Abstract
Objective
To investigate Leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5) expressions in gastric cancer and to evaluate its clinical significance.
Methods
LGR5 expression was assessed by immunohistochemistry in 257 gastric cancer patients after surgery. The relationships between LGR5 expression and clinicopathological features and patients prognosis were statistically analyzed.
Results
The expression of LGR5 was significantly higher in gastric cancers as a cancer stem cell marker than in adjacent normal tissues (P<0.001), and more frequently in patients with intestinal type, well-moderate differentiation and stage I and II (P<0.05). Although we found gastric cancer patients with LGR5 positive expression had a poorer prognosis, it didn’t meet statistical significance (P>0.05). LGR5 negative expression was significantly related to the favorable overall survival in stage I and II gastric cancer patients (P<0.05). Furthermore, patients with high LGR5 expression tended to be more likely to get progression and have poorer progress-free survival (P<0.05). Multivariate Cox regression analysis revealed that LGR5 expression was an independent factor of overall survival for the patients with stage I and II gastric cancer (P<0.05).
Conclusions
Our results show that LGR5 may play an important role in tumorigenesis and progression and would be a powerful marker to predict the prognosis of patients with stage I and II gastric cancer.
Key Words: Leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5), gastric cancer, cancer stem cell, biomarker, prognosis
Introduction
Gastric cancer (GC) is the fourth most prevalent malignant tumor worldwide and the second most frequent cause of cancer death (1). Although the overall prognosis of gastric cancer has gradually improved over the past decades, the survival of gastric cancer patients is only 20-25% in China, USA and Europe generally for delayed first diagnosis, whereas Japanese patients have much better survival (>50%) because of its early screening programs (2). About 80-90% of all gastric cancer patients are either diagnosed at an advanced stage when the tumor is inoperable, or develop recurrence in few years after surgery, and the 5-year survival rate of these patients is very poor (3,4). One of the crucial problems is that the majority of present progression, metastatic disease at diagnosis and predicting markers remain the cornerstone for gastric cancer patients, especially in early stage. Therefore, it is necessary to develop new biomarkers with the potential to estimate the efficacy of individual therapeutic strategies and to make optimistic management.
Leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5), also known as GPR49, a target of Wnt signaling, is a potential marker of human intestinal stem cells and could also be a new cancer stem cell (CSC) marker (5,6). It has been demonstrated to be involved in different human cancer entities, including hepatocellular carcinoma, basal cell carcinoma, endometrial cancer, colon cancer, ovarian cancer and so on (7-10). LGR5 was identified to be expressed on crypt stem cells (precursor cells) as well as lesions which had progressed to cancer. It always appeared at the base of prospective corpus and pyloric glands in neonatal stomach, and predominantly restricted to the base of mature pyloric glands in the adult (11,12).
Although previous studies indicated that LGR5 plays a vital role in several cancers, there is little known about the relationship between LGR5 and clinical characters of gastric cancers. In our study, we analyzed LGR5 expression in gastric cancers after surgery by immunohistochemistry and evaluated the relationship with clinicopathological features and prognosis.
Materials and methods
Patients and tissue samples
Two hundred and fifty-seven gastric cancer patients who were diagnosed and underwent surgical treatment in Peking University Cancer Hospital between January, 2002 and December, 2007 were included in our study. A total of 236 patients had matched gastric cancer and adjacent normal tissues. None of the patients had been administered by chemotherapy or radiotherapy prior to undergoing surgical resection. None of them had synchronous cancers. Clinical stage of gastric cancer was assessed on the basis of the tumor node metastasis (TNM) classification system recommended by the American Joint Committee on Cancer stage (7th edition) and JGCA guidelines. For accurate N staging, more than 15 lymph nodes in one patient were collected by means of careful examinations. Demographic and clinicopathological characteristics of patients were collected from our hospital electronic patient records. All samples were taken from surgically resected materials, and immediately formalin-fixed for following experiments. Follow-up data were retrieved from hospital records by interview, telephone or letters. The follow-up time started from the day of primary tumor operation. The end point for the disease-associated overall survival analysis was the time of death of the patient or our last review. The end point for the progress-free survival analysis was the first recurrence, progression or death. This investigation was approved by the Ethics Committee of Peking University Cancer Hospital. Informed consent was obtained from each patient.
Immunohistochemical analysis
All of the cancer and adjacent normal tissues of gastric cancers were formalin-fixed and paraffin-embedded immediately. Each paraffin block was cut at 4 µm in thickness. Sections were mounted on poly-lysine-coated slides, deparaffinized in xylene, and rehydrated through descending concentrations of ethanol series and ultimately distilled water. Blocking of endogenous peroxidases was accomplished by incubating sections in 3% hydrogen peroxide for 10 min. Antigen retrieval was performed using EDTA buffer (Zhongshan Biotechnology Inc., Beijing, China) heated in a pressure cooker for 5 min and then cooled to room temperature. 10% goat blood serum was used to prevent non-specific binding. Thereafter, LGR5 purified rabbit polyclonal antibody (AP2745d, Abgent, San Diego, CA, USA; at 1:10 dilution, according to the manufacturer’s instructions) was incubated with sections overnight at 4 °C. Immunostaining was performed by using two-step diaminobenzidine visualization (GK500705, Dako, Glostrup, Denmark). Sections were counterstained with hematoxylin for 40 s, rinsed in water, dehydrated in ascending concentrations of ethanol followed by clearance with xylene, and cover slipped permanently for light microscopy. Negative controls were carried out with the same procedure without primary antibody. Repeatedly validated esophageal adenocarcinoma specimens with LGR5 positive served as positive controls.
