Abstract
Immature CD4+CD8+ (double-positive (DP)) thymocytes are signaled via T cell antigen receptors (TCRs) to undergo positive selection and become responsive to intrathymic cytokines such as interleukin 7 (IL-7). We report here that cytokine signaling is required for positively selected thymocytes to express the transcription factor Runx3, specify CD8 lineage choice and differentiate into cytotoxic-lineage T cells. In DP thymocytes genetically engineered to be cytokine responsive, IL-7 signaling induced TCR-unsignaled DP thymocytes to express Runx3 and to differentiate into mature CD8+ T cells, completely circumventing positive selection. We conclude that TCR-mediated positive selection converts DP cells into cytokine-responsive thymocytes, but it is subsequent signaling by intrathymic cytokines that specifies CD8 lineage choice and promotes differentiation into cytotoxic-lineage T cells.
The fate of T cells developing in the thymus is determined during positive selection by the specificity of their αβ T cell antigen receptors (TCRs)1. Thymocytes at the CD4+CD8+ (double-positive (DP)) stage of development are signaled by their TCR to undergo positive selection and to differentiate into either CD4+ helper T cells or CD8+ cytotoxic T cells2. However, most TCRs fail to signal in the thymus because they fail to engage intrathymic ligands, which causes most DP thymocytes to undergo death by neglect3. Consequently, only DP thymocytes that receive a TCR signal successfully complete their differentiation into mature T cells, which has the result that every mature T cell expresses a rigorously screened self-specific TCR.
Before receiving a TCR signal, DP thymocytes are unresponsive to intrathymic cytokines such as interleukin 7 (IL-7; A004205)4,5. Indeed, TCR-unsignaled DP thymocytes do not express IL-7 receptor-α (IL-7Rα; A001267)5 and do have uniquely high expression of suppressor of cytokine signaling 1 (SOCS1), which blocks signal transduction by all common γ-chain (γc) cytokines6. Consequently, despite their expression of IL-4Rα and γc proteins5, TCR-unsignaled DP thymocytes are unresponsive to both IL-7 and IL-4. Moreover, preselection DP thymocytes reside in the thymic cortex, which lacks IL-7-producing cells7, so they may not encounter IL-7 or other γc cytokines unless the cells migrate to other areas of the thymus8,9.
Because TCR signaling in DP thymocytes mediates positive selection and induces the generation of mature CD4+ and CD8+ T cells, TCR signaling is thought to specify both CD4 and CD8 lineage choices and to drive thymocyte maturation10. Experimentally, DP thymocytes can be induced to differentiate into mature T cells independently of TCR-ligand engagements through the use of agonistic antibodies to TCR11 and pharmacological or genetic mimics of TCR signaling11,12. Although these approaches avoid TCR-ligand engagements, they satisfy the TCR signaling requirement of DP thymocytes. Consequently, TCR-signaled positive selection is generally considered essential for the differentiation of DP thymocytes into mature T cells.
After DP thymocytes are signaled to undergo positive selection, CD4 or CD8 lineage specification is induced by a mechanism that is best explained at present by the kinetic signaling model of T cell development2,10,13. The kinetic signaling model proposes that TCR-mediated positive selection converts cytokine-unresponsive DP thymocytes into cytokine-responsive intermediate thymocytes that are transcriptionally Cd4+Cd8− and that lineage specification is then determined in intermediate thymocytes by whether TCR signaling persists or ceases. During positive selection, persistent TCR signaling drives intermediate thymocytes to differentiate into CD4+ T cells, whereas cessation of TCR signaling permits intermediate thymocytes to be signaled by intrathymic γc cytokines such as IL-7 and to differentiate into CD8+ T cells. However, key predictions of the kinetic signaling model have not been evaluated in vivo, including the hypothesis that cytokine signaling is required for both CD8 lineage specification and cytotoxic T cell differentiation in the thymus.
The transcription factors that specify CD4 or CD8 lineage choice in positively selected thymocytes have largely been identified14–18. CD4 lineage choice is specified by the zinc-finger transcription factors Th-POK and GATA-3 (refs. 15,18,19), whereas CD8 lineage choice is specified by the Runt-family transcription factor Runx3 (refs. 14,17,20). Th-POK and Runx3 negatively regulate each other’s expression and thus reinforce lineage choices16,17,21–23. In agreement with predictions of the kinetic signaling model, persistent TCR signaling in positively selected intermediate thymocytes can induce Th-POK expression and specify CD4 lineage choice16. However, the intrathymic signal that induces Runx3 expression and specifies CD8 lineage choice has not yet been identified.
The present study investigates whether CD8 lineage specification and CD8+ T cell differentiation in the thymus requires signaling by intrathymic cytokines. We show that in positively selected thymocytes, γc cytokine activation of intracellular signal transducer and activator of transcription (STAT) molecules was needed to induce Runx3 expression, specify CD8 lineage choice and promote the differentiation of cytotoxic-lineage T cells in vivo. We also show that IL-7 signaling of DP thymocytes genetically engineered to be cytokine responsive circumvented positive selection by inducing preselection DP thymocytes to differentiate into mature CD8+ T cells in the complete absence of TCR signaling. Our observations substantially enhance the understanding of positive selection, lineage specification and T cell differentiation in the thymus.
RESULTS
γc cytokine signaling is required for thymic CD8+ T cell generation
In the present study we assessed whether signals transduced by intrathymic γc cytokines are required to specify CD8 lineage choice and promote the differentiation of cytotoxic-lineage T cells in the thymus (Supplementary Fig. 1). Signaling by IL-7, the main cytokine expressed in the thymus, is transduced by STAT5a and STAT5b molecules, so we examined the effect of deleting Stat5a and Stat5b in positively selected thymocytes24. To avoid interfering with cytokine signal transduction in early CD4−CD8− double-negative (DN) thymocytes, we conditionally deleted Stat5a and Stat5b in thymocytes beyond the DN4 stage of differentiation. We used a Cre transgene construct (E8III-Cre) that uses the E8III enhancer and promoter elements from Cd8a to drive expression of Cre recombinase in preselection immature single-positive and DP thymocytes (Fig. 1a). To confirm the developmental timing of E8III-Cre-mediated deletion, we introduced the E8III-Cre transgene into Rosa26-loxP-STOP-loxP-green fluorescent protein (GFP) reporter mice and observed that most preselection DN thymocytes that were TCRβ−CD4−CD8− were GFP−, whereas DP thymocytes and their post-selection progeny (CD8+ or CD4+ single-positive (SP) thymocytes and TCR+ DN thymocytes) were GFP+ (Supplementary Fig. 2a). We then introduced the E8III-Cre transgene into mice carrying a loxP-flanked allele encoding STAT5 to generate E8III-Cre+Stat5fl/− mice (STAT5-cKO mice) and confirmed that STAT5 expression and IL-7-induced STAT5 phosphorylation were abrogated in DP thymocytes and their post-selection progeny (Fig. 1b and Supplementary Fig. 2b). Notably, conditional deletion of Stat5a and Stat5b by E8III-Cre in preselection DP thymocytes had no effect on overall thymocyte cellularity or on the generation of CD4+ T cells (Fig. 1c). In contrast, conditional deletion of Stat5a and Stat5b in preselection DP thymocytes resulted in a 50% lower frequency of CD8 SP (CD8SP) thymocytes in STAT5-cKO mice than that in wild-type mice (P < 0.005; Fig. 1c), which revealed that expression of Stat5a and Stat5b in DP thymocytes was important for their differentiation into CD8+ T cells.
Figure 1.
