Abstract
Menin acts as contextual a tumor suppressor and a tumor promoter, partly via epigenetic regulation of gene transcription. While menin is phosphorylated, it remains unclear whether wild type menin has other post-translational modifications. Here, we report that menin is SUMOylated by SUMO1 in vivo and in vitro, and the SUMOylation is reduced by a SUMO protease. Lysine 591 of menin was covalently modified by SUMO1 and K591R mutation in menin blocked SUMOylation of the C-terminal part of menin in transfected cells. Full-length menin with K591 mutation was still SUMOylated in vivo, suggesting the existence of multiple SUMOylation sites. Menin K591R mutant or menin-SUMO fusion protein still retains the ability to regulate cell proliferation and the expression of the examined menin target genes.
Keywords: Menin, SUMOylation, SUMO1, K591R
Introduction
Menin is encoded by the MEN1 gene, and mutations in the MEN1 gene leads to multiple endocrine neoplasia type 1 (MEN1), an inherited tumor syndrome, with development of tumors in several endocrine organs including pancreatic islets [1-4]. Menin missense mutants associated with MEN1 syndrome are rapidly degraded via the ubiquitin-mediated proteolysis [5]. Menin is phosphotylated at several serine residues and the phosphorylation is induced by DNA damage [6,7]. It is not yet known whether phosphorylation alters the function of menin and whether menin has other modifications.
Post-translational modifications (PTMs) including methylation, phosphorylation, acetylation, and ubiquitination are crucial for regulation of multiple biological processes as well as biological activities such as transcriptional regulation and protein degradation [8,9]. Another PTM is modified by the small ubiquitin-related modifier (SUMO), a modification referred to as SUMOylation [10,11].
SUMO is ~11kD in size and has the three-dimensional structure similar to ubiquitin [10]. In mammals three major SUMO isoforms, SUMO1–3, have been found. Like ubiquitin, SUMO covalently conjugated to substrates through the C-terminus Gly-Gly motif under the help of E1 activating enzyme (SAE1/SAE2), E2 conjugating enzyme (Ubc9) and E3 protein ligase [12,13]. SUMOylation is a reversible and highly transient modification and the de-conjugation of SUMO is mediated by isopeptidases, also known as sentrin-specific proteases (SENP) or SUMO de-conjugating enzymes. Six human SENP family proteins, SENPs 1–3 and 5-7 have been identified [14]. SUMOylation has been shown to be important for regulating many cell processes, including transcription, replication, chromosome segregation and DNA repair [11,15]. SUMO modification regulates gene transcription, mainly repressing gene transcription, but also in some cases inducing transcriptional activation, and the majority of SUMOylation targets are transcription factors [16-18]. Although recent studies have shown that regulation of SUMOylation contributes to the pathogenesis and development of various diseases [19-22], if it remains unclear whether menin is modified by SUMOylation. Here, we show that menin undergoes SUMO modification and Lysine 591 is one of the SUMOylation sites.
Materials and methods
In vitro SUMOylation assay
In vitro SUMOylation of menin was performed using a SUMOylation kit (Biomol). Briefly, deleted or full-length His-tagged menin was expressed in E. coli and purified as previously described [23], and suspended in a 20 μl reaction buffer containing SAE1, SAE2, Ubc9, SUMO1 and ATP. Following 1 hr incubation at 37°C, reactions were terminated by an SDS loading buffer. The products were analyzed by western blotting.
In vitro de-SUMOylation assay
The de-SUMOylation of menin by SUMO protease Ulp1 was carried out using a SUMO protease kit (Invitrogen). Briefly, immunoprecipitation generated by co-IP was incubated in a SUMO-protease buffer with or without Ulp1 at 25°C for different times. Reactions were terminated by adding SDS loading buffer and the products were analyzed by Western blotting with indicated antibodies.
Mass spectrometry analysis
Purified menin and SUMO1 (RGG) were suspended in a reaction buffer with SAE1, SAE2, Ubc9 and ATP at 37 °C for 1 hr. Reaction products were separated by SDS-PAGE and were analyzed by Coomassie Brilliant Blue (CBB) staining. The predicted bands were cut and digested with trypsin [24]. The peptides were analyzed with nanoLC/MS/MS at the Penn Proteomics Core and the data were analyzed with Sequest and Scaffold software packages.
Plasmids and cell culture
Flag-SUMO1, His-SUMO1 and pUBC9 were previously described [25]. The pUbc9/SUMO1 used for bacterial expression of His-SUMO1 was from Mutsuhiro Takekawa (Nagoya University) [26]. The SENP1 plasmid was purchased from Addgene. Retroviral plasmid expressing Flag-tagged menin was previously described [27]. Full-length Flag-menin and menin fragment Flag-F3, which encode amino acid residues 1–610 and 396–610, respectively, were amplified by PCR and cloned into the BamHI/Not I site of pCDNA3.1. Menin-SUMO fusion construct was generated by fusion of SUMO1 to the C-terminus of menin. His-SUMO1 (G97A), His-SUMO1 (RGG), menin fragment F3 (K591R), full-length menin (K591R), retroviral expressed menin (K591) and menin-SUMO fusion (K591R) were generated using the site-directed mutagenesis kit (Stratagene). Men1-null MEF cells complemented with wild type or mutant menin [27], WT MEFs and HEK293T cells were cultured in Dulbecco’s modified Eagle’s Medium (DMEM) supplemented with 10 % fetal bovine serum (FBS) and 1% Pen/Strep.
