Abstract
Water-soluble poly(L-serine)s bearing long side-chain with terminal charge groups were synthesized via ring-opening polymerization of O-pentenyl-L-serine N-carboxyanhydride followed by thiol-ene reactions. These side-chain modified poly(L-serine)s adopt β-sheet conformation in aqueous solution with excellent stability against changes in pH and temperature. These water-soluble poly(L-serine) derivatives with charged side-chain functional groups and stable β-sheet conformations showed membrane-penetrating capabilities in different cell lines with low cytotoxicity.
Introduction
The secondary structures of proteins facilitate the folding of proteins into unique three-dimensional structures and regulate their biological activities1. To better understand protein folding/unfolding and eventually mimic their biological functions, there has been growing interest in designing and synthesizing water-soluble, synthetic polypeptides that adopt stable secondary conformations2–7. A variety of water-soluble polypeptides bearing neutrally charged, hydrophilic functional moieties8–14 or groups15–21 have been prepared, which can adopt stable α-helical or β-sheet conformations against various denaturing conditions. However, these polypeptides lack electrostatic property, which dictates critical interactions regulating the folding/unfolding and controlling the biological activities of these polypeptides22.
Water-soluble polypeptides electrolytes, such as negatively charged poly-L-glutamate and positively charged poly-L-lysine, have been extensively studied in various applications. They have been used as the building blocks of self-assembly23, drug delivery vehicles24 and coating materials25. However, they adopt random coil conformations in water solution at physiological conditions due to the electrostatic repulsion of their charged side chains, which tends to destabilize the helical conformation26. Much effort has been devoted to achieve water-soluble helical peptides with side-chain charged amino acid residues. The α-helical conformation of such peptides can be stabilized by side-chain hydrophobic interactions27, salt bridges28, or tethering of the side-chain functional groups situated on the same helical surface29. However, these strategies require the design of peptides with specific sequences that involve time-consuming, stepwise synthesis and tedious chemistry to control the orientation of peptide side chains30.
Recently, we demonstrated that the helical structure of cationic polypeptides can be stabilized by increasing the hydrophobic interaction of the side chain and minimizing the effect of side chain charge repulsion31–32. These ionic polypeptides with unusual helical stability were realized by maintaining a minimum separation distance of 11 σ-bonds between the peptide backbone and the side chain charge. Further enhanced helicity and helical stability were achieved when the side chain charges are situated even further away from the peptide backbone33. Nevertheless, all these designs of polypeptides are based on glutamate, an amino acid with high helical propensity. Ionic polypeptide derivative from other amino-acid, especially those atypical helix-favoring amino acids (e.g., L-serine), has never been investigated. Doruker and Bahar have observed that the peptide with serine residue has higher unwinding rate of helical states than the peptides with alanine or valine residues34. However, in a separate study, Hwang and Deming designed and synthesized poly(L-serine) with pendant oligoethylene glycol moieties35. They observed a random coil to β-sheet transition with the addition of methanol to the aqueous polypeptide solution but did not observe the formation of helical conformation. Here, we report the design and synthesis of a poly(L-serine) bearing terminally charged long hydrocarbon side chains and evaluation of their physical and biological properties, aiming to expand our understanding of polypeptide electrolytes. These polypeptides adopt stable β-sheet conformation against external environmental changes (e.g., pH and temperature) and showed interesting cell membrane permeability in several cell-lines with low cytotoxicity. To our knowledge, this is the first demonstration of cell membrane penetration with a peptide containing a large portion of β-sheet instead of α-helical conformation.
Experimental section
Materials
Anhydrous dimethylformamide (DMF) was dried by columns packed with 4Å molecular sieves and stored in a glove-box. Tetrahydrofuran (THF), ethyl acetate (EtOAc) and hexane were dried by columns packed with alumina and stored in a glove-box. PEG112-NH2 (MW = 5000) was purchased from Laysan Bio Inc (Arab, AL, USA). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as received unless otherwise specified. Raw264.7 (mouse monocyte macrophage) and HeLa (human cervix adenocarcinoma) cells were purchased from the American Type Culture Collection (Rockville, MD, USA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS).
