Abstract
Objective
Registries are a well-established mechanism for obtaining high quality, disease-specific data, but are often highly project-specific in their design, implementation, and policies for data use. In contrast to the conventional model of centralized data contribution, warehousing, and control, we design a self-scaling registry technology for collaborative data sharing, based upon the widely adopted Integrating Biology & the Bedside (i2b2) data warehousing framework and the Shared Health Research Information Network (SHRINE) peer-to-peer networking software.
Materials and methods
Focusing our design around creation of a scalable solution for collaboration within multi-site disease registries, we leverage the i2b2 and SHRINE open source software to create a modular, ontology-based, federated infrastructure that provides research investigators full ownership and access to their contributed data while supporting permissioned yet robust data sharing. We accomplish these objectives via web services supporting peer-group overlays, group-aware data aggregation, and administrative functions.
Results
The 56-site Childhood Arthritis & Rheumatology Research Alliance (CARRA) Registry and 3-site Harvard Inflammatory Bowel Diseases Longitudinal Data Repository now utilize i2b2 self-scaling registry technology (i2b2-SSR). This platform, extensible to federation of multiple projects within and between research networks, encompasses >6000 subjects at sites throughout the USA.
Discussion
We utilize the i2b2-SSR platform to minimize technical barriers to collaboration while enabling fine-grained control over data sharing.
Conclusions
The implementation of i2b2-SSR for the multi-site, multi-stakeholder CARRA Registry has established a digital infrastructure for community-driven research data sharing in pediatric rheumatology in the USA. We envision i2b2-SSR as a scalable, reusable solution facilitating interdisciplinary research across diseases.
Keywords: Registries, information dissemination, information storage and retrieval, health services research, rheumatic diseases, informatics, registry, pediatric rheumatology, medical informatics, disease association and prediction, natural language processing, multi-site disease studies, pediatric, monitoring the health of populations, personal health records and self-care systems
Objective
Registries are a well-established mechanism for obtaining high quality, disease-specific data on distinct cohorts of subjects with preselected diseases, environmental exposures, and/or treatments of interest.1 We describe the development and implementation of a self-scaling, interoperable platform for collaborative data sharing based upon the widely adopted Informatics for Integrating Biology & the Bedside (i2b2) framework2 and report use of this i2b2 self-scaling registry technology (i2b2-SSR) for the 56-site Childhood Arthritis & Rheumatism Research Alliance (CARRA) Registry of pediatric rheumatic diseases and Harvard Inflammatory Bowel Disease Longitudinal Data Repository.
Background and significance
The potential of disease registries to collect high quality data and support multi-center studies, comparative effectiveness research, and post-marketing surveillance, has never been greater. Yet registry efforts are often highly project-specific in their design, implementation, and policies for data use.
Disease registries range from rare disease projects (where no single center can ever produce sufficient numbers for study),3 4 to single-investigator studies,5 6 and to large, multi-site, national public health efforts such as the $50+ million/year Centers for Disease Control National Program of Cancer Registries7 and the National Cancer Institute's Surveillance, Epidemiology, and End Results Registry (SEER) program. The landscape is one of isolated, autonomous, and often overlapping clinical data repositories with dissimilar data schemas.8 Historically, registries have conformed to a model of centralized data contribution, warehousing, and control.9 Further, considerations of authorship and academic credit10 11 may deter principal investigators from more widely sharing their datasets,1 12 producing a chilling effect on multicenter study.
The Institute of Medicine and others have persuasively argued that the development of less compartmentalized, multi-stakeholder strategies for data sharing is critical to the conduct of relevant and innovative clinical research analyses.2 13–16 Nonetheless, only a relatively few successful efforts for widespread, registry-based data sharing have been accomplished in the USA to date and no infrastructure has yet emerged as a recognized standard. Instead, registry data collection, warehousing, and use traditionally follow a self-contained model that, by design, offers neither modularity nor scalability: data are typically collected for registry-limited use cases, data elements may not be recorded or normalized to externally meaningful standards, and data providers usually surrender their data to a centrally administered repository to which they then have limited access. While serving individual study goals, such constraints adversely impact the reusability, cross-disciplinary generalizability, and return on investment of registries to the larger research enterprise.
Firmly grounded on the use case of a rare disease registry—the multi-site CARRA Registry of pediatric rheumatic diseases—we address the desiderata of a national scale infrastructure for chronic disease registries in a real world deployment of a modular, reusable, and readily extensible research data storage and sharing framework. Our efforts are based on two highly diffusible, open source technologies: the i2b2 informatics framework3 4 17 18 and the Shared Health Research Information Network (SHRINE).5 6 18 These platforms provide a well-tested foundation for cross-institutional data aggregation within the health information sector and have been used for multi-municipality syndromic biosurveillance (AEGIS),7 19 cohort identification across the Harvard-affiliated hospitals (SHRINE),8 18 and multi-institutional neuroscientific research (Biomedical Informatics Research Network (BIRN)).9 20 21
Materials and methods
In 2009, the CARRA—a consortium comprising pediatric rheumatology clinical research centers in North America—received critical infrastructure funding from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS, RC2AR058934) to establish the longitudinal CARRA Registry network (CARRAnet).
