Abstract
Objective
Xeroderma pigmentosum complementation group C (XPC) participates in the initial recognition of DNA damage during nucleotide excision repair process in global genomic repair. Our meta-analysis was performed to evaluate the association between three polymorphisms (Lys939Gln, PAT+/– and Ala499Val) of XPC gene and risk of digestive system cancers.
Methods
All the relevant case-control studies published to April 2011 were identified through searching PubMed. Digestive system cancer risk with the three polymorphisms was estimated for each study by odds ratio (OR) with its 95% confidence interval (95% CI).
Results
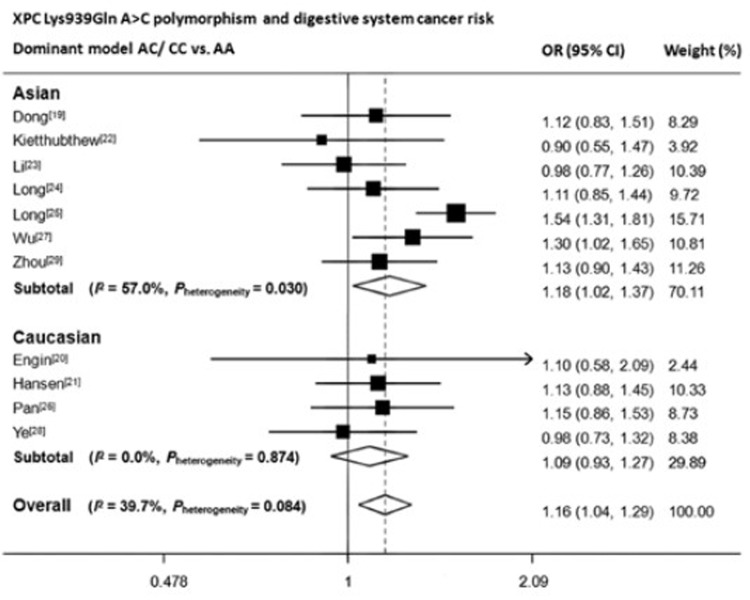
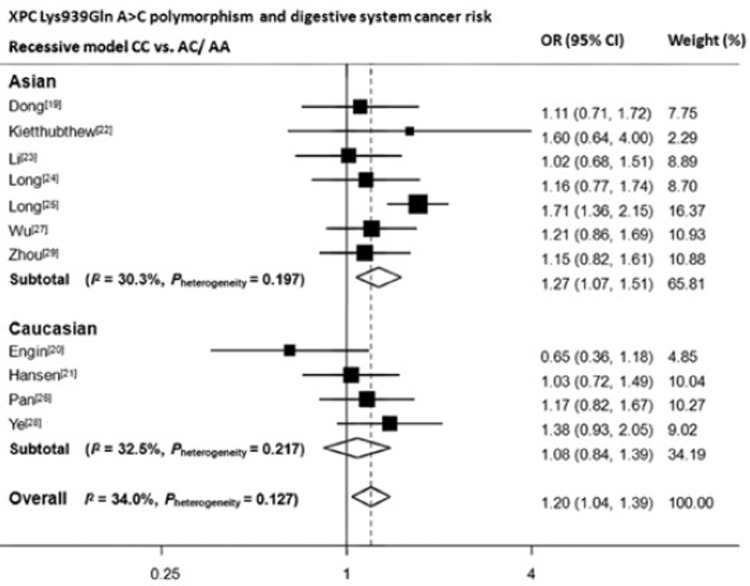
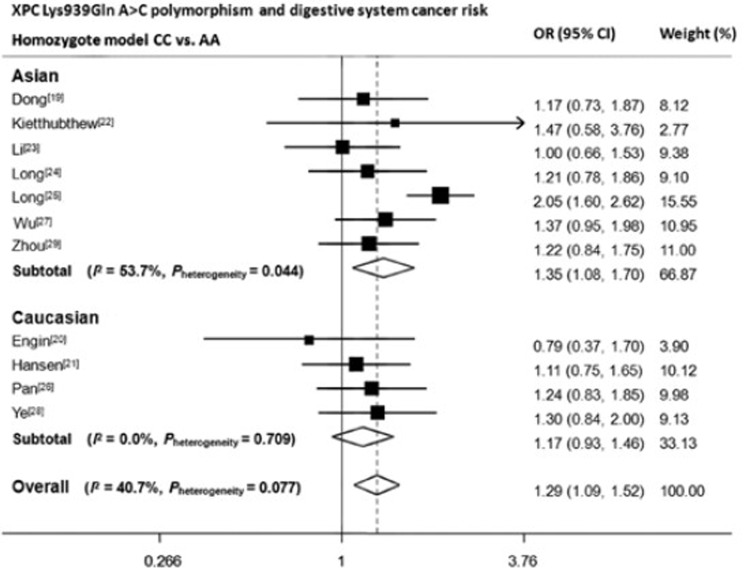
We found an increased overall risk for digestive system cancers in all three models of Lys939Gln A>C (AC/CC vs. AA: OR, 1.20; 95% CI, 1.11–1.30; CC vs. AC/AA: OR, 1.24; 95% CI, 1.11–1.39; CC vs. AA: OR, 1.36; 95% CI, 1.21–1.53). When stratified by ethnicity, results remained significant in Asian population (AC/CC vs. AA: OR, 1.18; 95% CI, 1.02–1.37; CC vs. AC/AA: OR, 1.32; 95% CI, 1.1–1.51; CC vs. AA: OR, 1.35; 95% CI, 1.08–1.70), but not for Caucasians. However for Ala499Val C>T, a significant protective effect of T allele was only observed in the dominant model. Otherwise, no significant results were observed for PAT+/–.
Conclusion
XPC Lys939Gln A>C polymorphism may play an important role in digestive system cancer susceptibility.