Instrumentation
1H and 13C NMR spectra were recorded on a Varian U500 MHz or UI500NB MHz spectrometer. Chemical shifts were reported in ppm and referenced to the solvent proton and 13C resonances. Size exclusion chromatography (SEC) experiments were performed on a system equipped with an isocratic pump (Model 1100, Agilent Technology, Santa Clara, CA, USA), a DAWN HELEOS multi-angle laser light scattering detector (MALLS) detector and an Optilab rEX refractive index detector (Wyatt Technology, Santa Barbara, CA, USA). The detection wavelength of HELEOS was set at 658 nm. Separations were performed using serially connected size exclusion columns (100 Å, 500 Å, 103 Å and 104 Å Phenogel columns, 5 μm, 300 × 7.8 mm, Phenomenex, Torrance, CA, USA) at 60 °C using DMF containing 0.1 M LiBr as the mobile phase. The MALLS detector is calibrated using pure toluene with no need for calibration using polymer standards and can be used for the determination of the absolute molecular weights (MWs). Infrared spectra were recorded on a Perkin Elmer 100 serial FTIR spectrophotometer calibrated with polystyrene film. Circular dichroism (CD) measurements were carried out on a JASCO J-700 CD spectrometer. The polymer samples for CD analysis were prepared at concentrations of 0.1 – 2.5 mg/mL in general unless otherwise specified. The solution was placed in a quartz cell with a path length of 0.2 cm. The mean residue molar ellipticity of each polymer was calculated based on the measured apparent ellipticity by following the literature reported formula: Ellipticity ([θ] in deg·cm2·dmol−1) = (millidegrees × mean residue weight)/(pathlength in millimeters × concentration of polypeptide in mg/ml). Lyophilization was performed on a FreeZone lyophilizer (Labconco, Kansas City, MO, USA). UV light was generated by an OmiCure S1000 UV lamp (EXFO, Mississauga, Canada).
Synthesis of O-pentenyl-L-serine (3)
In an ice-water bath, Boc-Ser-OH (6.7 g, 33.6 mmol) was dissolved in anhydrous DMF (100 mL) followed by slowly addition of sodium hydride (1.7 g, 95%, 67.2 mmol). Thirty minutes later, 5-bromo-1-pentene (10.0 g, 67.2 mmol) was added in one portion. The reaction mixture was stirred at room temperature for 16 h. DMF was distilled at 60 °C under vacuum. The residue was stirred in water (75 mL) for 16 h, followed by extracting with diethyl ether (30 mL × 2). The aqueous layer was acidified to pH 3 with 1N HCl and then extracted with ethyl acetate (50 mL × 3). The organic layer was dried over anhydrous MgSO4, collected by filtration and removed under vacuum to yield the product as a yellow oil (2, 7.0 g, 76% yield). The deprotection of 2 (7.0 g, 25.6 mmol) was performed with HCl/dioxane (4 M, 150 mL) by stirring the reaction solution at room temperature for 4 h. The solid precipitate was collected by centrifuge and purified by washing with diethyl ether (100 mL). The resulting product, O-pentenyl-L-serine (3, 4.0 g, 90% yield), was dried at room temperature under vacuum for 8 h before use. 1H NMR (500 MHz, D2O): δ 5.74 (m, 1H, CH2=CHCH2-), 4.88 (m, 2H, CH2=CHCH2-), 4.01 (m, 1H, -CHNH2), 3.90 (m, 2H, -OCH2CHNH2), 3.44 (m, 2H, CH2=CHCH2CH2CH2-), 1.97 (m, 2H, CH2=CHCH2CH2CH2-) and 1.54 (m, 2H, CH2=CHCH2CH2CH2-).
Synthesis of O-pentenyl-L-serine-based N-carboxylanhydride (PE-L-Ser NCA, 4)
O-pentenyl-L-serine 3 (1.0 g, 4.8 mmol) and triphosgene (1.1 g, 3.7 mmol) were mixed in anhydrous THF (15 mL) in a round-bottomed flask. The reaction was stirred at room temperature for 16 h. The solvent was distilled at room temperature under vacuum to yield a brown liquid residue which was purified by silica gel chromatography (Note: the silica gel was dried at 120 °C under vacuum for 8 h before use) with hexane as the eluent containing EtOAc gradient (from 0% to 40%). A light yellow liquid product was obtained (0.8 g, 83% yield). 1H NMR (500 MHz, CDCl3): δ 6.74 (s, 1H, -NH), 5.75 (m, 1H, CH2=CHCH2CH2CH2-), 5.22 (t, 2H, CH2=CHCH2CH2CH2-), 4.43 (t, 1H, -CHNH2), 3.73 (d, 2H, -OCH2CHNH), 3.46 (m, 2H, CH2=CHCH2CH2CH2-), 2.04 (m, 2H, CH2=CHCH2CH2CH2-) and 1.61 (m, 2H, CH2=CHCH2CH2CH2-). 13C NMR (125 MHz, CDCl3): δ 168.0, 153.0, 138.0, 115.3, 71.4, 68.7, 58.8, 30.2, 28.6.