The i2b2-based self-scaling registry platform (i2b2-SSR) developed for this purpose allows individual investigators and institutions to join a secure research data network by contributing a unique dataset and working with others to create larger, collaborative datasets that may be shared with the network as a whole or within specific subsets of sites and investigators. An administrative and auditing layer provides control of user permissions along with logging of user queries, assuring compliance with Institutional Review Board (IRB) and other regulatory protections for subjects, as well as allowing monitoring of data usage according to the research network's guidelines. A scenario illustrates the functionality:
Part 1. Dr Susan Smith, a clinician-researcher, is principal investigator (PI) of a small, multi-site trial demonstrating efficacy of the new biologic therapy ‘BioX’ for childhood-onset arthritis. Following regulatory approval of BioX therapy, she wants to extend her study to address the long-term safety of BioX and to engage more sites. Dr Smith discusses this with Dr Robert Rogers, who is wrapping up a similar post-marketing study for the earlier generation ‘BioA’ therapy.
Dr Rogers expresses concerns about the difficulties that Dr Smith will encounter in recruiting and retaining enough sites and subjects for another independent, long-term post-marketing surveillance study. Dr Smith, however, explains that rather than undertake another self-contained study, she intends to join her current study to the new, 60-site i2b2-SSR consolidated registry, which is already collecting 75% of the data elements she needs for the new study. She proposes a further collaboration in which Dr Rogers would be able to share his study's multi-site data by establishing his own i2b2-SSR repository. She explains to Dr Rogers—who is somewhat reluctant to share data widely prior to reaching his study's final endpoint—that he may first elect a limited collaboration with Dr Smith and make future decisions about data sharing on a study-by-study basis.
To join the i2b2-SSR network, Dr Smith uploads each participating site's data from her existing BioX trial to i2b2-SSR (one data warehouse per site). As study PI, Dr Smith is able to query all of the study sites as one virtual repository, and each site is also able to view the data it has contributed to the study. As an incentive for existing BioX study sites to continue their participation in the next, post-marketing surveillance phase, and to encourage new participation from additional sites, Dr Smith decides to grant appropriate permissions for any study site investigator to execute limited queries (counts of patients only, no subject-specific results) across all sites in this new BioX study.
Dr Rogers, now comfortable with sharing his BioA study's extensive data in a permissioned fashion exclusively with Dr Smith, uploads the BioA data to his own i2b2-SSR instance and provides Dr Smith with ‘counts-only’ query permission. Dr Smith provides Dr Rogers with reciprocal permissions, and they are now able to query summary data from both of their studies as a combined BioA/BioX virtual database.
Part 2. Dr Amy Allen, a junior clinician-researcher, believes that patients taking certain biologics, including BioX, improve when taking VitaG supplements. As a participating investigator in Dr Smith's BioX post-marketing surveillance study, she logs onto i2b2-SSR and defines a query for patients on BioX who are also taking vitamin supplements. Dr Allen discovers there are 80 subjects who might be candidates for testing, but wonders if there are too few control subjects in Dr Smith's cohort for her planned analyses to be meaningful. On contacting Dr Smith to discuss obtaining full, subject-level access to the BioX dataset, Dr Smith confirms an inadequate control population in the BioX cohort alone, but executes a query on the i2b2-SSR multi-site BioA/BioX database, returning aggregate counts that confirm adequate numbers for Dr Allen's research.
Dr Smith suggests that they both contact Dr Rogers with a request that Dr Allen be allowed to run a set of specific, subject-level queries on the BioA/BioX repository. The three researchers agree to collaborate and both Dr Rogers and Dr Smith grant Dr Allen permission to execute subject-level queries within the scope of her research on their respective datasets over a 2-month period.
This scenario illustrates our objectives for a modular, collaborative, self-scaling registry providing minimal barriers to participation. We have focused our i2b2-SSR development efforts around five design principles:
Provide data contributors with full ownership of and access to their own data
Minimize barriers for data owners to collaboratively contribute their data to new or existing datasets
Support a tiered sharing model which provides a granular, permissioned, and audit-capable data sharing framework
Enable near real-time access to data, supporting a virtuous cycle in which immediate data access promotes further data contribution and collaboration
Encourage ongoing incorporation of outside datasets from multiple sources.
