Abstract
Breakthroughs in molecular profiling technologies are enabling a new data-intensive approach to biomedical research, with the potential to revolutionize how we study, manage, and treat complex diseases. The next great challenge for clinical applications of these innovations will be to create scalable computational solutions for intelligently linking complex biomedical patient data to clinically actionable knowledge. Traditional database management systems (DBMS) are not well suited to representing complex syntactic and semantic relationships in unstructured biomedical information, introducing barriers to realizing such solutions. We propose a scalable computational framework for addressing this need, which leverages a hypergraph-based data model and query language that may be better suited for representing complex multi-lateral, multi-scalar, and multi-dimensional relationships. We also discuss how this framework can be used to create rapid learning knowledge base systems to intelligently capture and relate complex patient data to biomedical knowledge in order to automate the recovery of clinically actionable information.
Keywords: Database management systems (L01.224.900.280), access to information (L01.143.024), knowledge (L01.535), data, information, knowledge, bioinformatics, databases, research, computer, science, biomedical, informatics, trent1952, genomics, knowledge, hypergraph, cancer, database
Introduction
Next generation genomic profiling technologies are generating deep and detailed characterizations of patients and disease states. This data-intensive approach is providing unprecedented insights that can be used to resolve mechanistic complexity and clinical heterogeneity, thereby revolutionizing how we study, manage, and treat complex diseases. To support this revolution, bioinformatics tools are rapidly emerging to process and analyze large-scale complex molecular data sets for discovery research applications. Unfortunately, when it comes to clinical (n=1) applications of genomics, the data deluge is rapidly outpacing our capacity to interpret rich data sets to extract medically useful and meaningful knowledge. The next great challenge will be to address the manual interpretation bottleneck through the development of computational solutions for intelligently linking complex patient data to actionable biomedical knowledge. This illuminates a need to represent and query large-scale complex relationships distributed across disparate types of biomedical knowledge. A recent report states a goal for the community is to transition from traditional database management to managing potentially unstructured data across many repositories.1
We propose the key challenges for intelligently linking prior knowledge to partially automate genomic interpretation require: (a) a fundamentally different computational framework for storing and representing disparate data types with complex relationships, and (b) advanced software applications that leverage this framework to structure the representation of prior knowledge so that it can be intelligently linked to patient data. We propose a framework conceptually based on requirements and cognitive strategies for knowledge computing, previously introduced as the BioIntelligence Framework.2
Our framework is compatible with future directions toward computational intelligence. Since it can support the capturing and querying of multilateral and multi-scalar relations among genomes, phenotype, environment, lifestyle, medical history, and clinical outcome data, our platform can support systems with higher order functions such as inference and learning. This will ultimately allow genomic data to be intelligently repurposed beyond personalized medicine to support more sophisticated translational research and highly iterative knowledge discovery.
BioIntelligence Framework
Systems biology is concerned with emergent properties in complex interactions of systems of systems involving disparate data elements. Extracting useful information requires syntactic and semantic linking of data within and across large data sets. Systems modeled as networks based on binary graphs (where edges connect node pairs) are suited to capturing bilateral relationships and interactions. To represent multilateral relationships requires a fundamental change in how we model systems. We generalize the binary graph model to a hypergraph model, an approach which has been previously suggested,3 and introduce a hypergraph-based solution for representing multilateral relations and multi-scalar networks.
Biological systems may benefit from a flexible data model that supports nesting of data elements and concept abstraction in a more natural manner than functionally equivalent relational counterparts, and the ability to readily query across multiple systems and abstraction layers representing complex relationships, leading to systems compatible with learning, reasoning, and inferencing. Following a model for human intelligence, information lives in different levels of the neocortex: from highly variable data inputs, to patterns, to patterns of patterns, to invariant concepts.4 Inspired by this model of intelligence, we extend the notion of a hypergraph to allow links among edges to capture relationships that cross bounds of scale and dimension, and develop a novel generic framework for capturing information that can benefit systems biology and other areas.