Synthesis of polyethylene glycol-block-poly(O-pentenyl-L-serine) (PEG-b-PPELS, 5)
In a glove-box, PEG114-NH2 (22.5 mg, 4.5 μmol of amino groups) and 4 (90 mg, 450 mmol) were dissolved in DMF (2.0 mL). The polymerization was carried out at room temperature for 48 h. FTIR was used to monitor the polymerization until the conversion of 4 was above 99%. The product 5 in DMF solution was directly used for the next-step reaction. A small portion of 5 was isolated for characterization. 1H NMR (500 MHz, CDCl3): δ 5.76 (m, 1H, CH2=CHCH2CH2CH2O-), 4.95 (m, 2H, CH2=CHCH2CH2CH2O-), 4.43-4.24 (m, 3H, CHCH2-), 3.62 (s, 4H, -CH2CH2O- of PEG), 3.55 (m, 2H, CH2=CHCH2CH2CH2O-), 2.05 and 1.62 (m, 4H, CH2=CHCH2CH2CH2O-). It should be noted that 5 was characterized by NMR immediately after the polymerization solution was concentrated under vacuum. If 5 is stored in solid form for an extended period of time, it is no longer soluble in common organic solvents (e.g., DMF and CHCl3), presumably due to the inter-chain H-bonding of 5 that has a large portion of β-sheet conformation and side-chain hydrophobic interaction that makes it very difficult for solvent molecules to get back in once they are depleted (completely dried).
Synthesis of poly(ethylene glycol)-block-poly(O-pentenyl-L-serine-graft-cysteamine) (PEG-b-(PPELS-g-CA), 6)
In a quartz flask, cysteamine hydrochloride (100 mg, 0.75 mmol, 1.7 equiv. of the allyl groups) and photo-initiator (2,2-dimethoxy-2-phenyl acetophenone, DMPA, 5 mg) were added to the polymerization solution (5, 0.45 mmol in 2 mL DMF), followed by purging with N2 for 5 min. The quartz flask was sealed and irradiated by UV (365 nm, 16 mW) for 30 min. HCl (1N, 5 mL) was added to the DMF solution and the resulting mixture was dialyzed against DI water in a dialysis bag with a cutoff molecular weight of 1,000 g/mol. After lyophilization, 6 in a white solid form was obtained (43 mg, 36% yield). 1H NMR (500 MHz, D2O): δ 3.66 (br, 2H, CHCH2O-), 3.57 (br, 4H, -CH2CH2O- of PEG), 3.41 (br, 3H, CHCH2O- and -OCH2CH2CH2CH2CH2S-), 3.09 (br, 2H, -SCH2CH2NH3Cl), 2.72 (br, 2H,-SCH2CH2NH3Cl), 2.47 (br, 2H, -OCH2CH2CH2CH2CH2S-), 1.48 (br, 4H, -OCH2CH2CH2CH2CH2S-), 1.28 (br, 2H, -OCH2CH2CH2CH2CH2S-).
Synthesis of rhodamine-polypeptide conjugates
PEG-b-(PPELS-g-CA) (10 mg, 3 × 10−5 mol of primary amine) was dissolved in a NaHCO3 solution (4 mL, 0.2 M). Rhodamine B isothiocyanate (RhB-NCS, 0.83 mg, 1.5 μmol) in DMSO (83 μL) was added to the polymer solution. The reaction vial was wrapped with aluminum foil. The reaction solution was stirred at room temperature for 12 h followed by dialysis in DI H2O to yield rhodamine-labeled polypeptide (8.0 mg, 80% yield).
Cell uptake
Cells were seeded in a 96-well plate at a density of 10,000 cells/well. The medium was replaced with serum-free DMEM 24 h later followed by the addition of the rhodamine-polypeptide. After incubation for a pre-defined period of time, the cells were washed with cold PBS (3 × 200 μL, containing 20 U/mL heparin) to completely remove surface-bound cationic proteins from the cells36. Cells were then lysed with 100 μL RIPA buffer; the rhodamine content in the lysate was quantified by spectrofluorimetry. A series of rhodamine-polypeptide solutions were prepared through the dilution of the stock solution (1 mg/mL) with RIPA lysis buffer to the final concentrations of 100, 80, 60, 40, 20, 10 or 5 μg/mL. Regression curves were obtained by plotting the fluorescence intensity of the standard solution (Ex = 560 nm, Em = 590 nm) against concentration (μg/mL). The amount of rhodamine-polypeptide in the cell lysate was calculated based on the regression curve. The protein level in the cell lysate was determined by the BCA kit. The uptake level was expressed as μg of polypeptide relative to 1 mg of cellular protein.
