Abstract
Objective
(1) To evaluate a state-of-the-art natural language processing (NLP)-based approach to automatically de-identify a large set of diverse clinical notes. (2) To measure the impact of de-identification on the performance of information extraction algorithms on the de-identified documents.
Material and methods
A cross-sectional study that included 3503 stratified, randomly selected clinical notes (over 22 note types) from five million documents produced at one of the largest US pediatric hospitals. Sensitivity, precision, F value of two automated de-identification systems for removing all 18 HIPAA-defined protected health information elements were computed. Performance was assessed against a manually generated ‘gold standard’. Statistical significance was tested. The automated de-identification performance was also compared with that of two humans on a 10% subsample of the gold standard. The effect of de-identification on the performance of subsequent medication extraction was measured.
Results
The gold standard included 30 815 protected health information elements and more than one million tokens. The most accurate NLP method had 91.92% sensitivity (R) and 95.08% precision (P) overall. The performance of the system was indistinguishable from that of human annotators (annotators' performance was 92.15%(R)/93.95%(P) and 94.55%(R)/88.45%(P) overall while the best system obtained 92.91%(R)/95.73%(P) on same text). The impact of automated de-identification was minimal on the utility of the narrative notes for subsequent information extraction as measured by the sensitivity and precision of medication name extraction.
Discussion and conclusion
NLP-based de-identification shows excellent performance that rivals the performance of human annotators. Furthermore, unlike manual de-identification, the automated approach scales up to millions of documents quickly and inexpensively.
Keywords: Natural language processing, privacy of patient data, health insurance portability and accountability act, automated de-identification, protected health information, NLP, text mining, human-computer interaction and human-centered computing, providing just-in-time access to the biomedical literature and other health information, applications that link biomedical knowledge from diverse primary sources (includes automated indexing), linking the genotype and phenotype, discovery, bionlp, medical informatics, biomedical informatics, disease networks, translational medicine, drug repositioning, rare diseases
This paper studied automated de-identification of clinical narrative text using natural language processing (NLP)-based methods. The specific aims were (1) to evaluate a state-of-the-art NLP-based approach to automatically de-identify a large set of diverse clinical notes for all HIPAA (Health Insurance Portability and Accountability Act)-defined protected health information (PHI) elements and (2) to measure the impact of de-identification on the performance of information extraction (IE) algorithms executed on the de-identified documents. In addition, we hope that our study—by contrasting the performance of human and automated de-identification—will shape policy expectations.
Background and significance
The importance of information included in narrative clinical text of the electronic health record (EHR) is gaining increasing recognition as a critical component of computerized decision support, quality improvement, and patient safety.1 2 In an August, 2011 JAMA editorial, Jha discusses the promises of the EHR, emphasizing the importance of NLP as an enabling tool for accessing the vast information residing in EHR notes.3 NLP could extract information from clinical free-text to fashion decision rules or represent clinical knowledge in a standardized format.4–6 Patient safety and clinical research could also benefit from information stored in text that is not available in either structured EHR entries or administrative data.7–9
However, the 1996 HIPAA privacy rule requires that before clinical text can be used for research, either (1) all PHI should be removed through a process of de-identification, (2) a patient's consent must be obtained, or (3) the institutional review board should grant a waiver of consent.10 Studies have shown that requesting consent reduces participation rate, and is often infeasible when dealing with large populations.11 12 Even if a waiver is granted, documents that include PHI should be tracked to prevent unauthorized disclosure. On the other hand, de-identification removes the requirements for consent, waiver, and tracking and facilitates clinical NLP research, and consequently, the use of information stored in narrative EHR notes.
Several studies have used NLP for removing PHI from medical documents.13 Rule-based methods14–23 make use of dictionaries and manually designed rules to match PHI patterns in the texts. They often lack generalizability and require both time and skill for creating rules, but perform better for rare PHI elements. Machine-learning-based methods,24–34 on the other hand, automatically learn to detect PHI patterns based on a set of examples and are more generalizable, but require a large set of manually annotated examples. Systems using a combination of both approaches usually tend to obtain the best results.13 35 Overall, the best systems report high recall and precision, often >90%, and sometimes as high as 99%. Nevertheless, no study has evaluated the performance of automated de-identification for all PHI classes.13 Important items are often ignored—in particular, ages >89,15 16 18 24 25 geographic locations,15 16 24 26 institution and contact information,16 24 26 dates, and IDs.16 24 Furthermore, systems should ideally be evaluated on a large scale, including the diverse document types of the EHRs, to have a good idea of their accuracy and generalizability. However, most systems use only one or two document types for evaluation, such as pathology reports,16 17 19 20 26 discharge summaries,23 25 27–30 34 nursing progress notes,23 34 outpatient follow-up notes,22 or medical message boards.33 Some of them were only evaluated on documents with synthetic patient PHI (manually de-identified documents re-identified with fake PHI).27–30 Very few systems have been evaluated on more than two note types.14 15 24 32 Only a handful of studies provide details on over-scrubbing (non-PHI wrongly identified as PHI) and none of them investigate the effect of de-identification on subsequent IE tasks.13 It is indeed possible that de-identification has an adverse effect on IE accuracy.13 Over-scrubbing errors could overlap with useful information—for example, if a disease name is erroneously recognized as a person name it will be removed and lost to subsequent IE application. Second, NLP techniques such as part-of-speech tagging and parsing may be less effective on modified text.
In this paper, we examine some of the gaps of the literature and conduct de-identification experiments on a large set and wide variety of clinical notes (over 22 different types), using real PHI data (as opposed to resynthesized data), studying all classes of PHI and measuring the impact of de-identification on a subsequent IE task. We also illustrate the strength of automatic de-identification by comparing human and system performances.
Material and methods
Data
Three thousand five hundred and three clinical notes were selected by stratified random sampling from five million notes composed by Cincinnati Children's Hospital Medical Center clinicians during 2010. The study was conducted under an approved institutional review board protocol. The notes (see descriptive statistics in figure 1) belong to three broad categories (with the same proportional distribution as the five million notes):
Labeled (created within the EHR system and includes division origin (eg, emergency department, operating room))
Unlabeled (created within the EHR but no division)
External (written outside the EHR (eg, on a radiology system and transferred into the EHR through an interface)).
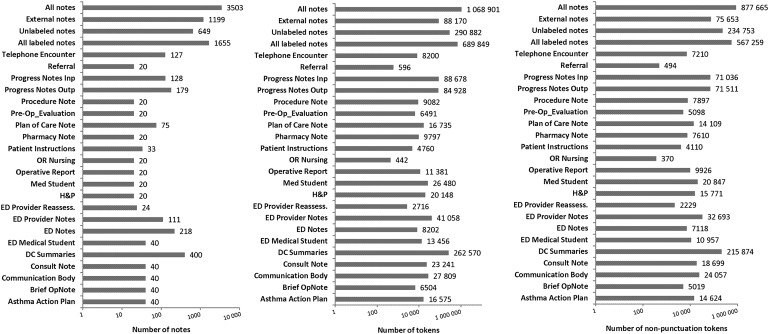
Figure 1.