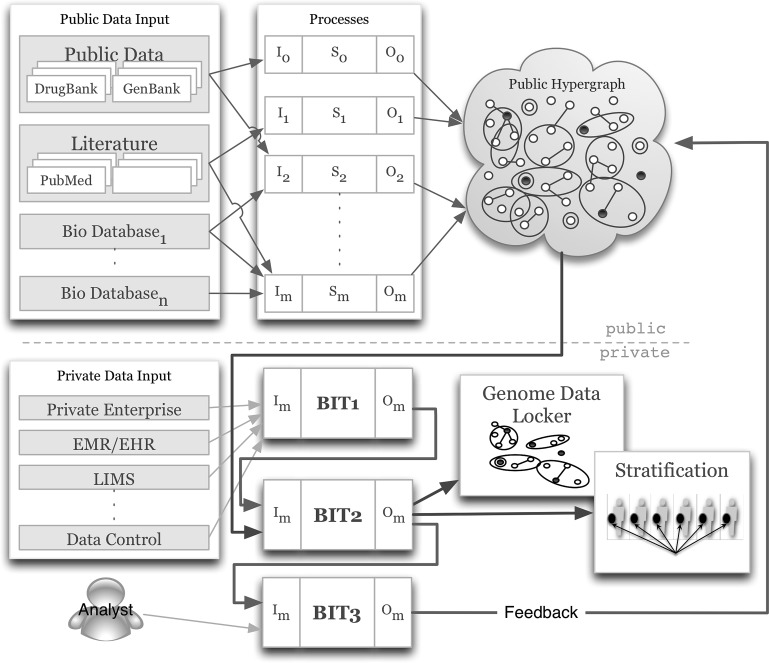
We desire a solution that is flexible to include various types of data from disparate sources, extensible to scale to massive stores of information, and accessible to permit the efficient extraction of patient-centric knowledge. Figure 1 outlines the architecture of our BioIntelligence Framework, the components of which are:
Figure 1.
A BioIntelligence Framework for creating a hypergraph-like store of public knowledge and using this, along with an individual's genomic and other patient information, to derive a personalized genome-based knowledge store for clinical translation and discovery research.
-
A public hypergraph-based network for representing knowledge, including
a. A scalable hypergraph-like model for representing a knowledge network
b. Processes to automate populating and updating the network with public domain knowledge from multiple sources
c. An efficient database solution for storing the network platform
-
A Patient Data Locker application built on top of the knowledge network, including:
d. An accessible web-based solution for storing patient-centric knowledge
e. Processes for structuring and formatting patient genomic and health data, inducing patient-centric subgraphs on the public hypergraph, and stratifying patients based on information in their lockers
A process for structuring and formatting analyst interpretation to facilitate feedback and rapid automated learning in the system.
Public hypergraph
A graph is defined G(V,E) where V is a set of vertices (nodes) and E is a set of edges (links) between two vertices. A hypergraph is a generalization of a graph in which an edge can connect any number of vertices. Biological networks have traditionally been modeled as graphs/networks. These graph models capture bilateral relationships among node pairs. Using hypergraphs as a modeling paradigm supports the characterization of multilateral relationships and processes.3 For example, in a general graph, an edge might represent a relationship between a gene and disease state. That relationship may change in the context of a drug, and a hypergraph can represent this contextual knowledge with an edge containing all three elements.
Hypergraphs are proving useful for capturing semantic and biomedical information in semantic web technologies for biological knowledge management and semantic knowledge networks.5 6 Approaches to extracting knowledge from biological research literature to store in a hypergraph have been proposed,7 8 with similar techniques used for population stratification.9
To support a data intelligent system, we wish for information to be stored not only explicitly in the data itself, but implicitly in how the data are linked, and capture this in our model. Abstraction is permitted by allowing hyperedges to contain other edges, forming a nested graph structure. A machine learning model called hierarchical temporal memory (HTM) mimics the human neocortex.4 10 Inspired by this design, we store knowledge at different levels of granularity, from single points of data, to collections of points, abstracting out to collections of collections. Thus, we perceive edges in lower levels of abstraction as nodes in higher levels, thereby permitting the network to be viewed and operated on within and across different scales of abstraction. Information is stored on the nodes and edges of the network and accessed via processes.
Populating the network platform requires processes to pull different types of information from multiple sources, such as the Cancer Biomedical Informatics Grid (caBIG) and BioWarehouse,11 12 and other tools and techniques.13–15 The processes are represented as S0… Sm in figure 1, and take unstructured, public domain knowledge as input and structures it as input to the public network.
