Abstract
Objective
De-identification allows faster and more collaborative clinical research while protecting patient confidentiality. Clinical narrative de-identification is a tedious process that can be alleviated by automated natural language processing methods. The goal of this research is the development of an automated text de-identification system for Veterans Health Administration (VHA) clinical documents.
Materials and methods
We devised a novel stepwise hybrid approach designed to improve the current strategies used for text de-identification. The proposed system is based on a previous study on the best de-identification methods for VHA documents. This best-of-breed automated clinical text de-identification system (aka BoB) tackles the problem as two separate tasks: (1) maximize patient confidentiality by redacting as much protected health information (PHI) as possible; and (2) leave de-identified documents in a usable state preserving as much clinical information as possible.
Results
We evaluated BoB with a manually annotated corpus of a variety of VHA clinical notes, as well as with the 2006 i2b2 de-identification challenge corpus. We present evaluations at the instance- and token-level, with detailed results for BoB's main components. Moreover, an existing text de-identification system was also included in our evaluation.
Discussion
BoB's design efficiently takes advantage of the methods implemented in its pipeline, resulting in high sensitivity values (especially for sensitive PHI categories) and a limited number of false positives.
Conclusions
Our system successfully addressed VHA clinical document de-identification, and its hybrid stepwise design demonstrates robustness and efficiency, prioritizing patient confidentiality while leaving most clinical information intact.
Introduction
Recent advances in health information technology promise considerable benefits to health care quality and clinical research. The widespread adoption of electronic health records (EHR) provides a unique framework for data-sharing, robust computational processing, and leading-edge research initiatives.1 However, it also comprises risks related to patient confidentiality. Medical identity theft is increasing, a risk exacerbated with the use of EHRs,2 and patients are concerned about unauthorized use of their personal health information.3
When clinical data are used for research purposes, patient informed consent is required unless the data are de-identified. In the USA, patient confidentiality is regulated by the Health Insurance Portability and Accountability Act (HIPAA; codified as 45 CFR §160 and 164) and the Common Rule.4 For clinical data to be considered de-identified, the HIPAA ‘Safe Harbor’ standard5 requires the removal of 18 different protected health information (PHI) elements, such as person names, social security numbers, dates, locations, etc. Therefore, since manual de-identification is a tedious and expensive process, the development of accurate software tools for automatically de-identifying documents has become an important need. Such tools will make clinical research easier across institutions, and facilitate the release of de-identified corpora, a critical resource for the development of scalable and generalizable clinical natural language processing (NLP) applications.
In this paper we introduce ‘BoB’, our Best-of-Breed automated clinical text de-identification system being developed within the Consortium for Healthcare Informatics Research at the Department of Veterans Affairs. In terms of de-identification, BoB is adopting a conservative approach, prioritizing patient confidentiality, as reflected in its design and the evaluation methodology we have followed.
Background and related work
Over the last decade, various methods for automated de-identification have been developed.1 This interest in de-identification has also been reflected in the successful organization of the 2006 i2b2 de-identification challenge.6
Automated text de-identification applications are focused on the removal of HIPAA PHI identifiers. Such applications must be exceptionally effective at removing PHI while keeping the resulting de-identified document usable. A system that excessively redacts documents, also redacting relevant clinical information, would compromise the interpretability of those documents. To accomplish this task, researchers have developed applications utilizing strategies based on rule-based and machine learning techniques. Rule-based approaches tackle the de-identification problem with pattern matching, regular expressions, and dictionary searches,7–10 but have limited generalizability that depends on the quality of the patterns and dictionaries. In contrast, machine learning-based approaches11–15 usually rely on supervised methods able to learn from training examples and predict PHI annotations. However, these methods require large annotated collections of representative documents. More details about de-identification approaches can be found in Meystre and colleagues’ review.1
Because of text de-identification's resemblance with traditional named entity recognition (NER),16 researchers have also experimented with pre-trained newswire NER tools.17 18 But even if they obtained decent performance with some PHI categories, NER approaches were always surpassed by systems specifically designed for de-identification. The special characteristics of clinical narratives such as fragmented and incomplete utterances, lack of punctuation marks and formatting, domain specific terminology, and the fact that the same entities can appear both as PHI and non-PHI (eg, ‘Mr Gilbert’ vs ‘Gilbert syndrome’), make the usage of pre-trained traditional newswire NER approaches complicated, and explain why de-identification of clinical texts is a challenging task.
The design and implementation of our de-identification system was based on a previous study on the suitability of existing de-identification tools used on Veterans Health Administration (VHA) documents19. In that study, we performed an ‘out-of-the-box’ evaluation of three rule-based de-identification systems7–9 and two systems based on machine learning.11 12 We observed that rule-based methods obtained better recall, while machine learning approaches addressed precision quite well. Although none of the evaluated systems reached sufficient ‘out-of-the-box’ performance to de-identify our VHA documents, that study gave us compelling insight into the best methods to use for the design and development of our best-of-breed (hence the name ‘BoB’) VHA clinical text de-identification system.