Descriptive statistics of the corpus. DC, discharge; ED, emergency department; H&P, history and physical; OR, operating room.
Within the labeled category, we included 22 note types in a randomly stratified sample. We selected a type only if the number of notes exceeded the subjective limit of 800 during the previous 12 months. We oversampled discharge summaries because of their richness in de-identification information,32 and some of the less common notes to have at least 20 notes for each type. Figure 1 shows the distribution of note types in our corpus. Including the unlabeled and external notes, the total number of note types was above 22.
All 18 HIPAA-defined PHI categories were included in the study.10 Some of them were collapsed into one category. In total we defined 12 classes:
NAME
DATE (eg, “12/29/2005”, “September 15th”)
AGE (any age, not only age >89)
EMAIL
INITIALS: person's initials
INSTITUTION: hospitals and other organizations
IP: internet provider addresses and URLs
LOCATION: geographic locations
PHONE: phone and fax numbers
SSN: social security number
ID: any identification number (medical record numbers, etc)
OTHER: all remaining identifiers.
To create a ‘gold standard’ for building and evaluating systems, clinical notes were manually annotated by two annotators (native English speakers with Bachelor degrees). All notes were double annotated and the final gold standard resulted from consensus seeking adjudication led by the annotators' supervisor. Before production annotation, the annotators were trained and the annotation guideline was iteratively developed. Double annotation is a standard method in NLP because it assures a strong gold standard. We will refer to the two annotators who created the gold standard as annotator 1 and annotator 2.
Additionally, the 1655 ‘labeled’ notes from the corpus were also double-annotated for medication names to test the impact of de-identification on the subsequent extraction of medication names.
De-identification systems
We studied the characteristics of two de-identification systems. One, MIST (MITRE Identification Scrubber Toolkit), is a prototype from MITRE.32 The other system was designed in-house based on the MALLET machine-learning package.36 Both systems are based on conditional random fields (CRFs),37 but implement the algorithm slightly differently. Using the MALLET package to build our system gave us access to the algorithm's source code (necessary to obtain probability scores for recall-bias experiments), while MIST's source code was not available.
We tested the MIST system in its default configuration, and with customizations (preprocessing and postprocessing steps and additional features for the CRF model). We also tested two configurations of the in-house system, one equivalent to the “out-of-the-box” MIST (ie, same feature generation process), and one with customizations.
Before training the customized systems, we performed two preprocessing steps: tokenization with an in-house tokenizer and part-of-speech tagging with the TreeTagger POS tagger (used with its downloadable English model).38 Features for the CRF models consisted of the default features generated by MIST: token-level properties (capitalization, punctuation, etc) and contextual features (token before, token after, etc). Additional features we used were token parts-of-speech and presence (or absence) of the tokens in a name lexicon (built using the US Census Bureau's dataset and the hospital's physician (employee) database).
We also added three postprocessing rules to the machine-learning algorithms, consisting of regular expressions to (1) identify EMAIL; (2) match strings to the entries of our name lexicon, with a match resulting in the assignment of a NAME label; and (3) label any string as a NAME if the algorithm tagged a matching string NAME in the document but missed the particular string somewhere else in the same document. Step (1) was necessary because of the rare frequency of EMAILs, which made it difficult for the system to learn their patterns. The presence of a word in a name lexicon was also used as a feature for machine learning, but adding step (2) as a postprocessing rule statistically significantly improved the performance.
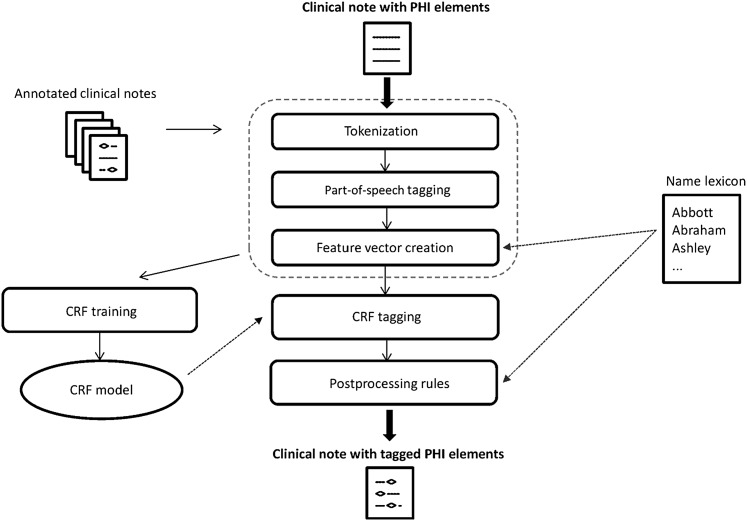
Figure 2 depicts the main steps of the de-identification process (identical for both customized systems).
Figure 2.
De-identification process. CRF, conditional random field; PHI, protected health information.
For convenience, we will refer to the four system versions as follows:
MIST1: original, “out-of-the-box” MIST system;
MIST2: customized MIST system (preprocessing, additional features and postprocessing);
MCRF1: in-house system with a configuration equivalent to MIST1;
MCRF2: configuration equivalent to MIST2.
Experiments
Evaluation metrics
We used three standard NLP metrics to measure performance: recall (sensitivity), precision (positive predictive value) and F value, which is the harmonic mean of recall (R) and precision (P) (F=(2*P*R)/(P+R)).39 40 We computed those metrics at span level (complete phrase is identified as PHI), token level (individual tokens are identified as PHI) and tag-blind token level (without taking into account the specific PHI tags). Span-level performance was computed for all performance tests. Token-level and tag-bling evaluations are provided only for the best performing system.
To rule out the possibility that the performance difference between two systems' outputs was due to chance, we also tested the statistical significance of the difference, using approximate randomization.41 42
Interannotator agreement (IAA)
IAA was calculated for the two annotators to define the strength of the gold standard,43 using the F value, after an initial 2-week training period. We required both span and tag to be the same for an annotated element to be counted as a match.
De-identification performance tests
We evaluated overall performance (all tags considered) and tag-based performance of the MIST and MCRF systems in a 10-fold cross-validation setting (the corpus was divided at the document level). In addition to the corpus-level test, we also measured the de-identification performance for document types.
A separate subset of 250 annotated documents (not part of either the training or testing) was manually examined during error analyses (development set).