Key words: XPC, Polymorphism, Digestive system cancer, Meta-analysis
INTRODUCTION
Xeroderma pigmentosum complementation group C gene (XPC) is one of the eight core genes (ERCC1, XPA, XPB, XPC, XPD, XPE, XPF and XPG) in the nuclear excision repair (NER) pathway. XPC binds to HR23B and forms XPC-HR23B complex, which is involved in DNA damage recognition and DNA repair initiation in the global genome of NER[1-3]. Binding of XPC to damaged DNA is the rate-limiting step for NER[4, 5]. The relationship between XPC and cancer starts with the observation of high incidence of skin cancers (approximately 1000-fold) in patients with mutations in XPC gene, which characterize the inherited disease xeroderma pigmentosum (XP)[6]. Three main single nucleotide polymorphisms (SNP) of XPC, Lys939Gln, Ala499Val and PAT+/–, have been identified and most commonly studied. The XPC- Lys939Gln poly morphism, with an A to C substitution in exon 15 that gives rise to a Lys to Gln substitution at position 939, was found to be associated with DNA repair capacity as measured chromatid aberrations[7]. The XPC Ala499Val is a non-synonymous substation of Ala for Val in codon 499, an interaction domain of XPC with hHRAD23, but its impact on the protein function was unknown, although it was in strong linkage disequilibrium with other two polymorphisms in the 30’-untranslated region (Exon 15−184 and Exon 15−177)[8]. PAT+/– was a novel variant in intron 9, and PAT+/+ carriers were demonstrated to have lower DNA repair capacity than PAT–/– carriers[9].
Numerous epidemiological studies have been conducted to explore the association of XPC poly-morphisms with cancer risk, covering different cancer types in diverse population, with results remained conflicting[10-14]. To date, four meta-analyses[15-18] have been performed to examine the modest effect of XPC variants on cancer risk that could not be achieved by single research. However, all of these meta-analyses focused their attention on cancers other than digestive system cancer. For example, Francisco, et al.[15] reported elevated risk for lung cancer in the recessive model of Lys939Gln (odds ratio: OR; confidence interval: CI. ORLys/Lys+Lys/Gln=1.30, 95% CI=1.11−1.53), as well as an increased risk for bladder cancer risk in both the recessive model and homozygote model of Ala499Val (ORAla/Ala+Ala/Val=1.32, 95% CI=1.06−1.63; ORAla/Ala=1.30, 95% CI=1.04−1.61). Two subsequently published meta-analyses written by Qiu, et al.[16], and Zhang, et al.[17] have drawn the same conclusions as Francisco, et al. Recently, Zheng, et al.[18] organized a meta-analysis on XPC polymorphism and breast cancer, and revealed that PAT+/– polymorphism may be a low-penetrant risk for developing breast cancer (OR+/– and +/+=1.41, 95% CI=1.05–1.89). However, they haven’t found any obvious associations for all genetic models of either Lys939Gln or Ala499Val. The reason why researchers neglected digestive system neoplasm might be due to the limited study numbers relevant to this topic (less than five in the above mentioned meta-analyses).
Through PubMed searching and literature reading, we noticed that a number of molecular epidemiologic studies have been performed to evaluate the role of XPC polymorphisms in digestive system neoplasm, with their results inconsistent[19-32]. To estimate the overall relationship of the three polymorphisms on digestive system cancer risk, as well as to quantify the potential between-study heterogeneity, a meta-analysis including 14 the most recently published and relevant articles was performed in present study.
MATERIALS AND METHODS
Identification and Eligibility of Relevant Studies
All the case-control studies published to date on the association between the three polymorphisms (Lys939Gln, Ala499Val, PAT+/–) of XPC gene and digestive system cancer risk were included in our analysis if they met the inclusion criteria. Eligible studies were identified by searching the electronic literature PubMed for relevant reports (last search update mid-April, 2011), using the search terms “Digestive System Neoplasms[Mesh] AND Poly-morphism, Genetic[Mesh] AND XPC, protein, Human[Mesh]”. A hand search of references from the retrieved studies or review articles was done to identify additional studies relevant to this topic. Literatures included in our meta-analysis need to satisfy all the following criteria: (1) published in English or Chinese. Abstracts, unpublished reports and articles written in languages other than English and Chinese were not considered; (2) study on human beings; (3) in a case-control study design (case-control study or nested case-control study); (4) had detailed genotype frequency of both cases and controls or can be calculated from the text of the articles; (5) excluded benign tumor, precancerous lesion and adenoma (e.g. colorectal adenoma[33]); (6) only the one with a larger sample size was selected if the case-control study was included by more than one articles using the same case serious; and (7) if the case-control study covered more than one cancer type using the same control subjects, we never separate them as several independent studies but combined the cancers of the same category together and consider them as one. In the current study, data for meta-analysis were available from 14 articles, including 4,562 cancer cases and 6,593 controls for Lys939Gln (11 studies from 11 articles)[19-29], 2,496 cases and 3,653 controls for Ala499Val (6 studies from 6 articles)[19, 23, 24, 26, 27,29], 1,429 cases and 2,724 controls for PAT+/– (7 studies from 7 articles)[19, 22, 26, 27, 30-32], respectively.
Data Extraction
Two investigators (X. Jiang and L. Zhou) carefully and independently extracted data, using “double data entry” command of Review Manager 4.2.10., to achieve the accuracy of data. If different results generated, they would check the data again to come to an agreement. The following items were included: first author’s name, year of publication, country of origin, ethnicity, source of control, genotyping methods, cancer types, numbers of cases and controls, and minor allele frequency (MAF) in controls. Different ethnicity was categorized as Asian, Caucasian and African. Colorectal cancer, gastric cancer, esophageal cancer, oral cancer, biliary tract cancer, gallbladder cancer and pancreatic cancer were categorized into digestive system neoplasm. Moreover, we also defined all the cancers according to different locations as upper digestive system cancer (including gastric cancer, esophageal cancer, oral cancer, biliary tract cancer, gallbladder cancer and pancreatic cancer) and lower digestive system cancer (colorectal cancer).
