Abstract
Signal transducers and activators of transcription (STAT) proteins function in the JAK/STAT signaling pathway and are activated by phosphorylation. As a result of this signaling event, they affect many cellular processes including cell growth, proliferation, differentiation, and survival. Increases in the expressions of STAT5A and STAT5B play a remarkable role in the development of leukemia in which leukemic cells gain uncontrolled proliferation and angiogenesis ability. At the same time, these cells acquire ability to escape from apoptosis and host immune system. In this study, we aimed to suppress STAT-5A and -5B genes in K562 CML cells by siRNA transfection and antisense oligonucleotides (ODN) targeting and then to evaluate apoptosis rate. Finally, we compared the transfection efficiencies of these approaches. Quantitative RT-PCR and Western blot results indicated that STAT expressions were downregulated at both mRNA and protein levels following siRNA transfection. However, electroporation mediated ODN transfection could only provide limited suppression rates at mRNA and protein levels. Moreover, it was displayed that apoptosis were significantly induced in siRNA treated leukemic cells as compared to ODN treated cells. As a conclusion, siRNA applications were found to be more effective in terms of gene silencing when compared to ODN treatment based on the higher apoptosis and mRNA suppression rates. siRNA application could be a new and alternative curative method as a supporting therapy in CML patients.
Keywords: Chronic myeloid leukemia, K562, STAT5, siRNA knockdown, antisense oligonucleotides, apoptosis
Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder of transformed, primitive hematopoietic progenitor cells and represents 15-20% of all adult leukemia cases. A reciprocal translocation between the ABL gene on chromosome 9 and the BCR gene on chromosome 22 [t(9;22)(q34;q11)] brings BCR and ABL genes together and BCR-ABL1 chimeric oncoprotein with constitutive tyrosine kinase activity is generated [1,2]. Dysregulated tyrosine kinase activity of BCR-ABL1 promotes cell proliferation and prevents apoptosis and this seems to be the main cause of CML phenotype [3]. The resultant BCR-ABL fusion protein activates many cytoplasmic and nuclear downstream signaling pathways that affect the proliferation and survival of hematopoietic cells [4].
Janus Kinase/Signal Transducers and Activators of Transcription (JAK/STAT) pathway is one of the most important signaling pathways in the regulation of cell proliferation, survival and apoptosis [5]. JAK/STAT pathway includes JAK tyrosine kinases, STATs and suppressers of cytokine signaling (CIS/SOCS) families [6]. Cytokines, hormones and growth factors are the ligands of JAK/STAT pathway and this pathway transduces signal from the cytokine receptor into the nucleus. JAKs are stimulated by activation of a cytokine receptor which in turn activates STAT transcription factor [7].
The STAT gene family is composed of seven proteins (from STAT1 to STAT6 including STAT5A and -5B) with a molecular weight between 75 to 96 kDa (748–851 aa). The structure of STAT family proteins is composed of an amino-terminal oligomerization domain, a DNA-binding domain, an src homology domain (SH2) and a transactivation domain. While the transactivation domain is responsible for the activation of transcription, there is a conserved tyrosine residue near the C-terminus of SH2 domain that is phosphorylated by the activated JAKs. As a result of this phosphorylation, STAT proteins interact with each other and become dimerized. STAT dimers migrate into the nucleus where they bind to specific regulatory DNA sequences of target genes and function as activator or repressor of these genes [8].
Overexpression of STATs, especially STAT5A, provides an efficient mechanism to translate an extracellular signal which results in posttranscriptional responses in hematological diseases [5,9]. It was indicated that mutant STAT5 had the ability to interact with other oncoproteins to transform primary hematopoietic cells and constitutively active mutant STAT5 promoted cell proliferation and triggered formation of leukemia [10-13]. STAT5 proteins are associated with the development of myeloproliferative diseases [14] and responsible for the antiapoptotic activity of Bcr-Abl [15,16].
STAT5 has become the initial target of emerging anti-cancer agents [17] since it was proved that suppression of STAT gene expressions caused the inhibition of leukemogenesis together with remarkable induction of tumor cell apoptosis [8]. Ideally, targeting STATs should cause preferential cancer cell death with minimal effects on normal cells, which can be achieved by post-transcriptional gene silencing mechanisms such as antisense oligonucleotides (ODN) and small interfering RNAs (siRNA). ODNs are synthetic short DNA or RNA segments which are designed based on sequence complementary to a specific target mRNA. After matching with the target mRNA, they cause either inhibition of translation or overexpression of the protein encoded by that mRNA. This is a novel gene regulatory mechanism in cancer therapy [18]. Intriguing siRNAs have the ability to silence/suppress a target gene by degrading its mRNA and their potential usage in the treatment of diseases have been paid increasing attention. siRNAs became promising therapeutic agents in the treatment of leukemias since varying expression profiles in leukemia affect important cellular events [19].
In the current study, we initially aimed to transfect K562 chronic myeloid leukemia cells by anti- STAT5A and -5B specific siRNAs using two different lipid based transfection reagents and ODNs either with incubation or electroporation methods and to compare their transfection efficiencies. Then, we focused on to determine suppression of STAT5A and -5B genes at mRNA and protein levels and apoptotic response of the leukemic cells following siRNA or ODN treatment. Finally, we made a general comparison of protocols and gene regulatory mechanisms for an in vitro leukemia model.