We build on two core open-source technologies in current use: (1) SHRINE, a peer-to-peer network designed for health informatics allowing construction of permissioned, well-defined data interchange topologies between data repositories10 11 18; and (2) the Informatics for Integrating Biology & the Bedside (i2b2) framework, a data warehousing and analytic platform that has been put into use at more than 60 medical centers, encompassing health data on an estimated 45 million subjects worldwide, and which is readily scalable via SHRINE.2
The i2b2-SSR system supports the following administrator actions:
Establishment of a trust relationship between a data contributor (i2b2-SSR node) and a data aggregator (i2b2-SSR Broadcast Aggregator) via exchange of digitally signed certificates distributed by mutually trusted certificate authorities (CAs)
Creation and modification of peer groups for collaboration, wherein one or more data contributors agree(s) to allow query access to a dataset
-
Mapping of fine-grained data query privileges to an i2b2-SSR end user, including designation of
User membership in one or more peer groups
One or more optional ‘My Home Node(s)’ for each user
-
User data access authorization, wherein queries will return either
Simple counts of subjects fulfilling a query, or
Detailed data resulting from the query, as well as counts of subjects
-
Data origin to display to a user, where returned counts and/or detailed data will be tagged with the identity of the data contributor in one of three permutations
Only peer group is identified (ie, no unique origin information is returned), or
Only ‘My Home Node’ data are identified (ie, ‘me’ versus all others in peer group), or
All data are fully identified by data origin
Display and monitoring of network health, including nodes not responding to queries
Generation of audit reports of user activity and queries submitted.
For the end-user, the i2b2-SSR system supports the following actions:
Display and selection of peer groups and data privileges for which a user is authorized
Ontology-based display of data elements and construction of complex queries via web interface
History of queries performed
Generation and display of aggregated query results, including a pluggable reporting system which supports BIRT22 and R-based23 24 data visualizations and exports.
Regulatory oversight of data use, most often in the form of IRB review, is a pre-condition for the release and sharing of health research data in the USA and elsewhere. Especially for multi-stakeholder collaborations where distinct data disclosure policies apply to different data contributors and consumers over time, a high cost-complexity penalty may exist which burdens and discourages productive information sharing. Cognizant of these concerns, we directly incorporate mechanisms for addressing third-party oversight of data sharing in our design. In addition to support for logging and auditing data access by users, the gatekeeper roles of regulatory agents are instantiated as digital certificate policies within i2b2-SSR, that is, an IRB is analogous to a certificate authority (CA). In this model, a data contributor must secure a digitally signed, time-limited certificate from a CA that is trusted by the data aggregator. Reciprocally, a data aggregator must present a certificate validated by a CA that the data contributor trusts, thereby establishing a bidirectional relationship.
Components of the system: access to data
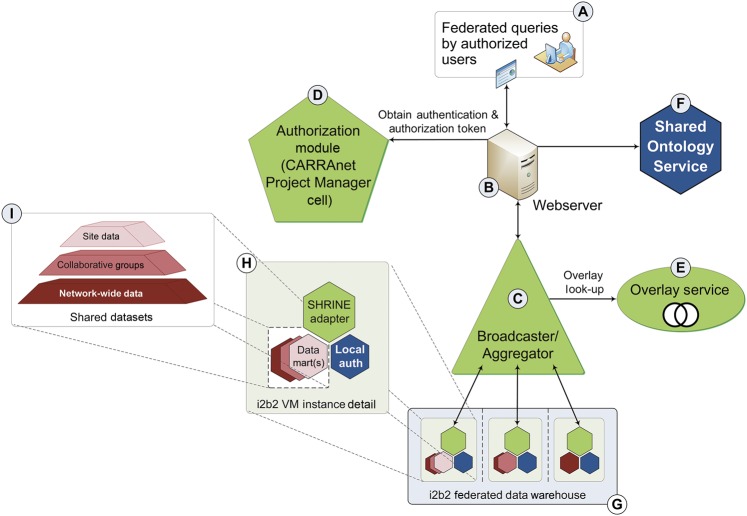
The i2b2-SSR architecture provides access to data via the components depicted in figure 1 and described below.
End user (A)
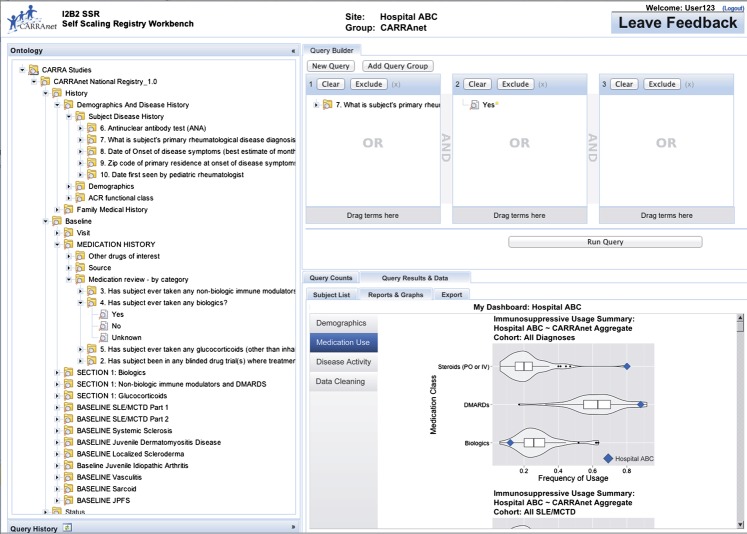
The end user, typically a research investigator, accesses the registry through a web-based query interface. Following secure log in, the user encounters a graphical query builder interface in which registry-specific ontologies may be browsed; search terms may be dragged and dropped to construct queries to define subject cohorts of potential interest. Selected cohorts are returned as patient sets, for which choices of pre-defined summary reports and visualizations may be generated in real time (figure 2). In this way, end users may iteratively refine their searches and are able, with appropriate authorizations, to download the resulting datasets for further analyses.