Evaluation of immunostaining
Histopathological sections were microscopically examined and scored by independent practiced pathologists who were blind to the clinical data pertaining to the patients. The evaluation was analyzed according to both the percentage of positive cells and the intensity of cytoplasmic staining. Intensity of staining was graded on a scale of 0 to 3, with 0 record as no staining, 1 as mild intensity, 2 as moderate intensity and 3 as severe intensity. The highest intensity score was assigned when a threshold of >10% of cells stained with that intensity. Percentage of stained cells was also scored on a scale from 0 to 3, where immunoreactivity was scored 0 if no carcinoma cells stained positive (negative, -), 1 if 1% to 25% were positive, 2 if 25% to 50% were positive, and 3 if >50% were positive. A composite expression score [0-6] was obtained by adding the intensity and percentage scores, with 1-2 recorded as weak positive [+], 3-4 as moderate positive [++], and 5-6 as strong positive [+++]. In the statistical analysis, 0 was ranked as negative, and 1-6 was recorded as positive; 0-2 was ranked as low expression, and 3-6 were recorded as high expression.
Statistical analysis
Statistical analyses were carried out using the SPSS software version 19.0 statistical package (SPSS Inc., Chicago, IL, USA). The differences of LGR5 expression between gastric cancer and adjacent normal tissues were analyzed by χ2 test or Fisher’s exact test. The interrelationship between gene expression level and patients’ clinicopathological characteristics was tested with the Spearman’s rank correlation analysis. Survival curves were fitted with the Kaplan-Meier method and the differences in survival assessed by the log rank test. The effect of different factors on patient survival was performed by multivariate analysis with Cox proportional hazards regression model. P-values <0.05 (two-sided) were considered as statistically significant.
Results
Demographics of patients with gastric cancer
A total of 257 patients with gastric cancer were eventually included in this study, including 185 males and 72 females. Age of patients at surgery ranged from 22 to 87 (Median age was 61) years old. As to Lauren type, there were 149 intestinal types, 78 diffuse type and 24 mixed type gastric cancers. According to TNM classification, 89 patients were classified into stage I and II, while 156 patients were classified into stage III and IV.
LGR5 expression in gastric cancer and adjacent normal tissues by immunohistochemical analysis
We examined LGR5 expression levels in 236 pair samples of primary gastric cancer and adjacent normal tissues using immunohistochemical analysis. The results showed that LGR5 protein was predominantly localized in the cytoplasm or on cell membrane. The LGR5 expression between gastric cancer and adjacent normal tissues were significantly different (P<0.001), for its staining percentage in carcinoma tissues was 50.0% (118/236), compared that the adjacent normal tissues were lower (37.3%, 88/236) (Figure 1A,B,C,D). And we also found that LGR5 positive cells were present at the base of normal human gastric crypts (Figure 1H).
Figure 1.
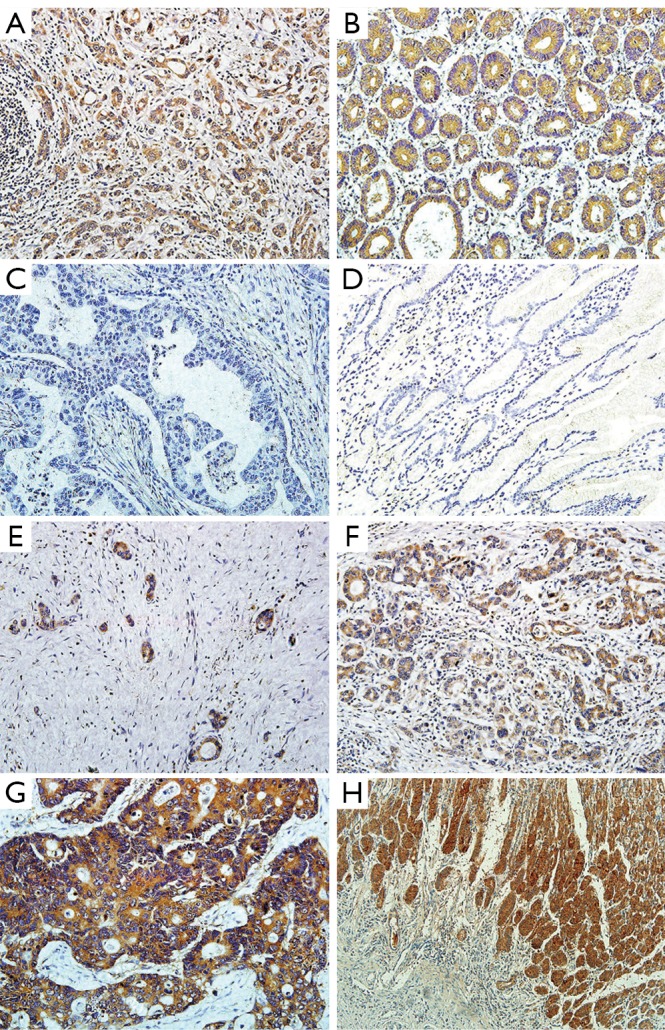
Representative immunohistochemical staining of LGR5 in gastric cancer and adjacent normal tissues. A and B show LGR5 positive staining in gastric cancer and adjacent normal tissues, respectively (magnification ×200). C and D show LGR5 negative staining in gastric cancer and adjacent normal tissues, respectively (magnification ×200). E, F and G show LGR5 staining in gastric cancer tissues with weak, moderate and strong expression (magnification ×200). H shows that LGR5+ cells are present at the base of normal human gastric crypts (magnification ×100)