Materials and methods
Culturing conditions of leukemia cells
Human K562 chronic myeloid leukemia cells were obtained from European Collection of Cell Cultures (ECACC). K562 cells were cultured in RPMI-1640 medium containing 10% (v/v) heat inactivated fetal calf serum, 100 units of penicillin-streptomycin/ml, 1% L-glutamine at 37°C in humidified air containing 5% CO2.
Confirmation of siRNA transfection
Initially, fluorescently labeled siRNA (siGLO RISC Free siRNA, Dharmacon) was transfected into leukemia cells with two different lipid based transfection reagents (TR); Dharmafect-I [DF-I, Dharmacon; part of Thermo Fisher Scientific] and Hiperfect [(HYP), Qiagen; Valencia CA and USA] in order to determine the most efficient one. Another transfection of siGLO siRNA having similar molecular weight as siRNA was designed for anti-sense oligonucleotides and performed by electroporation with the same conditions in order to have a general idea about their transfection efficiency.
Cell proliferation assay
In order to determine nontoxic effects of siRNA and ODN concentrations and also transfection reagents on K562 cells, XTT cell proliferation assay was performed (Cell Proliferation Kit II, Roche, Germany). Absorbance of each sample was measured spectrophotometrically by an ELISA reader (Thermo multiscan) for 24 and 48 hours.
Gene silencing by siRNA treatments
Each siRNA targeting STAT5A or -5B were composed of 4 different sequences (ON-TARGET plus set of 4 duplexes, Dharmacon) that match different parts of STAT5A and -5B mRNAs. The aim of using 4 different siRNA sequences for a target gene was to reduce off target effects and maintain high silencing potency. The final concentration of used siRNAs was 100 nM (4 siRNAs, 25nM for each) and siRNA transfection was performed in accordance with either HYP or DF-I manuals. Nucleotide sequences of siRNA are given in Table 1. To reach the optimal silencing effect, cells were incubated at 37°C in the presence of 5% CO2 for 96 hours. During this incubation period, particular number of cells were obtained at different time points to be used for real-time quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) (in LightCycler ver:2.0, at 24th and 48th hours), apoptotic assay (at 24th-96th hours) and Western Blot assay (at 72th-96th hours). Extra control groups were included in western blot analyses together with untreated control group, non-targeting siRNA treated cells as a negative control (NC) and “mock” group (transfected only with transfection reagent, no siRNA addition).
Table 1.
ON-TARGETplus set of 4 duplexes (25 nM each; totally 100 nM) specifically designed for STAT5A and STAT5B mRNA and negative control siRNA
| J-005169-11/12/13/14 STAT5A | |
|
| |
| SENSE | ANTISENSE 5’-P |
| CGG CUG GGA UCC UGG UUG AUU | UCA ACC AGG AUC CCA GCC GUU |
| GCG CUU UAG UGA CUC AGA AUU | UUC UGA GUC ACU AAA GCG CUU |
| GUA CUA CAC UCC UGU GCU GUU | CAG CAC AGG AGU GUA GUA CUU |
| GCA AUG AGC UUG UGU UCC AUU | UGG AAC ACA AGC UCA UUG CUU |
|
| |
| J-010539-06/07/08/09 STAT5B | |
|
| |
| SENSE | ANTISENSE 5’-P |
| GAA UUU ACC AGG ACG GAA UUU | AUU CCG UCC UGG UAA AUU CUU |
| GGA CAC AGA GAA UGA GUU AUU | UAA CUC AUU CUC UGU GUC CUU |
| GGG CAG AGU CGG UGA CAG AUU | UCU GUC ACC GAC UCU GCC CUU |
| UCU CAA GCC UCA UUG GAA UUU | AUU CCA AUG AGG CUU GAG AUU |
|
| |
| Negative control (siCONTROL Non-Targeting siRNA 2, D–001210–02–05) | |
|
| |
| 5’- UAAGGCUAUGAAGAGAUAC-3’ | |
Gene silencing by ODN applications
Anti-sense oligonucleotides specifically designed for STAT5A or STAT5B (as STAT5A ODN and STAT5B ODN) and negative control scrambled sequences (SCR ODN) were transfected into the leukemic cells for 24 hour as suggested before [20,21] or by electroporation method developed in our laboratory using BTX ECM 830 apparatus. Briefly, lyophilized ODNs were resuspended in 1ml of serum free Iscove’s Dulbecco Modification of Eagle’s Medium and stock solutions were prepared. 1x106 leukemic cells were incubated for 24 h with indicated ODNs (75 μg/ml as a low dose and 150 μg/ml as a high dose). Nucleotide sequences of ODN are given in Table 2. After incubation, the cells were collected and washed twice with Iscove’s Dulbecco Modification of Eagle’s Medium and total mRNAs and proteins were extracted. Simultaneously, the number of cells undergoing apoptosis was determined. The same ODN concentrations were used in electroporation-based transfection (1 pulse in 250 V for 10 msec in 4 mm cuvettes).
Table 2.