Retroviral infection and cell proliferation
Plasmids for retroviral packaging were cotransfected with psi-2 helper plasmid into 293T cells using the calcium chloride precipitation method. The resulting recombinant virus was collected and transduced into Men1 null MEFs, followed by selection in 2 μg/ml puromycin for 3 days. The above cells were seeded with 2×104 cells/well in 6-well plates, and cell numbers were counted with a hemacytometer, with dead cells excluded by Trypan blue staining.
RNA extraction and quantitative real time PCR (qRT-PCR)
Total RNA was extracted from cultured cells with Trizol and an RNeasy extraction kit from Qiagen. 1 μg RNA was used as template to synthesize cDNA for qRT-PCR using a 7500 fast real-time PCR from Applied Biosystems. The relative mRNA level was calculated by the ΔΔCt values calibrated with GAPDH, using SYBR Green dye from Qiagen for detection. The following primers were used for quantitative RT-PCR: Gas1-For, 5’-AGATGGTCGGGAACACTGAC-3’; Gas1-Rev, 5’-TCCCTTCTCCAAGTCCATTG Gli1-For, 5’-AAGGAATTCGTGTGCCATTGGG-3’;-3’; Gli1-Rev, 5’-ACATGTAAGGCTTCTCACCCGT-3’.
Purification of protein generated in E. coli
Briefly, E. coli BL21 transformed with full-length His-menin, internally deleted His-menin (460-510 AA) or pUbc9/SUMO1 (RGG) was incubated at 25°C, and then induced by isopropyl β-D-1-thiogalactopyranoside (0.25 mM) for 4 hrs. Cells were harvested and lysed with 1% Triton X-100 in PBS, 1.0 mM PMSF, 4 μg/mL bacterial protease inhibitor cocktail (Sigma), and 100 μg/mL lysozyme. Lysates were spun down and the supernatant was added to Ni-NTA beads (Qiagen) overnight at 4°C. After washed with 1 m Mimidazole in PBS, protein was eluted from Ni-NTA beads with 150 mM imidazole in PBS.
Western blotting
Briefly, cells were lysed in a RIPA buffer (Sigma) with protease inhibitors on ice for 30 min and sonicated to shear the genomic DNA. The extracted protein concentration was determined by BCA assay, and the samples were separated by SDS-PAGE, and then were transferred onto a PVDF membrane. Each of the antibodies was incubated against one of the following proteins: anti-menin (Bethyl), anti-SUMO1 (Abcam), anti-Flag (Sigma), anti-His (Clontech) and β-actin (Sigma). Membranes were further washed and incubated with an anti-rabbit or anti-mouse secondary antibody (BD). Detection of immunoreactive bands was performed using an ECL detection kit (GE). Equal protein loading was indicated by Ponceau-S staining of blotted membranes.
Co-immunoprecipitation
HEK293T cells were transiently transfected with indicated plasmids and were suspended in lysis buffer (50 mMTris-Cl, pH 7.4, 150 mMNaCl, 5 % glycerol, 1% mM NP-40, 1 mM EDTA) supplemented with protease inhibitors and N-ethylmaleimide (NEM) (20 mM). Cell lysates were incubated with anti-menin (Bethyl) antibody at 4°C for 2 hrs with rotation, and then bound to protein A-Sepharose (Invitrogen), or with Flag M2 beads (sigma) and washed four times with the lysis buffer. Proteins were separated by SDS-PAGE and immunoblotted with the indicated antibodies. To detect endogenous menin SUMOylation, MEFs were lysed with the lysis buffer and lysates were incubated with anti-menin antibody or control IgG (Abcam) at 4°C for 4 hrs. The immunoprecipitates were blotted with anti-menin or anti-SUMO1 antibodies.
Statistical analyses
Statistical analyses were performed using Graphpad Prism (version 5.0; Graphpad Software). The data are presented as the mean ±s.d. of n determinations unless noted otherwise. A two-tailed student’s t test was used for measuring statistical differences.
Results
Ectopic and endogenous menin are modified by SUMO1 in cells
During our routine detection of menin in Western blotting we occasionally observed a slow-migrating band (data not shown) and suspected that this band might be due to SUMOylation. To determine whether menin undergoes SUMOylation in cells, we transfected cDNAs for menin and Flag-tagged SUMO1 individually or in combination into HEK293T cells. The resulting cell lysates were immunoprecipitated with an anti-menin antibody and then immunoblotted with an anti-Flag antibody. A slow-migrating band was detected by the anti-Flag antibody in the immunoprecipitated products from cells co-transfected with both menin and F-SUMO1 (Figure 1A, lane 2), but not from cells transfected with either menin or SUMO1 alone (Figure 1A, Lanes 1 and 3). The apparent molecular weight is compatible with attachment of one SUMO molecule. We performed the reverse IP with an anti-Flag antibody to confirm menin modification with SUMO1. Again, a slow-migrating band was observed with Western blotting using an anti-menin antibody, in cells transfected with both menin and SUMO1 (Figure 1B, lane 2). This slow-migrating band was also detected by the anti-SUMO1 antibody (Figure 1C, lane 2). The asterisk denotes a likely unknown protein on beads that were crossreactive to the SUMO1 antibody.