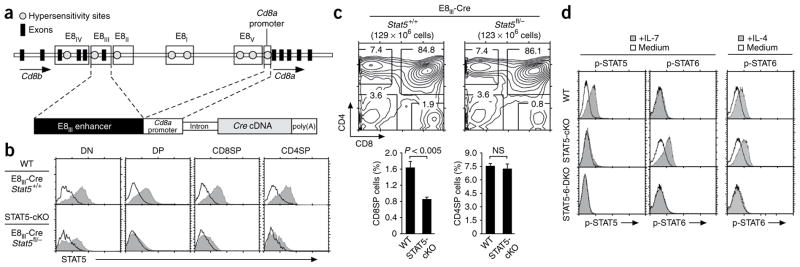
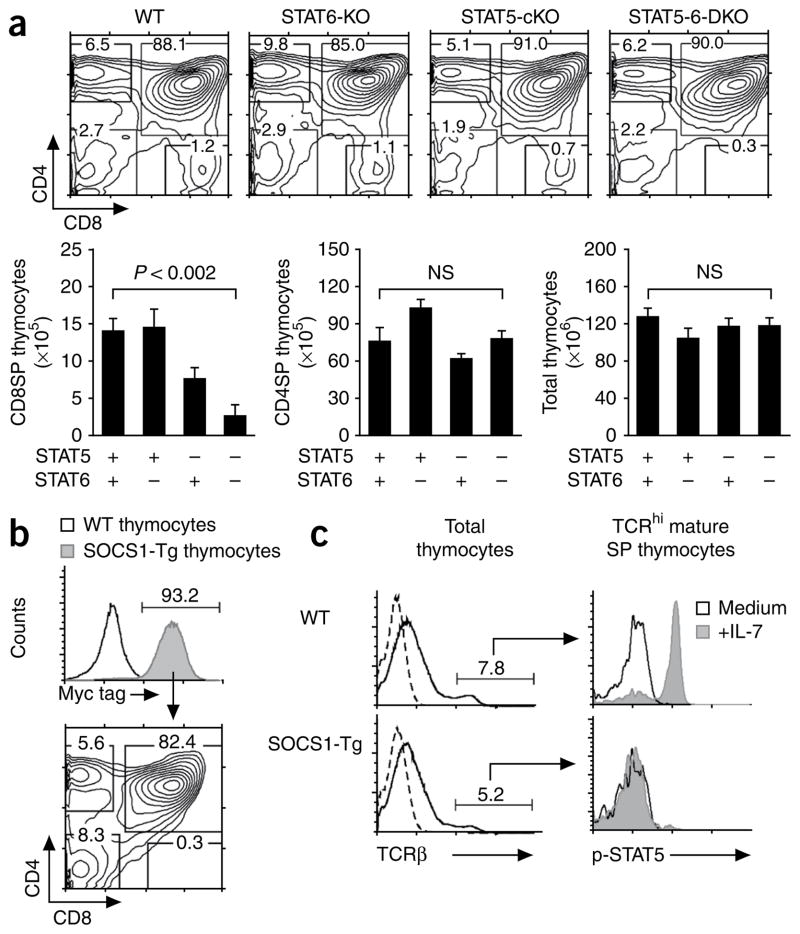
Impaired CD8+ T cell generation in Stat5a- and Stat5b-deficient mice. (a) The E8III-Cre transgene consists of enhancer and promoter elements from Cd8a that drive expression of Cre cDNA. (b) STAT5 protein content of thymocytes from wild-type (WT) and STAT5-cKO mice, assessed by intracellular staining with anti-STAT5 (shaded histograms) or control antibodies (open histograms). Data are representative of seven experiments. (c) Flow cytometry analysis (top) and frequencies of TCRhiCD4+ and TCRhiCD8+ SP thymocytes (bottom) in wild-type and STAT5-cKO mice. Numbers above plots indicate total thymocytes; numbers in outlined areas indicate percent cells in each. P value, Student’s two-tailed t-test; NS, not significant. Data are a summary of seven independent experiments with at least seven mice of each genotype (bottom; mean and s.e.m.). (d) Intracellular staining of phosphorylated STAT5 (p-STAT5) and phosphorylated STAT6 (p-STAT6) in lymph node T cells from wild-type, STAT5-cKO and STAT5-6-DKO mice after overnight stimulation with medium alone or with IL-7 (1 ng/ml) or IL-4 (10 ng/ml). Data are representative of three experiments.
Nevertheless, substantial numbers of CD8+ T cells were still present among STAT5-cKO thymocytes. One explanation could be that other cytokines, such as IL-4, might activate STAT proteins such as STAT6 to induce differentiation of CD8+ T cells in STAT5-cKO mice. Alternatively, IL-7 might signal through other STAT molecules in STAT5-deficient T cells, even though IL-7 signals are transduced exclusively by STAT5 in STAT5-sufficient T cells. For example, IL-7 stimulation of wild-type (C57BL/6) T cells phosphorylated STAT5 but not STAT6 (Fig. 1d). Nevertheless, we found that IL-7 stimulation of STAT5-deficient T cells from STAT5-cKO mice induced STAT6 phosphorylation (Fig. 1d), which shows that other STAT molecules can transduce IL-7 signals in the absence of STAT5 expression.
We investigated whether other STAT molecules, such as STAT6, were responsible for the generation of CD8+ T cells in STAT5-cKO mice as a result of signaling by IL-7 or another cytokine. To assess this possibility, we produced mice deficient in both STAT5 and STAT6 (STAT5-6-DKO mice) by mating germline Stat6−/− (STAT6-KO) mice25 with STAT5-cKO mice (Fig. 2a). STAT6 deficiency by itself had no apparent effect on thymocyte development, as thymocytes from STAT6-KO and wild-type mice were not markedly different25 (Fig. 2a). Nevertheless, double deficiency in STAT5 and STAT6 resulted in an 85% lower absolute number of CD8SP thymocytes than that in wild-type mice (P < 0.002), whereas the number of CD4SP thymocytes and overall thymocyte cellularity were unchanged (Fig. 2a). The CD8SP thymocytes detected in STAT5-6-DKO could have been the result of signaling by IL-7, or other intrathymic cytokines, through still other STAT molecules (for example, STAT1). In either case, these results document that STAT-mediated cytokine signaling is required for the thymic generation of nearly all, if not all, CD8SP cells.
Figure 2.
The generation of CD8+ T cells requires STAT-mediated cytokine signaling. (a) Thymic profiles of wild-type, STAT6-KO, STAT5-cKO and STAT5-6-DKO mice (top row) and absolute numbers of TCRhiCD8+ SP, TCRhiCD4+ SP and total thymocytes (bottom row). P value, Student’s two-tailed t-test. Data are from five independent experiments (mean and s.e.m.). (b) Thymic profile (below) of mice expressing a Myc-tagged SOCS1 transgene (tag expression, top plot). Number above bracketed line (top) indicates percent cells expressing Myc tag; numbers in outlined areas (below) indicate percent cells in each. Data are representative of two experiments. (c) Intracellular expression of phosphorylated STAT5 in TCRhi wild-type and TCRhi SOCS1-Tg thymocytes after 30 min of stimulation with IL-7 or medium (right). Left, TCRβ expression by wild-type and SOCS1-Tg thymocytes; dashed lines indicate control antibody staining; numbers above bracketed lines indicate percent TCRhi cells. Data are representative of three independent experiments.
Because it was not possible to generate mice lacking all STAT molecules, we used a different experimental strategy to confirm that cytokine signal transduction was necessary for thymic generation of CD8+ T cells. In mice expressing Myc-tagged SOCS1 under the control of the mouse proximal Lck promoter (SOCS1-Tg mice), γc cytokine signal transduction is essentially eliminated26. SOCS1 impairs γc cytokine signal transduction by binding to γc-associated Jak3 kinase molecules and preventing their phosphorylation27,28. As determined by intracellular staining for Myc, nearly all thymocytes in SOCS1-Tg mice expressed the Socs1 transgene (Fig. 2b). Mature TCRhi thymocytes were unable to phosphorylate STAT5 in response to IL-7 stimulation in SOCS1-Tg mice, in contrast to results obtained with wild-type cells (Fig. 2c). Developmentally, SOCS1-Tg mice were specifically devoid of CD8SP thymocytes (Fig. 2b and Supplementary Fig. 3), which confirmed that cytokine signal transduction was required for CD8+ T cell generation in the thymus. The absence of CD8SP thymocytes in SOCS1-Tg mice was caused by a block in CD8 lineage development and was not due to redirected differentiation of major histocompatibility complex (MHC) class I–selected thymocytes into the CD4 lineage, because MHC class II–deficient SOCS1-Tg mice were additionally devoid of mature CD4-lineage thymocytes (Supplementary Fig. 3), although they did contain the phenotypically CD4+CD8lo intermediate cells that are precursors of both CD4-lineage and CD8-lineage mature T cells13,29–32. Together these results demonstrate that STAT-mediated signaling by intrathymic γc cytokines is specifically required for CD8+ T cell generation in the thymus.