Figure 2.
End user query interface. The Site Investigator dashboard view is shown, illustrating a sample visualization of summary statistics for site ‘ABC’ versus the entire CARRAnet registry.
Figure 1.
The i2b2-SSR architecture. The diagram illustrates the interaction of i2b2-SSR core web services C and D, which are customized, i2b2-SSR ‘drop-in’ replacements for the standard SHRINE Broadcaster/Aggregator and i2b2 Project Manager Cell, respectively. In coordination with the i2b2-SSR Overlay Service (E), these modules support introduction of peer-group overlays for sharing of multiple datasets (I) using standard i2b2 nodes and SHRINE adapters (detail H). The authorized end-user (A) constructs a query based on shared ontologies that are pre-defined for the shared datasets. The Shared Ontology Service (F) may employ a standard i2b2 Ontology cell; alternatively, we provide an i2b2 Ontology module with the i2b2-SSR distribution that implements memory-based caching with ontology term search and autocomplete capabilities.
Webserver (B)
The webserver proxies all federated queries from the end user. It is a server-side component designed as a replacement to the standard SHRINE web client. The webserver assembles i2b2 query panels from the end user-defined queries and employs secure messaging between the various services it consumes. A reporting interface for providing summary data, data visualizations, analyses incorporating missing data, and exports is implemented using a business-intelligence layer (BIRT reports and/or R).
Broadcaster/Aggregator (C)
The Broadcaster/Aggregator is the i2b2-SSR component responsible for receiving end user i2b2 query panels, broadcasting them to various groupings of nodes that are sharing data, and then aggregating the results received. The Broadcaster/Aggregator uses XML digital signatures to encrypt and validate all incoming and outgoing messages. Using digitally signed certificates for coordinating trust relationships, the Broadcaster/Aggregator component verifies that data contributor nodes are trusted sources.
Authorization module (D)
The Authorization (Auth) module is a customized i2b2-SSR service that exposes an i2b2 Project Management cell interface to the Webserver and Broadcaster/Aggregator. It acts as the central, trusted authentication provider for the i2b2-SSR platform by issuing signed tokens that the Broadcaster/Aggregator subsequently validates. In addition, the Auth service functions as an identity provider or may proxy to an outside identity service such as Lightweight Directory Access Protocol or Microsoft Active Directory. In either case, once a user has been authenticated and properly logged-in, a session token is vended by the Auth service. This token encodes the combinations of query and data viewing privileges for which the user has been authorized, thereby determining which datasets a user can access and controlling the data granularity exposed.
Overlay Service (E)
The Overlay Service (OLS) is the i2b2-SSR directory service that is responsible for maintaining the distinct federated, collaborative topologies supported for a specific i2b2-SSR network. It uses a simple RESTful web service application programming interface (API) that allows users with administrative privileges to add or remove i2b2 data contributor nodes and define new peer groupings. The Broadcaster/Aggregator references these OLS groupings to determine which set of nodes to route a particular query broadcast to.
Shared Ontology service (F)
The Shared Ontology service provides web service access to hierarchical vocabularies that describe i2b2 data elements and provide term mappings for i2b2 query panels. This component functions identically to the standard i2b2 Ontology Cell for query panels, with the additional external policy requirement that at least one common vocabulary exists and is mapped at all data contributor nodes. In practice, as part of the Shared Ontology model, we additionally provide the requisite i2b2 concept dimension rows as a Shared Ontology service public table; however, these additional term mappings may be equivalently implemented at the SHRINE adapter translation layer.25 We also utilize a new, streamlined Shared Ontology module that incorporates Apache Lucene search capabilities.
i2b2 federated Data Warehouses (G)
The collection of site-specific i2b2 Data Warehouses defines the pool of potential data contributors to an i2b2-SSR network. For production use in CARRAnet, a hosted i2b2 Virtual Machine (i2b2VM) server farm has been established, within which each collaborating data contributor is provisioned a dedicated i2b2VM. Each site investigator is provided with full access to their contributed data, with capability for the hosting and sharing of multiple datasets across the entire registry network or only within specified subgroups of collaborating investigators.
i2b2 instance details (H)
i2b2 instances deployed within i2b2-SSR consist of a combination of three standard SHRINE and i2b2 components: a SHRINE adapter, an i2b2 Data Repository (also called the Clinical Research Chart, or CRC) cell, and a Local Authorization module (i2b2 Project Management cell). The SHRINE adapter receives query panels conveyed within SHRINE requests from a trusted Broadcaster/Aggregator and provides bidirectional translation between federated SHRINE requests and local CRC concept mappings. The CRC stores data in a set of star-schema data marts that comprises a local i2b2 data warehouse and interacts with the Local Authorization module to expose an ontology-aware query interface.