Figure 1.
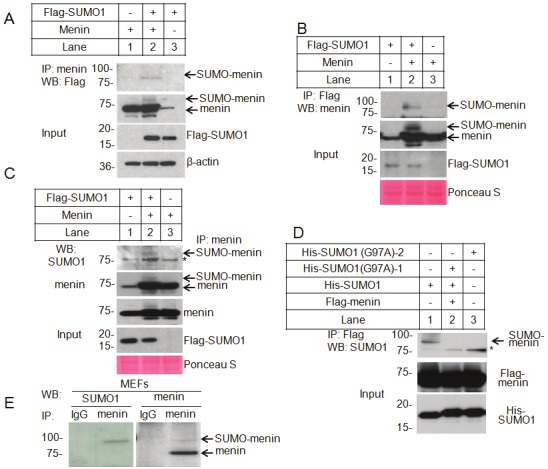
SUMOylation of menin by SUMO1 in vivo. A. HEK293T cells were co-transfected with menin and Flag-SUMO1. Immunoprecipitated menin was blotted with an anti-Flag antibody. The expression levels of SUMO1 and menin in the cell lysates are also shown. B. Menin and Flag-SUMO1 were co-transfected in HEK293T cells. SUMO1 was Immunoprecipitated, followed by western blotting with an anti-menin antibody. The total menin and SUMO1 proteins are also shown in the bottom. C. HEK293T cells were transfected to express menin along with Flag-SUMO1. Immunoprecipitated menin was blotted with an anti-menin antibody or was probed for SUMOylation using an anti-SUMO1 antibody. The total amounts of menin and SUMO1 are also shown. Asterisk indicates the non-specific binding. D. Flag-menin was expressed together with His–SUMO1 or two independent point mutants that cannot be attached to its substrate (His–SUMO1-G97A) into HEK293T cells. Menin was immunoprecipitated with Flag M2 beads, followed by western blotting for SUMOylation with anti-SUMO1 antibody. The total amounts of menin and SUMO1 are also shown; Asterisk indicates the non-specific binding. E. Immunoprecipitated endogenous menin from MEF cells was probed for SUMOylation with either anti-menin or anti-SUMO1 antibody.
To determine whether SUMO1 covalently links to menin, but not simply binds menin, we generated SUMO1 mutants (G97A) that lack the C-terminus G-G motif and thus cannot be conjugated to substrates by site-directed mutagenesis. Wild type (WT) or mutated SUMO1 cDNA were co-transfected into HEK293T cells, followed by immunoprecipitation with Flag M2 beads. The results showed that the G97A mutant failed to be conjugated to menin (Figure 1D, lane 2 and 3). These results suggest modification of transfected menin by SUMO1.
To verify whether endogenous menin is also SUMOylated, a large amount of mouse embryonic fibroblasts (MEFs) were used for immunoprecipitation with either anti-menin antibody or control IgG, followed by Western blotting with an anti-SMUO1 or anti-menin antibody. The results showed that a slow-migrating band was also detected by both the anti-SUMO1 and anti-menin antibodies (Figure 1E). To examine the possibility that other type of modification of menin, such as glucosylation, may also change the mobility, we treated the cells with o-glucosyltransferase inhibitor, but we failed to observe any change of menin in mobility in gel (data not shown). Together, these results indicate that endogenous menin is also SUMOylated by SUMO1 inside cells.
Removal of SUMO-modificaiton of menin by SUMO protease
SUMOylation modification is a dynamic process involving both conjugation and de-conjugation. The de-SUMOylation enzymes cleave the covalent bond between SUMO and the modified protein, removing the SUMO from the substrate, rendering it ready for the next round of modification [28]. To determine whether the slow-migration band can be removed by some of SENPs, we co-transfected Flag-tagged menin and His-tagged SUMO1 with or without increasing amounts of cDNA for human SENP1, one of several SUMO-specific proteases [14], into HEK293T cells. The resulting cell lysates were immunoprecipitated with Flag antibody-conjugated M2 beads and then immunoblotted with anti-His antibody. The slow-migrating band, or likely SUMOylated menin, was markedly reduced in SENP1-cotransfected cells (Figure 2A, lanes 4 and 5). These results further support the notion that menin is SUMOylated in cells. As a control, expression of menin and SUMO1 was as expected (Figure 2A, bottom panel).
Figure 2.