γc cytokine signaling induces Runx3 expression
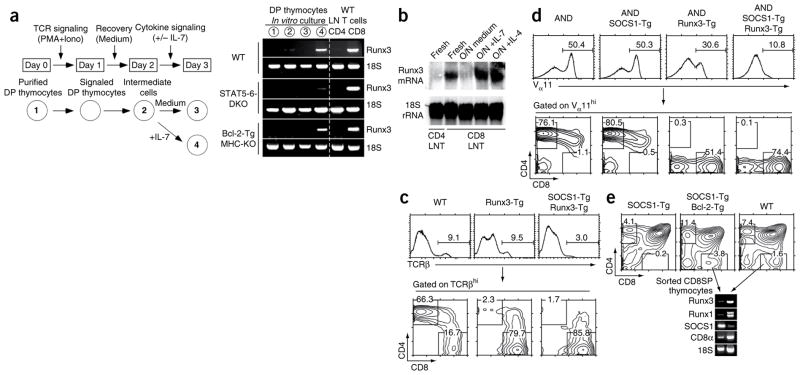
We next investigated whether cytokine signaling induced expression of Runx3, the transcription factor thought to specify CD8 lineage choice14,20,33. CD8 lineage choice occurs in intermediate thymocytes that are transcriptionally Cd4+Cd8− and that are the immediate progeny of TCR-signaled DP thymocytes13 (Supplementary Fig. 1). We used a well-established in vitro model of positive selection in which DP thymocytes were induced to differentiate into intermediate thymocytes through the use of phorbol 12-myristate 13-acetate and ionomycin and were subsequently stimulated with IL-7 to induce differentiation into CD8SP T cells13,34 (Fig. 3a). We assessed expression of Runx3 mRNA using primers specific for transcripts generated from the Runx3 distal promoter, which is expressed specifically in CD8+ T cells but not in CD4+ T cells14 (Fig. 3a). Analysis of wild-type thymocytes at various stages of in vitro differentiation revealed that Runx3 mRNA was not expressed in purified DP thymocytes, newly generated intermediate thymocytes or intermediate thymocytes cultured in medium (Fig. 3a). Runx3 mRNA expression was induced only in wild-type intermediate thymocytes stimulated with IL-7 (Fig. 3a). Runx3 induction required STAT5 and/or STAT6 because Runx3 mRNA was not expressed in STAT5-6-DKO intermediate thymocytes stimulated with IL-7 (Fig. 3a). Notably, transgenic expression of the prosurvival factor Bcl-2 did not circumvent the dependence of Runx3 expression on IL-7 stimulation (Fig. 3a).
Figure 3.
IL-7 signaling induces Runx3, which specifies CD8 lineage choice. (a) Experimental setup for the generation of intermediate thymocytes from preselection thymocytes (left): purified DP thymocytes (population 1) were stimulated overnight with phorbol 12-myristate 13-acetate plus ionomycin (PMA+Iono) and were allowed to differentiate into intermediate cells (population 2), which were further cultured in either medium alone (population 3) or with IL-7 (population 4). Right, RT-PCR analysis of cultured thymocytes with primers specific for Runx3 mRNA transcribed from the Runx3 distal promoter. WT LN T cells (far right), RNA from freshly isolated B6 CD4+ and CD8+ lymph node T cells (tissue specificity control). (b) RNA-hybridization analysis of Runx3 mRNA expression in CD8+ lymph node T cells cultured overnight (O/N) in medium, IL-7 or IL-4, as well as in freshly isolated CD8+ and CD4+ lymph node T cells (Fresh). (c) Expression of CD4 and CD8 in TCR cells from wild-type, Runx3-Tg and SOCS1-Tg–Runx3-Tg mice. (d) Expression of CD4 and CD8 in Vα11hi cells from MHC class II–specific AND–transgenic (AND), AND SOCS1-Tg, AND Runx3-Tg, and AND SOCS1-Tg–Runx3-Tg mice. (e) Thymic profiles of SOCS1-Tg, SOCS1-Tg–Bcl-2-Tg and wild-type mice (top), and RT-PCR analysis of Runx3 mRNA expression in CD8+ SP thymocytes sorted from SOCS1-Tg–Bcl-2-Tg and wild-type mice (bottom). Numbers above bracketed lines (top, c,d) indicate percent TCRhi cells (c) or Vα11hi cells (variable α-region 11; d); numbers in outlined areas (bottom, c,d; top, e) indicate frequency of cells in each subset. Data are representative of six (a), three (b,d) or two (c,e) experiments.
Following our observation that IL-7 induced Runx3 expression in TCR-signaled thymocytes, we investigated if γc cytokine signals also regulated Runx3 expression in mature CD8+ T cells in the periphery. Removal of CD8+ lymph node T cells from their cytokine-rich in vivo environment resulted in nearly complete loss of Runx3 mRNA expression (Fig. 3b). This effect was completely reversed by the addition of either IL-7 or IL-4 to overnight cultures (Fig. 3b), which demonstrated that signaling by IL-7 and other γc cytokines induces Runx3 gene expression in mature CD8+ T cells as well as positively selected thymocytes.
Because Runx3 expression was downstream of cytokine signaling, we tested if transgenic Runx3 expression was able to specify CD8 lineage choice in the absence of γc cytokine signals. To test this possibility, we generated mice that transgenically expressed Runx3 (Runx3-Tg mice) using human CD2 (CD2) control elements that are expressed in essentially all thymocytes. Consistent with the role of Runx3 in CD8 lineage specification, we found that Runx3-Tg mice contained only TCRhiCD8+ T cells (Fig. 3c), including mice transgenic for the MHC class II–restricted AND TCR, whose thymocytes normally differentiate into TCRhiCD4+ T cells (Fig. 3d). To assess if transgenic expression of Runx3 can specify CD8 lineage choice in the absence of γc cytokine signaling, we used the Socs1 transgene to block γc cytokine signaling in developing thymocytes (Fig. 3d and Supplementary Fig. 3). We crossed the SOCS1-Tg with Runx3-Tg mice to generate SOCS1-Tg–Runx3-Tg mice and found that despite the absence of γc cytokine signaling, SOCS1-Tg–Runx3-Tg mice contained mature TCRhi thymocytes at a lower frequency, almost all of which were TCRhiCD8+ T cells (Fig. 3c). AND SOCS1-Tg–Runx3-Tg mice had a similar phenotype (Fig. 3d). These results indicate that Runx3 is downstream of γc cytokine signaling and can induce CD8 lineage specification even in MHC class II–specific thymocytes and even in the absence of γc cytokine signaling. However, the Runx3 transgene did not quantitatively restore positive selection of TCRhiCD8+ T cells among SOCS1-Tg thymocytes (Fig. 3c), including those expressing the AND TCR (Fig. 3d), which suggests that signaling by intrathymic cytokines does more than induce expression of Runx3, such as upregulating Bcl2 gene expression to promote the survival of thymocytes during differentiation into CD8+ T cells.