Statistical Analysis
The risk of cancer associated with the polymorphisms of XPC gene was estimated for each study by OR together with its 95% CI, respectively. Cochran’s Q statistic test was performed to assess the between-study heterogeneity, and was considered significant for P<0.05. To combine values from studies, a fixed-effect model (Mantel-Haenszel method) and a random-effects model (DerSimonian and Laird method) were applied, respectively[34]. These two models provide similar results when heterogeneity between studies is absent; otherwise, random-effects model is more appropriate. We estimated the risks of combined variant homozygote and heterozygote versus wild-type homozygote, variant homozygote versus combined heterozygote and wild-type homozygote, assuming dominant and recessive effects of the variant allele, respectively. Variant homozygote compared with wild-type homozygote was also evaluated, as the homozygote model. We further performed stratification analyses on ethnicity, different sources of controls, genotyping methods and different locations of cancer. Meta-regression was performed to illustrate potential reasons of between-study heterogeneity. Egger’s test and funnel plots were used to examine the influence of publication bias (Linear regression asymmetry test). We checked deviation from the Hardy-Weinberg equilibrium (HWE) among controls for each polymorphism by χ2-test, with one degree of freedom. All P values were two-sided if not indicated, and all analyses were performed in the software Stata (version 11.0; STATA Corp, College Station, Texas) and Review Manager (version 4.2; Oxford, England).
RESULTS
Characteristics of Published Studies
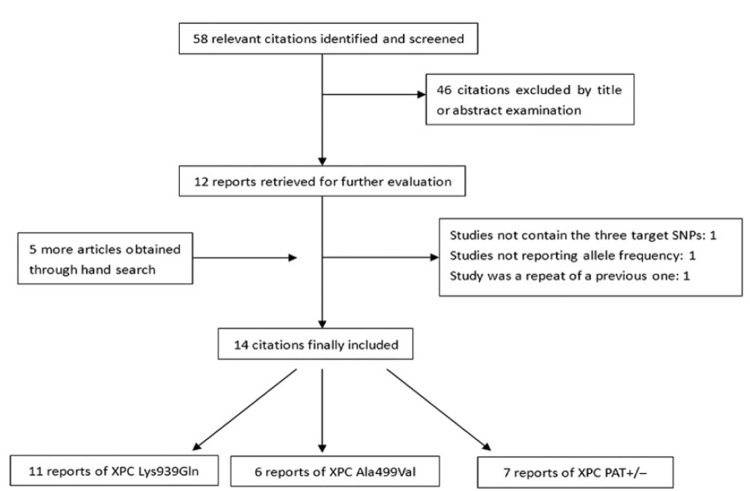
After initial screening, 58 relevant publications were identified, among which 12 publications appeared to meet the inclusion criteria. Five more articles were added through hand search. Altogether, 17 literatures were subjected to further examination. Three publications were excluded because they didn’t contain targeted SNPs[35], didn’t provide detailed genotype data[36], or was repeated data from one already-included study[37]. Therefore, our final data pooling consisted of 14 studies. Procedure of literature-picking is represented in Figure 1. Characteristics of the selected studies are summarized in Table 1. The distribution of genotypes in the controls was in accord with HWE for all selected studies. When the OR for an allelic genetic association was assumed to be 1.2 for Lys939Gln, 0.85 for both Ala499Val and PAT+/–, no study reached a statistical power greater than 80%. The more detailed information of all SNPs is shown in supplementary Table 1 (Table S1).
Figure 1.
This flow chat shows the procedure of literature selection.
Table 1. Characteristics of literatures included in the meta-analysis.
| SNP Author | Year | Country | Ethnictiy | Source of controls |
Genotyping method | Cancer types | Case | Control | HWE in control (P) | MAF* | Power | (%)** |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XPC Lys939Gln rs2228001 | ||||||||||||
| Kietthubthew[22] | 2006 | Thailand | Asian | Population-based | PCR-RFLP | OSCC | 106 | 164 | 0.547 | 0.27 | - | 9.7 |
| Zhou[29] | 2006 | China | Asian | Population-based | PCR-RFLP | ESCC and GCA | 580 | 612 | 0.699 | 0.36 | - | 33.3 |
| Ye[28] | 2006 | Sweden | Caucasian | Population-based | PCR-RFLP | ESCC, EA and GCA | 303 | 472 | 0.540 | 0.38 | - | 22.8 |
| Pan[26] | 2009 | USA | Causasian | Hospital-based | TaqMan Squence Detection System 7900HT | EC | 378 | 453 | 0.713 | 0.41 | - | 25.3 |
| Li[23] | 2010 | China | Asian | Hospital-based | TaqMan MGB real time PCR | HCC | 500 | 507 | 0.507 | 0.34 | - | 28.4 |
| Long[25] | 2010 | China | Asian | Hospital-based | TaqMan iQ5 Bio-Rad | HCC | 1,156 | 1,402 | 0.606 | 0.36 | - | 60.6 |
| Dong[19] | 2008 | China | Asian | Population-based | PCR-RFLP | GCA | 253 | 612 | 0.699 | 0.36 | - | 21.9 |
| Long[24] | 2010 | China | Asian | Hospital-based | PCR-RFLP | GAA | 361 | 616 | 0.445 | 0.35 | - | 26.4 |
| Hansen[21] | 2007 | Denmark | Caucasian | Population-based | TaqMan Squence Detection System ABI7500 | CRC | 395 | 797 | 0.112 | 0.37 | - | 30.4 |
| Engin[20] | 2010 | Turkey | Causasian | Hospital-based | PCR-RFLP | CRC | 110 | 116 | 0.642 | 0.53 | - | 10.0 |
| Wu[27] | 2011 | China | Asian | Population-based | PCR-RFLP | CRC | 420 | 842 | 0.639 | 0.36 | - | 31.6 |
| XPC cintron9 PAT+/- | ||||||||||||
| Casson[30] | 2005 | Canada | Caucasian | Population-based | PCR-RFLP | EA | 56 | 95 | 0.808 | 0.36 | 6.6 | - |
| Wang[32] | 2006 | China | Asian | Hospital-based | PCR-RFLP | PC | 101 | 337 | 0.564 | 0.36 | 10.0 | - |
| Sugimura[31] | 2006 | Japan | Asian | Hospital-based | PCR-RFLP | OSCC | 122 | 241 | 0.131 | 0.40 | 10.6 | - |