The most common database management system (DBMS) is based on the relational data model, which is best suited to capturing data structured in a predefined schema, and presents limitations in handling complexity and scalability.16 Our solution handles unstructured and semi-structured data, and is in the scope of non-relational (NoSQL) databases, which do not require pre-defined schemas.16 17 Such databases include those based on graph models and triplestores (eg, RDF), and are best suited to capturing binary relationships between two elements (a triplestore is even more restrictive as it is essentially a ‘directed’ binary graph). To effectively represent multilateral relationships and interactions present in biomedical data, hypergraph-based approaches have been suggested.3 18–21 The flexibility granted by allowing elements to contain elements allows processing knowledge at different levels of abstraction, in a less restrictive way than hierarchical models that restrict the network's topology. HyperGraphDB resembles our model, but uses a directed hypergraph and is built on a traditional database.22 Our model is more general, based on less restrictive undirected edges, and intended to be implemented natively, although a DBMS using our model may use other DB solutions as a persistent data store. In fact, other solutions can be built on our model as it is a generalization of other models.
Data model
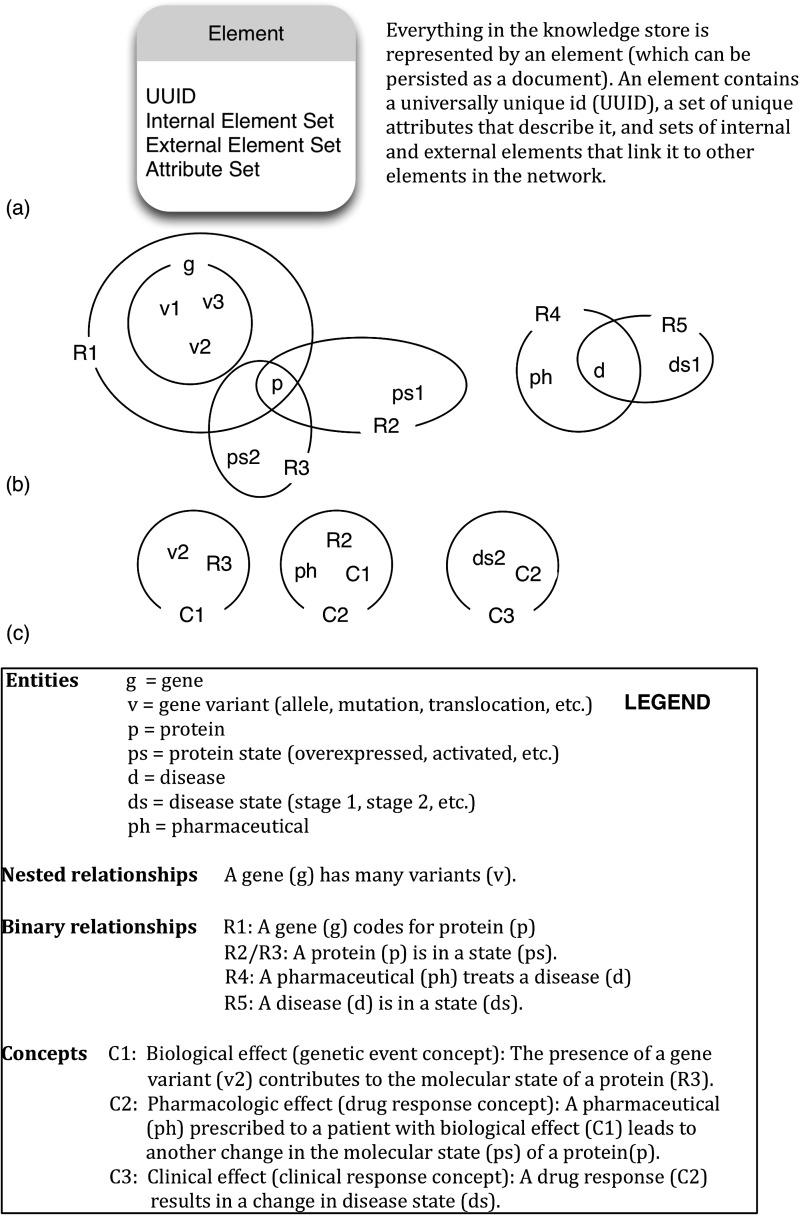
An example use of our model is shown in figure 2. This example captures multilateral, multiscalar, and multidimensional relationships using a general model (A), and viewing the network at different levels of abstraction (B and C). The elements shown in this solution are described in the legend of the image, and based on a real-world problem we are exploring. A possible schema model for a relational database capturing these same data can be found in the online supplementary material, and demonstrates the added effort involved in capturing these complex relationships in a SQL database.
Figure 2.