Cell uptake mechanisms
To explore the mechanism of polypeptide internalization, cells were pre-incubated with one of an endocytosis inhibitor (chlorpromazine (10 μg/mL), genistein (100 μg/mL), methyl-β-cyclodextrin (mβCD, 5 mM), wortmannin (10 μg/mL), or dynasore (80 μM)) for 30 min prior to the addition of polypeptide, which was present in the cell culture medium throughout the 2-h uptake experiment at 37 °C. Similar cell uptake experiments were also performed at 4 °C. Results were expressed as percentage uptake level of the control cells which were incubated with polypeptides at 37 °C for 2 h in the absence of endocytic inhibitors.
Cytotoxicity measurement
Cells were seeded in a 96-well plate at a density of 10,000 cells/well and incubated for 24 h at 37 °C. The medium was replaced with serum-free DMEM. Polypeptides were then added at 10, 5, 2, 1, and 0.5 μg/well, respectively. Cells were incubated for 4 h, and then the medium was replaced with serum-containing DMEM. After another 20-h incubation at 37 °C, cell viability was assessed using the standard MTT assay.
Results and Discussion
O-pentene-L-serine N-carboxylanhydride (PE-L-Ser NCA, 4) was synthesized in three steps from Boc-Ser-OH (1) with an overall yield of 50–60% (Scheme 1). Specifically, 4 was synthesized via the etherification of 1 with 5-bromo-1-pentene in the presence of sodium hydride to afford 2, followed by the deprotection of tert-butyloxycarbonyl group of 2 in HCl/dioxane and the cyclization of O-pentenyl-L-serine (3) with triphosgene. 4 is readily soluble in THF, ethyl acetate, ethyl ether, dichloromethane and chloroform, but insoluble in hexane. Purification of 4 via recrystallization was attempted but only resulted in a dark-colored oily residue. Therefore, a recently developed purification method by Deming and coworkers through the use of silica gel chromatography was used to purify 4,37 which yielded a colorless oil with satisfactory purity. The molecular structure of 4 was verified by NMR (Figure 1) and FTIR (Figure S1).
Scheme 1.
Synthesis of poly(ethylene glycol)-block-poly(O-pentenyl-L-serine-graft-cysteamine) [PEG-b-(PPELS-g-CA), 6]
Figure 1.
(a) 1H and (b) 13C NMR spectra of O-pentenyl-L-serine N-carboxylanhydrides (PE-L-Ser NCA, 4) in CDCl3.
The NCAs can be polymerized using primary amines and hexamethyldisilazane (HMDS)38–39. However, the resulting polymers showed poor solubility in common organic solvents, such as DMF, chloroform, dichloromethane and THF, which made it difficult to assess the molecular weights (MWs) of these polymers. PEG112-NH2 was then used to initiate the ring-opening polymerization of 4 in DMF, aiming at obtaining polymers with improved solubility yet undistorted polypeptide conformation via terminal grafting of PEG that has excellent solubility in common organic solvents and water. The MWs of the PEG-poly(4) (denoted as PEGn-b-PPELSm) was determined by 1H NMR and SEC. PEGn-b-PPELSm with a PPELS segment up to 70 mer (m = degree of polymerization (DP) = 70) was prepared, which was verified by SEC (Figure S2) and 1H NMR (Figure S3). A prominent peak shift was observed by comparing the SEC traces of PEG112-NH2 and PEG112-b-PPELS70 (Figure S2), indicating the successful synthesis of PEGn-b-PPELSm. The PPELS with a DP larger than 70 in the PEGn-b-PPELSm was also attempted. But precipitation was observed in the DMF polymerization solution. The precipitates, presumably aggregated PEGn-b-PPELSm with a PPELS block larger than ca. 70 repeating units, was insoluble in most common organic solvents including THF and chloroform.
Even for a PEGn-b-PPELSm prepared with a DP < 70 that is soluble in the DMF polymerization solution, it becomes insoluble in DMF after it is dried and stored for an extended period of time. Because PEGn-b-PPELSm has very hydrophobic side chains, it should be very difficult for the polar organic solvent molecules, such as DMF, to return to the non-polar, hydrophobic side chain areas and solvate (re-dissolve) the aggregated polypeptide side chains once the organic solvent molecules are depleted during the drying process. Therefore, the polymerization solutions of PEGn-b-PPELSm were directly used for the thiol-ene “click” reaction which was performed inside a quartz flask under N2 with catalytic amount of DMPA, the photo initiator, and purified by dialysis. Highly water-soluble (solubility > 5 mg/mL) ionic poly(L-serine) analogues (6) was obtained after freeze-drying. The molecular structure of 6 was verified by 1H NMR (Figure 2). A complete disappearance of the characteristic peaks of vinyl groups (CH2=CH-) (Figure 2 and Figure S3) indicates the quantitative grafting efficiency.