The antisense oligonucleotides specifically designed for STAT5A and STAT5B mRNAs
| ODN TYPE (5’→3’) | Sequence |
|---|---|
| STAT5A ODN | CsTsGsGsGsCsTsGsGsAsTsCsCsAsGsCsCsCsGsCsCsAsT |
| STAT5B ODN | CsTsGsGsCsTsTsGsTsAsTsCsCsAsCsAsCsAsGsCsCsAsT |
| SCR ODN | TsGsTsAsCsCsTsGsAsCsGsGsGsTsAsCsGsTsAsCsGsTsGsT |
Quantitative assessment of STAT5 mRNA expression levels by qRT-PCR
Following gene specific siRNA treatment for 24- and 48 hours, mRNA levels of STAT5A and -5B genes were assessed by qRT-PCR [22]. Gene silencing rate was determined by comparing the relative expression values of siRNA applied groups (either with HYP or DF-I) with untreated control group and repression values were identified for each target gene. By this way, a comparison of silencing efficiencies between two lipid based transfection reagents, HYP or DF-I, was obtained. Moreover, relative STAT5A and -5B mRNA levels of ODN applied groups were compared with scrambled group’s expressions and, then suppression rates were identified for 24 hour by qRT-PCR. Gene silencing efficiencies of ODNs were determined in terms of their concentrations and usage of two different protocols, incubation or electroporation.
Determination of STAT protein expression levels by western blot analyses
Protein extraction from siRNA treated, untreated (UT), non-targeting negative control (NC) and Mock control cells at 72- and 96 hours and the cells treated with low or high doses of ODN and SCR control cells at 24 hour was performed with “Proteojet Mammalian Cell Lysis Reagent” (Thermo Scientific, Fermentas, USA) according to manufacturer instructions.
Protein concentrations were determined by Bradford assay using bovine serum albumin (BSA) standards. Finally, 13 μg of each protein extract was resolved at 8% SDS-PAGE gel and transferred to PVDF membranes using wet transfer system (Bio-Rad, USA). The primer antibody concentrations used in the study were as follows: 1:1000 diluted β-Actin (Cell Signaling Technology, USA), 1:2000 diluted STAT5A (Upstate, USA) and STAT5B (Upstate, USA). Transfer was carried out overnight at 4°C and the protein levels of target genes in each fraction were detected using the enhanced colorimetric detection kit (Amplified Alkaline Phosphatase Goat Anti-Rabbit Immune-Blot Assay Kit, Bio-Rad, USA) according to manufacturer’s instructions. The western blot results were evaluated with gel imaging system (Chemi Smart 2000).
Apoptosis assay
The apoptosis rates of siRNA or ODN treated cells and untreated or scrambled control group cells were determined by Ethidium Bromide/Acridine Orange staining under a fluorescent microscope and the number of alive and apoptotic cells (early and late stages) was evaluated. Propidium Iodide (PI) staining was also done for confirmation.
Statistical analyses
siRNA or ODN mediated gene silencing experiments were performed in triplicate and the average of relative expression values was calculated together with standard deviations. Statistical analyses were performed by using GraphPad Prism (V.5.01) software. Two-way ANOVA and Bonferroni post-tests were used to compare groups and p<0.05 was defined as significant.
Results
Verification of siRNA transfection and cytotoxicity assay
Transfection efficiencies of the methods were determined under fluorescent microscope by counting the transfected cells. The results demonstrated that HYP, DF-I and electroporation based transfections resulted in 100-, 87-, and 75% of cell populations were transfected, respectively (Figure 1). The results of cell proliferation assay indicated that gene silencing could be achieved due to the post transcriptional modification of genes and it is not related with the cytotoxic effect of siRNA or ODN concentrations and trasfection reagent (TR) amounts. XTT assay results showed that siRNA, ODN and transfection reagents were not cytotoxic; thereby 100 nM siRNA, 75 μg/mL ODN and 6 μl of HYP/DF-1 doses were appropriate for the transfection of cells. However, 150 μg/mL ODN exhibited a lower optic density ratio with 0.83 when compared to control SCR (150 μg/mL) as given in Figure 2.
Figure 1.
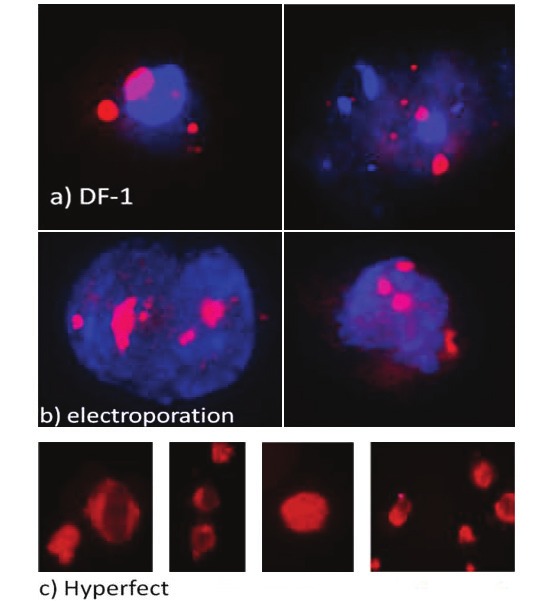
Visualization of K562 cells transfected with siRNA. The images were taken after 1 h of transfection under a fluorescence microscope transfected via DF-1, X40 (A), HYP, X100 (B), Electroporation; X40 (C) magnificated. The red spots indicate that siRNA was uptaken by the cell.
Figure 2.
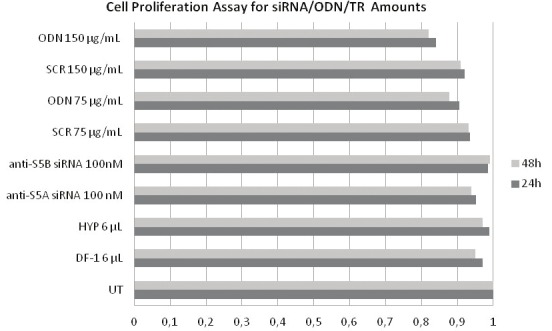
Cytotoxic effects of transfection reagents and siRNA/ODN concentration on K562 cells. The absorbances of siRNAs targeting STAT5A and -5B (100 nM each), transfection reagents (DF-1 or HYP) (6 μl each) and 75 μg/mL and 150 μg/mL ODN/SCR were measured spectrophotometrically. OD ratio of untreated (UT) control group was defined as 1.