Consequently, we asked if transgenic expression of Bcl2 would permit the generation of CD8+ T cells in the absence of γc cytokine signaling among Socs1-transgenic thymocytes. Introduction of the Bcl2 transgene in SOCS1-Tg mice resulted in a higher CD4SP thymocyte frequency and, more importantly, restored the generation of CD8SP thymocytes (Fig. 3e). Notably, Bcl2 transgene expression is known to generate unconventional CD8SP thymocytes that arise independently of MHC class I recognition35, fail to attain functional competence35 and may be the direct progeny of negatively selected DN thymocytes rather than positively selected DP thymocytes (data not shown). Notably, analysis of CD8SP thymocytes from Bcl-2-Tg–SOCS1-Tg mice revealed that they did not express either Runx3 mRNA or Runx1 mRNA (both of which encode Runx proteins with similar DNA-binding sites and similar downstream target genes; Fig. 3e). These results demonstrate that positively selected thymocytes do not express Runx3 in the absence of γc cytokine signaling, despite the appearance of unconventional CD8SP thymocytes induced by transgenic Bcl2 expression.
CD8 lineage specification in the absence of TCR signaling
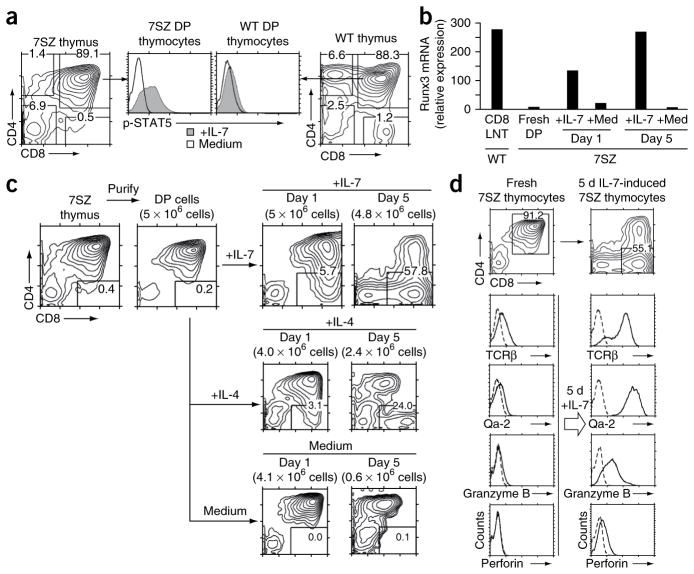
Next we investigated whether cytokine signaling could induce CD8 lineage specification in the absence of TCR-mediated positive selection signals. By genetic manipulation, we generated mice in which preselection DP thymocytes were responsive to IL-7 signaling (Fig. 4a and Supplementary Fig. 4). First, we introduced an Il7r transgene into Socs1−/− mice to generate IL-7Rα-Tg–SOCS1-KO mice. Then, to eliminate TCR signaling, we bred IL-7Rα-Tg–SOCS1-KO mice with mice deficient in the tyrosine kinase Zap70 (Zap70−/− mice) to generate IL-7Rα-Tg–SOCS1-KO–Zap70-KO mice (7SZ mice). Consistent with the absence of Zap70 expression, 7SZ thymocytes were arrested at the DP stage and lacked positively selected SP T cells (Fig. 4a). But unlike normal DP thymocytes, 7SZ DP thymocytes were IL-7 responsive, as exogenous IL-7 strongly induced STAT5 phosphorylation in vitro (Fig. 4a).
Figure 4.
Effect of in vitro IL-7 signaling on cytokine-responsive preselection DP thymocytes. (a) Intracellular content of phosphorylated STAT5 in preselection DP thymocytes from 7SZ and wild-type mice stimulated for 30 min with IL-7 (1 ng/ml) or medium. (b) Quantification of Runx3 mRNA expression in 7SZ DP thymocytes freshly isolated or stimulated for 1 or 5 d with IL-7 or medium alone, presented in arbitrary units relative to endogenous HPRT mRNA expression (encoding hypoxanthine guanine phosphoribosyl transferase). CD8 LNT (far left), purified wild-type CD8+ lymph node T cells (control). (c) Expression of CD4 and CD8 by 7SZ DP thymocytes purified by anti-CD8 panning and cultured for 1 or 5 d with IL-7 (10 ng/ml), IL-4 (10 ng/ml) or medium alone. Numbers in parentheses above plots indicate number of cells at end of culture. (d) Expression of TCRβ and Qa-2, assessed by surface staining, and expression of granzyme B and perforin, assessed by intracellular staining, in freshly purified 7SZ DP thymocytes and after 5 d of stimulation with IL-7 (10 ng/ml). Dashed lines indicate control antibody staining. Top, expression of CD4 and CD8 before and after IL-7 stimulation. Data are representative of four (a,c,d), or two (b) experiments.
Because 7SZ DP thymocytes phosphorylated STAT5 in response to IL-7 stimulation, we assessed whether IL-7 signaling would induce Runx3 expression. Purified fresh 7SZ DP thymocytes did not have detectable expression of Runx3 mRNA, but culture of purified 7SZ DP thymocytes with IL-7 markedly upregulated their expression of Runx3 mRNA, which eventually reached an amount equivalent to that in peripheral CD8+ lymph node T cells (Fig. 4b). These results demonstrate that cytokines such as IL-7 can induce Runx3 expression even in TCR-unsignaled DP thymocytes, as long as the DP thymocytes are capable of transducing cytokine signals.
To assess the consequences of cytokine signaling in TCR-unsignaled DP thymocytes, we assessed the developmental progression of 7SZ DP thymocytes in in vitro cultures supplemented with IL-7 or IL-4 (Fig. 4c). Cell recovery of 7SZ DP thymocytes was essentially 100% after 5 d in culture with IL-7 but was much lower with IL-4 and even lower in medium-alone conditions (Fig. 4c). Notably, we observed that after 5 d in IL-7-supplemented cultures, almost all (>95 %) 7SZ DP thymocytes had terminated CD4 expression and most had differentiated into CD8SP T cells (Fig. 4c), both events concordant with their Runx3 expression (Fig. 4b). As expected, cytokine-unresponsive DP thymocytes from wild-type and Zap70-KO mice remained DP after 5 d in IL-7-supplemented cultures (Supplementary Fig. 5a,b). IL-7 was far more potent than IL-4 in promoting the survival of 7SZ DP thymocytes and promoting CD8+ T cell differentiation (Fig. 4c), even though DP thymocytes express surface IL-4R5.
IL-7-induced CD8SP T cells were mature CD8+ T cells by phenotypic criteria, including expression of TCR, Qa-2, granzyme B and perforin (Fig. 4d). Most IL-7-induced 7SZ CD8+ T cells were TCRβhi and resembled in vivo–generated mature CD8+ T cells from wild-type mice. However, about 30% of IL-7-induced 7SZ CD8+ T cells were surface TCRβ−, which indicated that they lacked TCRα proteins, consistent with their TCR-independent differentiation (Fig. 4d). The Qa-2 marker is expressed on mature T cells that have acquired functional competence36, and all IL-7-induced 7SZ CD8+ T cells, including those that were TCRβ−, were Qa-2+ and also contained granzyme B and perforin proteins (Fig. 4d), which revealed that they had differentiated into functional cytotoxic-lineage T cells. However, because 7SZ T cells lack Zap70 expression, we could not use cytotoxic assays to assess the cytotoxic function of these CD8+ 7SZ T cells. The kinase Syk could potentially transduce TCR signals in the absence of Zap70 expression, but neither DP nor SP 7SZ thymocytes expressed Syk (Supplementary Fig. 6). We conclude that in vitro IL-7 signaling in cytokine-responsive preselection DP thymocytes induces Runx3 expression, specifies CD8 lineage choice and promotes their differentiation into phenotypically mature cytotoxic-lineage T cells, despite the absence of TCR-mediated positive selection signals.