Materials and methods
De-identification corpus
We generated a manually annotated reference standard with a variety of VHA clinical documents. We used a stratified random sampling approach to select clinical documents with more than 500 words authored between April 1, 2008 and March 31, 2009. The 100 most frequent note types (addendum excluded), from about 180 different types, were used as strata for sampling. They included consult notes from different specialties, nursing notes, discharge summaries, emergency room notes, progress notes, preventive health notes, surgical pathology reports, psychiatry notes, history and physical reports, informed consent, operation reports, and other less common note types. We then randomly selected eight documents in each stratum, reaching a total of 800 clinical documents. Each document was independently annotated by two reviewers, disagreements were adjudicated by a third reviewer, and a fourth reviewer eventually examined ambiguous and difficult adjudicated cases. The overall inter-annotator agreement (IAA) was 0.83 when considering exact agreement, and 0.91 for inexact agreement. As reported in other works about manual annotation of PHI,20 the IAA varies depending on the PHI category; in our case, Social Security Numbers, Patient Names, and Dates were the categories with high agreement (see more details in supplementary online appendix A).
The annotation schema was designed in accordance with the HIPAA ‘Safe Harbor’ legislation.5 We adopted a more conservative approach, considering states and countries as PHI, as well as the year in all date annotations, and other identifiers such as organizations (Other Organization Name), armed forces-specific information (Deployment), and specific and generic mentions of health care facilities (Healthcare Unit Name).
The additional complexity of recognizing these PHI categories makes successful automated text de-identification more difficult, but this conservative perspective is justified by our main concern: maximize patient confidentiality.
Further details about our annotation schema are available in online appendix A.
Best-of-breed automatic clinical text de-identification system (aka BoB) description
The architectural design of BoB was focused on the following goals:
Take advantage of rule-based and machine learning-based methods that have been previously exploited for de-identification.
Prioritize patient confidentiality. Since recall (equivalent to sensitivity here) is the most important measure for de-identification—patient data cannot be disclosed at any rate—we decided to focus our efforts on high recall, even if somehow compromising on precision.
Tackle the issue of scarcity of training examples. Large manually de-identified corpora are difficult to create, costly, and always a tedious task for human annotators. With our system, we want to alleviate this need, creating accurate techniques able to work satisfactorily with fewer training examples.
Make the system platform-independent and easily configurable and reusable.
BoB's architecture is based on the UIMA framework,21 which provides platform independency and makes it easily customizable. We designed BoB's architecture based on two main components:
A high-sensitivity extraction component, prioritizing patient confidentiality and implementing methods specifically tailored to obtain high sensitivity. It will detect all candidates that could possibly be considered PHI.
A false positives filtering component to mitigate the large amount of false positives produced by the previous component. This independent component is intended to improve overall precision (equivalent to positive predictive value) and integrates techniques that allow the system to disambiguate the candidate PHI annotations and classify them as true or false positives.
This stepwise processing enables us to separately design and implement methods focused exclusively on recall or precision, and then tackle the task as two independent problems. In our previous study, we identified that rule-based methods achieved higher recall, while machine learning approaches obtained better precision. We therefore decided to implement our first component—the high-sensitivity extraction—mainly using rule-based and pattern matching techniques, and mainly use machine learning algorithms for the second component. This truly hybrid architectural design differs from other systems that confront the de-identification task as a whole. Moreover, considering two independent steps allows us to take advantage of the strong points of both rule-based and machine learning methods, unlike other systems that base their predictions on one of them, or on a limited combination of techniques.
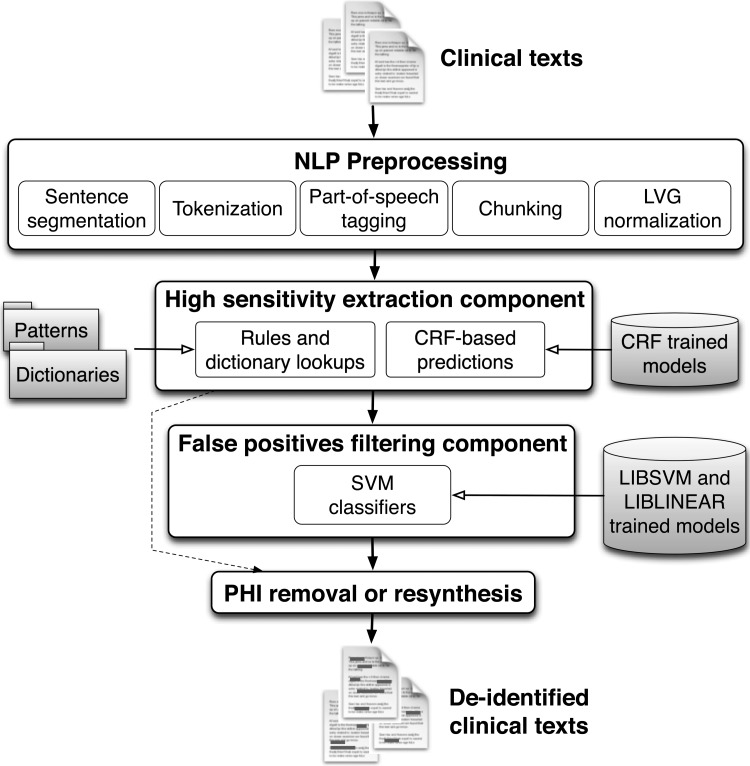
Figure 1 depicts an overview of our system's architecture. As shown in the figure, the last step could be PHI removal or its resynthesis (ie, replacing PHI identifiers with realistic surrogates).