| Kietthubthew[22] | 2006 | Thailand | Asian | Population-based | PCR-RFLP | OSCC | 106 | 164 | 0.472 | 0.26 | 8.1 | - |
| Pan[26] | 2009 | USA | Caucasian | Hospital-based | PCR-RFLP | EC | 379 | 443 | 0.362 | 0.37 | 19.7 | - |
| Dong[19] | 2008 | China | Asian | Population-based | PCR-RFLP | GCA | 253 | 612 | 0.786 | 0.35 | 17.5 | - |
| Wu[27] | 2011 | China | Asian | Population-based | PCR-RFLP | CRC | 412 | 832 | 0.689 | 0.35 | 24.5 | - |
| XPC Ala499Val rs2228000 | ||||||||||||
| Zhou[29] | 2006 | China | Asian | Population-based | PCR-RFLP | ESCC and GCA | 580 | 612 | 0.217 | 0.31 | 24.7 | - |
| Pan[26] | 2009 | USA | Caucasian | Hospital-based | TaqMan Squence Detection System 7900HT | EC | 383 | 450 | 0.133 | 0.24 | 16.3 | - |
| Dong[19] | 2008 | China | Asian | Population-based | PCR-RFLP | GCA | 253 | 612 | 0.169 | 0.31 | 16.6 | - |
| Long[24] | 2010 | China | Asian | Hospital-based | PCR-RFLP | GAA | 361 | 616 | 0.673 | 0.32 | 20.2 | - |
| Li[23] | 2010 | China | Asian | Hospital-based | TaqMan MGB real time PCR | HCC | 500 | 507 | 0.787 | 0.42 | 24.3 | - |
| Wu[27] | 2011 | China | Asian | Population-based | PCR-RFLP | CRC | 419 | 838 | 0.447 | 0.37 | 25.3 | - |
HWE, Hardy-weinberg equilibrium; MAF, Minor allele frequency; PAT, Poly AT; *for Lys939Gln rs2228001, MAF refers to the frequency of C allele; for Ala499Val rs2228000, it refers to frequency of T allele; for PAT, it refers to poly AT deletion; **Power was computed by the DSTPLAN4.3 software with MAF in controls as the frequency of risk factor and OR was selected with 1.20 and 1.85 as relative risk, respectively. OSCC, oral squamous cell carcinoma; ESCC, esophageal squamous cell carcinoma; GCA, gastric cardiac adenocarcinama; EC, esophageal cancer; HCC, hapatocellular carcinoma; PCR-RFLP, polymerase chain reaction fragment length polymorpaism
Quantitative Synthesis
Evaluation of the associations between the three polymorphisms and digestive system cancer risk is presented in Table 2. For Lys939Gln A>C, C allele was observed to be positively associated with overall digestive system cancer risk in all three models (for dominant model AC/CC vs. AA: OR, 1.20; 95% CI, 1.11−1.30; Pheterogeneity=0.08; for recessive model CC vs. AC/AA: OR, 1.24; 95% CI, 1.11−1.39; Pheterogeneity=0.13; for homozygote comparison CC vs. AA: OR, 1.36; 95% CI, 1.21−1.53; Pheterogeneity=0.08). However for Ala499Val C>T, a significant protective effect of T allele was only observed in the dominant model (TT/TC vs. CC: OR, 0.84; 95% CI, 0.76–0.94; Pheterogeneity=0.20). Otherwise, no statistically significant associations were found for PAT+/–.
Table 2. Summary ORs of the XPC polymorphism and cancer risk by ethnicity.
| SNP | Ethnicity | Studies | Case/control | Dominant model |
Recessive model |
Homozygote model |
|||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P* | OR (95% CI) | P* | OR (95% CI) | P* | ||||
| XPC Lys939Gln rs2228001 | Overall | 11 | 4,562/6,593 | 1.20 (1.11-1.30) | 0.08 | 1.24 (1.11-1.39) | 0.13 | 1.36 (1.21-1.53) | 0.08 |
| Asian | 7 | 3,376/4,755 | 1.18 (1.02-1.37) | 0.03† | 1.32 (1.15-1.51) | 0.20 | 1.35 (1.08-1.70) | 0.04† | |
| Caucasian | 4 | 1,186/1,838 | 1.09 (0.93-1.27) | 0.87 | 1.10 (0.90-1.34) | 0.22 | 1.17 (0.93-1.46) | 0.71 | |
| Overall | 7 | 1,429/2,724 | 0.95 (0.83-1.08) | 0.45 | 1.09 (0.90-1.33) | 0.24 | 1.04 (0.85-1.28) | 0.24 | |
| XPC intron9 PAT | Asian | 5 | 994/2,186 | 0.99 (0.85-1.16) | 0.55 | 1.05 (0.83-1.32) | 0.10 | 1.04 (0.81-1.34) | 0.10 |
| Caucasian | 2 | 435/538 | 0.83 (0.64-1.07) | 0.27 | 1.20 (0.85-1.69) | 0.83 | 1.04 (0.72-1.51) | 0.54 | |
| XPC Ala499Val rs2228000 | Overall | 6 | 2,496/3,635 | 0.84 (0.76-0.94) | 0.20 | 1.01 (0.86-1.19) | 0.74 | 0.94 (0.79-1.12) | 0.66 |
| Asian | 5 | 2,113/3,185 | 0.84 (0.75-0.94) | 0.12 | 0.98 (0.82-1.16) | 0.91 | 0.90 (0.75-1.09) | 0.81 | |
Test for Heterogeneity; †Random-effects model was used when P value for het erogeneity test <0.05; otherwise, fixed-effect model was used
We then assessed the effect of the three polymorphisms by ethnicity. As shown in Table 2, Figure 2, Figure 3 and Figure 4, we found that C variant of Lys939Gln significantly increased digestive system cancer risk in Asians (for dominant model AC/CC vs. AA: OR, 1.18; 95% CI, 1.02−1.37; Pheterogeneity=0.03; for recessive model CC vs. AC/AA: OR, 1.32; 95% CI, 1.15−1.51; Pheterogeneity=0.20; for homozygote comparison CC vs. AA: OR, 1.35; 95% CI, 1.08–1.70; Pheterogeneity=0.04) but not in Caucasians. For Ala499Val, five out of six articles adopted Chinese as study population, therefore, only one subgroup was generated. The decreased risk was only seen in dominant model (TT/TC vs. CC: OR, 0.84; 95% CI, 0.75−0.94; Pheterogeneity=0.12). For PAT+/–, no results reached statistically significant level.
Figure 2.
Forest plot of XPC Lys939Gln polymorphism and digestive system cancer risk in dominant model.
Figure 3.