Association of immunohistochemical expression of LGR5 with clinicopathological features
Among these 257 patients, LGR5 positive tumor cells were found in 133 (51.8%) patients, with 124 (48.2%) tumors lacked LGR5 immunoreactivity. Strong cytoplasmic or membranous immunoreaction was observed in 17 (6.6%) case, moderate staining in 43 (16.8%) cases and weak staining in 73 (28.4%) cases (Figure 1E,F,G).
In order to investigate the clinical significance of LGR5 expression, the relationship between LGR5 expression and clinicopathological features in gastric cancers was further assessed. As shown in Table 1, LGR5 expression appeared to be significantly associated with gender (P=0.022), age (P=0.001), Lauren type (P=0.015), differentiation (P=0.001) and TNM stage (I+II vs. III+IV, P=0.033) in correlation analysis, but not correlated with tumor site, size, histology, lymphovascular invasion, depth of invasion, lymph node metastasis, or distant metastasis statistically (P>0.05).
Table 1. Association of LGR5 expression with clinicopathological features.
| Clinicopathological features | LGR5 expression |
P | |
|---|---|---|---|
| Positive (%) | Negative (%) | ||
| Gender | |||
| Male | 104 (56.2%) | 81 (43.8%) | 0.022 |
| Female | 29 (40.3%) | 43 (59.7%) | |
| Age, year | |||
| ≤60 | 52 (40.9%) | 75 (59.1%) | 0.001 |
| >60 | 81 (62.3%) | 49 (37.7%) | |
| Tumor site | |||
| Upper | 33 (60.0%) | 22 (40.0%) | 0.194 |
| Middle | 25 (48.1%) | 27 (51.9%) | |
| Low | 66 (51.2%) | 63 (48.8%) | |
| Tumor size | |||
| ≤4 cm | 61 (56.5%) | 47 (43.5%) | 0.352 |
| >4 cm | 50 (50.0%) | 50 (50.0%) | |
| Lauren | |||
| Intestinal | 88 (59.1%) | 61 (40.9%) | 0.015 |
| diffuse | 27 (34.6%) | 51 (65.4%) | |
| mixed | 14 (58.3%) | 10 (41.7%) | |
| Differentiation | |||
| Well-Moderate | 73 (63.2%) | 42 (36.8%) | 0.001 |
| Poor | 53 (41.4%) | 75 (58.6%) | |
| Histology | |||
| Adenocarcinoma | 104 (53.9%) | 89 (46.1%) | 0.191 |
| Other types | 27 (44.3%) | 34 (55.7%) | |
| Lymphovascular invasion | |||
| Absent | 74 (54.8%) | 61 (45.2%) | 0.273 |
| Present | 57 (47.9%) | 62 (52.1%) | |
| Depth of invasion | |||
| T1+T2+T3 | 80 (57.1%) | 60 (42.9%) | 0.068 |
| T4 | 52 (45.6%) | 62 (54.4%) | |
| Lymph node metastasis | |||
| N0 | 28 (53.8%) | 24 (46.2%) | 0.744 |
| N1+N2+N3 | 100 (51.3%) | 95 (48.7%) | |
| Distant metastasis | |||
| No | 111 (52.4%) | 101 (47.6%) | 0.570 |
| Yes | 17 (47.2%) | 19 (52.8%) | |
| TNM stage | |||
| I+II | 54 (60.7%) | 35 (39.3%) | 0.033 |
| III+IV | 74 (46.5%) | 85 (53.5%) | |
Correlation of LGR5 expression with prognosis of gastric cancer patients after surgery
Survival analysis showed that patients with LGR5 positive expression had a poorer prognosis than those with LGR5 negative expression, but didn’t meet statistical significance (5-year survival rate, 50.8% vs. 39.0%, P=0.891). Similar result was found in different scores of LGR5 expression (P>0.05). For the other features, survival status was significantly correlated with whether cardiac cancer (P=0.029), Lauren type (P=0.015), differentiation (P=0.046), lymphovascular invasion (P=0.000), depth of invasion (P=0.000), lymph node metastasis (P=0.002), distant metastasis (P=0.000) and TNM stage (P=0.000), but not the others. Detailed data are shown in Table 2. Although in Cox univariate analysis, whether cardiac cancer (P=0.031), differentiation (P=0.049), lymphovascular invasion (P=0.000), depth of invasion (P=0.001), lymph node metastasis (P=0.003), distant metastasis (P=0.000) and TNM stage (P=0.000) were associated with overall survival, but in multivariate analysis, only lymph node metastasis, distant metastasis and TNM stage were independent prognostic factors (P=0.020, 0.001 and 0.000, respectively), as shown in Table 3.