Figure 1.
BoB's architecture. CRF, conditional random fields; LVG, lexical variant generation; NLP, natural language processing; PHI, protected health information; SVM, support vector machine.
NLP preprocessing
The workflow of the system starts with several NLP steps that prepare documents. They include sentence segmentation, tokenization, part-of-speech tagging, phrase chunking, and word normalization using lexical variant generation (LVG).22
We adapted several cTAKES modules,23 which implement wrappers for OpenNLP tools.24 Additionally, we added a regular expression-based annotator prior to the detection of the sentences. This annotator was developed to better handle special tabulations and whitespaces formatting, splitting these portions of the document into different chunks that can then be processed for sentence segmentation.
The high-sensitivity extraction component
The aim of this component is to obtain high recall (sensitivity), and the following methods dedicated to this objective are therefore used:
Rule-based module: This integrates pattern matching techniques, dictionary lookups, and several heuristics. Our patterns were adapted from patterns implemented previously7–9 and new patterns were developed to cover the different PHI formats present in VHA documents (eg, to support date-time formats such as ‘09/09/09@1200’). A total of about 130 regular expressions were considered. We used our training corpus for creating and adapting the patterns. We also implemented dictionary lookups using Lucene,25 with keyword and fuzzy searches on dictionaries of first and last names (from the 1990 US census, as in Neamatullah et al8), US states, cities, and counties, countries, companies (from Wikipedia, usps.com, and other web resources), common words (from Neamatullah et al8), and clinical eponyms and healthcare clinics extracted from our VHA training corpus. Fuzzy dictionary searches were implemented for person names with a similarity threshold (based on the Levenshtein edit distance) higher than 0.74. Our dictionary matches are not case sensitive, and we integrated a simple disambiguation procedure based on a list of common words and the capitalization of the token, as well as some heuristics based on part-of-speech tagging (eg, ‘a_DT brown_JJ spot_NN’ vs ‘by_IN mr._NN brown_NN’, brown_NN is considered PHI while brown_JJ would not).
The rule-based module maximizes recall, even if precision is altered. However, it is also dependent on the quality of the patterns and on the completeness of dictionaries. Thus, if unusual PHI formats or instances not supported by our patterns and dictionaries appear in the documents, they will be missed. To solve this issue, we added another module based on machine learning predictions:
CRF module: Machine learning classifiers are more generalizable and can detect annotations based on contextual and morphological features rather than using a fixed set of patterns. Therefore, to further enhance recall, we added machine learning classifiers, as commonly used in NER tasks.26 27 We used the conditional random fields (CRF) classifier provided by the Stanford NLP group.28 We trained the classifiers using our training corpus for: (1) person names, including Patient Names, Relative Names, Healthcare Provider Names, and Other Person Names; (2) Street City; (3) State Country; (4) Healthcare Units; (5) Other Organizations; and (6) Dates. We also integrated the default models provided with the Stanford NER. See online appendix B for details about the learning features and their selection.
The false positives filtering component
The high-sensitivity component maximizes recall but also produces numerous false positives. We therefore designed the next component of our pipeline to filter out as many false positives as possible. For this task, we built machine learning classifiers trained to differentiate PHI candidate annotations as true or false positives. Unlike other machine learning-based de-identification systems, our classifiers were trained with the annotations derived from the high-sensitivity extraction component, and then instead of tagging every token as PHI or non-PHI, our classifiers decide if an actual annotation is a false or true positive. This design allows us to create more accurate classifiers with less training examples (especially for PHI categories with few instances in our corpus), to use more discriminative features, and to reduce the decision dimensionality. We only created classifiers for PHI categories that needed improved precision. ZIP code, Age >89, Electronic Address and SSN categories already achieved about 100% precision by processing the high-sensitivity extraction component. We therefore did not generate classifiers for these PHI categories. We experimented with the following combinations of classifiers.
Individual classifiers
We created one support vector machine (SVM) classifier for each PHI category (using LIBSVM29), with all person name categories considered as one training class, and also one classifier for clinical eponyms. We then filter annotations from the high-sensitivity extraction component using the corresponding PHI category SVM classifier.
Multi-class classifier
For this configuration, we created one multi-class SVM classifier to filter out false positives. This classifier then decides to which PHI category (if any) each annotation corresponds. We also included clinical eponyms as a class in this classifier.
Best configuration
This configuration obtained the highest sensitivity for all PHI categories. Instead of creating one multi-class classifier, or creating individual classifiers that could not have enough specific training examples for some categories, we created several classifiers for similar PHI categories:
Three SVM classifiers: (1) person names (ie, Patient Name, Relative Name, Healthcare Provider Name, Other Person Name); (2) numerical PHI identifiers (ie, Date, Phone Number, Other ID Number); and (3) clinical eponyms.