An illustrative example of storing biomedical information in our proposed knowledge base: a component of the BioIntelligence Framework. A shows our data model, and describes its components. B and C show the elements described in the legend (at the bottom of the figure) at two different levels of abstraction.
The entities in figure 2 are represented as elements in our database. An attribute is a key/value pair, and a list of attributes (Attribute Set) is stored with each element. In this way, we can arbitrarily add any type of attribute to an element without changing the database structure. In the relational model, each entity type requires its own table, with pre-defined fields for attributes, and adding an attribute requires adding a field to the table, and migrating the database. A characteristic of the relational model is that all entities of the same type are stored in the same table. Our model is flexible in that it does not require pre-structuring data, but we can certainly mimic this behavior if desired by enforcing a rule that requires all elements to have an attribute with key=‘type.’ Using our model, this decision is left to the database designer, and not enforced by the model itself.
An element in our model can contain an arbitrary number of other elements (allowing ‘has-a’ and ‘has-many’ relationships). These elements are referenced in the ‘Internal Element Set’ of the element. For example, in figure 2, element g is a gene and contains three gene variant elements (v1, v2, v3) in its internal element set. This is an example of a multi-lateral relationship as g can be viewed as a ‘hyperedge’ in a hypergraph, connecting three ‘nodes.’ While this behavior is easy to model in a relational database, using a one-to-many relation, it becomes more complex when an element contains an arbitrary number of arbitrary types of elements. For example, the element R2 represents a molecular state, which in this case is a protein p associated with a protein state ps1. In a relational database, we can capture these relationships in a ‘molecular state’ table with foreign key fields pointing to a ‘protein’ table and a ‘protein state’ table. Now consider a molecular state that exists in the context of a modifier drug (ie, the state of the protein is perturbed by a modifier drug in a laboratory experiment). To capture this in our model, we can simply create a new element with three internal elements: p, ps1 and the element representing the modifier drug. To capture this in the relational database, we would need to either add a field to the ‘molecular state’ table, which may be blank for many records, or create a new table with the three related fields. Both of the later options require a change to the underlying data structure, and migrating the database.
The elements shown in figure 2C represent higher-level concepts and recursive nesting of elements at different levels, illustrating the ability to flexibly and efficiently capture multi-scalar relationships among elements, the motivation behind our data model. For example, note that R1 captures the relationship that gene g codes for protein p. C1 captures the genetic event concept, where gene variant v2 changes the state of the protein (the molecular state represented by R3). C2 is a drug response concept representing the higher-level concept that C1, in the context of the pharmaceutical ph, leads to a changed molecular state, R2. These combined biological and pharmacologic effects lead to a change in disease state to ds2, captured by the clinical response concept C3. The internal (nested) element sets contain the topology of the network, that is, they define how entities are related, and capture meaning in those relationships. Further details to describe the nature of the relationships can always be stored in attributes of the elements.
The ‘External Element Set’ of an element is the inverse of internal element relations. For example, v1, v2, and v3 all have g in their external element set. While it is not necessary to store this information (the network of relationships among elements can be constructed via the internal element sets alone), it does aid in querying the database. We have defined three new types of queries associated with our model: recover, context, and expand. The expand and context queries retrieve all internal and external elements of an element, respectively, optionally limited by modifiers presented with the query. Recover is a combination of these, and returns both internal and external elements. All three query actions have an optional level constraint, defining how deep to traverse the graph when retrieving related elements. For instance, expand n will retrieve all internal elements, and their internal elements, recursively up to n times. For instance, we can view C3 as an abstraction; a clinical effect that we can relate to patients and other concepts. Expanding C3 by one level shows us that it represents a disease state ds2, triggered by a pharmacologic effect C2. Expanding C3 by two levels shows us the details of the pharmacologic effect, and so on. In this way, we can choose which level of abstraction we wish to view and compute over, and we can create new concepts that cross these layers of abstraction (multi-scalar relationships).
By the definition of the internal and external element sets, it follows that our model naturally handles many-to-many relationships as well. In summary, we do not argue that a relational database is incapable of capturing the types of relationships we discuss here, rather that it requires more work, and added layers of complexity to the underlying structure of the database, which makes capturing and querying complex biomedical relationships more difficult. Our model is an abstraction of other models, including relational, graph, hierarchical, and object-oriented, and can therefore be used to model data represented using any and all of these models at once. The potential benefit of our model is that it provides levels of scalability and flexibility that are difficult to achieve with existing models. We are currently developing a solution based on this model and will present additional details of the model and related query language in future publications.