Table 2. Overall survivals related to several clinicopathological characteristics with gastric cancer patients.
| Variables | Patients | Events | Median survival, months (95% CI) | Log rank test |
|---|---|---|---|---|
| Gender | ||||
| Male | 184 | 78 | 38.0±6.5 (25.3-50.7) | 0.400 |
| Female | 72 | 26 | NA | |
| Age, year | ||||
| ≤60 | 127 | 46 | 47.3 | 0.349 |
| >60 | 129 | 48 | 36.0±5.1 (25.9-46.1) | |
| Tumor site | ||||
| (I) Upper | 55 | 26 | 38.0±8.9 (20.6-55.4) | 0.537 |
| Middle | 52 | 20 | NA | |
| Low | 129 | 52 | NA | |
| (II) Cardiac cancer | 49 | 26 | 26.5±4.8 (17.2-36.0) | 0.029 |
| Non-cardiac cancer | 207 | 78 | NA | |
| Tumor size | ||||
| ≤4 cm | 108 | 37 | NA | 0.062 |
| >4 cm | 100 | 52 | 31.1±3.1 (25.1-37.1) | |
| Lauren | ||||
| Intestinal | 148 | 56 | 41.5±5.5 (30.8-52.2) | 0.015 |
| diffuse | 78 | 39 | 28.0±8.1 (12.1-43.9) | |
| mixed | 24 | 7 | NA | |
| Differentiation | ||||
| Well-Moderate | 114 | 41 | 47.3 | 0.046 |
| Poor | 127 | 56 | 34.3 | |
| Histology | ||||
| Adenocarcinoma | 192 | 83 | 38.0±5.6 (26.9-49.1) | 0.479 |
| Other types | 61 | 20 | NA | |
| Lymphovascular invasion | ||||
| Absent | 135 | 40 | NA | 0.000 |
| Present | 118 | 63 | 25.0±3.2 (18.6-31.4) | |
| Depth of invasion | ||||
| T1+T2+T3 | 139 | 45 | NA | 0.000 |
| T4 | 114 | 59 | 28.0±2.8 (22.5-33.5) | |
| Lymph node metastasis | ||||
| N0 | 52 | 10 | NA | 0.002 |
| N1+N2+N3 | 194 | 92 | 31.8±3.0 (25.9-37.7) | |
| Distant metastasis | ||||
| No | 211 | 73 | NA | 0.000 |
| Yes | 36 | 30 | 12.0±1.5 (9.1-14.9) | |
| TNM stage | ||||
| I+II | 89 | 12 | NA | 0.000 |
| III+IV | 158 | 91 | 25.0±3.3 (18.5-31.5) | |
| LGR5 | ||||
| (I) Negative (-) | 123 | 50 | NA | 0.872 |
| Weak positive (+) | 73 | 29 | 47.3±11.6 (24.5-70.1) | |
| Moderate positive (++) | 43 | 19 | 38.0±5.0 (28.3-47.7) | |
| Strong positive (+++) | 17 | 6 | NA | |
| (II) Negative | 123 | 50 | NA | 0.891 |
| Positive | 133 | 54 | 38.0±4.0 (30.2-45.8) | |
| (III) Low expression | 196 | 79 | 47.3 | 0.947 |
| High expression | 60 | 25 | 38.0±4.5 (29.1-46.9) |
NA, because the overall survival rates of these patients were more than 50%, we didn’t get the median survival time
Table 3. Cox proportional hazard regression model analysis of all the patients.