Additionally, we also measured the performance of MCRF2i on two publicly available datasets: the i2b2 corpus,35 which consists of de-identified discharge summaries (669 reports for training and 220 reports for testing) that have been re-synthetized with fake PHI; and the PhysioNet corpus,23 44 which consists of 2483 nursing notes, with very sparse PHI elements (1779 in total). We report performance using a cross-validation setting for this corpus.
Humans versus systems performance tests
We conducted an experiment to compare the performance of the automated systems with that of humans. Two native English speakers (with Masters and Bachelor degrees) who had not previously taken part in the project annotated (independently) a random subset of 10% of the corpus (350 documents). We evaluated their individual performance against our gold standard. We will refer to the two additional annotators as annotator 3 and annotator 4.
Recall bias
In de-identification processes, recall is usually more important than precision, so we experimented with infusing recall bias into both systems.45 For MIST, we used the built-in command line parameter that implements Minkov's algorithm.45 For the MCRF system, we increased recall by selecting tokens labeled non-PHI and changing their label to the PHI label with the next highest probability suggested by the system. We selected non-PHI labels only if their system-generated probability score was less than or equal to a given threshold (eg, if we set the probability threshold at 0.95, every non-PHI label with a score >0.95 retained the original label). The threshold was varied between 0.85 and 0.99. In general, the higher we set the threshold, the more non-PHI tokens we selected and replaced, leading to higher recall.
Impact of de-identification on subsequent IE
The impact was tested by measuring the performance of automated IE on medication names (a subset of the corpus was annotated for medication names, as mentioned in the ‘Data’ subsection). We extracted medication names from clinical notes (1) before removing PHI (system trained and tested on original corpus), (2) after removing and replacing PHI with asterisks (system trained and tested on the corpus with asterisks), and (3) after removing and replacing PHI with synthetically generated PHI surrogates (system trained and tested on corpus with synthetic PHI). In the evaluation of medication IE—for example, if the medication name “aspirin” was erroneously tagged as NAME and removed from the corpus, then it was counted as false negative for IE.
We used MIST's built-in functionality to replace the original PHI with synthetic PHI. For medication name extraction, we used an automated system being developed in-house.46 47
Results
Corpus descriptive statistics
The corpus included at least 22 different note types, and more than one million tokens (see figure 1). Figure 3 shows the number of annotated PHI elements. Almost 50% are located in discharge summaries and progress notes. This lopsided distribution is due to the fact that these note types generally are the longest. More than 30% of all PHI was found in discharge summaries, confirming findings of Aberdeen et al.32
Figure 3.
Number of annotated protected health information (PHI) elements for each document type. DC, discharge; ED, emergency department; H&P, history and physical; OR, operating room.
DATE comprised more than one-third of all PHI, and NAME about a quarter. The third largest category was the mixed group of OTHER. Not shown in the figures are categories with extremely low frequencies: EMAIL (frequency: 14), INITIALS (16), IP (10), and SSN (1).
Interannotator agreement
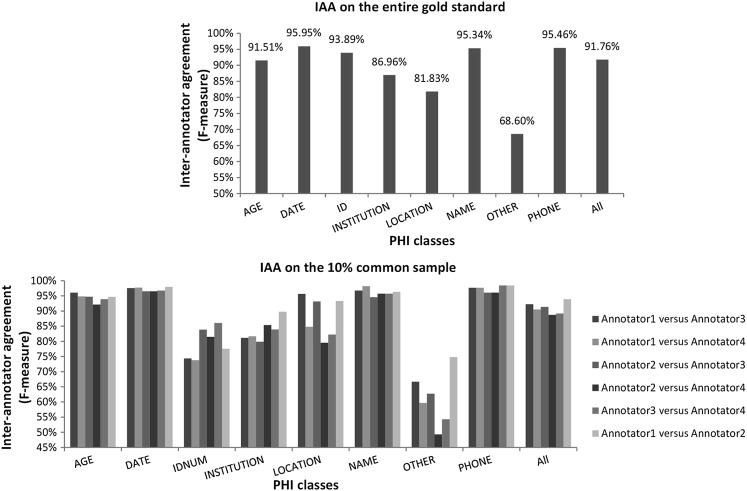
The overall F value of IAA was 91.76 for manual de-identification between annotators 1 and 2 (see top part of figure 4). The IAA for manual medication name annotation was 93.51 (1655 “Labeled” notes were annotated for medications). These values indicate good agreement for both the de-identification and the subsequent medication name extraction annotations.
Figure 4.
Inter-annotator agreement (IAA; F value) for each protected health information (PHI) class on the entire gold standard (annotators 1 and 2) and on the 10% common sample (annotators 1, 2, 3, and 4).
Automated de-identification performance
Table 1 (upper section) presents the performance of the de-identification systems for each tag type and overall, for the “out-of-the box” systems (MIST1 and MCRF1) and customized systems (MIST2 and MCRF2). In five cases, of the eight PHI tags shown, and for overall F value, MCRF2 achieved the highest performance. The difference between the two customized systems was found to be statistically significant for AGE, OTHER, ID, NAME, and overall F values (see lower section of table 1). For each tag level and overall F value, the customizations increased performance of both systems. This increase was statistically significant for NAME and overall F values for MCRF2 and for AGE, PHONE, DATE, NAME, and overall F values for MIST2.
Table 1.