Exogenous IL-7 signaling of DP thymocytes in vivo
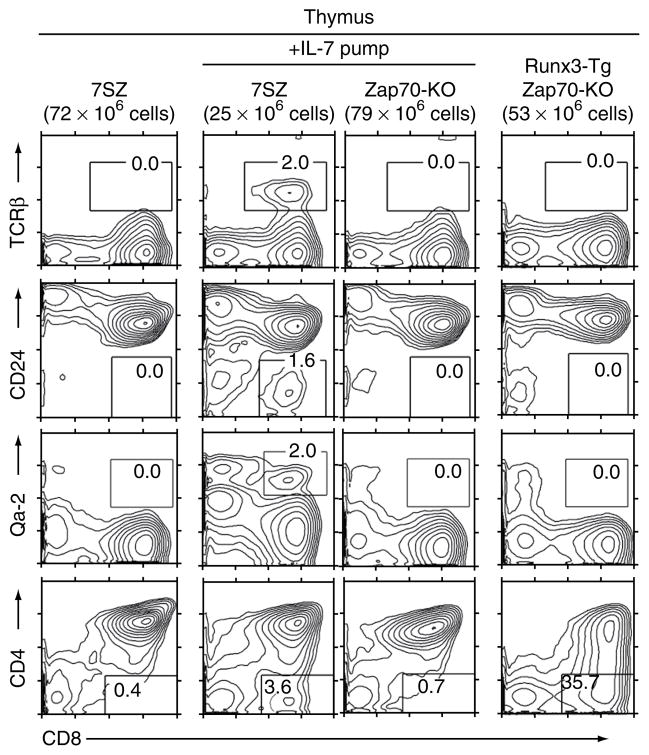
The 7SZ thymus contained cytokine-responsive DP thymocytes but no CD8SP cells, which raised the possibility that 7SZ DP thymocytes do not have access to endogenous γc cytokine in sufficient amounts to be signaled in the thymic cortex. Consequently, we tested if exogenous IL-7 could induce cytokine-responsive 7SZ DP thymocytes to differentiate into CD8+ T cells in vivo. Systemic IL-7 administration by a subcutaneously implanted osmotic pump did induce the differentiation of modest numbers of TCRβhi, CD24lo, Qa-2+ and CD4− mature CD8+ T cells in 7SZ mice (Fig. 5). Thus, exogenously administered IL-7 induces cytokine-responsive DP thymocytes to differentiate in vivo into phenotypically mature CD8+ T cells in the absence of TCR-signaled positive selection.
Figure 5.
In vivo IL-7 signaling of 7SZ thymocytes induces their differentiation into mature CD8+ T cells. Expression of TCRβ, CD24, Qa-2 and CD4 on CD8+ thymocytes from 7SZ and Zap70-KO mice after 14 d of in vivo administration of recombinant mouse IL-7 (10 μg/day) via a subcutaneously implanted osmotic pump (middle columns) or thymocytes from unstimulated 7SZ and Runx3-Tg–Zap70-KO mice (far left and far right). Numbers in outlined areas indicate percent cells in each. Data are representative of four experiments.
Because Runx3 is a downstream target of IL-7, we also asked if transgenic Runx3 expression was itself sufficient to induce TCR-unsignaled DP thymocytes to differentiate into mature CD8+ T cells in vivo. However, the thymus of Runx3-Tg–Zap70-KO mice lacked mature CD8+ T cells (Fig. 5), which confirmed that Runx3 transgene expression does not fully replace cytokine signaling in generating CD8+ T cells.
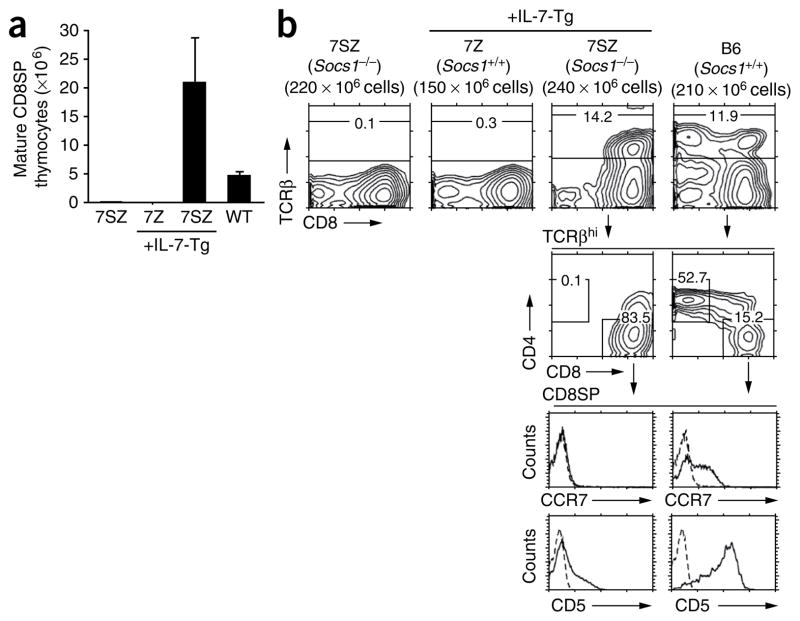
Given the modest effect of systemic IL-7 administration on CD8+ T cell generation in 7SZ mice, we next used a mouse Il7 transgene driven by the proximal Lck promoter37 as an intrathymic source of IL-7. We introduced the Il7 transgene into 7SZ mice with cytokine-responsive DP thymocytes and into IL-7Rα-Tg–Zap70-KO mice (7Z mice) with cytokine-unresponsive DP thymocytes that express endogenous SOCS1 (Fig. 6). Total thymocyte numbers were roughly equivalent in all experimental mice; however, the expression of transgenic Il7 resulted in an average of 21 × 106 TCRβhiCD8SP thymocytes in 7SZ mice, compared with 4 × 106 TCRβhiCD8SP thymocytes in wild-type mice (Fig. 6a). Transgenic Il7 had no discernible effect in the 7Z thymus because IL-7-Tg 7Z thymi lacked mature CD8SP thymocytes (Fig. 6a) and contained no TCRβhi thymocytes (Fig. 6b).
Figure 6.
Transgenic IL-7 expression induces efficient CD8+ T cell differentiation in 7SZ mice. (a) Absolute number of mature TCRhiCD8+ thymocytes in 7SZ, IL-7-Tg 7Z, IL-7-Tg 7SZ and wild-type mice. Data are the summary of two independent experiments with two mice of each genotype (error bars, s.e.m.). (b) Expression of CD4 and CD8 in TCRβhi thymocytes from 7SZ, IL-7-Tg 7Z, IL-7-Tg 7SZ and wild-type (B6) mice (above), and expression of CCR7 and CD5 in mature TCRβhiCD8+ thymocytes (below). Numbers in outlined areas indicate frequency of cells in each gate. Dashed lines indicate control antibody staining. Summary of developmental implications of these results, Supplementary Figure 8. Data are representative of two experiments.
In wild-type mice, TCR signaling in DP thymocytes induced differentiation into CD8+ T cells that were CD5hi and expressed CCR7, the chemokine receptor that promotes the migration of thymocytes into the thymic medulla (Fig. 6b). In contrast, IL-7-generated CD8+ T cells in the 7SZ thymus were CD5− to CD5lo, which confirmed that they had not been TCR signaled. We found that IL-7-generated 7SZ CD8+ T cells were also CCR7− (Fig. 6b), which revealed that cytokine signaling does not induce CCR7 expression. Nevertheless, mature thymocytes can emigrate into the periphery directly from the thymic cortex38, and we found that IL-7-generated 7SZ thymocytes did migrate into the periphery, because mature TCRβhiCD24−Qa-2+ CD8+ T cells were present in the lymph nodes of IL-7-Tg 7SZ mice (Supplementary Fig. 7). However, because mature CD8+ T cells cannot survive in the periphery in the absence of homeostatic TCR signaling, CD8+ T cells were present in the lymph nodes of IL-7-Tg 7SZ mice at much lower frequencies (Supplementary Fig. 7).