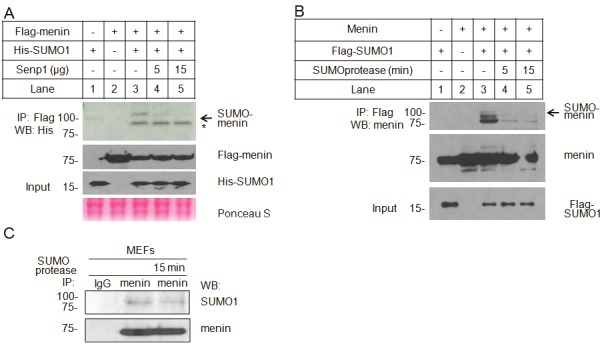
SUMOylation of menin is removed by a SUMO protease. A. Flag-menin and His-SUMO1 were co-transfected into HEK293T cells with or without different amount of SENP1 plasmid. Menin immunoprecipitated with Flag M2 beads was probed with an anti-His antibody. Over-expression of menin and SUMO1 were determined. B. HEK293T cells were co-transfected with menin and Flag-SUMO1. SUMO1 was pull-down with Flag M2 beads. The immunoprecipitated products were incubated with purified SUMO protease at different times, followed by western blotting with anti-menin antibody. The total menin and SUMO1 proteins are also shown in the bottom. C. Immunoprecipitated endogenous menin from MEF cells was incubated with purified SUMO protease for 15 min. The SUMOylated and WT menin were detected with anti-SUMO1 or anti-menin, respectively.
To further verify the above results, Flag-tagged SUMO1 and menin were co-expressed into HEK293T cells, followed by immunoprecipitation with Flag M2 beads. The immunoprecipitated products were treated with recombinant SUMO protease for different times, followed by Western blotting with an anti-menin antibody. We found that the slow-migrating band was substantially reduced after the immunoprecipitated products were treated with SUMO protease (Figure 2B, lanes 4 and 5). It was unclear what constituted the menin-antibody-detected materials above the SUMO-menin band (Figure 2B, lane 3), but it was possibly due to other types of modificaiton or protein degradation. Further experiments showed that immunoprecipitated SUMOylated endogenous menin from MEFs was also removed by incubation with SUMO protease (Figure 2C, lanes 2 and 3). Together, these results indicate that menin is modified by SUMO1.
Purified menin protein is SUMOylated by SUMO1 and SUMO2/3 in vitro
SUMOylation is a multiple-step process involving the use of three enzymes: SUMO-activating enzymes (E1), SUMO-conjugating enzyme Ubc9 (E2) and E3 ligase [12,13]. To determine whether menin can be sumoylated in vitro, we incubated purified full-length or internally deleted menin (460-510 AA) protein, with purified SUMO-activating enzymes (E1), SUMO-conjugating enzyme Ubc9 (E2), together with His-tagged SUMO1. The reactions were separated in SDS-PAGE and immunoblotted with anti-SUMO1 or anti-His antibodies. We found that in the presence of ATP, both the deleted menin and full-length menin protein were shifted to slow-migration bands detected by both anti-SUMO1 antibody (Figure 3A, top panel, lanes 4 and 6) and anti-His antibody (Figure 3A, bottom panel, lanes 4 and 6), when SUMO1 was included in the presence of E1 and E2 enzymes. In the absence of ATP, no SUMOylated menin bands were observed (Figure 3A, top panel, lanes 3 and 5), consistent with the prediction that SUMOylation process requires ATP as a source of energy. These results indicate that SUMO1 can SUMOylate menin in vitro in the presence of E1 and E2. It is noteworthy that more than one slow-migrating bands were detected in the gel (Figure 3A, lanes 4 and 6), suggesting that the robust in vitro SUMOylation system may SUMOylate multiple sites of menin. As a positive control, RanGAP1, which has previously been shown to be a substrate for SUMOylation, was also SUMOylated (Figure 3A, lane 2). We also tested SUMO2/3 in vitro SUMOylation assay, and showed that menin could also be modified with SUMO2/3 (Figure 3B, Lane 3 and 4).
Figure 3.
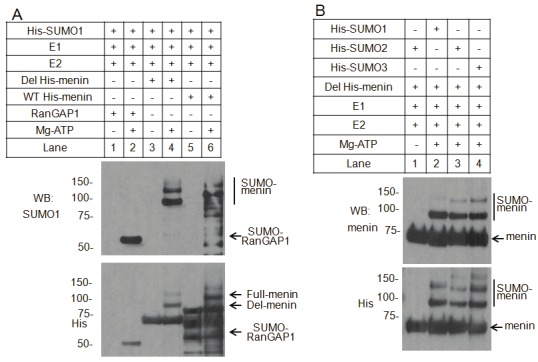
In vitro SUMOylation of menin. A. Affinity-purified RanGAP1, Deleted or Full length His-tagged menin were incubated with SAE1/SAE2 (E1), Ubc9 (E2), and His-tagged SUMO1 (SUMO1) in the presence or absence of ATP, as indicated. Reaction products were analyzed by western blotting using an anti-SUMO1 or anti-His antibodies. B. Purified Deleted His-tagged menin was incubated with SAE1/SAE2 (E1), Ubc9 (E2), and one of the His-tagged SUMO isoforms, as indicated. Reaction products were detected by western blotting with anti-menin or anti-His antibodies.