Performance of systems (per-tag and overall precision (P), recall (R) and F value (F)) and statistical significance tests
| Performance of systems (10-fold cross-validation) | ||||||||||||
| MIST1 | MCRF1 | MIST2 | MCRF2 | |||||||||
| P | R | F | P | R | F | P | R | F | P | R | F | |
| AGE | 94.47 | 92.31 | 93.38 | 96.7 | 90.42 | 93.45 | 95.87 | 92.46 | 94.13 | 96.69 | 90 | 93.22 |
| DATE | 95.77 | 97.12 | 96.44 | 97.97 | 96.61 | 97.29 | 97.25 | 97.76 | 97.5 | 97.95 | 96.98 | 97.46 |
| ID | 90.58 | 92.64 | 91.6 | 97.23 | 95.64 | 96.43 | 91.17 | 93.38 | 92.26 | 97.27 | 95.7 | 96.48 |
| INST | 90.59 | 86.41 | 88.45 | 93.18 | 85.01 | 88.91 | 90.61 | 87.06 | 88.8 | 93.25 | 85.26 | 89.08 |
| LOC | 79.82 | 67.93 | 73.4 | 86.12 | 68.94 | 76.58 | 78.92 | 69.95 | 74.16 | 87.38 | 69.95 | 77.7 |
| NAME | 93.19 | 88.64 | 90.86 | 95.62 | 86.99 | 91.1 | 92.48 | 94.16 | 93.31 | 94.47 | 94.56 | 94.52 |
| OTH | 77.21 | 77.39 | 77.3 | 83.94 | 74.17 | 78.76 | 78.17 | 77.13 | 77.65 | 84.68 | 73.87 | 78.91 |
| PH | 90.06 | 93.04 | 91.52 | 94.08 | 90.64 | 92.33 | 91.44 | 93.95 | 92.68 | 94.42 | 90.87 | 92.61 |
| All | 92.05 | 91.02 | 91.54 | 95.25 | 89.86 | 92.48 | 92.79 | 92.81 | 92.8 | 95.08 | 91.92 | 93.48 |
| Token-level performance for best system (MCRF2) | ||||||
| Token-level | Token-level + tag-blind | |||||
| P | R | F | P | R | F | |
| AGE | 98.21 | 93.40 | 95.75 | 93.42 | ||
| DATE | 98.17 | 96.87 | 97.52 | 96.98 | ||
| ID | 97.57 | 95.49 | 96.52 | 96.43 | ||
| INST | 97.48 | 92.74 | 95.05 | 94.79 | ||
| LOC | 97.95 | 93.92 | 95.89 | 96.03 | ||
| NAME | 97.26 | 97.38 | 97.32 | 97.53 | ||
| OTH | 86.81 | 76.45 | 81.30 | 78.31 | ||
| PH | 97.13 | 93.40 | 95.23 | 94.85 | ||
| All | 96.68 | 93.77 | 95.20 | 97.42 | 94.49 | 95.93 |
| Statistical significance tests between F values obtained by systems (cross-validation evaluation) | |||||
| MCRF1 vs MIST1 | MCRF2 vs MIST2 | MIST1 vs MIST2 | MCRF2 vs MCRF1 | MCRF2 vs gold standard | |
| p Value | p Value | p Value | p Value | p Value | |
| AGE | 0.8490 | *0.0087 | *0.0389 | 0.4922 | *0.0001 |
| DATE | *0.0001 | 0.7650 | *0.0001 | 0.2470 | *0.0001 |
| ID | *0.0001 | *0.0001 | 0.0996 | 0.8856 | *0.0001 |
| INST | 0.3572 | 0.6087 | 0.2738 | 0.6623 | *0.0001 |
| LOC | 0.0777 | 0.0553 | 0.5897 | 0.3812 | *0.0001 |
| NAME | 0.4897 | *0.0001 | *0.0001 | *0.0001 | *0.0001 |
| OTH | *0.0071 | *0.0180 | 0.3676 | 0.6118 | *0.0001 |
| PH | 0.2936 | 0.9248 | *0.0458 | 0.6208 | *0.0001 |
| All | *0.0001 | *0.0001 | *0.0001 | *0.0001 | *0.0001 |
*Indicates statistical significance (p<0.05).
INST, institution; LOC, location; OTH, other; PH, phone.
Table 1 also shows token-level performance for the best system (MCRF2). Compared with span level, the token-level performance gains range from <0.1% (DATE) to approximately 18% (LOCATION). Tag-blind token-level performance is even higher, with an overall F value of 95.93.
Table 2 gives the F values obtained by MCRF2 for each document type. Performance varies between the different note types, although high performance (>90%) is achieved for the majority of notes.
Table 2.
Best system (MCRF2) performance (F values) per document type
| AGE | DATE | ID | INST | LOC | NAME | OTH | PH | All PHI | |
| Asthma action plan | 100 | 96.59 | 80 | 96.47 | 100 | 95.17 | 98.11 | 69.72 | 92.19 |
| Brief OpNote | 100 | 99.12 | 98.51 | 80 | 100 | 94.56 | 35.29 | 100 | 95.17 |
| Communication body | 95.24 | 93.07 | 75 | 82.35 | 80 | 97.02 | 80.99 | 50 | 92.11 |
| Consult note | 92.96 | 98.5 | 80 | 93.62 | 75 | 98.35 | 90 | 80 | 95.94 |
| DC summaries | 92.89 | 98.12 | 98.26 | 94.54 | 77.27 | 96.37 | 75.62 | 97.96 | 94.55 |
| ED medical student | 96.77 | 69.23 | 100 | 66.67 | 0 | 95.45 | 96.7 | 100 | 92.13 |
| ED notes | 85.71 | 80 | 100 | 72.73 | 100 | 72.73 | 45.83 | 50 | 65.84 |
| ED provider notes | 96.71 | 90.48 | 100 | 76.19 | 0 | 80 | 92.35 | 50 | 92.47 |
| ED provider reassess. | 100 | 100 | 100 | 100 | 100 | 85.71 | 92.31 | 100 | 92.86 |
| H&P | 92.75 | 97.31 | 80 | 87.1 | 0 | 94.51 | 83.72 | 50 | 92.55 |
| Med student | 88.1 | 98.22 | 0 | 69.44 | 50 | 95.87 | 76.06 | 100 | 92.93 |
| Operative report | 90.91 | 98.68 | 100 | 100 | 100 | 93.94 | 94.31 | 100 | 96.48 |
| OR nursing | 100 | 100 | 100 | 100 | 100 | 88.89 | 0 | 100 | 81.82 |
| Patient instructions | 100 | 94.74 | 100 | 62.5 | 66.67 | 87.91 | 55 | 90.62 | 81.51 |
| Pharmacy note | 50 | 91.2 | 100 | 100 | 100 | 72 | 65.31 | 100 | 88.01 |
| Plan of care note | 100 | 96.97 | 100 | 50 | 0 | 91.09 | 78.12 | 66.67 | 87.35 |
| Pre-Op_evaluation | 100 | 99.37 | 100 | 100 | 100 | 94.49 | 100 | 100 | 98.6 |
| Procedure note | 100 | 98.06 | 100 | 96.3 | 0 | 93.51 | 91.36 | 100 | 94.96 |
| Progress notes Outp | 88.08 | 95.53 | 94.12 | 73.3 | 71.79 | 93.8 | 70.29 | 81.25 | 90.9 |
| Progress notes Inp | 93.23 | 98.19 | 93.88 | 87.91 | 54.55 | 93.32 | 76.64 | 98.41 | 94.75 |
| Referral | 100 | 100 | 100 | 90.91 | 0 | 100 | 100 | 100 | 93.75 |
| Telephone encounter | 100 | 91.94 | 100 | 56.25 | 40 | 88.21 | 47.62 | 76.36 | 82.16 |
| All labeled notes | 92.9 | 97.51 | 97.04 | 90.17 | 73.02 | 94.75 | 78.68 | 91.54 | 93.54 |
| Unlabeled notes | 92.44 | 97.44 | 97.07 | 87.68 | 83.65 | 94.68 | 80.09 | 95.92 | 93.58 |
| External notes | 94.87 | 96.79 | 69.57 | 63.89 | 70.59 | 85.31 | 60.71 | 82.22 | 91.88 |
| All notes | 93.22 | 97.46 | 96.48 | 89.08 | 77.7 | 94.52 | 78.91 | 92.61 | 93.48 |
Zero F value is the consequence of insufficient representation of a particular PHI type in that particular note category (eg, if there was one Location PHI element in 20 notes and it was missed then the F value was zero).