These results suggest that the thymic cortex contains insufficient amounts of endogenous cytokines to signal cytokine-responsive 7SZ DP thymocytes. Exogenously provided IL-7 was able to signal 7SZ DP thymocytes in vivo, efficiently inducing their differentiation into phenotypically mature cytotoxic-lineage T cells (Supplementary Fig. 8a). In addition, these results reveal that TCR signaling was required for expression of CCR7, the chemokine receptor that promotes the migration of thymocytes from the cortex to the cortico-medullary junction and thymic medulla, which are relatively cytokine-rich thymic regions (Supplementary Fig. 8b).
DISCUSSION
Our present study fundamentally changes the understanding of CD8+ T cell positive selection and differentiation by demonstrating that TCR signaling is dispensable, whereas cytokine signaling is required, for CD8-lineage specification and cytotoxic-lineage T cell differentiation in the thymus. In positively selected thymocytes, STAT-mediated γc cytokine signaling induced expression of Runx3, a transcription factor essential for CD8 lineage specification, and promoted the differentiation of DP thymocytes into phenotypically mature, cytotoxic-lineage T cells. In preselection DP thymocytes genetically engineered to be cytokine responsive, IL-7 signaling was sufficient to induce Runx3 expression and differentiation into mature CD8+ T cells, circumventing positive selection altogether. Notably, cytokine-responsive DP thymocytes were not signaled by endogenous intrathymic cytokines in vivo, which suggests that preselection DP thymocytes reside in a region of the thymic cortex with low expression of endogenous γc cytokines. In the presence of exogenous IL-7, however, cytokine-responsive DP thymocytes differentiated into CD8+ T cells that lacked expression of CCR7, a chemokine receptor that promotes the migration of thymocytes into the cytokine-rich cortico-medullary and medullary regions38,39. On the basis of these results, we propose that TCR-mediated positive selection converts DP thymocytes into cytokine-responsive thymocytes and induces their migration into cytokine-rich thymic regions. Signaling by endogenous intrathymic cytokines subsequently induces Runx3 expression, specifies CD8 lineage choice and promotes thymocyte differentiation into phenotypically mature CD8+ T cells.
TCR-mediated positive selection signals have been classically thought to specify CD4-CD8 lineage choice and to drive the differentiation of DP thymocytes into mature T cells. However, our present study has documented that TCR-mediated positive selection signals do not specify CD8 lineage choice and are not sufficient to drive the differentiation of DP thymocytes into mature CD8+ T cells. Instead, TCR-mediated positive selection signals make it possible for DP thymocytes and/or their immediate progeny to be signaled by intrathymic cytokines, which is accomplished by TCR-mediated termination of Cd8a and Socs1 expression and by TCR-mediated induction of Il7r and Ccr7 expression. Termination of Cd8a and Socs1 expression in TCR-signaled DP thymocytes disrupts MHC class I–specific TCR signaling and permits γc cytokine signaling, whereas persistent TCR signaling (such as that mediated by MHC class II–specific TCRs) prevents the transduction of γc cytokine signals in both developing CD4+ thymocytes5,13 and mature T cells40,41. Induction of Il7r and Ccr7 expression permits TCR-signaled DP thymocytes to migrate into cytokine-rich areas of the thymus (such as the cortico-medullary junction and thymic medulla) and to bind IL-7.
CD8 lineage specification is thought to be mediated in positively selected thymocytes by the transcription factor Runx3 (refs. 14,17,20). In DP thymocytes signaled by their TCR to undergo positive selection, Runx3 is not expressed until DP thymocytes have converted transcriptionally into Cd4+Cd8− intermediate thymocytes and have received STAT-mediated cytokine signals. Runx3 may be a direct STAT target, as sequence analysis has revealed three potential STAT-binding sites in the Runx3 distal promoter (data not shown). When expressed in IL-7-signaled intermediate thymocytes, Runx3 mediates the transcriptional events referred to as ‘coreceptor reversal’, activating the Cd4 silencer to extinguish Cd4 expression and activating the E8I Cd8a transcriptional enhancer to reactivate Cd8 expression13,20,42, which converts intermediate thymocytes into Cd4−Cd8+ (that is, CD8 lineage) thymocytes. Notably, in addition to mediating coreceptor reversal, Runx3 silences expression of Zbtb7b, the gene that encodes Th-POK, a transcription factor important for CD4 lineage specification15,16,18,19,21,43. Thus, STAT-mediated induction of Runx3 results in CD8 lineage specification because Runx3 converts positively selected thymocytes into Cd4−Cd8+ (that is, CD8 lineage) cells and suppresses Th-POK to extinguish CD4 lineage potential.
Our findings provide an additional explanation for why it is critical to prevent DP thymocytes from being signaled by intrathymic cytokines. It is now believed that IL-7 signaling would interfere with preselection DP thymocytes undergoing death by neglect5. Our present study has additionally suggested that IL-7 signaling in preselection DP thymocytes can circumvent positive selection and drive DP thymocytes to differentiate into CD8 lineage T cells expressing unscreened TCRs.
Notably, conditional deletion of the genes encoding the E protein transcription factors HEB and E2A results in intrathymic generation of mature CD8+ T cells even in the absence of TCR signaling44, similar to the intrathymic generation of mature CD8+ T cells in IL-7-Tg 7SZ mice reported here. However, the effects of deficiency in HEB and E2A on thymocyte development were ascribed to a proposed ‘gatekeeper’ function for HEB and E2A in preventing DP thymocyte differentiation until a functional αβTCR was produced and to a proposed ‘default pathway’ for CD8+ T cell differentiation for thymocytes not instructed to become CD4+ T cells. Our present study offers an alternative explanation for the phenotype observed in HEB-E2A–deficient thymocytes. We suggest that in the absence of HEB and E2A transcription factors, preselection DP thymocytes are responsive to intrathymic cytokines such as IL-7 and express CCR7 despite absent TCR signaling44, which causes HEB-E2A–deficient preselection thymocytes to migrate to cytokine-rich thymic areas, where they are signaled by cytokines to express Runx3 and to differentiate into phenotypically mature CD8+ T cells. In fact, in support of this perspective, HEB-E2A–deficient preselection DP thymocytes are IL-7Rα+CCR7+ (ref. 44. Thus, we propose that HEB and E2A contribute to the cytokine-unresponsive phenotype of preselection DP thymocytes in wild-type mice by downregulating expression of IL-7Rα and CCR7 and by upregulating expression of SOCS1 in these cells.
Although our present study has documented that CD8 lineage choice and CD8+ T cell differentiation require cytokine signaling, it has also documented that CD4 lineage choice and CD4+ T cell differentiation are γc cytokine independent and are unaffected by the ability of positively selected thymocytes to transduce γc cytokine signals. Because IL-7 is produced by thymic stromal cells and is the most prevalent intrathymic cytokine, we expected that conditional deletion of Stat5a and Sta5b in preselection thymocytes would be sufficient to abrogate CD8+ T cell generation. However, conditional deletion of Stat5a and Stat5b in preselection DP thymocytes diminished but did not abrogate CD8+ T cell generation. One possibility was that other cytokines such as IL-4 contribute to CD8+ T cell generation. However, IL-4 was much less effective than IL-7 in supporting thymocyte survival and differentiation, which suggested that IL-7 signaling might be mediated by other STAT molecules, such as STAT6, in STAT5-deficient cells. Although STAT5 deficiency does affect the activity of other STAT molecules in nonlymphoid cells45, STAT redundancy, as observed in our present study, has not been documented before in T cells, to our knowledge. We think the mechanism underlying our observation is that STAT6 and other STAT proteins can be recruited to phosphorylated Tyr449 in the IL-7Rα cytosolic tail and can then be phosphorylated. In wild-type cells, other STAT proteins are probably outcompeted by STAT5, which presumably binds to phosphorylated Tyr449 with the highest affinity. Indeed, mice deficient in both STAT5 and STAT6 contained considerably fewer CD8+ T cells in the thymus than did either wild-type mice or mice deficient in only one or the other STAT protein, whereas CD4+ T cell numbers and overall thymocyte numbers were equivalent, which demonstrates that CD8+ T cells require STAT-mediated cytokine signals for their generation in the thymus. Blocking γc cytokine signaling in developing thymocytes using a Socs1 trans-gene confirmed the critical importance of cytokine signaling for the generation of CD8+ T cells but not CD4+ T cells.
