [PubMed] [Google Scholar]
- 9.Hajishengallis G. Porphyromonas gingivalis–host interactions: open war or intelligent guerilla tactics? Microbes Infect. 2009;11(6–7):637–645. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 11.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 1998;62(4):1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia RI, Henshaw MM, Krall EA. Relationship between periodontal disease and systemic health. Periodontol. 2000. 2001;25:21–36. doi: 10.1034/j.1600-0757.2001.22250103.x. [DOI] [PubMed] [Google Scholar]
- 13.Foster PL. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 2007;42(5):373–397. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain V, Saleem-Batcha R, Chatterji D. Synthesis and hydrolysis of pppGpp in mycobacteria: a ligand mediated conformational switch in Rel. Biophys. Chem. 2007;127(1–2):41–50. doi: 10.1016/j.bpc.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Wang P, Bouwman FG, Mariman EC. Generally detected proteins in comparative proteomics–a matter of cellular stress response? Proteomics. 2009;9(11):2955–2966. doi: 10.1002/pmic.200800826. [DOI] [PubMed] [Google Scholar]
- 16.Petrak J, Ivanek R, Toman O, et al. Déjà vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8(9):1744–1749. doi: 10.1002/pmic.200700919. [DOI] [PubMed] [Google Scholar]
- 17.Boles BR, Thoendel M, Singh PK. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl Acad. Sci. USA. 2004;101(47):16630–16635. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes CS, Low DA. Signals of growth regulation in bacteria. Curr. Opin. Microbiol. 2009;12(6):667–673. doi: 10.1016/j.mib.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3(5):371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 20. Kuboniwa M, Hendrickson EL, Xia Q, et al. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 2009;9:98. doi: 10.1186/1471-2180-9-98. • Identifies and establishes the effectiveness of proteomics for investigating a simple model oral microbial community consisting of Porphyromonas gingivalis, Fusobacterium nucleatum and Streptococcus gordonii.
- 21.Mitchell HL, Dashper SG, Catmull DV, et al. Treponema denticola biofilm-induced expression of a bacteriophage, toxin-antitoxin systems and transposases. Microbiology. 2010;156(Pt 3):774–788. doi: 10.1099/mic.0.033654-0. [DOI] [PubMed] [Google Scholar]
- 22.Frees D, Savijoki K, Varmanen P, Ingmer H. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 2007;63(5):1285–1295. doi: 10.1111/j.1365-2958.2007.05598.x. [DOI] [PubMed] [Google Scholar]
- 23.Butler SM, Festa RA, Pearce MJ, Darwin KH. Self-compartmentalized bacterial proteases and pathogenesis. Mol. Microbiol. 2006;60(3):553–562. doi: 10.1111/j.1365-2958.2006.05128.x. [DOI] [PubMed] [Google Scholar]
- 24.Capestany CA, Tribble GD, Maeda K, Demuth DR, Lamont RJ. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J. Bacteriol. 2008;190(4):1436–1446. doi: 10.1128/JB.01632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chattoraj P, Banerjee A, Biswas S, Biswas I. ClpP of Streptococcus mutans differentially regulates expression of genomic islands, mutacin production, and antibiotic tolerance. J. Bacteriol. 2010;192(5):1312–1323. doi: 10.1128/JB.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kung RT, Ochs B, Goodson JM. Temperature as a periodontal diagnostic. J. Clin. Periodontol. 1990;17(8):557–563. [PubMed] [Google Scholar]
- 27.Takahashi N. Acid-neutralizing activity during amino acid fermentation by Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol. Immunol. 2003;18(2):109–113. doi: 10.1034/j.1399-302x.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 28.Zilm PS, Bagley CJ, Rogers AH, Milne IR, Gully NJ. The proteomic profile of Fusobacterium nucleatum is regulated by growth pH. Microbiology. 2007;153(Pt 1):148–159. doi: 10.1099/mic.0.2006/001040-0. [DOI] [PubMed] [Google Scholar]
- 29.Welin J, Wilkins JC, Beighton D, Svensäter G. Protein expression by Streptococcus mutansduring initial stage of biofilm formation. Appl. Environ. Microbiol. 2004;70(6):3736–3741. doi: 10.1128/AEM.70.6.3736-3741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welin J, Wilkins JC, Beighton D, et al. Effect of acid shock on protein expression by biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 2003;227(2):287–293. doi: 10.1016/S0378-1097(03)00693-1. [DOI] [PubMed] [Google Scholar]
- 31.Martinez AR, Abranches J, Kajfasz JK, Lemos JA. Characterization of the Streptococcus sobrinus acid-stress response by interspecies microarrays and proteomics. Mol. Oral Microbiol. 2010;25(5):331–342. doi: 10.1111/j.2041-1014.2010.00580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steeves CH, Potrykus J, Barnett DA, Bearne SL. Oxidative stress response in the opportunistic oral pathogen Fusobacterium nucleatum. Proteomics. 2011;11(10):2027–2037. doi: 10.1002/pmic.201000631. [DOI] [PubMed] [Google Scholar]