One linear classifier for narrative text PHI categories (ie, Street City, State Country, Deployment, Healthcare Units, Other Organizations). In this case we experimented with SVM, but linear classification (LIBLINEAR30) works well when the number of learning features is much larger than the number of training instances, and indeed performed better than SVM.
More details about the machine learning models can be found in online appendix B.
Results
To evaluate BoB's performance, we randomly split our annotated corpus in two subsets of 500 documents for training and 300 for blind testing. The selection was carried out without stratification of document types. The size of the testing corpus was estimated to allow for the demonstration of a difference of 2% or more in patient names’ recall (two-tailed, significance level of 0.05, and power of 0.8). Details about the PHI distribution can be found in online appendix A.
We present results in terms of precision (positive predictive value), recall (sensitivity), and F measure (harmonic mean of recall and precision31). To emphasize sensitivity, we also provide the F2 measure results in addition to the traditional F1 measure, which weighs recall (twice) higher than precision, as described in the equation:
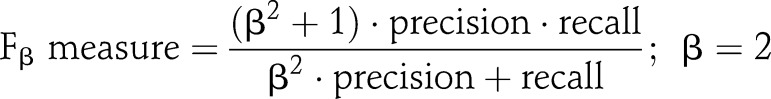 |
Statistical analysis of differences between systems and configurations performance was realized with the unpaired Student t test (two-tailed; level of significance 0.05).
In addition, we considered two different levels of evaluation:
Instance-level evaluation (table 1) considers the entire PHI annotation as the unit of evaluation. To emphasize sensitivity, we propose ‘fully contained’ matches, which consider the predictions as true positives when they at least overlap with the entire PHI annotation in the reference. We observed that exact matches are sometimes excessively strict (eg, when annotations include non-functional words or word delimiters) and that partial matches could leave fragments which could uniquely establish a link to the patient. Fully contained matches prioritize sensitivity by assuring complete redaction, but also relax the exact matching strategy.
Token-level evaluation (table 2) considers each token (eg, word) as the unit of analysis. We split all text in tokens separated by whitespace (eg, space, carriage return).
Table 1.
Instance-level evaluation results considering fully contained matches
| VHA PHI categories | MIT deid | One-step CRF | BoB rules | BoB CRF | BoB rules+CRF | BoB full | |
|---|---|---|---|---|---|---|---|
| R | R | R | R | R | R | P | |
| Patient Name | 0.590 | 0.949 | 0.972 | 0.953 | 0.992 | 0.980 | 0.707* |
| Relative Name | 0.600 | 0.920 | 0.960 | 0.960 | 0.960 | 0.920 | |
| Healthcare Provider Name | 0.319 | 0.898 | 0.920 | 0.898 | 0.963 | 0.943 | |
| Other Person Name | 0.111 | 0.667 | 1 | 0.667 | 1 | 0.888 | |
| Street City | 0.828 | 0.802 | 0.962 | 0.872 | 0.974 | 0.943 | 0.679 |
| State Country | 0.689 | 0.824 | 0.953 | 0.757 | 0.973 | 0.878 | 0.751 |
| Deployment | 0.057 | 0.887 | 1 | – | 1 | 0.887 | 0.859 |
| ZIP Code | 1 | 1 | 1 | – | 1 | 1 | 1 |
| Healthcare Units | 0.008 | 0.732 | 0.832 | 0.755 | 0.914 | 0.811 | 0.836 |
| Other Organizations | 0.033 | 0.483 | 0.824 | 0.549 | 0.912 | 0.725 | 0.578 |
| Date | 0.399 | 0.892 | 0.963 | 0.917 | 0.977 | 0.971 | 0.934 |
| Age>89 | 0.250 | 0.500 | 1 | – | 1 | 1 | 0.8 |
| Phone Number | 0.494 | 0.835 | 0.989 | – | 0.989 | 0.956 | 1 |
| Electronic Address | 1 | 0.500 | 1 | – | 1 | 1 | 1 |
| SSN | 1 | 0.407 | 1 | – | 1 | 1 | 0.964 |
| Other ID Number | 0.117 | 0.822 | 0.978 | – | 0.978 | 0.917 | 0.831 |
| Overall macro-averaged | 0.468 | 0.757 | 0.960 | – | 0.977 | 0.926 | 0.841 |
| Overall micro-averaged | |||||||
| Precision Recall F1 measure F2 measure | 0.311 | 0.920 | 0.362 | – | 0.346 | 0.836 | |
| 0.350 | 0.842 | 0.928 | – | 0.961 | 0.922 | ||
| 0.329 | 0.879 | 0.521 | – | 0.509 | 0.877 | ||
| 0.341 | 0.856 | 0.707 | – | 0.709 | 0.904 | ||
*BoB annotates all person names as one PHI category.
CRF, conditional random fields; P, precision; PHI, protected health information; R, recall; VHA, Veterans Health Administration.
Table 2.