| Variables | Univariate analysis |
Multivariate analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Relative risk | 95% CI | P | Relative risk | 95% CI | P | |||||
| Gender | ||||||||||
| Male vs. female | 0.827 | 0.530-1.289 | 0.402 | |||||||
| Age, years | ||||||||||
| ≤60 vs. >60 | 1.243 | 0.816-1.771 | 0.351 | |||||||
| Site | ||||||||||
| Non-cardiac cancer vs. cardiac cancer | 1.630 | 1.045-2.542 | 0.031 | |||||||
| Size | ||||||||||
| ≤4 cm vs. >4 cm | 1.489 | 0.977-2.270 | 0.064 | |||||||
| Lauren | ||||||||||
| Intestinal | 1.606 | 0.732-3.526 | 0.237 | |||||||
| diffuse | 2.582 | 1.154-5.777 | 0.021 | |||||||
| mixed | 1.000 | 0.018 | ||||||||
| Differentiation | ||||||||||
| Well-moderate vs. poor | 1.501 | 1.002-2.247 | 0.049 | |||||||
| Lymphovascular invasion | ||||||||||
| Absent vs. present | 2.370 | 1.593-3.527 | 0.000 | |||||||
| Depth of invasion | ||||||||||
| T1+T2 vs. T3+T4 | 3.935 | 1.725-8.974 | 0.001 | |||||||
| Lymph node metastasis | ||||||||||
| N0 vs. N1+N2+N3 | 2.737 | 1.425-5.258 | 0.003 | 0.280 | 0.098-0.801 | 0.020 | ||||
| Distant metastasis | ||||||||||
| No vs. Yes | 5.008 | 3.235-7.754 | 0.000 | 2.701 | 1.602-4.554 | 0.001 | ||||
| TNM stage | ||||||||||
| I+II vs. III+IV | 6.215 | 3.393-11.384 | 0.000 | 11.364 | 3.980-34.012 | 0.000 | ||||
| LGR5 expression | ||||||||||
| Negative vs. positive | 0.974 | 0.662-1.431 | 0.892 | |||||||
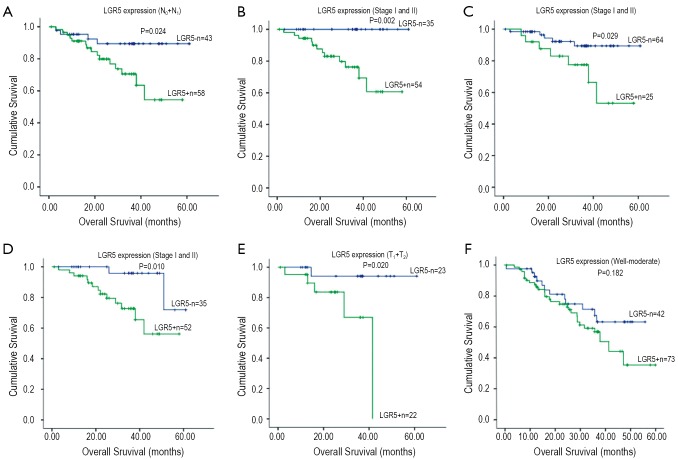
Furthermore, we investigated the subgroups of gastric cancer patients. In N0-1 group, the overall survival time between LGR5 positive and negative was significantly different (5-year survival rate, 54.4% vs. 89.4%, P=0.024, Figure 2A). In TNM stage I-II group, a detailed analysis of prognoses further revealed that patients with LGR5 positive lived shorter after surgery compared with negative ones (5-year survival rate, 100% vs. 60.6%, P=0.002, Figure 2B). The similar result was found in patients with high LGR5 expression (P=0.029, Figure 2C). Meanwhile, the expression of LGR5 was correlated with progression after surgery (P=0.011), and the rates increased with the score of LGR5 expression. Moreover, LGR5 positive patients had shorter progress-free survival time than the negative ones since surgery (P=0.010, Figure 2D). In T1-2 group, patients had dramatically different prognosis according to different LGR5 expression (LGR5- vs. LGR5+, 94.1% vs. 0%, P=0.020, Figure 2E). In well-moderate differentiation group, the overall survival time of LGR5 positive patients was shorter than negative patients (63.2% vs. 36.2%), but didn’t reach statistical difference (P>0.05, Figure 2F). The other different subgroups may have similar survival results (P>0.05). In Cox regression univariate analysis, LGR5 expression was associated with overall survival (P=0.039), and lymph node metastasis and whether cardiac cancer nearly get statistically significant (P=0.088 and 0.094, respectively). However, LGR5 expression and lymph node metastasis were independent factors for stage I and II gastric cancer patients in multivariate analysis (P=0.020 and 0.038, respectively). Detailed data are shown in Table 4.
Figure 2.
A. Kaplan-Meier overall survival analysis of gastric cancer patients with less than 3 lymph node metastasis (N0 + N1). Survival analysis was performed according to the expression status of LGR5; B,C. Kaplan-Meier overall survival analysis of gastric cancer patients in stage I and II. Survival analysis was performed according to the expression status of LGR5: B (Negative vs. Positive), C (Low vs. High expression); D. Kaplan-Meier progress-free survival analysis of gastric cancer patients in stage I and II. Survival analysis was performed according to the expression status of LGR5 (Negative vs. Positive); E. Kaplan-Meier overall survival analysis of gastric cancer patients with under muscularis propria invasion (T1 + T2). Survival analysis was performed according to the expression status of LGR5; F. Kaplan-Meier overall survival analysis of gastric cancer patients with well to moderate differentiation (well-moderate). Survival analysis was performed according to the expression status of LGR5
Table 4. Cox proportional hazard regression model analysis of gastric cancer patients with stage I and II.