DC, discharge; ED, emergency department; H&P, history and physical; OR, operating room; PHI, protected health information.
Overall token-level performance of MCRF2 on the i2b2 corpus was 96.68% F value (99.18% precision, 94.26% recall) with our default configuration and 97.44% F value (97.89% precision, 97.01% recall) using our recall bias method (threshold of 0.91). These results are similar to those obtained by the top systems in the i2b2 challenge and slightly lower than the performance of MIST (98.1% F value, 98.7% precision, 97.5% recall, as reported in Uzuner et al 35; however, our system was not customized for the i2b2 dataset). Performance on the PhysioNet corpus was much lower: 70.60 F value (89.02 precision, 58.49 recall) with our regular MCRF2 set-up and 74.61 F value (74.93 precision, 74.28 recall) using the recall bias method (0.97 threshold). This is explained by the very low frequency of PHI (1779) in the PhysioNet corpus, which makes this corpus ill-suited for machine-learning methods (there are not enough training instances). In that case, a rule-based method such as the one used by the providers of the corpus23 will have higher performance (74.9% precision, 96.7% recall and 84.41% F value). Gardner et al 34 also evaluated their CRF algorithm on the PhysioNet corpus and observed a large performance drop: they obtained a 25.5% precision for a 97.2% recall (40.04 F value) and a 70% precision for an 80% recall (74.66 F value).
Human de-identification performance
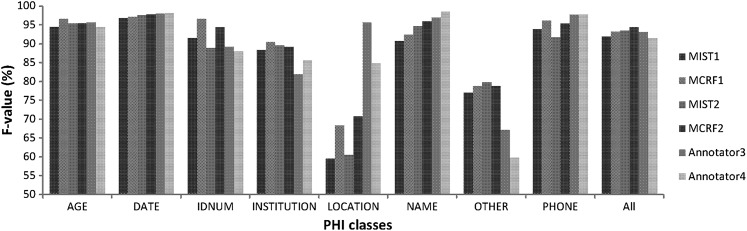
Table 3 shows the performance of the humans compared with that of the customized systems on the 10% random subset. Both humans performed worst when identifying PHI in the OTHER category. Performance of humans and systems are close, especially for AGE, DATE, and ID, where statistical tests found no significant difference (lower part of table 3). Both systems performed significantly better than the two humans on OTHER and better than annotator 3 on INSTITUTION. They both performed worse on LOCATION. Humans achieved better performance than the systems on NAME and better than MIST2 on PHONE. Both humans obtained a lower overall F value than the systems, but the highest recall was obtained by annotator 4. Figure 5 visualizes the F values obtained by the four systems and the two annotators.
Table 3.
Performance of humans versus automated systems (per-tag and overall precision (P), recall (R) and F value (F))
| Performance of humans versus automated systems | ||||||||||||
| Annotator 3 | Annotator 4 | MIST2 | MCRF2 | |||||||||
| P | R | F | P | R | F | P | R | F | P | R | F | |
| AGE | 99.51 | 91.93 | 95.57 | 94.59 | 94.17 | 94.38 | 95.50 | 95.07 | 95.28 | 97.21 | 93.72 | 95.43 |
| DATE | 98.17 | 97.78 | 97.97 | 98.56 | 97.62 | 98.09 | 96.73 | 98.57 | 97.65 | 97.86 | 97.78 | 97.82 |
| ID | 93.67 | 85.06 | 89.16 | 88.37 | 87.36 | 87.86 | 88.89 | 88.89 | 88.89 | 95.45 | 93.33 | 94.38 |
| INST | 78.33 | 85.98 | 81.98 | 84.52 | 86.59 | 85.54 | 90.12 | 89.02 | 89.57 | 93.33 | 85.37 | 89.17 |
| LOC | 97.78 | 93.62 | 95.65 | 86.67 | 82.98 | 84.78 | 66.67 | 55.32 | 60.47 | 82.86 | 61.70 | 70.73 |
| NAME | 98.92 | 94.93 | 96.88 | 99.08 | 97.92 | 98.50 | 93.52 | 95.71 | 94.60 | 95.49 | 96.36 | 95.92 |
| OTH | 68.77 | 65.55 | 67.12 | 47.28 | 81.27 | 59.78 | 83.27 | 76.59 | 79.79 | 87.70 | 71.57 | 78.82 |
| PH | 96.92 | 98.44 | 97.67 | 96.92 | 98.44 | 97.67 | 88.41 | 95.31 | 91.73 | 95.31 | 95.31 | 95.31 |
| All | 93.95 | 92.15 | 93.04 | 88.45 | 94.55 | 91.40 | 93.31 | 93.66 | 93.49 | 95.73 | 92.91 | 94.30 |
| Anno3 vs Anno4 | Anno3 vs MCRF2 | Anno4 vs MCRF2 | Anno3 vs MIST2 | Anno4 vs MIST2 | Anno3 vs gold standard | Anno4 vs gold standard | MCRF2 vs gold standard | MIST2 vs gold standard | |
| p Value | p Value | p Value | p Value | p Value | p Value | p Value | p Value | p Value | |
| AGE | 0.5226 | 0.9628 | 0.6493 | 0.9368 | 0.6516 | *0.0001 | *0.0001 | *0.0001 | *0.0001 |
| DATE | 0.881 | 0.7514 | 0.7 | 0.4288 | 0.555 | *0.0001 | *0.0001 | *0.0001 | *0.0001 |
| ID | 0.7526 | 0.1885 | 0.1428 | 0.9377 | 0.8021 | *0.0001 | *0.0001 | *0.0001 | *0.0001 |
| INST | 0.2785 | *0.0107 | 0.3347 | *0.0078 | 0.2949 | *0.0001 | *0.0001 | *0.0001 | *0.0001 |
| LOC | 0.1579 | *0.0078 | 0.1891 | *0.0001 | *0.021 | *0.0001 | *0.0001 | *0.0001 | *0.0001 |
| NAME | *0.0221 | 0.2246 | *0.0026 | *0.0093 | *0.0002 | *0.0001 | *0.0001 | *0.0001 | *0.0001 |
| OTH | 0.0506 | *0.0029 | *0.0001 | *0.0006 | *0.0001 | *0.0001 | *0.0001 | *0.0001 | *0.0001 |
| PH | 1 | 0.1299 | 0.1231 | *0.0414 | *0.0492 | *0.0001 | *0.0001 | *0.0001 | *0.0001 |
| All | *0.0169 | 0.054 | *0.0002 | 0.5088 | *0.0105 | *0.0001 | *0.0001 | *0.0001 | *0.0001 |
Statistical significance tests between F values obtained by humans versus systems (*indicates statistical significance (p<0.05), Anno3=annotator 3, Anno4=annotator 4).