Figure 2.
1H NMR spectrum of PEG112-b-(PPELS70-g-CA) (6) in D2O.
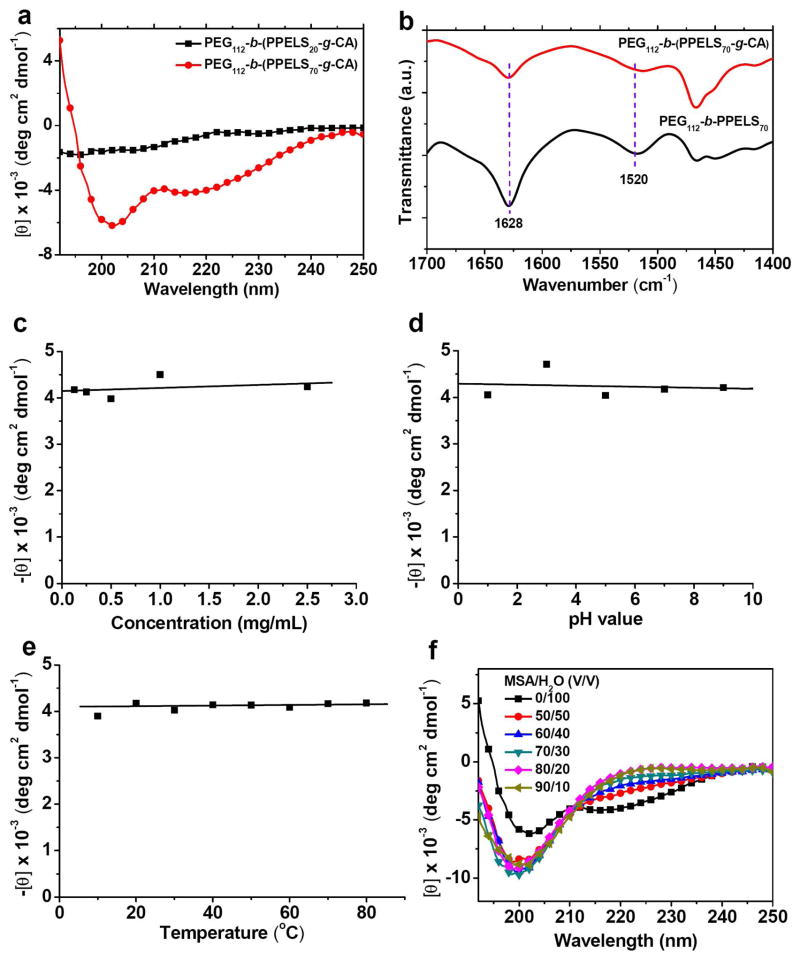
The solution conformations of PEG-b-(PPELS-g-CA)s were studied by CD spectroscopy. PEG112-b-(PPELS20-g-CA) showed no Cotton effect in the scanning range between 190–250 nm while PEG112-b-(PPELS70-g-CA) showed two negative dichroic peaks at 202 nm and 216 nm (Figure 3a). CDPro (CONTIN method) was used to determine the secondary structure of PEG112-b-(PPELS70-g-CA), which indicated that PEG112-b-(PPELS70-g-CA) adopts 45% of β-sheet and significant portion of random coil. FTIR results revealed an amide I peak at 1628 cm−1 and amide II peak at 1520 cm−1 for PEG112-b-(PPELS70-g-CA), confirming that PEG112-b-(PPELS70-g-CA) adopts β-sheet conformation in the solid state (Figure 3b), which is consistent with the conformation determined by CD. While PEG112-b-(PPELS70-g-CA) has comprehensive β-sheet conformations in aqueous solution, it showed concentration independence of its –[θ]216 values (Figure 3c) and remarkable stability against pH changes (from pH 1 to pH 9, Figure 3d). These properties of PEG112-b-(PPELS70-g-CA) resembles the properties of ionic poly(L-glutamate)s bearing elongated and charged side-chains.27 Trifluoroethanol (TFE) and methanol are well known solvents to induce the formation and enhance the stability of secondary structures of peptides35,40. For example, Hwang and Deming have reported a random coil to β-sheet transition by adding methanol to the aqueous solution of PEGylated poly(L-serine)s.35 We did similar experiments by using these solvents to trigger the conformation transitions of PEG112-b-(PPELS70-g-CA) in the aqueous solution, but observed an unusual stability of the β-sheet conformation of PEG112-b-(PPELS70-g-CA) by varying the fraction of MeOH or TFE (Figure S4) in water. PEG112-b-(PPELS70-g-CA) also showed excellent stability against elevated temperatures (Figure 3e), with negligible change of –[θ]216 value at a temperature ranging from 10 °C to 80 °C. A typical secondary structure transition was observed when increasing the volume fraction of methanesulfonic acid (MSA), a very strong peptide denaturing agent (Figure 3f), presumably due to the destruction of the hydrogen bonding. By comparing our results with those reported by Deming,35 it is apparent that the hydrophobic side chains play an important role in stabilizing the β-sheet conformation of PEG112-b-(PPELS70-g-CA). We also noticed that PEG112-b-(PPELS70-g-CA) in aqueous solution showed some degree of aggregation based on the analysis of dynamic light scattering. It is unclear, however, how exactly the hydrophobicity of side chains improves the β-sheet stability, which is subjected to further study.