Lysine 591 is one of menin SUMOylation sites
The process of SUMO attachment to the substrates forms an isopeptide bond between the C-terminal G-G motif of SUMO and the lysine residue on the substrate [11-13,15]. It is important to identify the amino acid residues in menin that is SUMOlylated in cells. However, it turned out to be extremely difficult to isolate adequate amount of the endogenous menin with SUMO modification due to the small quantity of the SUMOylated menin as well as the transient nature of this modification. As an alternative, we sought to identify the amino acid residues of menin that was modified by SUMO in vitro, as many proteins SUMOylated in vitro and in vivo at similar sites [26,29]. We expressed and purified a SUMO1 mutant protein (R95GG vs wild type T95GG) using a bacterial reconstitution system (Figure 4A), as previously reported [26]. The reason to use the R95GG mutant of SUMO1 is that “GG” in the mutated SUMO can be cleaved and released by an arginine(R)-specific protease from SUMO for mass spectrometry analysis.
Figure 4.
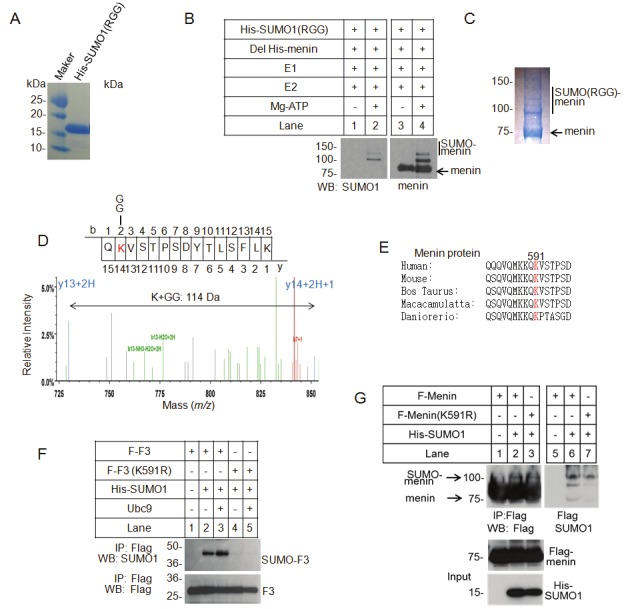
Identification of a menin SUMOylation site by mass spectrometric analysis. A. Bacterial cells were co-transformed with His-tagged SUMO1 (RGG), a T95R mutant, which can be digested by trypsin and generates a 114 Da peptide with a GG-tag attached to the SUMOylated lysine. Purified His-SUMO1 (RGG) protein was separated by SDS–PAGE and was analyzed by Coomassie Brilliant Blue (CBB) staining. B. Purified Deleted His-tagged menin was incubated with SAE1-SAE2 (E1), Ubc9 (E2), and SUMO1 (RGG) with or without ATP, as indicated. Reaction products were detected by western blotting with anti-menin or anti-SUMO1 antibody. C. A large amount of purified Deleted His-tagged menin was incubated with SAE1-SAE2 (E1), Ubc9 (E2), and SUMO1 (RGG). Reaction products for mass-spectrometric analysis were separated by SDS–PAGE and were analyzed by Coomassie Brilliant Blue (CBB) staining. D. The structure of the SUMO1 (RGG)-conjugated menin peptide generated by tryptic digestion was analyzed by tandem mass-spectrometry. E. Alignment of various menin sequences with SUMOylation sites (red). F. K591R mutant of menin C-teminal fragment (F3) was generated by site direct point mutagenesis. WT or K591R mutated F3 was transfected together with His-SUMO1 in the presence or absence of UBC9 into HEK293T cells, as indicated. Immunoprecipitated F3 by Flag M2 beads was separated by SDS–PAGE and was blotted with anti-SUMO1 or anti Flag antibody. G. WT or K591R mutated full-length menin was transfected into HEK392T cells with His-SUMO1. Menin was immunoprecipitated with flag M2 beads, followed by western blotting with anti-Flag (upper left) or anti-SUMO1 (upper right) antibody. The total amounts of menin and SUMO1 are also shown (lower).
We performed in vitro SUMOylation assay, and found that menin was conjugated by the SUMO (RGG) mutant efficiently (Figure 4B, lanes 2 and 4). The SUMOylated (RGG) menin protein was separated by SDS–PAGE, stained by CBB staining, and excised for mass spectrometry (Figure 4C). Following trypsin digestion, the SUMOylated peptide of menin could be detected as a new peptide with covalently attached G-G sequence. The mass-spectrometric analysis identified Lysine 591 as a SUMOylation site, which is conserved among diverse organisms (Figure 4D, E).
Considering that multiple SUMOylation sites of menin may exist, we first substitute lysine 591 by arginine on the C-terminal fragment of menin (F3), which has less lysine residues. To determine whether K591 is also crucial for SUMOylation of menin inside cells. K591R mutated F3 and SUMO1 were co-transfected into HEK293T cells with or without cDNA for E2 ligase, UBC9. Immunoprecipitation and Western blotting analysis revealed that the WT menin fragment, but not the K591R mutant was efficiently conjugated by SUMO1 (Figure 4F, lane 2), and this SUMOylation was further increased in cells co-transfected with UBC9 cDNA (Figure 4F, lane 3). This finding suggests that Lysine 591 is a critical site for in vivo SUMOylation of menin. Next, we mutated the Lysine 591 to arginine on full-length menin and performed the immunoprecipitation. However, the K591R mutant was still modified with SUMO1 in SUMO1 co-transfected cells, suggesting that other SUMO modification sites also exist in full-length menin (Figure 4G, lane 3).