INST, institution; LOC, location; OTH, other; PH, phone.
Figure 5.
F values obtained by the systems and the humans. MCRF, Mallet conditional random field; MIST, MITRE Identification Scrubber Toolkit.
For each tag level and overall F value, the difference between each human and the gold standard was statistically significant (lower section of table 1), as was the difference between each system and the gold standard.
We also computed IAA between the four humans on the 350 documents they all annotated (bottom part of figure 4). IAA is high between all annotator pairs for AGE, DATE, NAME, PHONE categories, and overall. It is low for OTHER, and fluctuates between the various pairs for IDNUM, INSTITUTION, and LOCATION.
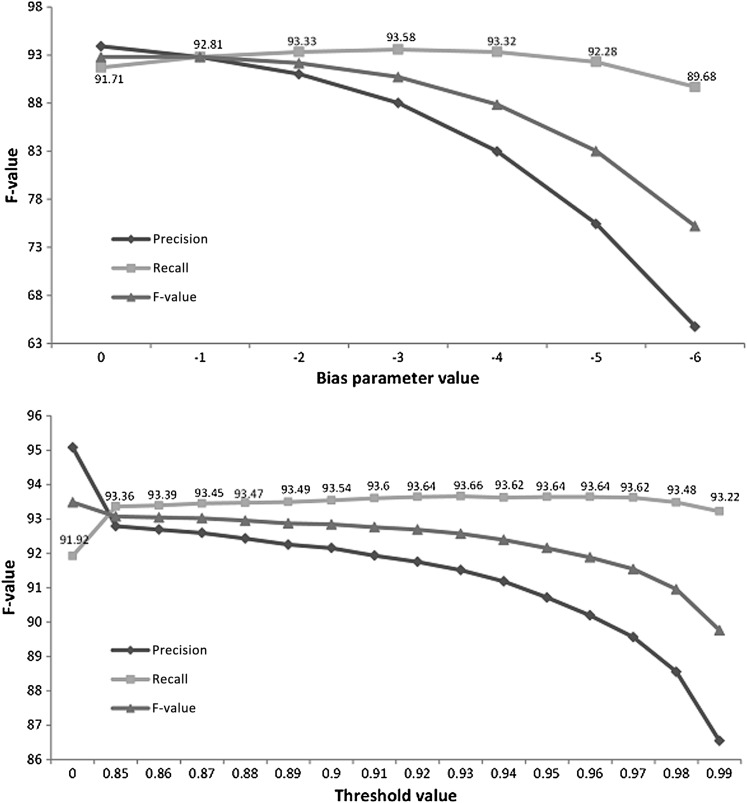
Recall bias
Changing the command line value parameter (MIST)ii and the threshold of non-PHI labels (in-house system) resulted in varying levels of recall changes. Figure 6 shows the results of the experiments for overall performance. The recall variation is rather limited on both systems. After a certain point, it reaches its maximum and then even decreases slightly, owing to the increasing number of non-PHI elements that are erroneously collapsed with true PHI. The maximum recall is 93.58 for MIST2 (bias parameter value of −3) and 93.66 for MCRF2 (0.93 threshold).
Figure 6.
Recall variations obtained by adjusting MIST's bias parameter and using thresholds for Mallet CRF probability scores (customized systems). CRF, conditional random field; MIST, MITRE Identification Scrubber Toolkit.
Impact of de-identification on subsequent IE
The impact of de-identification on the subsequent extraction of medication names is negligible. Results are shown in table 4, with statistical significance tests. The performance is slightly higher on de-identified text (including manually de-identified), but the difference is significant on the p<0.05 level only for two de-identified corpora. If Bonferroni correction is considered (because of the multiple comparisons), then none of the differences are significant.
Table 4.
Impact of de-identification on subsequent medication extraction task
| Original corpus with PHI | (1) Corpus de-identified with MCRF2 (PHI replaced with fake PHI) | (2) Corpus de-identified with MCRF2 (PHI replaced with *****) | (3) Corpus de-identified with MIST2 (PHI replaced with *****) | (4) Corpus de-identified with MCRF2 (configuration with best recall = 0.93 threshold; PHI replaced with *****) | (5) Corpus de-identified with MIST2 (configuration with best recall = −3 bias parameter; PHI replaced with *****) | (6) Corpus manually de-identified (PHI replaced with *****) | |||||||||||||||
| P | R | F | P | R | F | P | R | F | P | R | F | P | R | F | P | R | F | P | R | F | |
| Medication extraction performance | 96.28 | 89.27 | 92.64 | 96.44 | 88.94 | 92.54 | 96.31 | 89.50 | 92.78 | 96.33 | 89.56 | 92.82 | 96.34 | 89.26 | 92.66 | 96.49 | 88.88 | 92.53 | 96.39 | 89.53 | 92.83 |
| Statistical significance tests | ||||||
| Original vs (1) | Original vs (2) | Original vs (3) | Original vs (4) | Original vs (5) | Original vs (6) | |
| p Value | 0.1265 | 0.0802 | 0.0122* | 0.8275 | 0.2187 | 0.0137* |
PHI, protected health information.
Discussion
We performed error analysis for the best system on the development set (350 documents with 3845 PHI). The system made 476 errors. Of these, 13% (62) were boundary detection errors (partially tagged PHI (eg, only “5/12” in “Monday 5/12”) or PHI including extra tokens (eg, in “Fax 513-555-6666” Fax was also tagged)), 24.2% (115) were false positives, although 26.1% (30) of them were actually PHI but were labeled as the wrong category (eg, “Rochester NY” tagged as NAME instead of LOCATION). Ten of the false-positive results were true positives missing from the gold standard (missed by annotators 1 and 2). This happened for the NAME, ID, DATE, and OTHER categories. For NAME, a majority of false positives were device names (eg, “Sheehy” in “Sheehy tube”) or capitalized words (eg, “Status Asthmaticus”). For DATE, scores and measurements that looked like dates (eg, pain scores such as “2/10”) were often wrongly tagged. Finally, 62.82% (299) of the errors were missing PHI, although 9% (27) of those had been tagged but with the wrong category. Not counting the mislabeled elements, the system missed 38 NAMEs (out of 952), 3 of 124 IDs, 32 of 1744 DATEs, 8 of 164 PHONEs, 27 of 209 AGEs, 23 of 186 INSTITUTIONs, 3 of 56 LOCATIONs, and 138 of 410 OTHERs. The majority of false negatives (58.9%) were single-token elements (eg, single first names were more often missed by the system than first names followed by last names).