- 33.Smalley JW, Silver J, Marsh PJ, Birss AJ. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the mu-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem. J. 1998;331(Pt 3):681–685. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dashper SG, Ang CS, Veith PD, et al. Response of Porphyromonas gingivalis to heme limitation in continuous culture. J. Bacteriol. 2009;191(3):1044–1055. doi: 10.1128/JB.01270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baughn AD, Malamy MH. The essential role of fumarate reductase in haemdependent growth stimulation of Bacteroides fragilis. Microbiology. 2003;149(Pt 6):1551–1558. doi: 10.1099/mic.0.26247-0. [DOI] [PubMed] [Google Scholar]
- 36.Guan SM, Shu L, Fu SM, Liu B, Xu XL, Wu JZ. Prevotella intermedia upregulates MMP-1 and MMP-8 expression in human periodontal ligament cells. FEMS Microbiol. Lett. 2009;299(2):214–222. doi: 10.1111/j.1574-6968.2009.01748.x. [DOI] [PubMed] [Google Scholar]
- 37.Kulik EM, Lenkeit K, Chenaux S, Meyer J. Antimicrobial susceptibility of periodontopathogenic bacteria. J. Antimicrob. Chemother. 2008;61(5):1087–1091. doi: 10.1093/jac/dkn079. [DOI] [PubMed] [Google Scholar]
- 38.van Winkelhoff AJ, Winkel EG, Barendregt D, Dellemijn-Kippuw N, Stijne A, van der Velden U. Beta-lactamase producing bacteria in adult periodontitis. J. Clin. Periodontol. 1997;24(8):538–543. doi: 10.1111/j.1600-051x.1997.tb00226.x. [DOI] [PubMed] [Google Scholar]
- 39.Yu F, Anaya C, Lewis JP. Outer membrane proteome of Prevotella intermedia 17: identification of thioredoxin and ironrepressible hemin uptake loci. Proteomics. 2007;7(3):403–412. doi: 10.1002/pmic.200600441. [DOI] [PubMed] [Google Scholar]
- 40.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15(2):167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ang CS, Veith PD, Dashper SG, Reynolds EC. Application of 16O/18O reverse proteolytic labeling to determine the effect of biofilm culture on the cell envelope proteome of Porphyromonas gingivalis W50. Proteomics. 2008;8(8):1645–1660. doi: 10.1002/pmic.200700557. [DOI] [PubMed] [Google Scholar]
- 42.Seers CA, Slakeski N, Veith PD, et al. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 2006;188(17):6376–6386. doi: 10.1128/JB.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998;25(2):134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 44.Veith PD, O’Brien-Simpson NM, Tan Y, Djatmiko DC, Dashper SG, Reynolds EC. Outer membrane proteome and antigens of Tannerella forsythia. J. Proteome Res. 2009;8(9):4279–4292. doi: 10.1021/pr900372c. [DOI] [PubMed] [Google Scholar]
- 45.Pham TK, Roy S, Noirel J, Douglas I, Wright PC, Stafford GP. A quantitative proteomic analysis of biofilm adaptation by the periodontal pathogen Tannerella forsythia. Proteomics. 2010;10(17):3130–3141. doi: 10.1002/pmic.200900448. [DOI] [PubMed] [Google Scholar]
- 46.Veith PD, Dashper SG, O’Brien-Simpson NM, et al. Major proteins and antigens of Treponema denticola. Biochim. Biophys. Acta. 2009;1794(10):1421–1432. doi: 10.1016/j.bbapap.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 47.Amarasinghe JJ, Scannapieco FA, Haase EM. Transcriptional and translational analysis of biofilm determinants of Aggregatibacter actinomycetemcomitans in response to environmental perturbation. Infect. Immun. 2009;77(7):2896–2907. doi: 10.1128/IAI.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fletcher JM, Nair SP, Ward JM, Henderson B, Wilson M. Analysis of the effect of changing environmental conditions on the expression patterns of exported surface-associated proteins of the oral pathogen Actinobacillus actinomycetemcomitans. Microb. Pathog. 2001;30(6):359–368. doi: 10.1006/mpat.2000.0439. [DOI] [PubMed] [Google Scholar]
- 49.Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ., Jr Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 2002;66(3):486–505. doi: 10.1128/MMBR.66.3.486-505.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marsh PD. Dental plaque as a microbial biofilm. Caries Res. 2004;38(3):204–211. doi: 10.1159/000077756. [DOI] [PubMed] [Google Scholar]
- 51.Simionato MR, Tucker CM, Kuboniwa M, et al. Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infect. Immun. 2006;74(11):6419–6428. doi: 10.1128/IAI.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maeda K, Tribble GD, Tucker CM, et al. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol. Microbiol. 2008;69(5):1153–1164. doi: 10.1111/j.1365-2958.2008.06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bradshaw DJ, Marsh PD, Watson GK, Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect. Immun. 1998;66(10):4729–4732. doi: 10.1128/iai.66.10.4729-4732.1998. • Provides direct demonstrations of some key concepts in oral microbial communities. The paper shows that F. nucleatum facilitates coaggregation of normally non-coaggregating species and provides support for anaerobic community members against aeration.