Forest plot of XPC Lys939Gln A>C polymorphism and digestive system cancer risk in recessive model. CC vs. AC/AA. OR: odds ratio; CI: confident interval. Data were calculated in random-effect models for Asian subgroup, Caucasian subgroup and overall.
Figure 4.
Forest plot of XPC Lys939Gln A>C polymorphism and digestive system cancer risk in homozygote model. CC vs. AA. OR: odds ratio; CI: confident interval. Data were calculated in random-effect models for Asian subgroup, Caucasian subgroup and overall.
Finally, the data were further analyzed and stratified by different control groups (hospital based or population based), genotyping methods [polymerase chain reaction-restriction fragment length poly-morphism (PCR-RFLP) or TaqMan], and tumor locations (upper digestive system or lower digestive system), as shown in Table 3. For Lys939Gln, only marginally significant result was found in homozygote model of those studies with population based controls. Moreover, C allele significantly elevated the risk of upper digestive system cancers (recessive model CC vs. AC/AA: OR, 1.29; 95% CI, 1.11−1.50; Pheterogeneity=0.26; for homozygote comparison CC vs. AA: OR, 1.33; 95% CI, 1.09–1.63; Pheterogeneity=0.06). For Ala499Val, the protective effect of T allele was particularly prominent in studies adopting population based controls (TT/TC vs. CC: OR, 0.76; 95% CI, 0.65−0.89; Pheterogeneity=0.28), genotyping through PCR-RFLP (TT/TC vs. CC: OR, 0.80; 95% CI, 0.68−0.93; Pheterogeneity=0.23), and in upper digestive system cancer (TT/TC vs. CC: OR, 0.84; 95% CI, 0.71−0.98; Pheterogeneity=0.13). However, the decreased risk was only restricted to dominant model. Otherwise, no significant gene effects were observed in subgroup analyses.
Table 3. Summary ORs of the XPC polymorphism and cancer risk by different control groups, genotyping methods and tumor locations.
| SNP | Variables | Classification | Studies | Case/control | Dominant model |
Recessive model |
Homozygote model |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P* | OR (95% CI) | P* | OR (95% CI) | P* | |||||
| XPC Lys939Gln rs2228001 | Source of control | Hospital-based | 5 | 2,505/3,094 | 1.19 (0.97-1.46) | 0.02† | 1.15 (0.86-1.56) | 0.01** | 1.27 (0.90-1.80) | 0.007** |
| Population-based | 6 | 2,057/3,499 | 1.13 (1.00-1.26) | 0.71 | 1.18 (1.00-1.39) | 0.89 | 1.24 (1.04-1.48) | 0.970 | ||
| Genotyping method | PCR-RFLP | 7 | 2,133/3,434 | 1.12 (1.00-1.26) | 0.81 | 1.16 (0.99-1.36) | 0.56 | 1.24 (1.04-1.47) | 0.930 | |
| TaqMan | 4 | 2,429/3,159 | 1.20 (0.96-1.50) | 0.01† | 1.24 (0.93-1.64) | 0.03** | 1.33 (0.92-1.92) | 0.005** | ||
| Tumor location | Upper digestive system | 8 | 3,637/4,838 | 1.14 (0.99-1.31) | 0.03 | 1.29 (1.11-1.50) | 0.26 | 1.33 (1.09-1.63) | 0.060 | |
| Lower digestive system | 3 | 925/1,755 | 1.21 (1.02-1.42) | 0.71 | 1.01 (0.75-1.35) | 0.22 | 1.18 (0.92-1.53) | 0.410 | ||
| XPC intron9 PAT | Source of control | Hospital-based | 3 | 602/1,021 | 1.21 (0.66-2.23) | 0.05 | 1.21 (0.98-1.50) | 0.82 | 1.28 (0.76-2.17) | 0.140 |
| Population-based | 4 | 827/1,703 | 0.82 (0.64-1.06) | 0.77 | 0.97 (0.82-1.15) | 0.45 | 0.82 (0.62-1.07) | 0.670 | ||
| Genotyping method | PCR-RFLP/TaqMan‡ | NA | NA | NA | NA | NA | NA | NA | NA | |
| Tumor location | Upper digestive system | 6 | 1,017/1,892 | 0.96 (0.70-1.31) | 0.19 | 1.16 (0.99-1.36) | 0.92 | 1.04 (0.78-1.39) | 0.300 | |
| Lower digestive system§ | 1 | NA | NA | NA | NA | NA | NA | NA | ||
| XPC Ala499Val rs2228000 | Source of control | Hospital-based | 3 | 1,244/1,573 | 0.94 (0.81-1.10) | 0.63 | 1.07 (0.84-1.36) | 0.48 | 1.06 (0.82-1.37) | 0.610 |
| Population-based | 3 | 1,252/2,062 | 0.76 (0.65-0.89) | 0.28 | 0.96 (0.76-1.21) | 0.66 | 0.84 (0.66-1.07) | 0.730 | ||
| Genotyping method | PCR-RFLP | 4 | 1,613/2,678 | 0.80 (0.68-0.93) | 0.23 | 0.96 (0.78-1.18) | 0.84 | 0.86 (0.70-1.07) | 0.860 | |
| TaqMan | 2 | 883/957 | 0.95 (0.78-1.14) | 0.34 | 1.14 (0.83-1.56) | 0.29 | 1.13 (0.82-1.54) | 0.470 | ||
| Tumor location | Upper digestive system | 5 | 2,077/2,797 | 0.84 (0.71-0.98) | 0.13 | 1.06 (0.87-1.27) | 0.76 | 0.98 (0.80-1.19) | 0.640 | |
| Lower digestive system§ | 1 | NA | NA | NA | NA | NA | NA | NA | ||
*Test for Heterogeneity; †Random-effects model was used when P value for het erogeneity test <0.05; otherwise, fixed-effect model was used. **All the articles were genotyping by PCR-RFLP; §, only one article available for lower digestive system
Publication Bias
Funnel plot and Egger’s test were used to address potential publication bias in the available literatures. For Lys939Gln, publication bias could be graphically judged from the shape of funnel plots in all the three models for both overall population and Asian subgroup. Egger’s tests were performed to provide statistical evidence for funnel plot asymmetry (P=0.02, 0.05 and 0.02 for Lys939Gln in the dominant model, recessive model and homozygote model of overall cancer, respectively; P=0.03, 0.24 and 0.14 of the three models in Asian population).