RNAi mediated silencing of STATs decreased mRNA and protein levels of STAT5A and 5B
siRNA-mediated suppression rates of STAT5A and -5B genes were determined by comparing the expressions of them with untreated group’s expression. STAT5A or -5B suppression rates were calculated by qRT-PCR as “suppression percentage” or “fold reduction” at mRNA level and by Western Blot analysis at protein level.
While siRNAs specifically designed for STAT5A mRNA caused 46.4% suppression (1.88 fold, p=0.06) at 24 hour with DF-1, transfection performed with HYP caused 76.6% (3.78 fold, p=0.002) suppression at the same time point. The results obtained after 48 hours were as follows: siRNAs transfected with DF-1 inhibited STAT5A mRNA expression by 65.4% (2.76 fold, p=0.04) whereas HYP caused 80.5% (4.68 fold, p=0.0003) downregulation. When HYP and DF-1-mediated silencing efficacies were compared, HYP mediated STAT5A suppression was 1.68 fold (p<0.01) and 2.08 fold (p<0.001) higher than DF-1-mediated one at 24 and 48 hours, respectively. A comparison between DF-1 treated anti-STAT5A siRNA applied group displayed that 48 hour silencing rate was higher than that of 24 hour with 1.53 fold increase (p<0.05) for STAT5A (Figure 3A). Since STAT5A mRNA expressions were inhibited more effectively with HYP, further analyses were performed by using this transfection reagent. As for western blot results, time-dependent decreases at STAT5A protein expression levels via anti-STAT5A siRNA were detected with a highest rate of suppression at 96th hour of transfection with HYP (Figure 3B).
Figure 3.
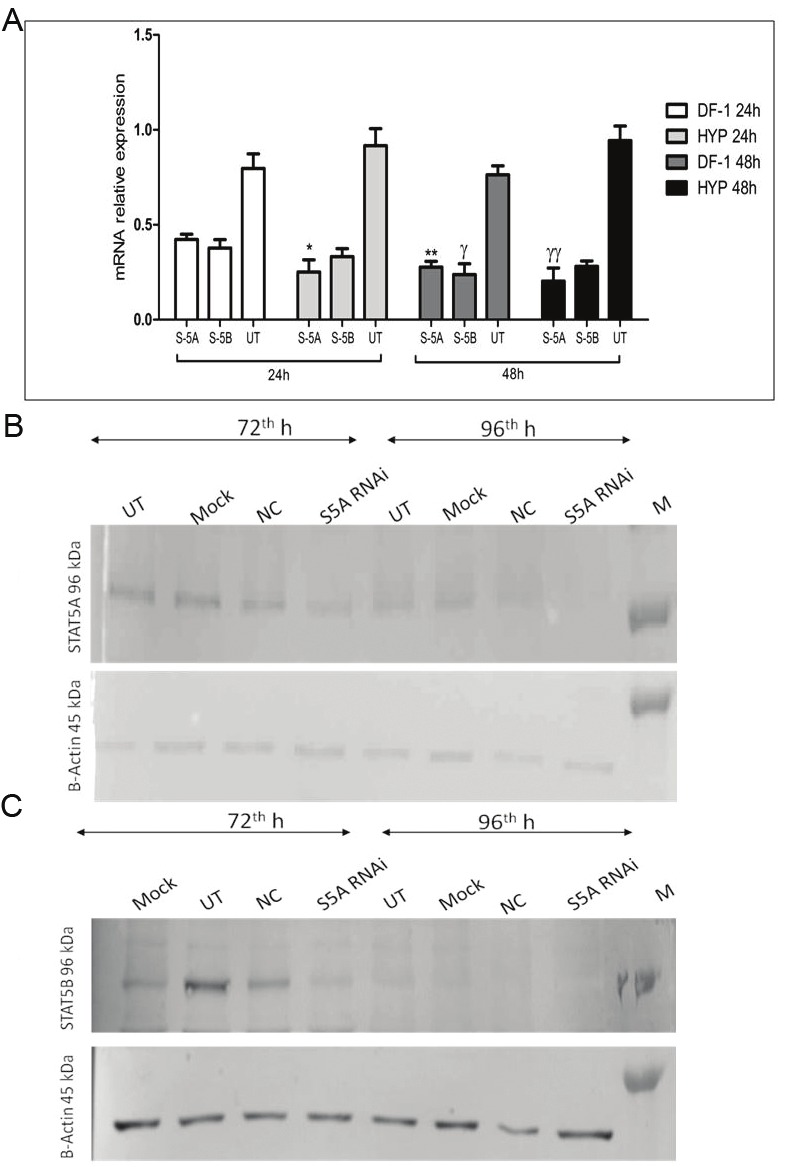
Effects of RNAi Mediated Silencing of STATs at mRNA and Protein Expression Levels. A: STAT5A and STAT5B mRNA relative expression results following siRNA treatment with DF-1 or HYP transfection reagents at 24th and 48th hours detected by qRT-PCR. S-5A and S-5B stands for anti-STAT5A and anti-STAT5B siRNA treated groups; whereas UT refers untreated group. *, γγ: HYP vs DF-1 STAT5A silencing efficacy for 24th and 48th hour differed statistically [1.68 fold (p<0.01) and 2.08 fold (p<0.001)] respectively. **, γ: DF-1 24th h vs DF-1 48th h: Significant increase was detected in STAT5A suppression at 48th h [1.53 fold (p<0.05)], STAT5B expressional inhibiton was 1.59 fold [p<0.05] higher. B-C: Stat5a and Stat5b protein expression results at the 72th and 96th hours of transfection revealed by western blot after HYP mediated siRNA transfection. NC: M: Protein Marker.