Figure 3.
(a) CD spectra of PEG112-b-(PPELS20-g-CA) and PEG112-b-(PPELS70-g-CA) in water. (b) FTIR spectra of PEG112-b-PPELS70 and PEG112-b-(PPELS70-g-CA) in the solid state. (c) The concentration dependence of the residue molar ellipticity at 216 nm for PEG112-b-(PPELS70-g-CA). (d) The pH dependence of the residue molar ellipticity at 216 nm for PEG112-b-(PPELS70-g-CA). (e) The temperature dependence of the molar ellipticity at 216 nm for PEG112-b-(PPELS70-g-CA). (f) CD spectra of PEG112-b-(PPELS70-g-CA) in a mixed solvent of MSA and H2O.
Cationic polypeptides with rigid molecular structures, mostly α-helical conformations, have been previously demonstrated to show membrane activities41–43. Report of using a β-sheet containing cationic polypeptide to mediate cell membrane permeation has not been reported in literature to the best of our knowledge. We were motivated to explore the cell penetration potentials of the resulting ionic poly(L-serine)s with rich β-sheet conformation using rhodamine-labeled polypeptide. HeLa, a cervical cancer cell line, and Raw 264.7, a macrophage cell line were selected for the cell penetration assessment. As shown in Figure 4a, PEG112-b-(PPELS70-g-CA) showed interesting cell penetrating properties and outperformed polyarginine (5000~15000 Da), a well-known membrane active polypeptide, with respect to the cell uptake23. The cell uptake level increased with incubation time and the feeding amount, and the maximal cell uptake level was noted 2 h post polypeptide treatment (Figure 4b and 4c). To further understand the mechanisms underlying the polypeptide-mediated cell penetration, we performed the cell uptake study at lower temperature (4 °C) or in the presence of various endocytic inhibitors. The cell uptake level was inhibited by 40–70% at 4 °C when energy-dependent endocytosis was completely blocked (Figure 4d), suggesting that part of the polypeptide was internalized through endocytosis while the remaining entered the cells via energy-independent transduction. Genistein and mβCD inhibited the cell uptake process while chlorpromazine showed unappreciable inhibitory effect, suggesting that the endocytosis was related to the caveolae- rather than clathrin-mediated pathway. Furthermore, the cell uptake level was decreased in the presence of dynasore which inhibited both the clathrin- and caveolar-mediated pathways by inhibiting dynamins, further substantiating the above observed results. Unappreciable inhibitory effect was observed for wortmannin, indicating that macropinocytosis was not involved. Despite their desired cell penetrating properties, the resulting polypeptides showed good cell tolerability as determined by the MTT assay, as evidenced by slight toxicity at high concentrations (10 and 5 μg/well) and unappreciable toxicity at concentrations ≤ 2 μg/well (Figure 4e). Such results further supported that the cell penetration was achieved without compromising the cell viability and biological functions.
Figure 4.
(a) Uptake level of rhodamine-labeled PEG112-b-(PPELS70-g-CA) in Raw 264.7 and HeLa cells. Cells in 96-well plates were treated with polypeptides (2 μg/well) for 2 h, and the uptake level was expressed as μg polypeptide associated with 1 mg cellular protein (n = 3). (b) Concentration-dependent uptake level of rhodamine-labeled PEG112-b-(PPELS70-g-CA)s in Raw 264.7 and HeLa cells. Cells in 96-well plates were treated with polypeptides (0.5, 1, 2, and 5 μg/well) for 2 h (n = 3). (c) Time-dependent uptake level of rhodamine-labeled PEG112-b-(PPELS70-g-CA)s in Raw 264.7 and HeLa cells. Cells in 96-well plates were treated with polypeptides (2 μg/well) for 0.4, 1, 2, and 4 h, respectively (n = 3). (d) Elucidation of the mechanisms underlying cell uptake of polypeptides. Cells were treated with rhodamine-labeled PEG112-b-(PPELS70-g-CA)s (2 μg/well) for 2 h in the presence of various endocytic inhibitors, and results were expressed as percentage uptake (%) of control cells which were incubated in the absence of endocytic inhibitors (n = 3). (e) Cytotoxicity of PEG112-b-(PPELS70-g-CA)s as determined by the MTT assay. Cells in 96-well plates were incubated with polypeptides at different concentrations for 4 h in serum-free media, and were further cultured in serum-containing media for another 20 h before the cell viability assessment. Results were expressed as percentage viability of control cells which did not receive polypeptide treatment (n = 3).