| Variables | Univariate analysis |
Multivariate analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Relative risk | 95% CI | P | Relative risk | 95% CI | P | |||||
| Gender | ||||||||||
| Male vs. female | 0.663 | 0.144-3.047 | 0.597 | |||||||
| Age, years | ||||||||||
| ≤60 vs. >60 | 1.509 | 0.454-5.021 | 0.502 | |||||||
| Site | ||||||||||
| Non-cardiac cancer vs. cardiac cancer | 2.718 | 0.862-8.576 | 0.088 | |||||||
| Lauren type | ||||||||||
| Intestinal vs. other types | 0.421 | 0.092-1.931 | 0.266 | |||||||
| Differentiation | ||||||||||
| Well-Moderatevs. poor | 1.449 | 0.466-4.502 | 0.522 | |||||||
| Lymphovascular invasion | ||||||||||
| Absent vs. present | 0.770 | 0.168-3.525 | 0.736 | |||||||
| Depth of invasion | ||||||||||
| T1+T2 vs. T3+T4 | 1.344 | 0.402-4.489 | 0.631 | |||||||
| Lymph node metastasis | ||||||||||
| Absent vs. present | 0.327 | 0.088-1.212 | 0.094 | 0.274 | 0.073-1.028 | 0.038 | ||||
| TNM stage | ||||||||||
| I vs. II | 1.758 | 0.385-8.039 | 0.467 | |||||||
| LGR5 expression | ||||||||||
| Low expression vs. high expression | 3.346 | 1.062-10.544 | 0.039 | 3.971 | 1.242-12.699 | 0.020 | ||||
Discussion
To develop sensitive and specific molecular biomarkers for tailored management of cancers is a major challenge in clinical oncology, including gastric cancer for its poor outcome (13,14). Using standard immunostaining, we have demonstrated that LGR5 expression was increased in gastric cancers compared with adjacent normal tissues, suggesting that LGR5 may serve as an important biomarker for higher risk of tumor genesis, supported by Fan et al. in colorectal cancers (15). And we also found that LGR5 positive cells were present at the base of normal human gastric crypts, while this distribution is similar to the report in mice and consistent with the known location of the stem cell niche (16-18).
However, Yamamoto et al. found no significant association between expression of LGR5 mRNA and clinicopathologic features in hepatocellular carcinomas (7). We investigated that LGR5 expression was significantly correlated with Lauren types, differentiation and TNM stage and were more frequent in well to moderate differentiation, intestinal type and lower-staged (I-II). Similar reports demonstrated that the expression of LGR5 in colorectal and ovarian carcinomas was higher in stage I-II and decreased in stage III-IV (10). In mouse endometrial cancer model, LGR5 is highly expressed in the epithelium during the initial stages of tumorigenesis, but is dramatically down-regulated in full-grown tumors (9). These hypotheses correspond with our finding that the overexpression of LGR5 may be an early event in tumorigenesis. Therefore, we presume that LGR5 expression occurs early in progression because of fine growth characteristics and favorable proliferations. When normal gastric mucosa changes into intestinal metaplasia or cancer, the LGR5 positive cancer cell may have powerful capability to efficiently generate, differentiate, develop and progress like cancer stem cells. However, when cancer cells get full grown-up, the reproductive activity apparently decline following lower LGR5 expression as the records. Although it happens almost like this, the exact functions with mechanism remain to be determined.
Although we didn’t find LGR5 expression have a statistically prognostic impact for all gastric cancer patients as Eva Simon (19) shown, we still investigated that LGR5 positive ones appeared to live shorter after surgery. It may due to much more advanced gastric cancers in our cohort which express LGR5 much lower. Furthermore, LGR5 negative expression tended to be more favorable prognostic factor for the patients with N0-1, T1-2 and stage I-II. For stage I and II, LGR5 negative patients were even found no death, and the similar result was found in progress-free survival. Based on our immunohistochemical assessment, analysis of LGR5 expression may provide more accurate information for predicting progression and prognosis for stage I and II gastric cancer patients after surgery. High expression of LGR5 had shorter progress-free survival in lower-staged gastric cancer patients (stage I and II), and it could be an independent factor of prognosis for these patients. In accordance with our results, the expression of LGR5 was significantly associated with poor prognosis for disease-free survival in colorectal cancer patients (20). These data suggest that LGR5 expression may be associated with the malignant potential of gastric cancer, including the appearance of progression in stage I and II patients, and would offer a clue to predict the prognosis of these people, but it will push towards our following cohort-based study.