SUMOylation of menin maintains the ability to regulate MEFs
To determine if SUMOylation of menin plays a role in cell regulation, we ectopically expressed cDNAs for WT or K591R mutant menin or menin-SUMO fusion protein (K519R) into Men1-null MEFs. Expression of menin in the cells was confirmed by Western blotting (Figure 5A). Both K591R mutant and menin-SUMO fusion (K519R) retained the function in inhibiting the proliferation of MEFs (Figure 5B). Our earlier data showed that menin repressed the expression of Gas1 and Gli1, two crucial factors for Hedgehog signaling pathway in MEFs. However, we found no differences of the WT or mutant menin or menin-SUMO1 fusion protein in regulating either Gas1 or Gli1mRNA expression in MEFs (Figure 5A, B), indicating that alteration of K591 or SUMOylation of menin does not have obvious functional consequences on proliferation of MEFs and expression of the examined menin target genes.
Figure 5.
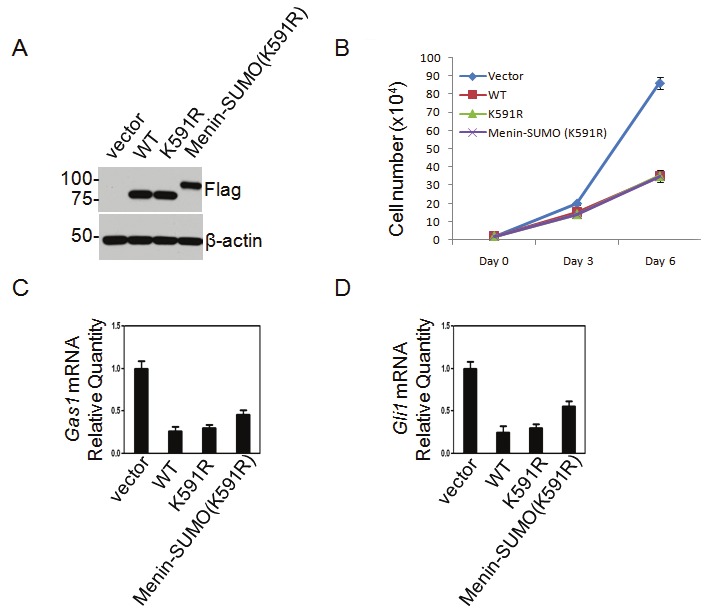
Mutation of a SUMOylation site or tagging SUMO1 to menin does not seem to affect MEFs. A, B. Western blotting (A) and growth curve (B) for MEFs expressing WT menin, K591R mutant and menin-SUMO fusion, and the indicated constructs were transduced to Men1-null MEFs. C, D. Quantitative RT-PCR (qRT-PCR) showing expression of Gas1 and Gli1 mRNA levels in menin-null MEFs complemented with WT menin, K591R mutant or menin-SUMO fusion.
Discussion
Menin is a scaffold protein that recruits transcription factors and epigenetically regulates gene expression [23,30]. However, whether PTMs of menin regulates its function remains unclear. Protein function is often tightly regulated by PTMs such as SUMOylation and dysregulation of PTMs is associated with various diseases such as cancer. It has been reported that menin is phosphorylated at several serine residues [6,7]; but the impact of menin phosphorylation on cell function remains to be elucidated. Our findings suggest that menin is SUMOylated in vivo and in vitro. SUMO1 catalyzes mono-SUMOylation, and SUMO2/3 is capable of forming a poly-SUMO chain. In vitro SUMOylation studies suggest that menin can be SUMOylated at more than one site (Figure 3A). However, in the cellular context, we only observed one SUMO-menin band (Figure 1A, B, C). It is possible that only one of these sites is SUMOylated under physiological conditions. Alternatively, it is possible that there are multiple SUMOylation sites, but the low level of SUMOylation in cells makes it difficult to detect menin with more than one SUMO modification. The SUMOylation band detected by in vivo system is much weaker compared to the in vitro system. This is consistent with the fact that SUMOylation is a transient and dynamic process, mediated by ligases as well as proteases like SENPs.
Through mass spectrometry analysis, we found that lysine 591 of menin is conjugated by SUMO1. The C-terminal part of menin with K591R mutation lost the ability to be SUMOylated by SUMO1 in transfected cells, suggesting that K591 is indeed a SUMOylation site. However, full-length menin with K591R mutation could still be conjugated by SUMO1, implying existence of multiple SUMOylation sites. Most SUMO-modified proteins identified contain an acceptor Lysine within a ψKX(D/E) consensus motif, where ψ is a large hydrophobic residue [31]. The menin SUMOylation site (QKVS) is not consistent with the consensus SUMO interacted motif, which implies the possible existence of a menin-specific E3 ligase [28].