Conclusions
In summary, PEG112-b-(PPELS70-g-CA), a water-soluble, poly(L-serine) analogue with elongated, positively charged side chains were synthesized through mPEG-NH2 initiated ring-opening polymerization of O-pentenyl-L-serine N-carboxyanhydrides followed by thiol-ene reactions to introduce the charged groups. PEG112-b-(PPELS70-g-CA) adopted a mixed β-sheet and random coil conformation, and the β-sheet conformation was stable against changes in pH and temperature. The resulting polypeptides also showed cell-penetrating properties. We are currently investigating whether these materials can be used as gene delivery vehicles.
Supplementary Material
Acknowledgments
This work is supported by NIH (R21 1R21EB013379 and Director’s New Innovator Award 1DP2OD007246). We thank Professor Martin Gruebele for allowing us to use their CD spectrometer.
Footnotes
Supporting Information Available
FTIR spectrum of PE-L-Ser NCA (4) (Figure S1), SEC traces of PEG112-NH2 and PEG112-b-PPELS70 (Figure S2), 1H NMR spectrum of PEG112-b-PPELS70 (5) in CDCl3 (Figure S3). The MW of PEG-b-PPELS can be calculated from the integrations of Hb and Hg (Figure S3). MeOH and TFE concentration dependence of the molar ellipticity at 216 nm for PEG112-b-(PPELS70-g-CA) (Figure S4). CD spectra of PEG112-b-(PPELS70-g-CA) in aqueous solutions against various changes (Figrue S5). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Stryer L. Biochemistry. W.H. Freeman and Company; 1998. [Google Scholar]
- 2.Robertson DE, Farid RS, Moser CC, Urbauer JL, Mulholland SE, Pidikiti R, Lear JD, Wand AJ, Degrado WF, Dutton PL. Nature. 1994;368:425–431. doi: 10.1038/368425a0. [DOI] [PubMed] [Google Scholar]
- 3.Goodman CM, Choi S, Shandler S, DeGrado WF. Nat Chem Biol. 2007;3:252–262. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiori WR, Lundberg KM, Millhauser GL. Nat Struct Biol. 1994;1:374–377. doi: 10.1038/nsb0694-374. [DOI] [PubMed] [Google Scholar]
- 5.Fezoui Y, Hartley DM, Walsh DM, Selkoe DJ, Osterhout JJ, Teplow DB. Nat Struct Biol. 2000;7:1095–1099. doi: 10.1038/81937. [DOI] [PubMed] [Google Scholar]
- 6.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 7.Bryson JW, Betz SF, Lu HS, Suich DJ, Zhou HXX, Oneil KT, Degrado WF. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- 8.Engler AC, Lee HI, Hammond PT. Angew Chem, Int Ed. 2009;48:9334–9338. doi: 10.1002/anie.200904070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang H, Li Y, Lahasky SH, Sheiko SS, Zhang D. Macromolecules. 2011;44:1491–1499. [Google Scholar]
- 10.Chen C, Wang Z, Li Z. Biomacromolecules. 2011;12:2859–2863. doi: 10.1021/bm200849m. [DOI] [PubMed] [Google Scholar]
- 11.Yu M, Nowak AP, Deming TJ, Pochan DJ. J Am Chem Soc. 1999;121:12210–12211. [Google Scholar]
- 12.Liu Y, Chen P, Li Z. Macromol Rapid Commun. 2012;33:287–295. doi: 10.1002/marc.201100649. [DOI] [PubMed] [Google Scholar]
- 13.Ding J, Xiao C, Zhao L, Cheng Y, Ma L, Tang Z, Zhuang X, Chen X. J Polym Sci Part A: Polym Chem. 2011;49:2665–2676. [Google Scholar]
- 14.Ding J, Xiao C, Tang Z, Zhuang X, Chen X. Macromol Biosci. 2011;11:192–198. doi: 10.1002/mabi.201000238. [DOI] [PubMed] [Google Scholar]
- 15.Lotan N, Berger A, Katchals E, Ingwall RT, Scheraga HA. Biopolymers. 1966;4:239. doi: 10.1002/bip.1966.360040211. [DOI] [PubMed] [Google Scholar]
- 16.Xiao C, Zhao C, He P, Tang Z, Chen X, Jing X. Macromol Rapid Commun. 2010;31:991–997. doi: 10.1002/marc.200900821. [DOI] [PubMed] [Google Scholar]