Token-level evaluation results considering exact matches
| VHA PHI categories | MIT deid | One-step CRF | BoB rules | BoB CRF | BoB rules+CRF | BoB full | |
|---|---|---|---|---|---|---|---|
| R | R | R | R | R | R | P | |
| Patient Name | 0.724 | 0.956 | 0.977 | 0.962 | 0.994 | 0.985 | 0.642* |
| Relative Name | 0.909 | 0.939 | 0.970 | 0.970 | 0.970 | 0.939 | |
| Healthcare Provider Name | 0.747 | 0.925 | 0.938 | 0.916 | 0.965 | 0.943 | |
| Other Person Name | 0.867 | 0.800 | 1 | 0.800 | 1 | 0.933 | |
| Street City | 0.765 | 0.728 | 0.878 | 0.798 | 0.929 | 0.887 | 0.682 |
| State Country | 0.656 | 0.812 | 0.944 | 0.744 | 0.956 | 0.869 | 0.839 |
| Deployment | 0.177 | 0.859 | 0.934 | – | 0.934 | 0.869 | 0.915 |
| ZIP Code | 1 | 1 | 1 | – | 1 | 1 | 1 |
| Healthcare Units | 0.080 | 0.716 | 0.834 | 0.748 | 0.902 | 0.798 | 0.779 |
| Other Organizations | 0.098 | 0.503 | 0.798 | 0.596 | 0.880 | 0.721 | 0.606 |
| Date | 0.617 | 0.922 | 0.972 | 0.938 | 0.978 | 0.972 | 0.935 |
| Age>89 | 0.250 | 0.500 | 1 | – | 1 | 1 | 0.8 |
| Phone Number | 0.565 | 0.810 | 0.991 | – | 0.991 | 0.939 | 1 |
| Electronic Address | 1 | 0.500 | 1 | – | 1 | 1 | 1 |
| SSN | 1 | 0.407 | 1 | – | 1 | 1 | 0.964 |
| Other ID number | 0.094 | 0.855 | 0.983 | – | 0.983 | 0.936 | 0.82 |
| Overall macro-averaged | 0.597 | 0.764 | 0.951 | – | 0.968 | 0.925 | 0.845 |
| Overall micro-averaged | |||||||
| Precision | 0.734 | 0.931 | 0.420 | – | 0.392 | 0.815 | |
| Recall | 0.489 | 0.859 | 0.933 | – | 0.957 | 0.921 | |
| F1 measure | 0.587 | 0.893 | 0.579 | – | 0.556 | 0.864 | |
| F2 measure | 0.524 | 0.872 | 0.749 | – | 0.743 | 0.897 | |
*BoB annotates all person names as one PHI category.
CRF, conditional random fields; P, precision; PHI, protected health information; R, recall; VHA, Veterans Health Administration.
To select the best false positives filtering component configuration, we compared BoB's performance with the three configurations described in the previous section. As shown in table 3, the configuration with the best recall (sensitivity) is BoB best configuration. Since BoB's main goal is to protect patient confidentiality, we chose this configuration for the rest of the results presented below. Nonetheless, we also observed that BoB achieved more balanced results in terms of recall and precision (ie, 89% in both recall and precision, for the BoB individual classifiers configuration), and BoB multi-class configuration offered the best precision (95%). BoB could therefore be tailored to preserve more clinical content with a slight impact on de-identification, depending on the final purpose of the de-identified documents and the legal agreements that could be imposed to avoid re-identification.
Table 3.
Overall results of the different configurations of BoB's false positives filtering component: instance-level results with fully contained matches and using the testing corpus
| BoB's false positives filtering component | BoB individual classifiers | BoB multi-class | BoB best configuration |
|---|---|---|---|
| Overall micro-averaged (PHI level) | |||
| Precision | 0.895 | 0.952 | 0.836 |
| Recall | 0.895 | 0.886 | 0.922 |
| F1 measure | 0.895 | 0.918 | 0.877 |
| F2 measure | 0.895 | 0.898 | 0.904 |
PHI, protected health information.
To gain insight into the strengths of BoB's components, we present BoB's performance at four processing steps: (1) after the rule-based module from the high-sensitivity extraction component (‘BoB rules’ in the tables) only; (2) after processing BoB's CRF models (‘BoB CRF’; these models were generated using our training corpus, and only for some PHI categories); (3) after the complete high-sensitivity extraction component (‘BoB rules+CRF’); and (4) after running the entire pipeline (ie, the high-sensitivity extraction and false positive filtering components; ‘BoB full’).
Additionally, we also present the results achieved by the Stanford CRF classifier trained using our 500 document training corpus, and detecting all PHI categories at the overall PHI level, that is, PHI versus non-PHI (‘one-step CRF’ column). For this experiment, we used the same learning features as in BoB's CRF module (see online appendix B). Furthermore, in order to have a reference point for available text de-identification systems, we also ran the MIT deid system8 ‘out-of-the-box’ in our evaluation. This system bases the de-identification on pattern matching techniques and dictionary searches. To coherently run this system, we had to map the MIT deid output to our categories (eg, the Medical Record Number category in the MIT deid system was mapped to Other ID Number).