There are many take-home messages in our experiments that we believe should influence the decisions of institutional review boards about whether to accept the output of automated de-identification systems as comparable to manual de-identification. First, no single manual de-identification is 100% accurate. Even the results of double manual de-identification are not perfect. We found statistically significant differences between the gold standard that was the result of an adjudicated double de-identification and the output of the individual annotators. Consequently, evaluations that are based on single-annotated standard could misjudge the automated system's performance. Second, different note types have a different density of PHI (and potentially different context of the same PHI), and a de-identification system that is trained on a mix of note types will show varying performance on these note types. As a result, the de-identification performance of machine-learning systems will depend on the frequency of PHI types in the training data. High performance was achieved for most note types in our corpus, so we believe a single system can work for multiple note types if the training corpus includes the particular note type in sufficient number or if the PHI elements of a note type are expressed in similar ways as in other note types. Finally, installing a high-performance MIST-based prototype automated de-identification system is straightforward. It involves a few hours setup. Annotating the gold standard requires additional effort and its extent depends on multiple factors (eg, frequency of PHI in notes). The amount of annotations required to achieve high performance varies among the different PHI classes, depending on the variability of their form and context. For instance, we observed that PHONEs (which have regular patterns) and IDs (which occurred in easily identifiable contexts, eg, following “MRN:”) only required a couple of hundred annotations to achieve good performance (≥90% F values), while the mixed category of OTHER could not reach such high performance even with a couple of thousand annotations.
In addition, of interest for the translational research community, we found that automated de-identification did not reduce the accuracy of subsequent IE. The performances of the automated de-identification systems were sufficiently high that over-scrubbing errors did not affect the value of the de-identified corpus for extracting medical information.
Some of the limitations of our results are the de-identification performance for the LOCATION and OTHER categories, which should be improved; for proper performance evaluation, a larger sample size is necessary for EMAIL, IP, SSN, INITIALS; the corpus was obtained from only one institution, though it did include over 22 different note types selected from more than five million notes; we should experiment with at least one more subsequent NLP task to measure the impact of de-identification as results might be different with another task. Finally, the prototype needs to be transferred to a production environment to adequately estimate the cost of setting up a hospital's automated de-identification system.
Conclusion
In this paper, we presented a large-scale study on automated de-identification of clinical text, including over 3500 notes from a variety of types (>22). We showed that two automated systems, an existing system (MIST)32 and an in-house system, could obtain high performance (93.48% span-level and 95.20% token-level overall F values for the best system). We also compared results of the systems with those obtained by two human annotators and found that the performance of the systems rivaled that of the humans, with the humans even performing slightly worse on a couple of PHI categories and overall. Furthermore, unlike manual de-identification, the automated approach scales up to millions of documents quickly and inexpensively. Finally, this study also goes beyond de-identification performance testing by looking at the effect of de-identification on a subsequent IE task (medication extraction), for which no decrease in performance was seen.
Footnotes
Contributors: LD supervised the development of the ‘gold standard’, ran about 50% of the experiments, analyzed the results, created the tables and figures, and contributed to the draft and final manuscript. KM implemented about 40% of the experiments. GS consulted on the medication extraction experiments and contributed to the final manuscript. FX consulted on the Mallet package and conditional random fields algorithm, and contributed to the final manuscript. TL ran 6% of the experiments, annotated 10% of the corpus, and contributed to the final manuscript. QL ran 4% of the experiments and contributed to the final manuscript. KM extracted the data, consulted on data quality and cleaning, and contributed to the final manuscript. AJ consulted on machine learning and data quality. MK analyzed the errors, contributed to calculating statistics, and annotated the medication extraction and de-identification corpora. LS annotated 10% of the corpus. IS coordinated the work, supervised the experiments, data cleaning, analysis of the results, wrote the first draft, and contributed to the final manuscript. LD and IS had full access to all of the data in the study and take responsibility for its integrity and the accuracy of the data analysis. From the MITRE Corporation, John Aberdeen, Samuel Bayer, Lynette Hirschman, and Benjamin Wellner provided suggestions for MIST (MITRE Identification Scrubber Toolkit) optimization and answered numerous questions about the MIST infrastructure. We appreciate the suggestions of the anonymous institutional review board (IRB) members who approved our IRB protocol, and of the reviewers and editors of an earlier version of the manuscript. The i2b2 de-identified clinical records used in this research were provided by the i2b2 National Center for Biomedical Computing funded by U54LM008748 and were originally prepared for the shared tasks for challenges in natural language processing for clinical data organized by Dr Ozlem Uzuner, i2b2, and SUNY. The PhysioNet de-identified records used in this research were provided by PhysioNet (http://www.physionet.org/physiotools/deid/).
Funding: IS and LD were partially supported by 5R00LM010227-04. Internal funds from Cincinnati Children's Hospital Medical Center supported the annotators, and partially KM.
Competing interests: None.
Ethics approval: Ethics approval was provided by the institutional review board of Cincinnati Children's Hospital Medical Center.
Provenance and peer review: Not commissioned; externally peer reviewed.
We did not evaluate MIST on those corpora because (1) the two systems are very similar and (2) MIST was already evaluated on the i2b2 corpus (its F value ranked first in the i2b2 challenge).
MIST is set to have a slight recall bias (−1) out-of-the-box.