Test of Heterogeneity
For Lys939Gln, we noticed that heterogeneity existed in the both dominant model (P=0.03) and homozygote model (P=0.04). Meta-regression analysis was employed to assess the source of heterogeneity. Results showed that sample size might be a source of heterogeneity (P=0.016). After careful examination of each individual study, we found that the study by Long, et al.[25] was the major cause of the heterogeneity. Therefore, we did sensitivity analysis to identify its impact. The estimations of between-study variance I-square reduced greatly when deselecting this article, from 0.57 to 0.00 for Asian population in dominant model and from 0.53 to 0.00 in homozygote model, respectively.
DISCUSSION
In this meta-analysis, containing a total of 4,841 cancer cases and 7,266 controls from 14 independent publications, associations of three well-characterized polymorphisms (Lys939Gln rs2228001, Ala499Val rs2228000 and PAT+/–) of XPC gene on digestive system cancer susceptibility were examined. We demonstrated that the C allele of Lys939Gln rs2228001 was associated with a significantly increased risk in digestive system cancers, especially among Asian population.
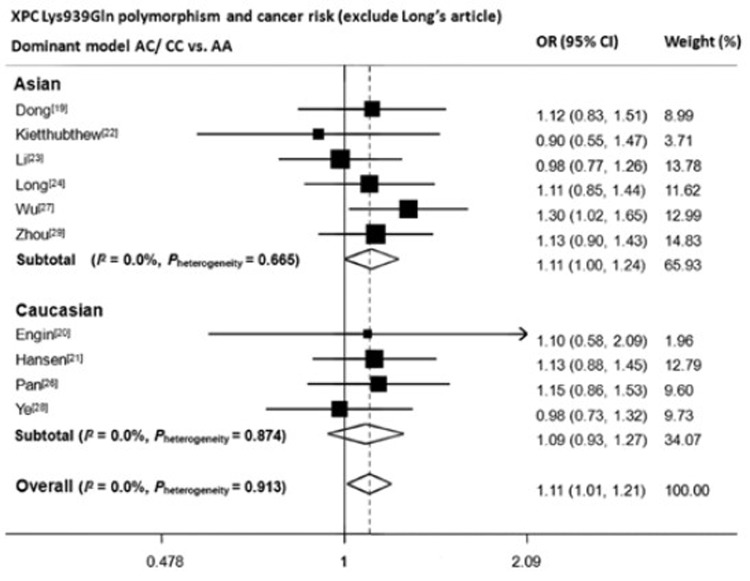
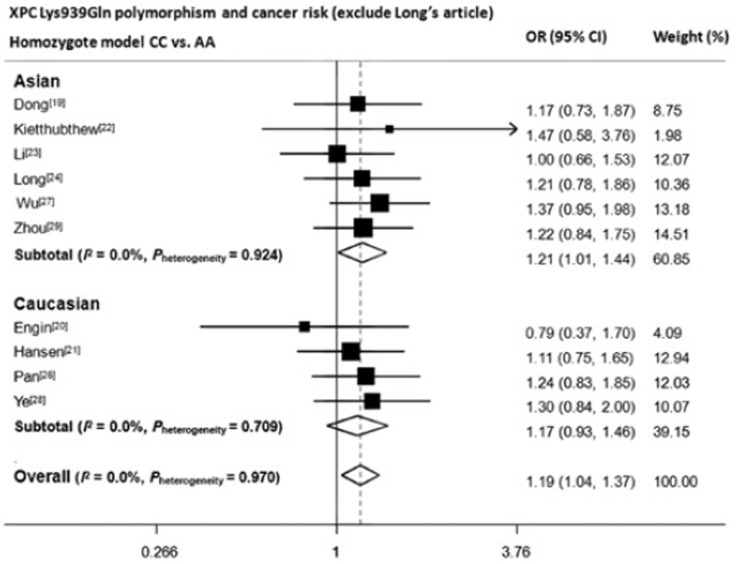
Our conclusion regarding with rs2228001 can be reliable due to following reasons. Primarily, the sample size was large enough to achieve sufficient study power. With 4,562 cases and 6,593 controls, this meta-analysis reached an overall power of 99.6% to detect an OR of 1.2 at α=0.05 level, and in Asian subgroup, the power was 97.5%. Moreover, three previous published meta-analyses[15-17], although did not explore the relationship between rs2228001 polymorphism and digestive system cancers, obtained quite similar results that C allele elevated lung cancer risk, suggesting the possibility of C allele to be the potential risk allele to cancers. In addition, we have noticed that the article by Long, et al.[25] was the major cause for heterogeneity of both dominant and homozygote model. Results still remained significant for both two models even when this article was excluded (overall: AC/CC vs. AA OR=1.11, 95% CI=1.01–1.21; CC vs. AA OR=1.19, 95% CI=1.04–1.37. Asian population: AC/CC vs. AA OR=1.11, 95% CI= 1.00−1.24; CC vs. AA OR=1.21, 95% CI=1.01–1.44) (Figure S1 and Figure S2). Finally, stratification analyses were performed to identify the confounding effects which might be caused by different source of controls, different genotyping methods and different cancer locations. Only marginally significant gene effects can be seen in homozygote model of those studies adopting population-based controls and those studies genotyping through PCR-RFLP. Therefore, we believe genotyping methods and source of control didn’t affect our results. We also noticed that gene effects were significant for upper digestive system cancer in both recessive and homozygote models (AC/CC vs. AA OR=1.29, 95% CI= 1.11−1.50; CC vs. AA OR=1.33, 95% CI=1.09−1.63). After further scrutiny, we found 6 out of the selected 8 articles were from Asian. When the rest two articles with Caucasian population were analyzed separately, no significant effects were observed.
PAT+/– polymorphism was first reported by Khan et al.[38] and found to be in strong linkage disequilibrium with Lys939Gln (Lewontin’s LD=1.0), which was also indicated by several other studies[10, 39]. However, in our meta-analysis, we did not observe any significant associations of PAT+/– polymorphism on digestive system cancer risk. Reason for this could be quite obvious, because in our present study PAT+/– and Lys939Gln only shared an overlapping of 4 articles. Through further examination on the data of the 4 articles[19,22,26,27], we discovered that Pan’s paper[26] badly challenged the LD rule (MAF, PAT+/– vs. Lys939Gln: 0.37 vs. 0.41) while the other three[19, 22, 27] obeyed it well, which means the possibility of not finding any significant effect could exist.