Finally, in order to test the generalizability of BoB's design and methods, we evaluated our system with the 2006 i2b2 de-identification corpus. This corpus differs in type and structure from our documents, and was resynthesized with surrogates that, in most cases, could not be found in dictionaries, making their de-identification difficult for techniques based on dictionaries (see further details in Uzuner et al6). Nevertheless, this experiment is a good challenge for our hybrid approach, and provides useful information about the generalizability of our methods. To test BoB with this corpus, we trained BoB's machine learning classifiers (CRF and SVM models) with the i2b2 training corpus, and mapped our PHI categories to the i2b2 ones (see table 4). We removed the annotators for PHI categories not supported by i2b2 annotations (ie, Deployment, Other Organizations, and Electronic Address) and performed two different experiments: (1) running BoB without any modification or adaptation of our rule-based techniques or dictionaries (‘BoB full’ in the table), and (2) as we realized that BoB's recall for ID, Phone Numbers, and Ages was low in comparison with our VHA-based results, we created a few new patterns for some formats of these PHI categories found in the i2b2 training corpus (ie, ‘BoB new patterns’). In more detail, we built one pattern for Ages like ‘98y’, another pattern for Phones with whitespaces between the parenthesis and area code (eg, “( 800 )” 000-000’), and finally three patterns for IDs such as ‘000-00-00-00 Abc123’ and ‘AZ12 ABC123/123Abc’.
Table 4.
Evaluation with the 2006 i2b2 de-identification challenge testing corpus: instance-level results with fully contained matches
| VHA PHI categories | i2b2 PHI categories | #inst. train | #inst. test | BoB full | BoB new patterns | ||
|---|---|---|---|---|---|---|---|
| R | P | R | P | ||||
| Patient Name | Patient | 684 | 245 | 0.975 | 0.834* | 0.975 | 0.834* |
| Relative Name | |||||||
| Other Person Name | |||||||
| Healthcare Provider Name | Doctor | 2681 | 1070 | 0.980 | 0.980 | ||
| Street City | Location | 144 | 119 | 0.613 | 0.767 | 0.613 | 0.767 |
| State Country | |||||||
| ZIP Code | |||||||
| Healthcare Units | Hospital | 1724 | 676 | 0.910 | 0.790 | 0.910 | 0.790 |
| Date | Date | 5167 | 1931 | 0.990 | 0.942 | 0.990 | 0.942 |
| Age>89 | Age | 13 | 3 | 0 | 0 | 1 | 1 |
| Phone Number | Phone number | 174 | 58 | 0.810 | 0.978 | 0.914 | 0.981 |
| SSN Other ID Number | ID | 3666 | 1143 | 0.784 | 0.964 | 0.980 | 0.805 |
| Overall macro-averaged | 0.758 | 0.753 | 0.920 | 0.874 | |||
| Overall micro-averaged | Precision | 0.878 | 0.846 | ||||
| Recall | 0.921 | 0.965 | |||||
| F1 measure | 0.899 | 0.902 | |||||
| F2 measure | 0.912 | 0.939 | |||||
*BoB annotates all person names as one PHI category.
P, precision; PHI, protected health information; R, recall; VHA, Veterans Health Administration.
Discussion
Our novel text de-identification system achieved very competitive performance. More importantly, it demonstrated that our choice of tackling the de-identification problem as two separate tasks allowed us to make the most of each method (ie, rule-based techniques for high sensitivity and machine learning algorithms for improved precision).
Sensitivity analysis
BoB's rule-based module in the high-sensitivity extraction component obtained high sensitivity for almost all PHI categories (instance-level macro-averaged recall of 96%). Most missed PHI were person names initials, or names, healthcare facilities, and organizations that were not included in our dictionaries, for example, ‘XXX Motorsports’ or ‘PT’ (physical therapy). Fortunately, adding the CRF module (ie, ‘BoB rules+CRF’) lessened this issue by increasing the sensitivity of all non-numerical PHI categories (eg, achieving 99% recall of Patient Names, which is one of the most sensitive categories). It indicates that, although the recall achieved by BoB's CRF models is always lower than considering BoB's rules and dictionaries, the CRF models predict annotations that were missed by the rules and dictionaries, successfully supporting the purpose of BoB's high-sensitivity extraction component.
BoB's token-level sensitivity is quite similar. Only PHI categories that often include many tokens within one annotation, such as Street City, Healthcare Units, and Other Organizations, had slightly decreased sensitivity. However, both macro- and micro-averaged recall reached similar values as instance-level measurements, at about 97% macro-averaged recall when considering BoB's complete high-sensitivity extraction component.
Overall performance analysis
Although BoB's high-sensitivity extraction component sensitivity is very high, it can compromise the usefulness of the documents by redacting many non-PHI tokens (instance-level micro-averaged precision of 34.6%). However, as anticipated, with BoB's false positive filtering component, precision was very significantly increased to 83.6% (highly significant difference with p<0.001), and sensitivity remained high at 92.2% (non-significant difference with p=0.349). Such an increase in precision demonstrates the efficient design of BoB's architecture, and the effective training strategy of our false positives filtering classifiers. However, these classifiers also filtered out some true positives, mostly PHI overlapping with common words such as ‘bill’ or ‘max’ for person names, and ‘IN’ for Indiana.