References
- 1. Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008:128–44 [PubMed] [Google Scholar]
- 2. Hicks J. The Potential of Claims Data to Support the Measurement of Health Care Quality. Santa Monica, CA: RAND Corporation, 2003 [Google Scholar]
- 3. Jha AK. The promise of electronic records: around the corner or down the road? JAMA 2011;306:880–1 [DOI] [PubMed] [Google Scholar]
- 4. Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform 2009;42:760–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Warner JL, Anick P, Hong P, et al. Natural language processing and the oncologic history: is there a match? J Oncol Pract 2011;7:e15–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhan C, Miller MR. Administrative data based patient safety research: a critical review. Qual Saf Health Care 2003;12(Suppl 2):ii58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc 2005;12:448–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA 2011;306:848–55 [DOI] [PubMed] [Google Scholar]
- 10. Health Insurance Portability and Accountability Act. P.L. 104–191, 42 U.S.C. 1996 [Google Scholar]
- 11. Wolf MS, Bennett CL. Local perspective of the impact of the HIPAA privacy rule on research. Cancer 2006;106:474–9 [DOI] [PubMed] [Google Scholar]
- 12. Dunlop AL, Graham T, Leroy Z, et al. The impact of HIPAA authorization on willingness to participate in clinical research. Ann Epidemiol 2007;17:899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meystre SM, Friedlin FJ, South BR, et al. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Methodol 2010;10:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sweeney L. Replacing personally-identifying information in medical records, the Scrub system. Proc AMIA Annu Fall Symp 1996:333–7 [PMC free article] [PubMed] [Google Scholar]
- 15. Ruch P, Baud RH, Rassinoux AM, et al. Medical document anonymization with a semantic lexicon. Proc AMIA Symp 2000:729–33 [PMC free article] [PubMed] [Google Scholar]
- 16. Thomas SM, Mamlin B, Schadow G, et al. A successful technique for removing names in pathology reports using an augmented search and replace method. Proc AMIA Symp 2002:777–81 [PMC free article] [PubMed] [Google Scholar]
- 17. Berman JJ. Concept-match medical data scrubbing. How pathology text can be used in research. Arch Pathol Lab Med 2003;127:680–6 [DOI] [PubMed] [Google Scholar]
- 18. Fielstein EM, Brown SH, Speroff T, eds. Algorithmic De-identification of VA Medical Exam Text for HIPAA Privacy Compliance: Preliminary Findings. Medinfo. 2004 [Google Scholar]
- 19. Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am J Clin Pathol 2004;121:176–86 [DOI] [PubMed] [Google Scholar]
- 20. Beckwith BA, Mahaadevan R, Balis UJ, et al. Development and evaluation of an open source software tool for deidentification of pathology reports. BMC Med Inform Decis Mak 2006;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedlin FJ, McDonald CJ. A software tool for removing patient identifying information from clinical documents. J Am Med Inform Assoc 2008;15:601–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morrison FP, Li L, Lai AM, et al. Repurposing the clinical record: can an existing natural language processing system de-identify clinical notes? J Am Med Inform Assoc 2009;16:37–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neamatullah I, Douglass MM, Lehman LW, et al. Automated de-identification of free-text medical records. BMC Med Inform Decis Mak 2008;8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taira RK, Bui AA, Kangarloo H. Identification of patient name references within medical documents using semantic selectional restrictions. Proc AMIA Symp 2002:757–61 [PMC free article] [PubMed] [Google Scholar]
- 25. Uzuner O, Sibanda TC, Luo Y, et al. A de-identifier for medical discharge summaries. Artif Intell Med 2008;42:13–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gardner J, Xiong L. HIDE: an integrated system for health information DE-identification. Comp Med Sy 2008:254–9 [Google Scholar]
- 27. Arakami E, ed. Automatic Deidentification by Using Sentence Features and Label Consistency. I2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. 2006 [Google Scholar]
- 28. Guo Y, Gaizauskas R, Roberts I, et al., eds. Identifying Personal Health Information Using Support Vector Machines. I2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. 2006 [Google Scholar]
- 29. Hara K, ed. Applying a SVM Based Chunker and a Text Classifier to the Deid Challenge. I2b2 Workshop on Challenges in Natural Language Processing for Clinical Data. 2006 [Google Scholar]
- 30. Szarvas G, Farkas R, Busa-Fekete R. State-of-the-art anonymization of medical records using an iterative machine learning framework. J Am Med Inform Assoc 2007;14:574–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wellner B, Huyck M, Mardis S, et al. Rapidly retargetable approaches to de-identification in medical records. J Am Med Inform Assoc 2007;14:564–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aberdeen J, Bayer S, Yeniterzi R, et al. The MITRE Identification Scrubber Toolkit: design, training, and assessment. Int J Med Inform 2010;79:849–59 [DOI] [PubMed] [Google Scholar]
- 33. Benton A, Hill S, Ungar L, et al. A system for de-identifying medical message board text. BMC Bioinformatics 2011;12(Suppl 3):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gardner J, Xiong L, Wang F, et al. An evaluation of feature sets and sampling techniques for de-identification of medical records. In: Veinot T, ed. Proceedings of the 1st ACM International Health Informatics Symposium (IHI '10). New York, NY, USA: ACM, 2010:183–90 [Google Scholar]
- 35. Uzuner O, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007;14:550–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCallum AC. MALLET: a Machine Learning for Language Toolkit. 2002 [Google Scholar]
- 37. Lafferty J, McCallum A, Pereira F. Conditional random fields: probabilistic models for segmenting and labeling sequence data. Proc. 18th International Conf. on Machine Learning. Morgan Kaufmann San Francisco, CA, USA, 2001:282–9 [Google Scholar]
- 38. Schmid H. Probabilistic part-of-speech tagging using decision trees. Proceedings of International Conference on New Methods in Language Processing. Manchester, UK, 1994 [Google Scholar]
- 39. Friedman C, Hripcsak G. Evaluating natural language processors in the clinical domain. Methods Inf Med 1998;37:334–44 [PubMed] [Google Scholar]
- 40. Hripcsak G, Rothschild AS. Agreement, the f-measure, and reliability in information retrieval. J Am Med Inform Assoc 2005;12:296–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Noreen EW. Computer-Intensive Methods for Testing Hypotheses: An Introduction. New-York: Wiley, 1989 [Google Scholar]
- 42. Cinchor N. The statistical significance of MUC4 results. MUC4 '92 Proceedings of the 4th Conference on Message Understanding; 1992. Association for Computational Linguistics, Stroudsburg, PA, USA [Google Scholar]
- 43. Ogren P, Savova G, Chute C. Constructing evaluation corpora for automated clinical named entity recognition. Proceedings of the Sixth International Conference on Language Resources and Evaluation (LREC'08), European Language Resources Association (ELRA). Marrakech, Morocco, 2008 [Google Scholar]
- 44. Goldberger AL, Amaral LA, Glass L, et al. PhysioBank, PhysioToolkit, and Physionet: components of a new research resource for complex physiologic signals. Circulation 2000;101:E215–20 [DOI] [PubMed] [Google Scholar]
- 45. Minkov E, Wang R, Tomasic A, et al. NER Systems that Suit User's Preferences: Adjusting the Recall-Precision Trade-off for Entity Extraction. Proceedings of the Human Language Technology Conference of the NAACL, Companion Volume: Short Papers 2006:93–6 [Google Scholar]
- 46. Halgrim S, Xia F, Solti I, et al. Extracting medication information from discharge summaries. Proceedings of the NAACL HLT 2010 Second Louhi Workshop on Text and Data Mining of Health Documents. Association for Computational Linguistics, Stroudsburg, PA, USA; 2010:61–7 [Google Scholar]
- 47. Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]