Shared datasets (I)
A single i2b2 instance may host multiple datasets. One dataset may be federated with many other datasets within and across i2b2 data warehouses that share common data ontologies. In this way, large, interrelated datasets may be added or removed incrementally in a self-scaling, modular fashion.
Components of the system: addition of network nodes
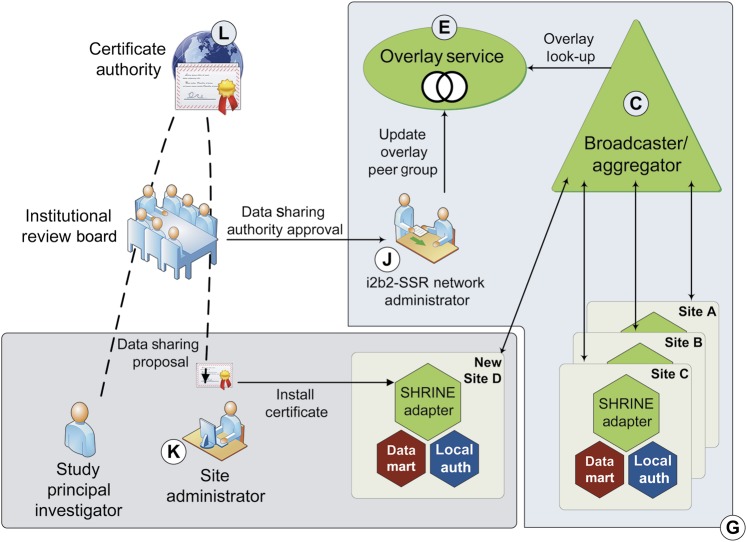
The self-scaling design of i2b2-SSR the enables addition of network nodes via the components depicted in figure 3 and described below.
Network Administrative User (J)
The Administrative User interface offers system management capabilities for i2b2-SSR. It enables the network operator to manage users, user permissions, Overlay Service peer groups, and trust relationships and provides a dashboard to monitor the overall health of the network.
Figure 3.
Self-scaling architecture—adding new sites (network nodes) and/or studies to an i2b2-SSR network. With appropriate approvals, a Site Administrator (K) configures the local SHRINE adapter to communicate with a particular registry Broadcaster/Aggregator endpoint (C) and installs a digital certificate distributed by a certificate authority (L) that is mutually trusted by the site and the i2b2-SSR Network Administrator (J).
i2b2 Data Contributor Administrative User (K)
Local i2b2 data repository administrative users enable their i2b2 instance to trust one or more Broadcaster/Aggregators via configuration of i2b2 and one or more local SHRINE adapters. This enables end users hosting their own i2b2 instances to flexibly join i2b2-SSR data sharing networks ad hoc. Trust relationships between an i2b2 instance data contributor and the network are established via digital certificate exchange, typically using a mutually trusted CA.
Certificate authority (L)
Our design accommodates use of one or more CAs to establish mutual trust relationships and provide gatekeeper functionality for data sharing within i2b2-SSR networks. A trusted third party, such as an IRB or other institutional regulatory body, may provide signed digital certificates via the CA; these are used by the Broadcaster/Aggregator and local SHRINE adapter to validate requests and securely encode responses exchanged between components. Certificates distributed by CAs may incorporate temporal constraints (eg, certificate expirations) and also support certificate revocation policies specified by a data sharing authority, such as an IRB.
Codebase
The i2b2-SSR code is available as open source software, licensed under LGPL version 3 for i2b2-SSR components; constituent components and dependencies are available under their respective open source licenses (repository and links at https://open.med.harvard.edu/display/CARRANET). The Webserver package is available from Cincinnati Children's Hospital Medical Center at https://bmi.cchmc.org/svn/i2b2/i2b2/public/.
Results
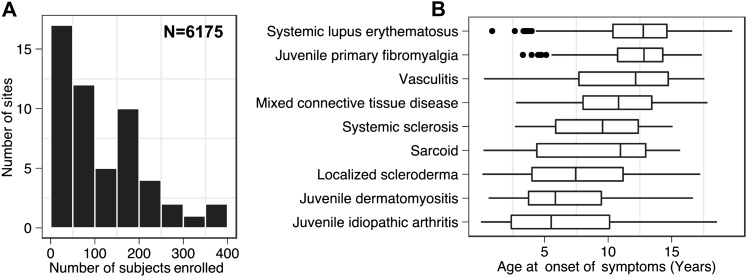
As of February 2012, the CARRA Registry network comprised 56 actively recruiting sites with 237 investigators and over 6000 subjects enrolled, including data on more than 11 000 registry visits gathered since activation of the first site in May 2010; this represents the largest pediatric rheumatic diseases study cohort in the USA to date and one of the largest worldwide.26 The i2b2-SSR platform provides secure, granularly permissioned, real-time access to registry data for CARRAnet investigators and has been adopted as the primary mechanism for return of results for the pediatric rheumatology research community. Data sharing is governed by well-defined policies27 intended to facilitate access to data while fostering collaborative research in areas of scientific priority and inclusion of early stage investigators. The dataset continues to grow as researchers enter information daily, with a current target enrollment of 10 000 subjects at 60 sites and regular longitudinal follow-up. Table 1 and figure 4 provide a brief cross-sectional overview of the characteristics of registry subjects at initial enrollment visit, obtained via i2b2-SSR federated query across all sites.