- 54.Diaz PI, Zilm PS, Rogers AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 2002;148(Pt 2):467–472. doi: 10.1099/00221287-148-2-467. [DOI] [PubMed] [Google Scholar]
- 55.Lo AW, Seers CA, Boyce JD, et al. Comparative transcriptomic analysis of Porphyromonas gingivalis biofilm and planktonic cells. BMC Microbiol. 2009;9:18. doi: 10.1186/1471-2180-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelson KE, Fleischmann RD, DeBoy RT, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 2003;185(18):5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mikx FH, Van der Hoeven JS. Symbiosis of Streptococcus mutans and Veillonella alcalescens in mixed continuous cultures. Arch. Oral Biol. 1975;20(7):407–410. doi: 10.1016/0003-9969(75)90224-1. [DOI] [PubMed] [Google Scholar]
- 58.Lewis JP, Iyer D, Anaya-Bergman C. Adaptation of Porphyromonas gingivalis to microaerophilic conditions involves increased consumption of formate and reduced utilization of lactate. Microbiology. 2009;155(Pt 11):3758–3774. doi: 10.1099/mic.0.027953-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rudney JD, Xie H, Rhodus NL, Ondrey FG, Griffin TJ. A metaproteomic analysis of the human salivary microbiota by three-dimensional peptide fractionation and tandem mass spectrometry. Mol. Oral Microbiol. 2010;25(1):38–49. doi: 10.1111/j.2041-1014.2009.00558.x. • This work demonstrates continued development of metaproteomics as a tool for understanding the diversity and metabolic activities of oral microbial communities.
- 60.Xie H, Onsongo G, Popko J, et al. Proteomics analysis of cells in whole saliva from oral cancer patients via value-added three-dimensional peptide fractionation and tandem mass spectrometry. Mol. Cell Proteomics. 2008;7(3):486–498. doi: 10.1074/mcp.M700146-MCP200. [DOI] [PubMed] [Google Scholar]
- 61.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17(3):377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tatusov RL, Fedorova ND, Jackson JD, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nandakumar R, Madayiputhiya N, Fouad AF. Proteomic analysis of endodontic infections by liquid chromatographytandem mass spectrometry. Oral Microbiol. Immunol. 2009;24(4):347–352. doi: 10.1111/j.1399-302X.2009.00520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tribble GD, Lamont RJ. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol. 2000. 2010;52(1):68–83. doi: 10.1111/j.1600-0757.2009.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Wang T, Chen W, et al. Differential protein expression by Porphyromonas gingivalis in response to secreted epithelial cell components. Proteomics. 2005;5(1):198–211. doi: 10.1002/pmic.200400922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hasegawa Y, Tribble GD, Baker HV, Mans JJ, Handfield M, Lamont RJ. Role of Porphyromonas gingivalis SerB in gingival epithelial cell cytoskeletal remodeling and cytokine production. Infect. Immun. 2008;76(6):2420–2427. doi: 10.1128/IAI.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tribble GD, Mao S, James CE, Lamont RJ. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc. Natl Acad. Sci. USA. 2006;103(29):11027–11032. doi: 10.1073/pnas.0509813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Darveau RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 1998;66(4):1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang GT, Zhang HB, Dang HN, Haake SK. Differential regulation of cytokine genes in gingival epithelial cells challenged by Fusobacterium nucleatum and Porphyromonas gingivalis. Microb. Pathog. 2004;37(6):303–312. doi: 10.1016/j.micpath.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 70.Ji S, Kim Y, Min BM, Han SH, Choi Y. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J. Periodont. Res. 2007;42(6):503–510. doi: 10.1111/j.1600-0765.2007.00974.x. [DOI] [PubMed] [Google Scholar]
- 71.Vankeerberghen A, Nuytten H, Dierickx K, Quirynen M, Cassiman JJ, Cuppens H. Differential induction of human beta-defensin expression by periodontal commensals and pathogens in periodontal pocket epithelial cells. J. Periodontol. 2005;76(8):1293–1303. doi: 10.1902/jop.2005.76.8.1293. [DOI] [PubMed] [Google Scholar]
- 72. Hendrickson EL, Xia Q, Wang T, Lamont RJ, Hackett M. Pathway analysis for intracellular Porphyromonas gingivalis using a strain ATCC 33277 specific database. BMC Microbiol. 2009;9:185. doi: 10.1186/1471-2180-9-185. • Uses proteomics to provide a direct measure of pathway-level changes that occur in P. gingivalis after internalization, an important, but difficult to measure environment.