The relative expression of STAT5B mRNA decreased by 51.2% (2.11 fold, p=0.05) with DF-1 at 24 hour and HYP caused 67.5% inhibition (2.86 fold, p=0.004) at this time point. While 68.7% suppression was detected (3.22 fold, p=0.002) with DF-1 at 48 hour; similar results were obtained with HYP with the inhibition rate of 70.1% [(3.37 fold, p=0.003), Figure 3A]. A comparison between DF-1 treated anti-STAT5B siRNA applied group showed that 48 hour silencing rate was higher than that of 24 hour with 1.59 fold increase for STAT5B [(p<0.05); Figure 3A]. As a result, STAT5B mRNA expression was downregulated with each transfection reagent at 48 hour. However, considering both STAT5A and -5B inhibition rates, HYP was accepted as a better reagent and further experiments were carried out with HYP. When STAT5B protein levels were evaluated, it was observed that HYP mediated siRNA applications silenced STAT5B efficiently both at 72- and 96 hours with an acceleration of suppression especially at 96 hour (Figure 3C).
Antisense oligonucleotide mediated silencing of STATs at mRNA and protein levels
Antisense oligonucleotides targeting STAT5A and STAT5B were transfected into the cells via the incubation method as described elsewhere and via electroporation method by applying low (75 μg/mL) and high (150 μg/mL) concentrations of ODNs for 24 hour. The inhibition of ODN mediated STAT5A and -5B gene expressions was determined by comparing the expression levels of groups treated with high/low concentrations of ODN and SCR (scrambled, ) group. STAT5A or -5B suppression rates were determined by qRT-PCR as “suppression percentage” or “fold reduction” at mRNA level and by Western Blot analysis at protein level at 24 hour for either incubation or electroporation method.
Based on the results of incubation method, the relative expression of STAT5A decreased by 12.2% (1.15 fold, p>0.05) and 37.5% (1.63 fold, p>0.05) at low and high concentrations of ODNs, respectively, as compared to their own SCR control groups (Figure 4A). On the other hand, low concentration of ODN caused 43.4% (1.77 fold, p>0.05) inhibition in STAT5A mRNA expression and high concentration of it achieved 74.9% suppression (3.95 fold, p<0.001) using electroporation method (Figure 4A). When STAT5A protein expressions were evaluated, it was observed that while incubation method didn’t cause efficient suppression at both low and high concentrations of ODNs, highER concentration of ODNs incorparated into the cells via electroporation highly suppressed STAT5A protein level (Figure 4B).
Figure 4.
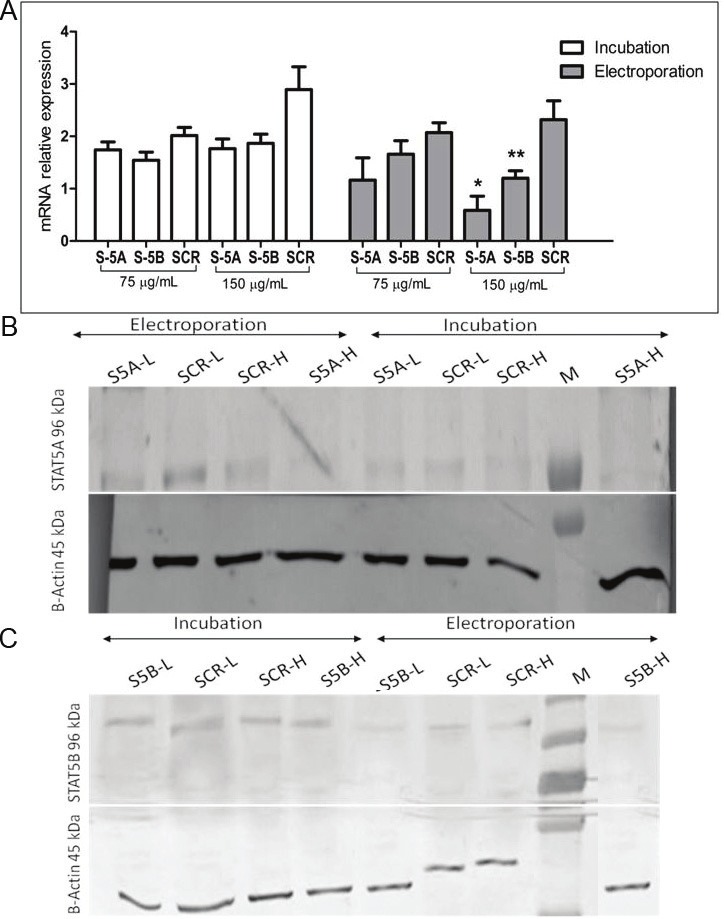
Antisense Oligonucleotide Mediated Silencing of STATs at mRNA and Protein Levels. The relative expression levels of STAT5A and STAT5B mRNAs following transfection of 75 μg/mL and 150 μg/mL of ODNs either with incubation or electroporation methods at 24th h were detected by qRT-PCR. S-5A and S-5B stands for AS-STAT5A and AS-STAT5B ODN treated groups whereas SCR refers negative control group. Significant inhibition of STAT5A and STAT5B mRNAs was detected in electroporation group (*, **: p<0.001 and p<0.05 respectively) (A). Stat5a and Stat5b protein expressions were detected by western blot after 24 hours of incubation/electroporation mediated ODN transfection (B and C). S5A-L/S5B-L: Low antisense STAT5A or STAT5B oligonucleotides concentration (75 μg/mL), S5A-H/S5B-H: High antisense STAT5A or STAT5B oligonucleotides concentration (150 μg/mL) and SCR-L/SCR-H: Low and high concentrations of Scrambled controls, M: Protein Marker.