LGR5/GPR49 is localized to banded DNA in chromosomal 12q22-23, originally isolated as a leucine-rich, orphan G-protein-coupled, seven-transmembrane receptor, and is a member of the G-protein-coupled receptor (GPCR) subfamily, including thyroid-stimulating hormone (TSH), follicle-stimulating hormone (FSH) and luteinizing hormone (LH) receptors (21,22). In adult mice, LGR5 expression is restricted to rare, scattered cells in the intestine, eye, brain, stomach, mammary gland and reproductive organs (23). More and more studies revealed the function of LGR5 both during development and cancer progression. LGR5 is often co-expressed in Wnt- driven proliferative compartments. Conditional deletion of LGR5 gene in the mouse gut impairs Wnt target gene expression and results in the rapid demise of intestinal crypts, thus phenocopying Wnt pathway inhibition (24). Colon cancer cells with high Wnt activity express stem cell markers, whereas cells with low Wnt activity express higher levels of differentiation markers and are unable to give rise to new tumors (25). In addition, tumorigenesis in mice is much more effective in LGR5 positive stem cells as compared to more differentiated cells (26). In this light, it is interesting to note that LGR5 is a Wnt target gene and could also be a cancer stem cell marker (6). By now, we had already discovered these cancer stem cells in hematological malignancies and several solid tumors, like breast, lung, ovarian, liver, prostate, pancreas, skin, brain and colon cancers (27-35). Some studies have already focused on the effects of different LGR5 expression in the context of tumor development and progression, and it was identified to be expressed on crypt stem cells (precursor cells) which had progressed to cancer (11). Many of the Wnt response genes modulated by LGR5 expression are linked to EMT, MMPs, collagens, fibronectin, wnt5a and FGF4 (36). These changes in expression pattern are associated with alterations in anchorage-independent proliferation, invasion, migration, cell adhesion, tumorigenicity and tumor morphology with opposing phenotypes of LGR5 expression.
Moreover, growing body of literatures detected the correlation between LGR5 expression and clinicopatological features of several cancer patients. In colorectal cancers, increases of LGR5 expression in putative stem cells occur early during colorectal tumorigenesis, shifts in their distribution towards the crypt bottom and/or invasive tumor front might play a role in the development and progression of colorectal cancer and up-regulation of LGR5 was found in the early stage of colorectal tumorigenesis (37,38). Persistent activation of LGR5 in intestinal metaplasia and esophageal adenocarcinomas (EACs) may sustain multistep carcinogenesis and high LGR5 expressions in EACs were associated with worse survival (39,40). The LGR5 overexpressions in hepatocellular carcinomas with well to moderate differentiation were more frequent than in poor differentiation, although there was no statistical significance (7). These results highlight the importance of LGR5 as a potential marker for many cancer studies.
Although we have identified the LGR5 expression in gastric cancers and find some useful information especially in lower-staged cancers, our cohort size is small and randomly selected from cases spanning six years, during which time changes in therapies may have introduced bias in the survival data. Because of only assessed gastric cancer tissues after surgery without previous therapy, we omitted individuals with complete response following neoadjuvant chemotherapy and radiotherapy. It is necessary to plan and perform a larger prospective further validated study with more sensibility and accuracy. However, LGR5 is not simply as a marker of tumor cells, but as a regulator of Wnt responses, cell motility and cell-cell adhesion. It will be a promising biomarker for the management of premalignant and malignant lesions especially for stage I and II gastric cancer patients.
In conclusion, we have already analyzed the expression of a promising cancer stem cell gene, LGR5 in gastric caners and the relationship with characteristic features. The LGR5 may play an important role in tumorigenesis and progression, and would be a powerful marker to predict the prognosis of stage I and II gastric cancer patients. Further studies are needed to verify the impact of LGR5 expression on gastric cancers, including prospective cohorts, multicenter studies, and functional experiments, and find more proper techniques for detection. Therefore, LGR5 could be a candidate target for future diagnosis and tailored therapy, if its potential clinical utilization is confirmed.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90 [DOI] [PubMed] [Google Scholar]
- 2.Hartgrink HH, Jansen EP, van Grieken NC, et al. Gastric cancer. Lancet 2009;374:477-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelley JR, Duggan JM. Gastric cancer epidemiology and risk factors. J Clin Epidemiol 2003;56:1-9 [DOI] [PubMed] [Google Scholar]
- 4.Wagner AD, Grothe W, Haerting J, et al. Chemotherapy in advanced gastric cancer: a systematic review and meta-analysis based on aggregate data. J Clin Oncol 2006;24:2903-9 [DOI] [PubMed] [Google Scholar]
- 5.Pinson KI, Brennan J, Monkley S, et al. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 2000;407:535-8 [DOI] [PubMed] [Google Scholar]
- 6.Van der Flier LG, Sabates-Bellver J, Oving I, et al. The Intestinal Wnt/TCF Signature. Gastroenterology 2007;132:628-32 [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto Y, Sakamoto M, Fujii G, et al. Overexpression of orphan G-protein-coupled receptor, Gpr49, in human hepatocellular carcinomas with beta-catenin mutations. Hepatology 2003;37:528-33 [DOI] [PubMed] [Google Scholar]