Similarly, with token-level analysis, false positives filtering also dramatically increased precision (highly significant difference with p<0.001) with a decrease in recall (significant difference with p=0.004). Also, the overall instance-level precision was about 2% higher than the token-level precision, indicating that only a few annotations by BoB included additional tokens that did not belong to the actual PHI identifier.
The performance of the single-stage CRF experiment reinforces our empirical evidence on machine learning methods to improve precision (instance-level micro-averaged precision of 92%). However, in terms of recall, and especially for some PHI categories, the de-identification achieved is not sufficient to guarantee high patient privacy rates (instance- and token-level micro-averaged recall of about 85%).
The ‘out-of-the-box’ results achieved by the MIT deid system indicate the strong need for adaptation of rule-based techniques to the type of target documents. This system reached significantly lower performance (instance-level recall and precision highly significantly different with p<0.001, token-level recall significantly different with p=0.004, and precision highly significantly different with p<0.001). This system had lower performance for several reasons: (1) it was not designed for some of our PHI categories (eg, Deployment, Other Organizations, and although it detects hospitals, it does not detect generic mentions of clinics or acronyms annotated as Healthcare Units in our reference); (2) it performed better at token-level for non-numerical PHI types such as Names (eg, detecting first names but missing last names). We believe this indicates a need for more general patterns that can lower the system dependency on dictionaries; and (3) it surprisingly missed Phone Numbers and Other ID Numbers, categories that should be well addressed with pattern matching. We believe it occurred because of missing patterns covering rare formats (eg, ‘000-CALLNOW’, ‘x9999’, ‘AzS45/56-0’ and ‘LS #0000’). The main objective of this comparison was to demonstrate that using a de-identification system ‘out-of-the-box’ would not work with our corpus, and that we successfully addressed the de-identification of VHA clinical documents with BoB.
Finally, our experiment with the 2006 i2b2 de-identification challenge corpus shows that BoB also performs quite well with other documents (table 4). Although most person names in the i2b2 corpus could not be found in dictionaries, BoB's patterns and CRF classifiers were able to detect them. On the other hand, BoB reports low recall for IDs, Phone Numbers and Ages. However, after a rapid analysis of the training subset and the addition of five new patterns for these categories, BoB was able to improve its performance dramatically. Overall, BoB's results with the i2b2 corpus also accomplishes our main goal satisfactorily (ie, prioritizing patient privacy; instance-level micro-averaged recall of 92% and 96%), while achieving competitive precision and preserving the interpretability of documents (instance-level micro-averaged precision of 87.8% and 84.6%).
Conclusion
We have developed an automated text de-identification system for VHA clinical documents. The novel design of our hybrid stepwise approach has demonstrated robustness and efficiency, prioritizing patient confidentiality while leaving most clinical information intact. Future efforts will include improvements of our strategies, such as adding patterns covering broader formats of PHI identifiers, and explorations of other learning features that could improve filtering. Finally, in a manner similar to previous efforts,32 33 we plan to estimate the risk of re-identification, as well as the impact on subsequent uses of automatically de-identified documents.
Acknowledgments
The authors would like to thank John Aberdeen, Lynette Hirschman, and Samuel Bayer at the MITRE Corporation, for helpful discussions and insightful comments. We also thank Matthew Maw from the SLC VA IDEAS Center for his assistance with the evaluation scripts.
Footnotes
Contributors: OF was the main developer of BoB, running the experiments and analyzing their performance, and also drafted the manuscript. BRS and SS led the development of the VHA documents’ reference standard. Both helped draft the manuscript, along with FJF. SMM conceived the design of BoB, analyzed the evaluation results, and drafted the manuscript. MHS revised the manuscript critically and in detail. All authors contributed with useful and critical comments during the development of the BoB de-identification tool. All authors read and approved the final manuscript.
Funding: This work was supported by the Consortium for Healthcare Informatics Research (CHIR), VA HSR HIR 08-374.
Competing interests: None.
Ethics approval: This study has been performed with the approval of the appropriate ethics committee (University of Utah IRB approval reference number IRB_00031374) and the VA Office of Research and Development (approval number 2011-06-007D).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Meystre SM, Friedlin FJ, South BR, et al. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Methodol 2010;10:70 http://www.biomedcentral.com/1471-2288/10/70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dixon P. Medical identity theft: the information crime that can kill you. 2006. http://www.worldprivacyforum.org/pdf/wpf_medicalidtheft2006.pdf (accessed April 2012).