On the other hand, low concentration of ODN suppressed the relative STAT5B mRNA levels by 22.3% (1.31 fold, p>0.05) while high concentration of ODN resulted in an inhibition of 35.7% (1.55 fold, p>0.05). However, in the electroporation method, low and high concentrations of ODN suppressed the relative STAT5B mRNA levels by 20.6% and 48.7% (1.24 fold, p>0.05 and 1.93 fold, p<0.05), respectively (Figure 4A). Based on the results of western blot analysis, it could be concluded that STAT5B expression was not inhibited by any of ODN via incubation method. On the other hand, an increase in the suppression rate of STAT5B was observed only at high concentration of ODN transfected with electroporation method (Figure 4C).
Triggered apoptosis of leukemic cells in response to STAT5 supression
The results of apoptotic analyses demonstrated that apoptosis was induced in K562 cells even at earlier periods of transfection due to siRNA or high dose ODN mediated (with electroporation method) gene silencing of STAT5A and STAT5B genes through both RNAi and antisense oligonucleotides. Apoptosis rate was found to be extremely high especially at 72- and 96 hours of the transfection in which the cells treated with HYP mediated anti-STAT5A siRNA by an average ratio of 72% and 82.5% apoptotic cells, respectively (p<0.001 and p<0.001). Apoptosis rates were 66.5- and 75.5% (p<0.001 and p<0.001) at 72 and 96 hours of the transfection, respectively, for anti-STAT5B siRNA-treated group (Figure 5A). Figure 5B and 5C showed the image of apoptotic cells treated with siRNA. As shown in Figure 5C, propidium iodide staining was also performed to verify apoptosis. The percentage of apoptotic cells in the group treated with high concentration of antisense STAT5A oligonucleotides was 45.3 ± 14.6% whereas antisense STAT5B caused 40.9 ± 4.8% apoptotic leukemic cells as shown in Table 3. Figure 5D showed Ethidium Bromide/Acridine Orange staining of the cells treated with high dose ODN by electroporation method.
Figure 5.
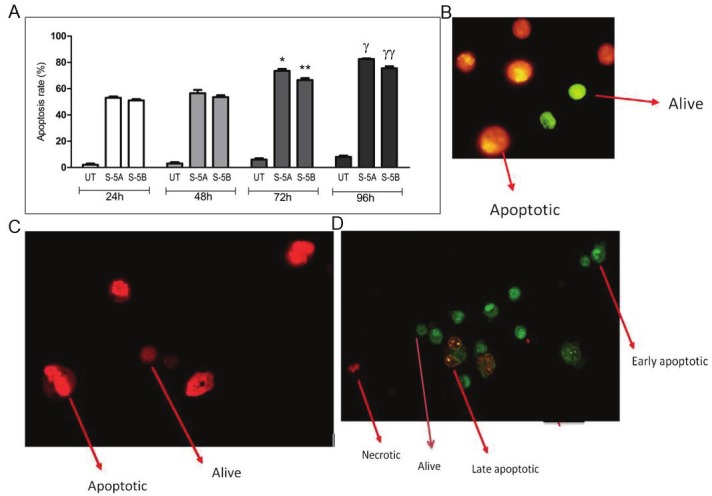
Induction of Apoptosis in Response to STAT5 Knockdown. High rates of apoptosis were detected in HYP mediated siRNA transfected cells and high concentration of ODN treated groups via Ethidium Bromide/Acridine Orange staining under a fluorescence microscope. STAT5A and STAT5B apoptosis rate ratios seen in siRNA applied groups compared to UT control group. *, **: 24h vs 72h: p*<0.001, p**<0.001. γ, γγ: 24h vs 96h: pγ<0.001, pγγ<0.001 (A). siRNA treated cell apoptosis. Green cells are alive and orange cells are indicated as apoptotic; X40 magnificated (B). PI staining of siRNA treated cells (C). High concentration of ODN treated cell apoptosis; red cells are necrotic, early apoptotic cells were also seen in green but the bright dots found in the nucleus differ them from alive cells, X40 magnification (D).
Table 3.