Patient data locker
Given a patient's data (genomic, health, etc), we wish to recover related knowledge from our network using BioIntelligence Tools (BIT). The first step (BIT1 in figure 1) is a process to structure data as input to a process for inducing patient-relevant subgraphs of the knowledge network (BIT2 in figure 1). BIT1 integrates many types of data sets across multiple databases to support electronic medical and health records (EMR/EHRs), and is designed as a modular based system to provide metadata and indexing for queries.
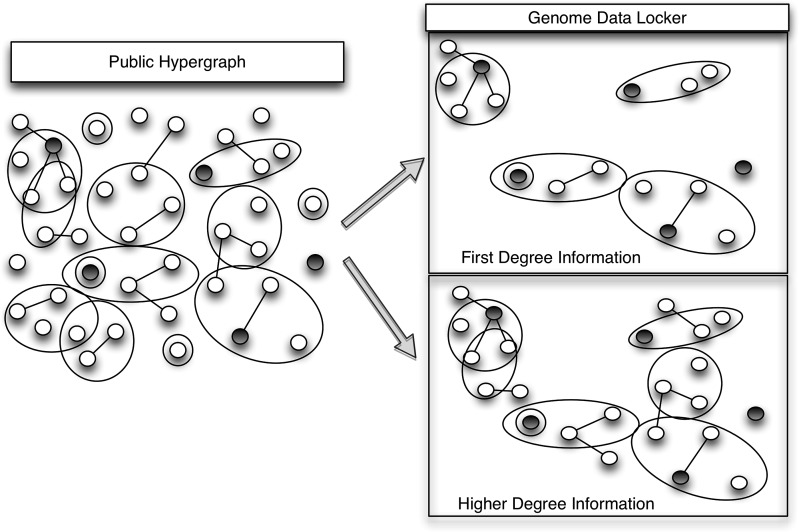
The next step (BIT2 in figure 1) is a process to extract relevant knowledge from the network based on individual patient information. An induced subgraph H(S,T) of network G(V,E) has the properties S⊂V, and for every vertex set Si of H, the set Si is an edge of H if and only if it is an edge in G. That is, H has the same edges that appear in G over the same set of nodes. We say that H is an induced subgraph of G, and H is induced by S. In our system, V is the set of nodes in the platform network G, and S is the set of nodes that map to an individual's genomic and health information. Thus, a subgraph is induced by an individual genome. The architecture in figure 1 shows an example public hypergraph, and private subgraphs stored in a data locker. These networks are detailed in figure 3, where the network on the left is a public knowledge network, and the darkened nodes are elements relevant to an individual's information (based on input patient data). The network on the top right is induced by this information, and contains all of the darkened nodes, and edges incident to them. Alternately, we can expand the information retrieved to include all connected components of the induced subgraph. An example of a connected-component induced subgraph is shown on the bottom right of figure 3.
Figure 3.
The network on the left is an example knowledge network platform. The darkened nodes represent gene variants present in an individual genome. The network on the top right is a genome-induced subgraph of the network. The network on the bottom right is a genome-induced subgraph, expanded out to include additional knowledge stored on edges in the connected-component each data element is contained in.
The most important characteristic of the data locker is that it contains all relevant knowledge to facilitate clinical translation. Induced subgraphs can be used to transform a large set of patient-relevant data to smaller, task-tailored formats void of extraneous detail. The patient data locker is linked to the public knowledge store, and automatically updated to contain only a subset of information related to the patient. Thus, an expert need not develop their own intricate search queries and perform the tedious task of progressively reducing the amount of irrelevant data returned by the query. Any query that can be run on the entire knowledge network, can be run on the subgraph in a patient's locker, leading to a more precise subset of knowledge returned, and potentially faster querying speeds as the search space is reduced.
The expert analyst is provided with knowledge tailored to a particular patient, partially automating the interpretation process, and a process (BIT3 in figure 1) allows the analyst to input new interpretation knowledge into the public network. We envision this type of feedback mechanism will support the inclusion of a learning model for our system, and allow the community to contribute to its growth. The system is diverse, providing framework and template libraries, allowing users to integrate their own tools for analysis, data collection, and beyond.
Conclusion
A deluge of biomedical data generated from next-generation sequencing (NGS) and clinical applications is overwhelming our ability to efficiently extract value from it. Existing bioinformatics tools were not developed to support clinical translation for an individual patient, causing an n=1 translation bottleneck. A new architecture for managing biomedical data is desired, and we present the BioIntelligence Framework as a genome-compatible biomedical knowledge representation platform. Our future efforts to achieve the goals outlined in this paper include ensuring that we develop algorithms on this framework that minimally meet the performance expectations of existing solutions in practice.