- 3.Harris Interactive Health information privacy (HIPAA) notices have improved public's confidence that their medical information is being handled properly. 2005. http://www.harrisinteractive.com/news/allnewsbydate.asp?NewsID=894 (accessed April 2012). [Google Scholar]
- 4.GPO US 45 C.F.R. § 46 Protection of Human Subjects. 2008. http://www.access.gpo.gov/nara/cfr/waisidx_08/45cfr46_08.html (accessed April 2012). [Google Scholar]
- 5.GPO US 45 C.F.R. § 164 Security and Privacy. 2008. http://www.access.gpo.gov/nara/cfr/waisidx_08/45cfr164_08.html (accessed April 2012). [Google Scholar]
- 6.Uzuner O, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007;14:550–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedlin FJ, McDonald CJ. A software tool for removing patient identifying information from clinical documents. J Am Med Inform Assoc 2008;15:601–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neamatullah I, Douglass MM, Lehman LW, et al. Automated de-identification of free-text medical records. BMC Med Inform Decis Mak 2008;8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beckwith BA, Mahaadevan R, Balis UJ, et al. Development and evaluation of an open source software tool for deidentification of pathology reports. BMC Med Inform Decis Mak 2006;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am J Clin Pathol 2004;121:176–86 [DOI] [PubMed] [Google Scholar]
- 11.Aberdeen J, Bayer S, Yeniterzi R, et al. The MITRE identification scrubber toolkit: design, training, and assessment. Inter J Med Inform 2010;79:849–59 [DOI] [PubMed] [Google Scholar]
- 12.Gardner J, Xiong L. An integrated framework for de-identifying unstructured medical data. Data Knowl Eng 2009;68:1441–51 [Google Scholar]
- 13.Uzuner O, Sibanda TC, Luo Y, et al. A de-identifier for medical discharge summaries. Artif Intell Med 2008;42:13–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szarvas G, Farkas R, Busa-Fekete R. State-of-the-art anonymization of medical records using an iterative machine learning framework. J Am Med Inform Assoc 2007;14:574–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, Gaizauskas R, Roberts I, et al. Identifying Personal Health Information using support vector machines training sub-system testing sub-system The Preprocessing sub-system . Proceeding of the i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006 [Google Scholar]
- 16.Grishman R, Sundheim B. Message understanding conference-6: a brief history. Proceedings of the Association for Computational Linguistics; 1996:466–71 [Google Scholar]
- 17.Benton A, Hill S, Ungar L, et al. A system for de-identifying medical message board text. BMC Bioinformatics 2011;12:S2 http://www.biomedcentral.com/1471-2105/12/S3/S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellner B, Huyck M, Mardis S, et al. Rapidly retargetable approaches to de-identification in medical records. J Am Med Inform Assoc 2007;14:564–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrández O, South BR, Shen S, et al. Evaluating current automatic de-identification methods with Veteran's health administration clinical documents. BMC Med Res Methodol 2012;12:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velupillai S, Dalianis H, Hassel M, et al. Developing a standard for de-identifying electronic patient records written in Swedish: precision, recall and F-measure in a manual and computerized annotation trial. Int J Med Inform 2009;78:e19–26 [DOI] [PubMed] [Google Scholar]
- 21.Apache UIMA 2008. http://uima.apache.org (accessed April 2012). [Google Scholar]
- 22.LVG (Lexical Variant Generation) 2011. http://lexsrv2.nlm.nih.gov/LexSysGroup/Projects/lvg/current/web/index.html (accessed Apr 2012). [Google Scholar]
- 23.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldridge J, Morton M, Bierner G. OpenNLP Maxent Package in Java. 2005 http://opennlp.apache.org/ (accessed Apr 2012).
- 25.The Apache Lucene project. http://lucene.apache.org/ (accessed Apr 2012).
- 26.Tjong Kim Sang EF, De Meulder F. Introduction to the CoNLL-2003 shared task: language-independent named entity recognition. Proceedings of the Seventh Conference on Natural language Learning; 2003:142–7 [Google Scholar]
- 27.Doddington G, Mitchell A, Przybocki M, et al. The Automatic Content Extraction (ACE) program—tasks, data, and evaluation. Evaluation. Proceedings of The International Conference on Language Resources and Evaluation; 2004:837–40 [Google Scholar]
- 28.Finkel JR, Grenager T, Manning C. Incorporating non-local information into information extraction systems by Gibbs sampling. Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics; 2005:363–70 [Google Scholar]
- 29.Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2011;2:1–27 [Google Scholar]
- 30.Fan R-E, Chang K-W, Hsieh C-J, et al. LIBLINEAR: a library for large linear classification. J Mach Lear Res 2008;9:1871–4 [Google Scholar]
- 31.van Rijsbergen CJ. Information retrieval. Newton, MA: Butterworth-Heinemann, 1979 [Google Scholar]
- 32.Hirschman L, Aberdeen J. Measuring risk and information preservation: toward new metrics for de-identification of clinical texts. Proceedings of the Second Louhi Workshop on Text and Data Mining of Health Documents; 2010:72–5 [Google Scholar]
- 33.Yeniterzi R, Aberdeen J, Bayer S, et al. Effects of personal identifier resynthesis on clinical text de-identification. J Am Med Inform Assoc 2010;17:159–68 [DOI] [PMC free article] [PubMed] [Google Scholar]