- 17.Tang H, Zhang D. Biomacromolecules. 2010;11:1585–1592. doi: 10.1021/bm1002174. [DOI] [PubMed] [Google Scholar]
- 18.Kramer JR, Deming TJ. J Am Chem Soc. 2010;132:15068–15071. doi: 10.1021/ja107425f. [DOI] [PubMed] [Google Scholar]
- 19.Tang H, Zhang D. Polym Chem. 2011;2:1542–1551. [Google Scholar]
- 20.Kramer JR, Deming TJ. J Am Chem Soc. 2012;134:4112–4115. doi: 10.1021/ja3007484. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, He C, Xiao C, Ding J, Zhuang X, Chen X. Polym Chem. 2011;2:2627–2634. [Google Scholar]
- 22.Whitford D. Proteins Structure and Function. John Wiley & Sons, Ltd; Chichester, West Sussex, England: 2005. [Google Scholar]
- 23.Holowka EP, Sun VZ, Kamei DT, Deming TJ. Nat Mater. 2007;6:52–57. doi: 10.1038/nmat1794. [DOI] [PubMed] [Google Scholar]
- 24.Bhatt RL, de Vries P, Tulinsky J, Bellamy G, Baker B, Singer JW, Klein P. J Med Chem. 2003;46:190–193. doi: 10.1021/jm020022r. [DOI] [PubMed] [Google Scholar]
- 25.Wong MS, Cha JN, Choi KS, Deming TJ, Stucky GD. Nano Lett. 2002;2:583–587. [Google Scholar]
- 26.Nagasawa M, Holtzer A. J Am Chem Soc. 1964;86:538. [Google Scholar]
- 27.Dill KA. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 28.Marqusee S, Baldwin RL. Proc Natl Acad Sci USA. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blackwell HE, Grubbs RH. Angew Chem, Int Ed. 1998;37:3281–3284. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 30.Atherton ES, RC . Solid phase peptide synthesis: A practical approach. IRL Press at Oxford University Press; Oxford, England: 1989. [Google Scholar]
- 31.Lu H, Wang J, Bai YG, Lang JW, Liu SY, Lin Y, Cheng JJ. Nat Commun. 2011:2. doi: 10.1038/ncomms1209. [DOI] [PubMed] [Google Scholar]
- 32.Lu H, Bai Y, Wang J, Gabrielson NP, Wang F, Lin Y, Cheng J. Macromolecules. 2011;44:6237–6240. doi: 10.1021/ma201164n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Lu H, Lin Y, Cheng J. Macromolecules. 2011;44:6641–6644. doi: 10.1021/ma201678r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doruker P, Bahar I. Biophys J. 1997;72:2445–2456. doi: 10.1016/S0006-3495(97)78889-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang J, Deming TJ. Biomacromolecules. 2001;2:17–21. doi: 10.1021/bm005597p. [DOI] [PubMed] [Google Scholar]
- 36.McNaughton BR, Cronican JJ, Thompson DB, Liu DR. Proc Natl Acad Sci USA. 2009;106:6111–6116. doi: 10.1073/pnas.0807883106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kramer JR, Deming TJ. Biomacromolecules. 2010;11:3668–3672. doi: 10.1021/bm101123k. [DOI] [PubMed] [Google Scholar]
- 38.Lu H, Cheng J. J Am Chem Soc. 2007;129:14114–14115. doi: 10.1021/ja074961q. [DOI] [PubMed] [Google Scholar]
- 39.Lu H, Cheng J. J Am Chem Soc. 2008;130:12562–12563. doi: 10.1021/ja803304x. [DOI] [PubMed] [Google Scholar]
- 40.Cammers-Goodwin A, Allen TJ, Oslick SL, McClure KF, Lee JH, Kemp DS. J Am Chem Soc. 1996;118:3082–3090. [Google Scholar]
- 41.Lättig-Tünnemann G, Prinz M, Hoffmann D, Behlke J, Palm-Apergi C, Morano I, Herce HD, Cardoso MC. Nat Commun. 2011;2:453. doi: 10.1038/ncomms1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabrielson NP, Lu H, Yin L, Li D, Wang F, Cheng J. Angew Chem, Int Ed. 2012;51:1143–1147. doi: 10.1002/anie.201104262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabrielson NP, Lu H, Yin L, Kim KH, Cheng J. Mol Ther. 2012 doi: 10.1038/mt.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.