SUMOylation has been shown to be important in many cell processes, including: transcription, replication, chromosome segregation and DNA repair [11,15]. SUMO modification of many transcription factors appears to correlate with transcriptional repression, which may reflect altered protein-protein or protein-DNA interaction. Menin recruits the trithorax group protein MLL to the Hox genes promoter and activates its expression, further increasing the proliferation of leukemia cells. Crystal structure of human menin displays a deep pocket that binds short peptides of MLL1 or JUND in the same manner, but that it can have opposite effects on transcription [23]. It is possible that SUMOylation may change the three-dimensional structure of menin and play a role in regulating the interaction between menin and MLL.
However, we failed to observe any menin K591R mutation or menin-SUMO (K591) fusion in menin function on regulating proliferation of MEFs or the expression of the target genes we examined. These findings do not necessarily mean that the SUMOylation of menin has no function or consequences. It is possible that the role of SUMOylation of menin is restricted by the tissue-specificity. On the other hand, the lack of phenotype of the sumolyation site mutant may also be due to existence of alternative SUMOylation site that can compensate for the mutation. SUMOylation modification is related to various cancers, such as breast cancer and lung cancer [32-34]. As a tumor suppressor, menin represses proliferation of lung cancer cells through inhibiting expression of PTN by binding to the promoter [35]. Alternatively, SUMOylation may only change the function of menin in regulating a select set of genes, and further work remains to explore this possibility.
Nevertheless, to our knowledge, our findings for the first time showed that menin has the SUMOylaiton post-translational modification and Lysine 591 is one of the SUMOylation sites. These findings form a basis for further investigation of the potential role of menin SUMOylation in various contexts of tissues and conditions.
Acknowledgements
This work was supported by AACR/Caring for Crcinoid Foundation grant (XH) and NIH R01-DK085121 (XH). Zijie Feng was supported partly by Chinese Consul Scholarship. We thank Prof. Mutsuhiro Takekawa (University of Tokyo) for pUbc9/SUMO1 plasmid and Chaoxing Yuan (University of Pennsylvania) for mass spectrometry analysis.
Conflict of interests
The authors have no conflict of interests for this manuscript.
References
- 1.Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, EmmertBuck MR, Debelenko LV, Zhuang ZP, Lubensky IA, Liotta LA, Crabtree JS, Wang YP, Roe BA, Weisemann J, Boguski MS, Agarwal SK, Kester MB, Kim YS, Heppner C, Dong QH, Spiegel AM, Burns AL, Marx SJ. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276:404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 2.Lemmens I, VandeVen WJM, Kas K, Zhang CX, Giraud S, Wautot V, Buisson N, DeWitte K, Salandre J, Lenoir G, Pugeat M, Calender A, Parente F, Quincey D, Gaudray P, DeWit MJ, Lips CJM, Hoppener JWM, Khodaei S, Grant AL, Weber G, Kytola S, Teh BT, Farnebo F, Phelan C, Hayward N, Larsson C, Pannett AAJ, Forbes SA, Bassett JHD, Thakker RV. Identification of the multiple endocrine neoplasia type 1 (MEN1) gene. Human Molecular Genetics. 1997;6:1177–1183. doi: 10.1093/hmg/6.7.1177. [DOI] [PubMed] [Google Scholar]
- 3.Marx SJ. Molecular genetics of multiple endocrine neoplasia types 1 and 2. Nat Rev Cancer. 2005;5:367–375. doi: 10.1038/nrc1610. [DOI] [PubMed] [Google Scholar]
- 4.Bertolino P, Tong WM, Galendo D, Wang ZQ, Zhang CX. Heterozygous Men1 mutant mice develop a range of endocrine tumors mimicking multiple endocrine neoplasia type 1. Mol Endocrinol. 2003;17:1880–1892. doi: 10.1210/me.2003-0154. [DOI] [PubMed] [Google Scholar]
- 5.Yaguchi H, Ohkura N, Takahashi M, Nagamura Y, Kitabayashi I, Tsukada T. Menin missense mutants associated with multiple endocrine neoplasia type 1 are rapidly degraded via the ubiquitin-proteasome pathway. Mol Cell Biol. 2004;24:6569–6580. doi: 10.1128/MCB.24.15.6569-6580.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis J, Lin WC, Rozenblatt-Rosen O, Meyerson M. The Menin Tumor Suppressor Protein Is Phosphorylated in Response to DNA Damage. Plos One. 2011;6:e16119. doi: 10.1371/journal.pone.0016119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacConaill LE, Hughes CM, Rozenblatt-Rosen O, Nannepaga S, Meyerson M. Phosphorylation of the menin tumor suppressor protein on serine 543 and serine 583. Mol Cancer Res. 2006;4:793–801. doi: 10.1158/1541-7786.MCR-06-0123. [DOI] [PubMed] [Google Scholar]