- 73. Xia Q, Wang T, Taub F, et al. Quantitative proteomics of intracellular Porphyromonas gingivalis. Proteomics. 2007;7(23):4323–4337. doi: 10.1002/pmic.200700543. •• The original paper describing the comprehensive, high-coverage proteome of an intact pathogen before and after internalization.
- 74.Kuboniwa M, Hasegawa Y, Mao S, et al. P. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 2008;10(2):122–128. doi: 10.1016/j.micinf.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mao S, Park Y, Hasegawa Y, et al. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell. Microbiol. 2007;9(8):1997–2007. doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Saba JA, McComb ME, Potts DL, Costello CE, Amar S. Proteomic mapping of stimulus-specific signaling pathways involved in THP-1 cells exposed to Porphyromonas gingivalis or its purified components. J. Proteome Res. 2007;6(6):2211–2221. doi: 10.1021/pr070031u. • An important goal in using proteomics to understand oral infections is measuring the host response. This paper uses proteomics to examine the response of human cells not just to known virulence factors (e.g., lipopolysaccharide and FimA) but also to whole and intact P. gingivalis cells.
- 77.Nares S, Moutsopoulos NM, Angelov N, et al. Rapid myeloid cell transcriptional and proteomic responses to periodontopathogenic Porphyromonas gingivalis. Am. J. Pathol. 2009;174(4):1400–1414. doi: 10.2353/ajpath.2009.080677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yoshimura M, Ohara N, Kondo Y, et al. Proteome analysis of Porphyromonas gingivalis cells placed in a subcutaneous chamber of mice. Oral Microbiol. Immunol. 2008;23(5):413–418. doi: 10.1111/j.1399-302X.2008.00444.x. • Proteomic studies of pathogens in vivo are rare. This paper identifies 18 proteins with altered levels in an in vivo infection and demonstrates that one is a previously unidentified virulence factor.
- 79.Bostanci N, Heywood W, Mills K, Parkar M, Nibali L, Donos N. Application of label-free absolute quantitative proteomics in human gingival crevicular fluid by LC/ MS E (gingival exudatome) J. Proteome Res. 2010;9(5):2191–2199. doi: 10.1021/pr900941z. [DOI] [PubMed] [Google Scholar]
- 80.Gibson FC, 3rd, Yumoto H, Takahashi Y, Chou HH, Genco CA. Innate immune signaling and Porphyromonas gingivalis accelerated atherosclerosis. J. Dent. Res. 2006;85(2):106–121. doi: 10.1177/154405910608500202. [DOI] [PubMed] [Google Scholar]
- 81. Madan M, Amar S. Toll-like receptor-2 mediates diet and/or pathogen associated atherosclerosis: proteomic findings. PLoS One. 2008;3(9):e3204. doi: 10.1371/journal.pone.0003204. •• Demonstrates support for the hypothesis that the oral pathogen P. gingivalis can contribute to the development of a systemic disease, atherosclerosis, through the stimulation of TLR2.
- 82.Lemos JA, Burne RA. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology. 2008;154(Pt 11):3247–3255. doi: 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nelson KE, Weinstock GM, Highlander SK, et al. Human Microbiome Jumpstart Reference Strains Consortium. A catalog of reference genomes from the human microbiome. Science. 2010;328(5981):994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hackett M. Science, marketing and wishful thinking in quantitative proteomics. Proteomics. 2008;8(22):4618–4623. doi: 10.1002/pmic.200800358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pekar T, 2nd, Blethrow JD, Schwartz JC, et al. Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal. Chem. 2009;81(18):7757–7765. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Afif H, Allali N, Couturier M, Van Melderen L. The ratio between CcdA and CcdB modulates the transcriptional repression of the ccd poison-antidote system. Mol. Microbiol. 2001;41(1):73–82. doi: 10.1046/j.1365-2958.2001.02492.x. [DOI] [PubMed] [Google Scholar]
- 87.Galvani C, Terry J, Ishiguro EE. Purification of the RelB and RelE proteins of Escherichia coli: RelE binds to RelB and to ribosomes. J. Bacteriol. 2001;183(8):2700–2703. doi: 10.1128/JB.183.8.2700-2703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 101.UniProt. www.uniprot.org.
- 102.BLAST®. http://blast.ncbi.nlm.nih.gov/Blast.cgi.
- 103.The Human Oral Microbiome Database. www.homd.org.
- 104.Clusters of Orthologous Groups of proteins. www.ncbi.nlm.nih.gov/COG.
- 105.MetaCyc database. http://metacyc.org.