- 8.Tanese K, Fukuma M, Yamada T, et al. G-protein-coupled receptor GPR49 is up-regulated in basal cell carcinoma and promotes cell proliferation and tumor formation. Am J Pathol 2008;173:835-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun X, Jackson L, Dey SK, et al. In pursuit of leucine-rich repeat-containing G protein-coupled receptor-5 regulation and function in the uterus. Endocrinology 2009;150:5065-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClanahan T, Koseoglu S, Smith K, et al. Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol Ther 2006;5:419-26 [DOI] [PubMed] [Google Scholar]
- 11.Brabletz S, Schmalhofer O, Brabletz T.Gastrointestinal stem cells in development and cancer. J Pathol 2009;217:307-17 [DOI] [PubMed] [Google Scholar]
- 12.Barker N, Huch M, Kujala P, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 2010;6:25-36 [DOI] [PubMed] [Google Scholar]
- 13.Filomena A, Saieva C, Lucchetti V, et al. Gastric cancer surveillance in a high-risk population in tuscany (Central Italy): preliminary results. Digestion 2011;84:70-7 [DOI] [PubMed] [Google Scholar]
- 14.McCune K, Bhat-Nakshatri P, Thorat MA, et al. Prognosis of hormone-dependent breast cancers: implications of the presence of dysfunctional transcriptional networks activated by insulin via the immune transcription factor T-bet. Cancer Res 2010;70:685-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan XS, Wu HY, Yu HP, et al. Expression of Lgr5 in human colorectal carcinogenesis and its potential correlation with beta-catenin. Int J Colorectal Dis 2010;25:583-90 [DOI] [PubMed] [Google Scholar]
- 16.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007;449:1003-7 [DOI] [PubMed] [Google Scholar]
- 17.Bjerknes M, Cheng H.Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology 1999;116:7-14 [DOI] [PubMed] [Google Scholar]
- 18.Mills JC, Gordon JI. The intestinal stem cell niche: there grows the neighborhood. Proc Natl Acad Sci U S A 2001;98:12334-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon E, Petke D, Böger C, et al. The spatial distribution of LGR5+ cells correlates with gastric cancer progression. PLoS One 2012;7:e35486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi H, Ishii H, Nishida N, et al. Significance of Lgr5(+ve) cancer stem cells in the colon and rectum. Ann Surg Oncol 2011;18:1166-74 [DOI] [PubMed] [Google Scholar]
- 21.Hsu SY, Liang SG, Hsueh AJ. Characterization of two LGR genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein-coupled, seven-transmembrane region. Mol Endocrinol 1998;12:1830-45 [DOI] [PubMed] [Google Scholar]
- 22.McDonald T, Wang R, Bailey W, et al. Identification and cloning of an orphan G protein-coupled receptor of the glycoprotein hormone receptor subfamily. Biochem Biophys Res Commun 1998;247:266-70 [DOI] [PubMed] [Google Scholar]
- 23.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature 2006;439:84-8 [DOI] [PubMed] [Google Scholar]
- 24.de Lau W, Barker N, Low TY, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 2011;476:293-7 [DOI] [PubMed] [Google Scholar]
- 25.Vermeulen L, De Sousa E. Melo F, van der Heijden M, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol 2010;12:468-76 [DOI] [PubMed] [Google Scholar]
- 26.Barker N, Ridgway RA, van Es JH, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 2009;457:608-11 [DOI] [PubMed] [Google Scholar]
- 27.Ponti D, Costa A, Zaffaroni N, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 2005;65:5506-11 [DOI] [PubMed] [Google Scholar]
- 28.Eramo A, Lotti F, Sette G, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ 2008;15:504-14 [DOI] [PubMed] [Google Scholar]
- 29.Curley MD, Therrien VA, Cummings CL, et al. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem Cells 2009;27:2875-83 [DOI] [PubMed] [Google Scholar]
- 30.Yang ZF, Ho DW, Ng MN, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 2008;13:153-66 [DOI] [PubMed] [Google Scholar]
- 31.Collins AT, Berry PA, Hyde C, et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005;65:10946-51 [DOI] [PubMed] [Google Scholar]
- 32.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res 2007;67:1030-7 [DOI] [PubMed] [Google Scholar]
- 33.Monzani E, Facchetti F, Galmozzi E, et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer 2007;43:935-46 [DOI] [PubMed] [Google Scholar]
- 34.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 2003;63:5821-8 [PubMed] [Google Scholar]
- 35.Vermeulen L, Todaro M, de Sousa Mello F, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A 2008;105:13427-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker F, Zhang HH, Odorizzi A, et al. LGR5 is a negative regulator of tumourigenicity, antagonizes Wnt signalling and regulates cell adhesion in colorectal cancer cell lines. PLoS One 2011;6:e22733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeda K, Kinoshita I, Shimizu Y, et al. Expression of LGR5, an intestinal stem cell marker, during each stage of colorectal tumorigenesis. Anticancer Res 2011;31:263-70 [PubMed] [Google Scholar]
- 38.Uchida H, Yamazaki K, Fukuma M, et al. Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in colorectal cancer. Cancer Sci 2010;101:1731-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Rahden BH, Kircher S, Lazariotou M, et al. LgR5 expression and cancer stem cell hypothesis: clue to define the true origin of esophageal adenocarcinomas with and without Barrett’s esophagus? J Exp Clin Cancer Res 2011;30:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker L, Huang Q, Mashimo H.Lgr5, an intestinal stem cell marker, is abnormally expressed in Barrett’s esophagus and esophageal adenocarcinoma. Dis Esophagus 2010;23:168-74 [DOI] [PubMed] [Google Scholar]