Interestingly, XPC Lys939Gln polymorphism was associated with increased risk of digestive system cancers only in Asian population. It might not be due to different genetic background because the two populations share very close minor allele frequency of the variant C allele (Asian vs. Caucasian: 0.35 vs. 0.39). We believed this discrepancy could be explained by sample size. When we assumed that the OR of the allelic genetic association for C variant was 1.2, the analyses on Caucasians (with 1,186 cases and 1,838 controls) only had a power of 67.6%, far from sufficient to detect such a risk, whereas Asians (with 3,376 cases and 4,755 controls) achieved statistical powers of 97.5%.
In interpreting our results of the current meta-analysis, some limitations need to be addressed. As revealed by the Egger’s test and funnel plot, considerable numbers of unpublished negative studies could not be included in this meta-analysis, suggesting the likelihood of potential publication bias, where positive rather than negative finding tend to be published. Moreover, selection bias might also exist because only studies written and published in English and Chinese were selected. Articles written in languages other than English and Chinese could not be indexed. In addition, the numbers of included studies were still insufficient for subgroup analysis. For example, altogether six articles were retrieved for Ala499Val polymorphism and only one study was available on Caucasian population. Limited study number leads to smaller sample size and lower power. Finally, our meta-analysis was based on unadjusted OR estimates. The authors tried to calculate ORs with adjustment, however, not all the studies presented adjusting their ORs by the same potential confounders. Given these results, our conclusions should be interpreted cautiously.
In summary, our meta-analysis provides evidence that C variant of XPC Lys939Gln rs2228001 is associated with increased risks of digestive system cancer, especially in Asian population. Further studies with larger sample size are needed to confirm our current findings.
Table S1. Polymorphic variants of DNA repair genes.
| Polymorphic variant | Chromosome | Intron/exon | Codon | Base substitution |
Amino acid change |
|---|---|---|---|---|---|
| XPC PAT | 3q25 | Intron9 | AT | Intron 9 of XPC contains an 83 bp poly(AT) insertion with a 5 bp deletion of GTAAC | |
| XPC A14187449C rs2228001 | 3p36.3 | Exon15 | 939 | A→C | Lys→Gln |
| XPC C14199887T rs2228000 | 3p37.3 | Exon8 | 499 | C→T | Val→Ala |
Figure S1.
Forest plot of XPC Lys939Gln A>C polymorphism and digestive system cancer risk in dominant model (exclude Long’s article)
Figure S2.
Forest plot of XPC Lys939Gln A>C polymorphism and digestive system cancer risk in homozygote model (exclude Long’s article)
REFERENCES
- 1.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem 1997; 272:23465-8 [DOI] [PubMed] [Google Scholar]
- 2.Sugasawa K, Ng JM, Masutani C, et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell 1998; 2:223-32 [DOI] [PubMed] [Google Scholar]
- 3.Thoma BS, Vasquez KM. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol Carcinog 2003; 38:1-13 [DOI] [PubMed] [Google Scholar]
- 4.Janićijević A, Sugasawa K, Shimizu Y, et al. DNA bending by the human damage recognition complex XPC-HR23B. DNA Repair (Amst) 2003; 2:325-36 [DOI] [PubMed] [Google Scholar]
- 5.Tapias A, Auriol J, Forget D, et al. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J Biol Chem 2004; 279:19074-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magnaldo T, Sarasin A.Xeroderma pigmentosum: from symptoms and genetics to gene-based skin therapy. Cells Tissues Organs 2004; 177:189-98 [DOI] [PubMed] [Google Scholar]
- 7.Vodicka P, Kumar R, Stetina R, et al. Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single-strand breaks in DNA. Carcinogenesis 2004; 25:757-63 [DOI] [PubMed] [Google Scholar]
- 8.Sak SC, Barrett JH, Paul AB, et al. Comprehensive analysis of 22 XPC polymorphisms and bladder cancer risk. Cancer Epidemiol Biomarkers Prev 2006; 15:2537-41 [DOI] [PubMed] [Google Scholar]
- 9.Qiao Y, Spitz MR, Shen H, et al. Modulation of repair of ultraviolet damage in the host-cell reactivation assay by polymorphic XPC and XPD/ERCC2 genotypes. Carcinogenesis 2002; 23:295-9 [DOI] [PubMed] [Google Scholar]
- 10.Blankenburg S, König IR, Moessner R, et al. Assessment of 3 xeroderma pigmentosum group C gene polymorphisms and risk of cutaneous melanoma: a case-control study. Carcinogenesis 2005; 26:1085-90 [DOI] [PubMed] [Google Scholar]
- 11.Broberg K, Bjork J, Paulsson K, et al. Constitutional short telomeres are strong genetic susceptibility markers for bladder cancer. Carcinogenesis 2005; 26:1263-71 [DOI] [PubMed] [Google Scholar]
- 12.Shen H, Sturgis EM, Khan SG, et al. An intronic poly (AT) polymorphism of the DNA repair gene XPC and risk of squamous cell carcinoma of the head and neck: a case-control study. Cancer Res 2001; 61:3321-5 [PubMed] [Google Scholar]
- 13.Vogel U, Overvad K, Wallin H, et al. Combinations of polymorphisms in XPD, XPC and XPA in relation to risk of lung cancer. Cancer Lett 2005; 222:67-74 [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Newcomb PA, Egan KM, et al. Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer. Cancer Epidemiol Biomarkers Prev 2006; 15:353-8 [DOI] [PubMed] [Google Scholar]
- 15.Francisco G, Menezes PR, Eluf-Neto J, et al. XPC polymorphisms play a role in tissue-specific carcinogenesis: a meta-analysis. Eur J Hum Genet 2008; 16:724–34 [DOI] [PubMed] [Google Scholar]