Percentage of apoptotic cells at 24 hour of electroporation mediated transfection of ODNs in K562 leukemic cells. K562 cells were exposed to 150 μg/mL antisense STAT5A, STAT5B and scrambled ODNs (mean ± SD; EtBr/Akridine Orange staining)
| K562 | SCR treated | STAT5A ODN treated | STAT5B ODN treated |
|---|---|---|---|
| Non-apoptotic cells | 83.4 ± 18.2 % | 50.9 ± 13.7 % | 52.8 ± 3.9 % |
| Apoptotic cells (early + late stage) | 11.7 ± 7.6 % | 45.3 ± 14.6 % | 40.9 ± 4.8 % |
Discussion
CML is known to be a clonal hematological disease that forms 15–20% of all adult leukemia cases. Increasing number of studies aim to define an efficient therapy for CML especially for patients with resistance to tyrosine kinase inhibitors and drug intolerance. Therefore, there is an urgent need to develop alternative therapeutic strategies such as post transcriptional gene silencing mechanisms for CML. In this study, we aimed to induce leukemic cell apoptosis via ODN or RNAi-mediated silencing of STAT5A and STAT5B genes which are dormant transcription factors in cytoplasm. When activated, they translocate into the nucleus where they increase the expression of their target genes and trigger leukemic development. The usage of antisense oligonucleotides for gene silencing studies has been started earlier and they have been played roles in the gene regulation [23]. However, the discovery of small double-stranded RNAs (siRNAs) has showed that they are very efficient molecules for the inhibition of gene expression in mammalian cells [24,25]. However, such gene silencing studies using either ODN or siRNA will be only achieved when they are introduced into the cells without being exposed to biodistribution and degradation by nucleases inside the cell [26] and inducing innate immune system [27]. For this purpose, we used two different lipid based transfection reagents in order to deliver siRNA and two different methods, incubation and electroporation, with low and high concentration of ODN in order to transfect leukemic cells. By conducting different strategies, we aimed to find the optimal gene silencing method which induces leukemic cell apoptosis. In addition, we aimed to trigger leukemic cell apoptosis by suppressing STAT5 expression at both mRNA and protein levels. Because, STAT5s are the key regulatory components of JAK/STAT pathway. Therefore, firstly we analyzed the uptake of fluorescently labeled siRNA by the cells treated with DF-1, HYP and electroporation and we concluded that siRNA was penetrated into every cell with HYP transfection reagent. Next, we checked whether the used TR, siRNA and ODN concentrations had any cytotoxic effect on cell proliferation. Eventually, we concluded that 6 μl TR and 100 nM siRNA were appropriate for the efficient transfection of K562 cells. This was the most important point of the study since the results showed that apoptosis of leukemic cells was due to siRNA or ODN mediated gene silencing. Another outcome of this assay was that high concentration of ODN (150 μg/mL) exhibited a lower OD ratio as compared to its SCR control and other agents. Therefore, we concluded that longer periods of treatment could decrease cell viability due to high concentration. This may also be the reason why the studies carried out with ODNs are usually last about 24 hours since ODNs have lower resistance to nuclease degradation [20,28]. However, longer periods of treatment like 12 days with chemically modified siRNAs have been shown in our previous study [22].
One of the most important aims of the study was to acquire high suppression rates of STAT5A and STAT5B at both mRNA and protein levels following siRNA or ODN treatment and to reveal the apoptotis induction in siRNA/ODN treated and untreated/SCR control leukemia cells. qRT-PCR results displayed that siRNA transfection performed using HYP inhibited STAT5A mRNA expression by 76.6- and 80.5% for 24- and 48 hours, respectively, and STAT5A protein levels was highly downregulated in a time dependent manner together with the induction of apoptosis (82.5% apoptotic leukemic cells). Moreover, STAT5B mRNA expression was suppressed by 67.5- and 70.1% at 24 and 48 hour, respectively, via siRNA applications mediated with HYP. DF-1 also exhibited an efficient silencing of STAT5B with suppression rate of 68.7% at 48 hour. STAT5B protein levels were efficiently downregulated at both 72 and 96 hours with an acceleration of suppression especially at 96 hour with 75.5% of apoptotic leukemic cells.
Although siRNA applications showed high mRNA and protein suppression rates and also an increase in leukemic cell apoptosis, ODN application had lower inhibition rates compared to siRNA even at high doses of ODN. ODN applications were carried out either with incubation method as reported before [20,21] or by an electroporation protocol generated by our group. We concluded that our revised electroporation protocol for high concentration of ODNs caused 74.9% suppression of STAT5A mRNA and also decreased protein levels together with the induction of apoptosis ( 45.3 ± 14.6 % apoptotic leukemic cells). The expression of STAT5B mRNA (decreased by 48.7%) was found to be more efficient using high concentration of ODNs by electroporation. Although incubation method caused no inhibition at protein level, an increase in the suppression rate of STAT5B protein was detected using high dose ODN transfected by electroporation method with 40.9 ± 4.8% of apoptosis in leukemic cells. Finally, we concluded that electroporation could be a quick and more efficient alternative method in ODN mediated gene silencing studies rather than incubation.
Taken together all these results, our data demonstrated that siRNA applications are more efficient gene silencing approach for suspension leukemic cells as compared to ODN treatment based on the higher apoptosis and mRNA suppression rates. Similar to our results, Bertrand J. et al reported that siRNA application was more efficient for targeting green fluorescent protein (GFP) rather than nuclease resistant antisense oligonucleotide in HeLa cells and in xenografted mice models. They displayed that siRNAs appeared to be quantitatively more efficient and their effect was lasted for a longer period in cell culture. Moreover, they observed an activity of siRNAs even in mice, but not of antisense oligonucleotides. They concluded that the absence of efficiency of ODN was probably due to their lower resistance to nuclease degradation. We also think that if the concentration of ODN is increased in order to perform a better gene silencing, this may also be cytotoxic for healthy cells [28].
In conclusion, RNAi process could be a powerful strategy for post transcriptional gene silencing that may open a new therapeutic era in the clinic. In the future, siRNA application could constitute a new and alternative curative method as a supporting therapy of CML diagnosed patients.