Table 1.
Diagnosis at baseline visit, Childhood Arthritis & Rheumatology Research Alliance (CARRA) registry population (as of February 2012)
| Diagnosis | N (%) |
| Juvenile idiopathic arthritis | 4510 (72%) |
| Pediatric systemic lupus erythematosus | 618 (10%) |
| Juvenile dermatomyositis | 433 (7%) |
| Localized scleroderma | 236 (4%) |
| Juvenile primary fibromyalgia | 122 (2%) |
| Vasculitis | 117 (2%) |
| Mixed connective tissue disease | 112 (2%) |
| Sarcoidosis | 38 (1%) |
| Systemic sclerosis | 36 (1%) |
Figure 4.
CARRA Registry, selected demographics (as of February 2012, data from 53 sites), see also table 1. (A) Distribution of subject enrollment by site. The majority of registry subjects (3647 out of 6175 total subjects enrolled, or ∼60%) are found at sites enrolling <200 subjects (N=44 sites), reflecting the broad collaboration needed within this research community to achieve sufficient populations to conduct significant research investigations; (B) age at onset of disease symptoms by disease diagnosis. Upper age distribution is right-censored due to pediatric-onset inclusion criteria.
As the registry has matured, CARRAnet has become the platform of choice for US investigators to initiate new and enhanced clinical research studies in this population. As of mid-2012, enhanced datasets from five newly funded disease-specific efforts studying the comparative effectiveness of various Consensus Treatment Plans28 in pediatric rheumatic diseases (pediatric systemic lupus erythematosus,29 juvenile dermatomyositis,30 31 systemic-onset and polyarticular juvenile idiopathic arthritis,32 33 localized scleroderma34) will be incorporated into the CARRAnet platform, with multiple additional efforts actively planned, including biospecimen collection for translational research applications. In addition, with the anticipated introduction of a public–private partnership for pharmaceutical post-marketing surveillance into CARRAnet,4 the i2b2-SSR infrastructure will provide a critical conduit for managing permissioned access by researchers to a dual, research and regulatory use dataset.
While initially deployed for within-network data sharing, the i2b2-SSR platform is equally extensible to interdisciplinary collaboration and data sharing. For certain crosscutting clinical questions, such as general studies in autoimmunity or determination of uncommon but serious adverse event signals of medications in pediatric patients, it is highly advantageous to combine data from disparate sources. This may involve federation of data from different studies or projects housed within a single node (intra-institutional), federation across different studies housed within nodes of different networks (inter-institutional), or even federation of queries between collections of distinct networks (trans-institutional). Within the context of an ontology-based data warehouse framework such as i2b2, extended topologies for federated queries are readily constructed by aligning local ontologies to external, standardized vocabularies such as SNOMED-CT and MedDRA. For example, Children's Hospital Boston is a contributing site for two multi-center registry studies housed in separate i2b2-SSR nodes and networks: the Harvard Inflammatory Bowel Diseases Longitudinal Repository and the CARRA Registry. With appropriate investigator and regulatory approvals, combined federation between the two local registry nodes can be accomplished via straightforward changes to i2b2-SSR peer-group configuration by network administrators. Likewise, with appropriate registry authorizations and existence of a shared, trans-network ontology, data federation between all sites of both registries becomes readily feasible.
Discussion
Broadly federating and aggregating clinical research data is a valuable capability pursued through a range of technological strategies by a number of projects, including the cancer Biomedical Informatics Grid (caBIG) caGrid platform 35 (which supports a Unified Modeling Language (UML) based, federated network architecture), the Mini-Sentinel Initiative, 36 37 the National Database for Autism Research (NDAR)38 and others,39 40 including efforts under active development such as Query Health. 41 Beyond purely technical considerations, however, there exist considerable operational complexities—societal, organizational, and economic—that must be simultaneously addressed in order to successfully bridge barriers to widespread data sharing in the healthcare enterprise.42–45
We believe that our two-tiered approach of (1) fostering grass-roots efforts for data sharing by allowing participating investigators full control of their contributed data while enabling them to flexibly join, or depart, data sharing networks in a dynamic, ad hoc fashion with fine-grained, transparent data access permissions, and (2) leveraging the large, established, open source infrastructure and installed base of i2b2-based data warehousing, is an attractive recipe for fostering multi-functional use of disease registries, enabling substantially greater economies of scale and resources than the traditional, centralized, data silo approaches of the past century.