- 16.Qiu L, Wang Z, Shi X, et al. Associations between XPC polymorphisms and risk of cancers: A meta-analysis. Eur J Cancer 2008; 44:2241-53 [DOI] [PubMed] [Google Scholar]
- 17.Zhang D, Chen C, Fu X, et al. A meta-analysis of DNA repair gene XPC polymorphisms and cancer risk. J Hum Genet 2008; 53:18-33 [DOI] [PubMed] [Google Scholar]
- 18.Zheng W, Cong XF, Cai WH, et al. Current evidences on XPC polymorphisms and breast cancer susceptibility: a meta-analysis. Breast Cancer Res Treat 2011; 128:811-5 [DOI] [PubMed] [Google Scholar]
- 19.Dong Z, Guo W, Zhou R, et al. Polymorphisms of the DNA repair gene XPA and XPC and its correlation with gastric cardiac adenocarcinoma in a high incidence population in North China. J Clin Gastroenterol 2008; 42:910-5 [DOI] [PubMed] [Google Scholar]
- 20.Engin AB, Karahalil B, Engin A, et al. Oxidative stress, Helicobacter pylori, and OGG1 Ser326Cys, XPC Lys939Gln, and XPD Lys751Gln polymorphisms in a Turkish population with colorectal carcinoma. Genet Test Mol Biomarkers 2010; 14:559-64 [DOI] [PubMed] [Google Scholar]
- 21.Hansen RD, Sørensen M, Tjønneland A, et al. XPA A23G, XPC Lys939Gln, XPD Lys751Gln and XPD Asp312Asn polymorphisms, interactions with smoking, alcohol and dietary factors, and risk of colorectal cancer. Mutat Res 2007; 619:68-80 [DOI] [PubMed] [Google Scholar]
- 22.Kietthubthew S, Sriplung H, Au WW, et al. Polymorphism in DNA repair genes and oral squamous cell carcinoma in Thailand. Int J Hyg Environ Health 2006; 209:21-9 [DOI] [PubMed] [Google Scholar]
- 23.Li LM, Zeng XY, Ji L, et al. Association of XPC and XPG polymorphisms with the risk of hepatocellular carcinoma. Zhonghua Gan Zang Bing Za Zhi (in Chinese)2010; 18:271-5 [DOI] [PubMed] [Google Scholar]
- 24.Long XD, Ma Y, Huang YZ, et al. Genetic polymorphisms in DNA repair genes XPC, XPD, and XRCC4, and susceptibility to Helicobacter pylori infection-related gastric antrum adenocarcinoma in Guangxi population, China. Mol Carcinog 2010; 49:611-8 [DOI] [PubMed] [Google Scholar]
- 25.Long XD, Ma Y, Zhou YF, et al. Polymorphism in xeroderma pigmentosum complementation group C codon 939 and aflatoxin B1-related hepatocellular carcinoma in the Guangxi population. Hepatology 2010; 52:1301-9 [DOI] [PubMed] [Google Scholar]
- 26.Pan J, Lin J, Izzo JG, et al. Genetic susceptibility to esophageal cancer: the role of the nucleotide excision repair pathway. Carcinogenesis 2009; 30:785-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Jin M, Liu B, et al. The association of XPC polymorphisms and tea drinking with colorectal cancer risk in a Chinese population. Mol Carcinog 2011; 50:189-98 [DOI] [PubMed] [Google Scholar]
- 28.Ye W, Kumar R, Bacova G, et al. The XPD 751Gln allele is associated with an increased risk for esophageal adenocarcinoma: a population-based case-control study in Sweden. Carcinogenesis 2006; 27:1835-41 [DOI] [PubMed] [Google Scholar]
- 29.Zhou RM, Li Y, Wang N, et al. Correlation of XPC Ala499Val and Lys939Gln polymorphisms to risks of esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma. Ai Zheng (in Chinese)2006; 25:1113-9 [PubMed] [Google Scholar]
- 30.Casson AG, Zheng Z, Evans SC, et al. Polymorphisms in DNA repair genes in the molecular pathogenesis of esophageal (Barrett) adenocarcinoma. Carcinogenesis 2005; 26:1536-41 [DOI] [PubMed] [Google Scholar]
- 31.Sugimura T, Kumimoto H, Tohnai I, et al. Gene-environment interaction involved in oral carcinogenesis: molecular epidemiological study for metabolic and DNA repair gene polymorphisms. J Oral Pathol Med 2006; 35:11-8 [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Lin DX, Lu XH, et al. Polymorphisms of the DNA repair genes XRCC1 and XPC: relationship to pancreatic cancer risk. Wei Sheng Yan Jiu (in Chinese)2006; 35:534-6 [PubMed] [Google Scholar]
- 33.Huang WY, Berndt SI, Kang D, et al. Nucleotide excision repair gene polymorphisms and risk of advanced colorectal adenoma: XPC polymorphisms modify smoking-related risk. Cancer Epidemiol Biomarkers Prev 2006; 15:306-11 [DOI] [PubMed] [Google Scholar]
- 34.DerSimonian R, Laird N.Meta-analysis in clinical trials. Control Clin Trials 1986; 7:177-88 [DOI] [PubMed] [Google Scholar]
- 35.Berndt SI, Platz EA, Fallin MD, et al. Genetic variation in the nucleotide excision repair pathway and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev 2006; 15:2263-9 [DOI] [PubMed] [Google Scholar]
- 36.Cai XL, Gao YH, Yu ZW, et al. A 1:1 matched case-control study on the interaction between HBV, HCV infection and DNA repair gene XPC Ala499Val, Lys939Gln for primary hepatocellular carcinoma. Zhonghua Liu Xing Bing Xue Za Zhi (in Chinese)2009; 30:942-5 [PubMed] [Google Scholar]
- 37.Guo W, Zhou RM, Wan LL, et al. Polymorphisms of the DNA repair gene xeroderma pigmentosum groups A and C and risk of esophageal squamous cell carcinoma in a population of high incidence region of North China. J Cancer Res Clin Oncol 2008; 134:263-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan SG, Muniz-Medina V, Shahlavi T, et al. The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function. Nucleic Acids Res 2002; 30:3624-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sak SC, Barrett JH, Paul AB, et al. The polyAT, intronic IVS11-6 and Lys939Gln XPC polymorphisms are not associated with transitional cell carcinoma of the bladder. Br J Cancer 2005; 92:2262-5 [DOI] [PMC free article] [PubMed] [Google Scholar]