- 8.Waby JS, Bingle CD, Corfe BM. Post-translational control of Sp-family transcription factors. Curr Genomics. 2008;9:301–311. doi: 10.2174/138920208785133244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 10.Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 11.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 12.Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr Opin Struct Biol. 2007;17:726–735. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 14.Mikolajczyk J, Drag M, Bekes M, Cao JT, Ronai Z, Salvesen GS. Small ubiquitin-related modifier (SUMO)-specific proteases - Profiling the specificities and activities of human SENPs. J Biol Chem. 2007;282:26217–26224. doi: 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- 15.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci U S A. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hay RT. SUMO: A history of modification. Molecular Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 18.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Sarge KD, Park-Sarge OK. Sumoylation and human disease pathogenesis. Trends Biochem Sci. 2009;34:200–205. doi: 10.1016/j.tibs.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YQ, Sarge KD. Sumoylation regulates lamin A function and is lost in lamin A mutants associated with familial cardiomyopathies. Journal of Cell Biology. 2008;182:35–39. doi: 10.1083/jcb.200712124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woo CH, Shishido T, McClain C, Lim JH, Li JD, Yang J, Yan C, Abe JI. Extracellular signal-regulated kinase 5 SUMOylation antagonizes shear stress-induced antiinflammatory response and endothelial nitric oxide synthase expression in endothelial cells. Circulation Research. 2008;102:538–545. doi: 10.1161/CIRCRESAHA.107.156877. [DOI] [PubMed] [Google Scholar]
- 22.Shishido T, Woo CH, Ding B, McClain C, Molina CA, Yan C, Yang J, Abe J. Effects of MEK5/ERK5 association on small ubiquitin-related modification of ERK5: Implications for diabetic ventricular dysfunction after myocardial infarction. Circulation Research. 2008;102:1416–1425. doi: 10.1161/CIRCRESAHA.107.168138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Gurung B, Wan BB, Matkar S, Veniaminova NA, Wan K, Merchant JL, Hua XX, Lei M. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482:542–U141. doi: 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strader MB, Tabb DL, Hervey WJ, Pan CL, Hurst GB. Efficient and specific trypsin digestion of microgram to nanogram quantities of proteins in organic-aqueous solvent systems. Anal Chem. 2006;78:125–134. doi: 10.1021/ac051348l. [DOI] [PubMed] [Google Scholar]
- 25.Chu Y, Yang X. SUMO E3 ligase activity of TRIM proteins. Oncogene. 2011;30:1108–1116. doi: 10.1038/onc.2010.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubota Y, O’Grady P, Saito H, Takekawa M. Oncogenic Ras abrogates MEK SUMOylation that suppresses the ERK pathway and cell transformation. Nat Cell Biol. 2011;13:282–U477. doi: 10.1038/ncb2169. [DOI] [PubMed] [Google Scholar]
- 27.Schnepp RW, Chen YX, Wang H, Cash T, Silva A, Diehl JA, Brown E, Hua X. Mutation of tumor suppressor gene Men1 acutely enhances proliferation of pancreatic islet cells. Cancer Res. 2006;66:5707–5715. doi: 10.1158/0008-5472.CAN-05-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melchior F, Schergaut M, Pichler A. SUMO: ligases, isopeptidases and nuclear pores. Trends in Biochemical Sciences. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Choi HK, Choi KC, Yoo JY, Song M, Ko SJ, Kim CH, Ahn JH, Chun KH, Yook JI, Yoon HG. Reversible SUMOylation of TBL1-TBLR1 regulates beta-catenin-mediated Wnt signaling. Molecular Cell. 2011;43:203–216. doi: 10.1016/j.molcel.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 30.La P, Desmond A, Hou Z, Silva AC, Schnepp RW, Hua X. Tumor suppressor menin: the essential role of nuclear localization signal domains in coordinating gene expression. Oncogene. 2006;25:3537–3546. doi: 10.1038/sj.onc.1209400. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 32.Dunnebier T, Bermejo JL, Haas S, Fischer HP, Pierl CB, Justenhoven C, Brauch H, Baisch C, Gilbert M, Harth V, Spickenheuer A, Rabstein S, Pesch B, Bruning T, Ko YD, Hamann U. Common variants in the UBC9 gene encoding the SUMO-conjugating enzyme are associated with breast tumor grade. Int J Cancer. 2009;125:596–602. doi: 10.1002/ijc.24286. [DOI] [PubMed] [Google Scholar]
- 33.Synowiec E, Krupa R, Morawiec Z, Wasylecka M, Dziki L, Morawiec J, Blasiak J, Wozniak K. Efficacy of DNA double-strand breaks repair in breast cancer is decreased in carriers of the variant allele of the UBC9 gene c 73G > A polymorphism. Mutat Res. 2010;694:31–38. doi: 10.1016/j.mrfmmm.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Kang JS, Saunier EF, Akhurst RJ, Derynck R. The type I TGF-beta receptor is covalently modified and regulated by sumoylation. Nat Cell Biol. 2008;10:654–664. doi: 10.1038/ncb1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao SB, Feng ZJ, Xu B, Wu Y, Yin P, Yang Y, Hua X, Jin GH. Suppression of lung adenocarcinoma through menin and polycomb gene-mediated repression of growth factor pleiotrophin. Oncogene. 2009;28:4095–4104. doi: 10.1038/onc.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]