Cytokine signaling is not strictly indispensable for the appearance of CD8SP cells in the thymus, as we detected such cells in SOCS1-Tg–Bcl-2-Tg mice despite the absence of γc cytokine signal transduction. Unconventional CD8SP thymocytes have been detected previously in Bcl-2-Tg mice and are highly unusual because they arise independently of MHC class I expression and fail to achieve functional maturity35. Our present study has further documented that these unconventional CD8SP thymocytes arose independently of γc cytokine signaling and, when they did not receive γc cytokine signals, did not express Runx3 (or Runx1). We think such unconventional CD8 thymocytes may not be the immediate progeny of DP thymocytes but instead may derive from DN thymocytes that, in the presence of the Bcl2 transgene, are signaled by their αβTCR to directly upregulate CD8 expression. Thus, we think that unconventional thymocytes, despite their CD8SP phenotype, do not require and have not undergone CD8 lineage specification because they do not arise from bipotent precursors (for example, DP thymocytes).
In conclusion, signaling by intrathymic cytokines is necessary for CD8 lineage specification and CD8+ T cell development. Because preselection DP thymocytes reside in a cytokine-insufficient region of the thymic cortex and are cytokine unresponsive, we propose that TCR signaling converts DP thymocytes into cytokine-responsive cells that migrate to cytokine-rich regions of the thymus and are signaled by endogenous cytokines. Thus, thymic generation of CD8+ T cells requires a carefully choreographed sequence initiated by TCR signaling and executed by cytokine signaling.
ONLINE METHODS
Animals
E8III-Cre-Tg mice, which express cDNA encoding Cre recombinase under the control of the mouse E8III-Cd8a enhancer-promoter elements, and Runx3-Tg mice, which express cDNA encoding hemagglutinin-tagged mouse Runx3 driven by the human CD2 promoter, were newly generated for this study. Other transgenic mice models used in this study were as follows: Bcl-2-Tg mice, which express cDNA encoding human Bcl-2 driven by the Lck proximal promoter46; SOCS1-Tg mice, which express cDNA encoding Myc-tagged mouse SOCS1 driven by the Lck proximal promoter26; IL-7-Tg mice, which express cDNA encoding mouse IL-7 driven by the Lck proximal promoter37; IL-7Rα-Tg mice, which express cDNA encoding mouse IL-7Rα driven by the human CD2 promoter47; and AND TCR–transgenic mice48.
Mice whose Stat5a and Stat5b loci are flanked with loxP sites have been reported24. Stat6−/− mice were from Taconic; Socs1+/−Ifng−/− mice were provided by J. Ihle49 and were bred to generate Socs1−/−Ifng−/− mice; Zap70-KO, MHC class II–KO and MHC-KO mice were bred in our own colony. C57BL/6 (wild-type) mice were from Charles River. Notably, all Socs1−/− mice were maintained as Socs1−/−Ifng−/− to avoid systemic inflammation. Animal experiments were approved by the National Cancer Institute Animal Care and Use Committee, and all mice were cared for in accordance with US National Institutes of Health guidelines.
Cell culture and cell purification
Single-cell suspensions were prepared from thymus and lymph nodes by gentle tweezing with forceps and were analyzed freshly or after overnight in vitro culture (5 × 106 cells per ml) in a 7.5% CO2 atmosphere in RPMI-1640 medium supplemented with 10% (vol/vol) FCS. Where indicated, cells were stimulated with mouse recombinant IL-7 (10 ng/ml) or IL-4 (40 ng/ml; Peprotech). CD4+ and CD8+ lymph node T cells were purified by depletion of immunoglobulin-positive cells plus either CD8+ or CD4+ cells, respectively, with antibody-mediated magnetic cell sorting.
Flow cytometry
Antibodies with the following specificities were used for staining: CD4 (GK1.5 and RM4.5), CD8α (53-6-7), CD8β (53-5.8), TCRβ (H57-597), IL-4Rα (M1), phosphorylated STAT5 (47), p-STAT6 (J71-773; all from BD Pharmingen); CD24 (30-F1), IL-7Rα (A7R34), Qa-2 (69H1-9-9; all from eBiosciences); STAT5 (9363) and phosphorylated STAT6 (9361; polyclonal antibodies; both from Cell Signaling Technology); and fluorescein isothiocyanate–conjugated anti-Myc (9E10; Sigma). Donkey secondary antibodies to rabbit IgG (7f11-096-152) were from Jackson ImmunoResearch Laboratories. Intracellular staining was done as described5. Fluorescein isothiocyanate–conjugated anti-Syk (5F5) was a gift from A. Weiss50.
For cell surface immunofluorescence analysis, cells were analyzed on a FACSVantage SEM (Becton Dickinson) with four-decade logarithmic amplification. Dead cells were excluded by forward light-scatter gating and propidium iodide staining. Data were analyzed using software designed by the Division of Computer Research and Technology, US National Institutes of Health.
ALZET osmotic pump installation
Infusion of recombinant mouse IL-7 (PeproTech) was achieved by subcutaneous implantation of ALZET osmotic pumps (Durect), which released 10 μg IL-7 per day for 2 weeks.
RNA hybridization and RT-PCR
Total RNA were isolated with TriZol (Invitrogen). RNA-hybridization analysis was done as described40. RT2 qPCR primers (SA Biosciences) were used for real-time PCR. Expression of Runx3 mRNA (distal isoform) was assessed by RT-PCR with the following primers: Runx3-433/down (5′-GGTCAGACCCACTTGGTTGG-3′) and Runx3-433/up (5′-GGTGAGCCTCGTTCATTCAT-3′). Expression of 18S rRNA was assessed with the following primers: 18S-S (5′-CCTGAGAAACGGCTACCACATC-3′) and 18S-AS (5′-CATCTAAGGGCATCACAGACCTG-3′).
Statistical analyses
Results of Student’s two-tailed t-tests with P values of less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank R. Bosselut, N. El Kassar, R. Hodes, D. Singer and N. Taylor for critical reading of the manuscript; S. Habu (Tokai University) for mouse Runx3 cDNA; A. Weiss (University of California San Francisco) for fluorescein isothiocyanate–conjugated monoclonal antibody to Syk (5F5); J. Ihle (St. Jude Children’s Research Hospital) for Socs1+/−Ifng−/− mice; and S. Sharrow, A. Adams and L. Granger for flow cytometry. Supported by the Intramural Research Program of the US National Institutes of Health, the National Cancer Institute and the Center for Cancer Research.
Footnotes
AUTHOR CONTRIBUTIONS
J.-H.P. did experiments, analyzed data and contributed to the writing of the manuscript; S.A., T.G., M.C., M.Y.K. and P.J.L. did experiments and analyzed data; B.E. and A.S.A. constructed some of the experimental mice; Y.C., R.E.G., M.K. and L.H. provided experimental mice; L.F. generated transgenic mice; and A.S. conceptualized the research, directed the study, analyzed data and wrote the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Accession codes. UCSD-Nature Signaling Gateway (http://www.signaling-gateway.org): A004205 and A001267.