Footnotes
Contributors: All authors contributed to the ideas behind the framework. TF, CC, and PL contributed computer science expertise. JK, DVH, JMT, and SM contributed expertise in clinical genomics and translational research.
Competing interests: None.
Provenance and peer review: Commissioned; externally peer reviewed.
References
- 1. Agrawal R, Ailamaki A, Bernstein PA, et al. The Claremont report on database research. ACM SIGMOD Record 2008;37:9–19 [Google Scholar]
- 2. Mousses S, Kiefer J, Von Hoff D, et al. Using biointelligence to search the cancer genome: an epistemological perspective on knowledge recovery strategies to enable precision medical genomics. Oncogene 2008;27:S58–66 [DOI] [PubMed] [Google Scholar]
- 3. Klamt S, Haus U, Theis F. Hypergraphs and cellular networks. PLoS Comput Biol 2009;5:e1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hawkins J, Blakeslee S. On Intelligence. New York: Times Books, 2004 [Google Scholar]
- 5. Antezana E, Kuiper M, Mironov V. Biological knowledge management: the emerging role of the semantic web technologies. Brief Bioinform 2009;10:392–407 [DOI] [PubMed] [Google Scholar]
- 6. Zhen L, Jiang Z. Hy-SN: hyper-graph based semantic network. Knowledge-Based Systems 2010;23:809–16 [Google Scholar]
- 7. Vailaya A, Bluvas P, Kincaid R, et al. An architecture for biological information extraction and representation. Bioinformatics 2005;21:430–8 [DOI] [PubMed] [Google Scholar]
- 8. Mukhopadhyay S, Palakal M, Maddu K. Multi-way association extraction and visualization from biological text documents using hyper-graphs: applications to genetic association studies for diseases. Artif Intell Med 2010;49:145–54 [DOI] [PubMed] [Google Scholar]
- 9. Vazquez A. Population stratification using a statistical model on hypergraphs. Phys Rev E Stat Nonlin Soft Matter Phys 2008;77:1–7 [DOI] [PubMed] [Google Scholar]
- 10. George D. How the Brain Might Work: a Hierarchical and Temporal Model for Learning and Recognition [dissertation]. Palo Alto, California: Stanford University, 2008 [Google Scholar]
- 11. NCI Cancer Biomedical Informatics Grid (caBIG). https://cabig.nci.nih.gov/ (accessed 16 Sep 2011).
- 12. Karp P. Biowarehouse database integration for bioinformatics. http://biowarehouse ai.sri.com/ (accessed 16 Sept 2011).
- 13. Chen H, Ding L, Wu Z, et al. Semantic web for integrated network analysis in biomedicine. Components 2009;10:177–92 [DOI] [PubMed] [Google Scholar]
- 14. Tudor CO, Schmidt CJ, Vijay-Shanker K. eGIFT: mining gene information from the literature. BMC Bioinformatics 2010;11:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valentin F, Squizzato S, Goujon M, et al. Fast and efficient searching of biological data resources–using EB-eye. Brief Bioinform 2010;11:375–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leavitt N. Will NoSQL databases live up to their promise? Computer 2010;43:12–14 [Google Scholar]
- 17. Angles R, Gutierrez C. Survey of graph database models. ACM Computing Surveys 2008;40:1–39 [Google Scholar]
- 18. Olken F. Graph data management for molecular biology. OMICS 2003;7:75–8 [DOI] [PubMed] [Google Scholar]
- 19. Hu Z, Mellor J, Wu J, et al. Towards zoomable multidimensional maps of the cell. Nat Biotechnol 2007;25:547–55 [DOI] [PubMed] [Google Scholar]
- 20. Spreckelsen C, Spitzer K. Formalising and acquiring model-based hypertext in medicine: an integrative approach. Methods Inform Med 1998;37:239–46 [PubMed] [Google Scholar]
- 21. Wu G, Li J, Hu J, et al. System: a native RDF repository based on the hypergraph representation for RDF data model. J Comput Sci Technol 2009;24:652–64 [Google Scholar]
- 22. Iordanov B. HyperGraphDB: a generalized graph database. Proceedings of the 2010 International Conference on Web-age Information Management 2010:25–36 [Google Scholar]