Federated queries over distributed i2b2 networks imply their own set of unique challenges, for example, proper accounting for same-patient data (when such facts are present in multiple nodes), imputation of missing data points when queries return null results, and the challenges of aggregating similar data referred to using different ontologies. The i2b2-SSR peer group-based trust and shared ontology approach provides a robust framework for addressing such complexities. The capability to implement heuristics on privileged, fact-level data while exposing only select, computed views of federated information to permissioned users enables innovative solutions to such challenges, thereby realizing the benefits of centralized data warehousing within a distributed, self-scaling system. Moreover, incentivizing the growth of networks using shared ontologies supports a much needed community-based, rather than data silo, approach to the clinical research enterprise.
Conclusion
Building upon prior successful efforts at clinical data federation using the i2b2 and SHRINE open source platforms,2 17–19 46 we have assembled a lightweight, reusable, peer-to-peer chronic disease registry framework that promotes investigator participation through robust mechanisms for local control over data ownership and sharing. The implementation of this framework for the multi-site, multi-stakeholder CARRA Registry has established a digital infrastructure for community-driven research data sharing in pediatric rheumatology in the USA. The future success of this technology will be measured by its real-world application to fostering new collaborations in comparative effectiveness and translational research within the network, as well as by its value as a model for fostering grass-roots efforts for data sharing in other investigator networks.
Our next steps for development of the i2b2-SSR infrastructure will focus upon use of registry data as a core, gold-standard reference by which to effectively leverage more voluminous ‘ambient’ health information, particularly patient-specific information from electronic health records and patient-reported outcomes. To accomplish this, we envision a self-scaling registry paradigm that encompasses secure data federation across multiple systems capable of patient-level linkage: i2b2-based registries, electronic health record warehouses, and patient-centric data systems (eg, personally controlled health record systems for direct patient report). In this way, i2b2-SSR will not only serve as infrastructure for data sharing, but will also fulfill the need for an incremental, highly scalable, and reusable solution to conduct interdisciplinary research across diseases.
Acknowledgments
The authors are indebted to the efforts of other key contributors to this project, including Kathleen Fox, Brian McCourt, and Jane Winsor at the Duke Clinical Research Institute (CARRAnet) and Lori Ashworth and Sarah Weber at Children's Hospital Boston (Harvard Longitudinal Inflammatory Bowel Diseases Repository). While it is not possible to individually acknowledge every contributor in a project of this size, we are especially appreciative of the efforts of the database and clinical research IT teams at Children's Hospital Boston; the informatics development teams at Cincinnati Children's Hospital Medical Center, Duke Clinical Research Institute, and Harvard Medical School; and co-investigators and research coordinators at the 56 CARRA Registry sites and three Harvard Inflammatory Bowel Diseases Longitudinal Repository sites.
Footnotes
Contributors: MDN provided the conceptual design in consultation with other authors and directed the implementation of the overall informatics framework. He is guarantor. KDM, with MDN, conceptualized the initial design and subsequent implementation of the i2b2-SSR framework. JQ and DMO are lead developers of the software and chief architects of the i2b2-SSR peer group design and subsequent reduction to software code. KM, AJM, and GMW provided critical software design contributions that were required for the reduction to practice of the overall i2b2-SSR design and, respectively, contributed the implementation of i2b2-SSR webserver interface, peer networking architecture, and data model and ETL design. CJI and RWW, along with MDN, conceived and designed the Shared Ontology concept for i2b2-SSR and implemented the CARRA Registry portion of the project. AB and MDN designed and implemented the Shared Ontology and research design for the Harvard IBD Longitudinal Repository portion of the project. NTI, CIS, LES, and CAW provided key contributions to the overall conceptual design, research design of the registry, and the design of Shared Ontology schemas, and contributed to the implementation and refinement of user-facing features and the reporting and business intelligence layer. KDM, CIS, and LES provided overall project leadership and directly contributed to each phase of i2b2-SSR development.
Funding: We gratefully acknowledge the support that has made this project possible. Funding support was received from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (RC2AR058934), the National Library of Medicine (1R01LM011185-01 and T15LM007092), Friends of CARRA, the Arthritis Foundation, the Rasmussen Fund, the National Institute of Diabetes and Digestive and Kidney Diseases (NIH Loan Repayment Program), and the Duke Clinical Research Institute.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The i2b2-SSR code is available as open source software, licensed under LGPL version 3 for i2b2-SSR components; constituent components and dependencies are available under their respective open source licenses (repository and links at https://open.med.harvard.edu/display/CARRANET). The Webserver package is available from Cincinnati Children's Hospital Medical Center at https://bmi.cchmc.org/svn/i2b2/i2b2/public/.
References
- 1. Gliklich R, Dreyer N, eds Registries for Evaluating Patient Outcomes: A User's Guide. (Prepared by Outcome DEcIDE Center [Outcome Sciences, Inc. d/b/a Outcome] under Contract No. HHSA29020050035I TO3). AHRQ Publication No. 07-EHC001-1. Rockville, MD: Agency for Healthcare Research and Quality, April 2007.