Acknowledgement
This work was supported by The Scientific and Technological Research Council of Turkey, (TUBITAK 105S459, to BK).
Conflict of interest statement
The authors declare no potential conflict of interest.
References
- 1.Nowell PC, Hungerford N. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497. [Google Scholar]
- 2.Koca E, Haznedaroglu IC. Imatinib mesylate and the management of chronic myeloid Leukemia (CML) Turk J Haematol. 2005;4:161–172. [PubMed] [Google Scholar]
- 3.Kantarjian H, Melo JV, Tura S, Giralt S, Talpaz M. Chronic myelogenous leukemia: disease biology and current and future therapeutic strategies. Hematol Am Soc Hematol Educ Program. 2000;1:90–109. doi: 10.1182/asheducation-2000.1.90. [DOI] [PubMed] [Google Scholar]
- 4.Kantarjian H, Talpaz M, Giles F, O’Brien S, Cortes J. New insights into the pathophysiology of chronic myeloid leukemia and imatinib resistance. Ann Intern Med. 2006;145:913–923. doi: 10.7326/0003-4819-145-12-200612190-00008. [DOI] [PubMed] [Google Scholar]
- 5.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 6.Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285:1–24. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 7.Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 8.Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, McCubrey JA. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18:189–218. doi: 10.1038/sj.leu.2403241. [DOI] [PubMed] [Google Scholar]
- 9.Valentino L, Pierre J. JAK/STAT signal transduction: regulators and implication in hematological malignancies. Biochem Pharmacol. 2006;71:713–721. doi: 10.1016/j.bcp.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 10.Onishi M, Nosaka T, Misawa K, Mui AL, Gorman D, McMahon M. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol Cell Biol. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nosaka T, Kitamura T. Janus kinases (JAKs) and signal transducers and activators of transcription (STATs) in hematopoietic cells. Int J Hematol. 2000;71:309–319. [PubMed] [Google Scholar]
- 12.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 13.Ilaria RL Jr, Van Etten RA. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem. 1996;271:31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- 14.Levy DE, Gilliland DG. Divergent roles of STAT1 and STAT5 in malignancy as revealed by gene disruptions in mice. Oncogene. 2000;19:2505–2510. doi: 10.1038/sj.onc.1203480. [DOI] [PubMed] [Google Scholar]
- 15.Donato NJ, Wu JY, Zhang L, Kantarjian H, Talpaz M. Down-regulation of interleukin-3/granulocyte-macrophage colonystimulating factor receptor beta-chain in BCR-ABL(+) human leukemic cells: association with loss of cytokine-mediated Stat-5 activation and protection from apoptosis after BCR-ABL inhibition. Blood. 2001;97:2846–2853. doi: 10.1182/blood.v97.9.2846. [DOI] [PubMed] [Google Scholar]
- 16.Nieborowska-Skorska M, Wasik MA, Slupianek A, Salomoni P, Kitamura T, Calabretta B, Skorski T. Signal transducer and activator of transcription (STAT)5 activation by BCR/ABL is dependent on intact Src homology (SH)3 and SH2 domains of BCR/ABL and is required for leukemogenesis. J Exp Med. 1999;189:1229–1242. doi: 10.1084/jem.189.8.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benekli M, Baumann H, Wetzler M. Targeting signal transducer and activator of transcription signaling pathway in leukemias. J. Clin. Oncol. 2009;27:4422–4432. doi: 10.1200/JCO.2008.21.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braasch DA, Corey DR. Novel antisense and peptide nucleic acid strategies for controlling gene expression. Biochemistry. 2002;41:4503–4510. doi: 10.1021/bi0122112. [DOI] [PubMed] [Google Scholar]
- 19.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baśkiewicz-Masiuk M, Masiuk M, Czajka R, Machaliński B. The role of STAT5 proteins in the regulation of normal hematopoiesis in a cord blood model. Cell Mol Biol Lett. 2003;8:317–331. [PubMed] [Google Scholar]
- 21.Baśkiewicz-Masiuk M, Masiuk M, Czajka R, Machaliński B. The influence of STAT5 antisense oligonucleotides on the proliferation and apoptosis of selected human leukaemic cell lines. Cell Prolif. 2003;36:265–278. doi: 10.1046/j.1365-2184.2003.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaymaz BT, Selvi N, Gündüz C, Aktan C, Dalmızrak A, Saydam G, Kosova B. Repression of STAT3, STAT5A, and STAT5B expressions in chronic myelogenous leukemia cell line K–562 with unmodified or chemically modified siRNAs and induction of apoptosis. Ann Hematol. 2013;92:152–162. doi: 10.1007/s00277-012-1575-2. [DOI] [PubMed] [Google Scholar]
- 23.Braasch DA, Corey DR. Novel antisense and peptide nucleic acid strategies for controlling gene expression. Biochemistry. 2002;41:4503–4510. doi: 10.1021/bi0122112. [DOI] [PubMed] [Google Scholar]
- 24.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 25.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crombez L, Aldrian-Herrada G, Konate K, Nguyen QN, McMaster GK, Brasseur R, Heitz F, Divita G. A new potent secondary amphipathic cell-penetrating peptide for siRNA delivery into mammalian cells. Mol Ther. 2009;17:95–103. doi: 10.1038/mt.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robbins M, Judge A, MacLachlan I. siRNA and innate Immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 28.Bertrand JR, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvy C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem Biophys Res Commun. 2002;296:1000–1004. doi: 10.1016/s0006-291x(02)02013-2. [DOI] [PubMed] [Google Scholar]