Note: Supplementary information is available on the Nature Immunology website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Jameson SC, Hogquist KA, Bevan MJ. Positive selection of thymocytes. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- 2.Singer A. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr Opin Immunol. 2002;14:207–215. doi: 10.1016/s0952-7915(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 3.Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. doi: 10.1038/372100a0. [DOI] [PubMed] [Google Scholar]
- 4.Van De Wiele CJ, et al. Thymocytes between the β-selection and positive selection checkpoints are nonresponsive to IL-7 as assessed by STAT-5 phosphorylation. J Immunol. 2004;172:4235–4244. doi: 10.4049/jimmunol.172.7.4235. [DOI] [PubMed] [Google Scholar]
- 5.Yu Q, et al. Cytokine signal transduction is suppressed in preselection double-positive thymocytes and restored by positive selection. J Exp Med. 2006;203:165–175. doi: 10.1084/jem.20051836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong MM, et al. Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation. Immunity. 2003;18:475–487. doi: 10.1016/s1074-7613(03)00078-5. [DOI] [PubMed] [Google Scholar]
- 7.Zamisch M, et al. Ontogeny and regulation of IL-7-expressing thymic epithelial cells. J Immunol. 2005;174:60–67. doi: 10.4049/jimmunol.174.1.60. [DOI] [PubMed] [Google Scholar]
- 8.Alves NL, et al. Characterization of the thymic IL-7 niche in vivo. Proc Natl Acad Sci USA. 2009;106:1512–1517. doi: 10.1073/pnas.0809559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Repass JF, et al. IL7-hCD25 and IL7-Cre BAC transgenic mouse lines: new tools for analysis of IL-7 expressing cells. Genesis. 2009;47:281–287. doi: 10.1002/dvg.20497. [DOI] [PubMed] [Google Scholar]
- 10.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahama Y, Suzuki H, Katz KS, Grusby MJ, Singer A. Positive selection of CD4+ T cells by TCR ligation without aggregation even in the absence of MHC. Nature. 1994;371:67–70. doi: 10.1038/371067a0. [DOI] [PubMed] [Google Scholar]
- 12.Ohoka Y, et al. In vitro differentiation and commitment of CD4+ CD8+ thymocytes to the CD4 lineage, without TCR engagement. Int Immunol. 1996;8:297–306. doi: 10.1093/intimm/8.3.297. [DOI] [PubMed] [Google Scholar]
- 13.Brugnera E, et al. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 2000;13:59–71. doi: 10.1016/s1074-7613(00)00008-x. [DOI] [PubMed] [Google Scholar]
- 14.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 16.He X, et al. CD4–CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–358. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Setoguchi R, et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 18.Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4+ T cells. Nat Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato T, et al. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity. 2005;22:317–328. doi: 10.1016/j.immuni.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egawa T, Taniuchi I. Antagonistic interplay between ThPOK and Runx in lineage choice of thymocytes. Blood Cells Mol Dis. 2009;43:27–29. doi: 10.1016/j.bcmd.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Bosselut R. CD4–CD8 lineage differentiation: Thpoking into the nucleus. J Immunol. 2009;183:2903–2910. doi: 10.4049/jimmunol.0901041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui Y, et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 26.Hanada T, et al. A mutant form of JAB/SOCS1 augments the cytokine-induced JAK/STAT pathway by accelerating degradation of wild-type JAB/CIS family proteins through the SOCS-box. J Biol Chem. 2001;276:40746–40754. doi: 10.1074/jbc.M106139200. [DOI] [PubMed] [Google Scholar]
- 27.Kubo M, Hanada T, Yoshimura A. Suppressors of cytokine signaling and immunity. Nat Immunol. 2003;4:1169–1176. doi: 10.1038/ni1012. [DOI] [PubMed] [Google Scholar]
- 28.Metcalf D. The SOCS-1 story. Exp Hematol. 1999;27:1715–1723. doi: 10.1016/s0301-472x(99)00120-4. [DOI] [PubMed] [Google Scholar]
- 29.Bosselut R, Guinter TI, Sharrow SO, Singer A. Unraveling a revealing paradox: Why major histocompatibility complex I-signaled thymocytes ‘paradoxically’ appear as CD4+8lo transitional cells during positive selection of CD8+ T cells. J Exp Med. 2003;197:1709–1719. doi: 10.1084/jem.20030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundberg K, Heath W, Kontgen F, Carbone FR, Shortman K. Intermediate steps in positive selection: differentiation of CD4+8int TCRint thymocytes into CD4−8+TCRhi thymocytes. J Exp Med. 1995;181:1643–1651. doi: 10.1084/jem.181.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merkenschlager M, et al. Centromeric repositioning of coreceptor loci predicts their stable silencing and the CD4/CD8 lineage choice. J Exp Med. 2004;200:1437–1444. doi: 10.1084/jem.20041127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki H, Punt JA, Granger LG, Singer A. Asymmetric signaling requirements for thymocyte commitment to the CD4+ versus CD8+ T cell lineages: a new perspective on thymic commitment and selection. Immunity. 1995;2:413–425. doi: 10.1016/1074-7613(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 33.Taniuchi I, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 34.Yu Q, Erman B, Bhandoola A, Sharrow SO, Singer A. In vitro evidence that cytokine receptor signals are required for differentiation of double positive thymocytes into functionally mature CD8+ T cells. J Exp Med. 2003;197:475–487. doi: 10.1084/jem.20021765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linette GP, et al. Bcl-2 is upregulated at the CD4+ CD8+ stage during positive selection and promotes thymocyte differentiation at several control points. Immunity. 1994;1:197–205. doi: 10.1016/1074-7613(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 36.Vernachio J, Li M, Donnenberg AD, Soloski MJ. Qa-2 expression in the adult murine thymus. A unique marker for a mature thymic subset. J Immunol. 1989;142:48–56. [PubMed] [Google Scholar]
- 37.El Kassar N, et al. A dose effect of IL-7 on thymocyte development. Blood. 2004;104:1419–1427. doi: 10.1182/blood-2004-01-0201. [DOI] [PubMed] [Google Scholar]
- 38.Ueno T, et al. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J Exp Med. 2004;200:493–505. doi: 10.1084/jem.20040643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwan J, Killeen N. CCR7 directs the migration of thymocytes into the thymic medulla. J Immunol. 2004;172:3999–4007. doi: 10.4049/jimmunol.172.7.3999. [DOI] [PubMed] [Google Scholar]
- 40.Park JH, et al. ‘Coreceptor tuning’: cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nat Immunol. 2007;8:1049–1059. doi: 10.1038/ni1512. [DOI] [PubMed] [Google Scholar]
- 41.Zhu J, et al. Transient inhibition of interleukin 4 signaling by T cell receptor ligation. J Exp Med. 2000;192:1125–1134. doi: 10.1084/jem.192.8.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellmeier W, Sunshine MJ, Losos K, Hatam F, Littman DR. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity. 1997;7:537–547. doi: 10.1016/s1074-7613(00)80375-1. [DOI] [PubMed] [Google Scholar]
- 43.Muroi S, et al. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 44.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosui A, et al. Loss of STAT5 causes liver fibrosis and cancer development through increased TGF-β and STAT3 activation. J Exp Med. 2009;206:819–831. doi: 10.1084/jem.20080003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sentman CL, Shutter JR, Hockenbery D, Kanagawa O, Korsmeyer SJ. Bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell. 1991;67:879–888. doi: 10.1016/0092-8674(91)90361-2. [DOI] [PubMed] [Google Scholar]
- 47.Yu Q, Erman B, Park JH, Feigenbaum L, Singer A. IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and ROR γt: impact on thymocyte development. J Exp Med. 2004;200:797–803. doi: 10.1084/jem.20032183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaye J, et al. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 1989;341:746–749. doi: 10.1038/341746a0. [DOI] [PubMed] [Google Scholar]
- 49.Marine JC, et al. SOCS1 deficiency causes a lymphocyte-dependent perinatal lethality. Cell. 1999;98:609–616. doi: 10.1016/s0092-8674(00)80048-3. [DOI] [PubMed] [Google Scholar]
- 50.Chu DH, et al. Pre-T cell receptor signals are responsible for the down-regulation of Syk protein tyrosine kinase expression. J Immunol. 1999;163:2610–2620. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.