- 2. Kohane IS, Churchill SE, Murphy SN. A translational engine at the national scale: informatics for integrating biology and the bedside. J Am Med Inform Assoc 2012;19:181–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith MY, Sobel RE, Wallace CA. Monitoring the long-term safety of therapies for children with juvenile idiopathic arthritis: time for a consolidated patient registry. Arthritis Care Res (Hoboken) 2010;62:800–4 [DOI] [PubMed] [Google Scholar]
- 4. United States, Food and Drug Administration Public Workshop on Developing a Consolidated Pediatric Rheumatology Observational Registry [Internet]. 2009. Report No.: FDA-2009-N-0145–0055. http://www.regulations.gov/#!documentDetail;D=FDA-2009-N-0145-0055 [Google Scholar]
- 5. United States, Department of Health and Human Services FAQ on Public Health Registries [Internet]. National Committee on Vital and Health Statistics. http://ncvhs.hhs.gov/9701138b.htm (accessed 20 Dec 2011).
- 6. United States, National Institutes of Health Catalog of NIH Funded Databases, Disease Registries, and Biomedical Information Resources, 2008–2009 [Internet]. Research Portfolio Online Reporting Tools (RePORT), 2010. http://report.nih.gov/FileLink.aspx?rid=630 (accessed 20 Dec 2011). [Google Scholar]
- 7. United States, Department of Health and Human Services Centers for Disease Control and Prevention, Justification of Estimates for Appropriation Committees [Internet]. Fiscal Year, 2011. http://www.cdc.gov/fmo/topic/Budget%20Information/appropriations_budget_form_pdf/FY2011_CDC_CJ_Final.pdf [Google Scholar]
- 8. Blackstone E, Lenat D, Ishwaran H. Methods that need to be developed. In: Olsen LA, Grossmann C, McGinnis JM. IOM (Institute of Medicine). Learning What Works: Infrastructure Required for Comparative Effectiveness Research: Workshop Summary. Washington (DC): National Academies Press (US), 2011:123–44 [PubMed] [Google Scholar]
- 9. United States, National Committee on Vital and Health Statistics, Subcommittee on Privacy and Confidentiality Roundtable Discussion: Health and Medical Registries [Transcript of Proceedings]. Washington, DC, 1998. http://ncvhs.hhs.gov/980129tr.htm [Google Scholar]
- 10. Wager E. Recognition, reward and responsibility: why the authorship of scientific papers matters. Maturitas 2009;62:109–12 [DOI] [PubMed] [Google Scholar]
- 11. Ross RG, Greco-Sanders L, Laudenslager M, et al. An institutional postdoctoral research training program: predictors of publication rate and federal funding success of its graduates. Acad Psychiatry 2009;33:234–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Y, Ascoli G. Rhyme and the Reason of Data Sharing: A Satellite Symposium of the 2007 Society for Neuroscience Annual Meeting. Bethesda, MD: National Institute of Neurological Disorders and Stroke (NINDS), 2007 [Google Scholar]
- 13. Freudenheim M. National Registry Is a Tool in the Fight on Cystic Fibrosis. New York: The New York Times [Internet], 2009. Sect. D:1. http://www.nytimes.com/2009/12/22/health/22cyst.html [Google Scholar]
- 14. Gawande A. Annals of medicine: the bell curve. The New Yorker [Internet] 6 December 2004. http://www.newyorker.com/archive/2004/12/06/041206fa_fact [Google Scholar]
- 15. Potash J, Toolan J, Steele J, et al. The bipolar disorder phenome database: a resource for genetic studies. Am J Psychiatry 2007;164:1229–37 [DOI] [PubMed] [Google Scholar]
- 16. Arenson AD, Bakhireva LN, Chambers CD, et al. Implementation of a shared data repository and common data dictionary for fetal alcohol spectrum disorders research. Alcohol 2010;44:643–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murphy S, Churchill S, Bry L, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res 2009;19:1675–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weber GM, Murphy SN, McMurry AJ, et al. The shared health research information network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc 2009;16:624–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reis BY, Kirby C, Hadden LE, et al. AEGIS: a robust and scalable real-time public health surveillance system. J Am Med Inform Assoc 2007;14:581–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Biomedical Informatics Research Network (BIRN) [Internet] History. Biomedical Informatics Research Network. http://www.birncommunity.org/about/history (accessed 21 Dec 2011).
- 21. Namini AH, Berkowicz DA, Kohane IS, et al. A submission model for use in the indexing, searching, and retrieval of distributed pathology case and tissue specimens. Stud Health Technol Inform 2004;107:1264–7 [PubMed] [Google Scholar]
- 22. BIRT Project [Internet] The Eclipse Foundation. 2012. http://www.eclipse.org/birt (accessed 14 Apr 2012). [Google Scholar]
- 23. Team R. R: A Language and Environment for Statistical Computing [Internet]. Vienna: R Foundation for Statistical Computing, 2012. http://www.R-project.org (accessed 14 Apr 2012). [Google Scholar]
- 24. Urbanek S. Rserve–a fast way to provide R functionality to applications. In: Hornik K, Leisch F, Zeileis A, eds. Proc. Of the 3rd International Workshop on Distributed Statistical Computing (DSC 2003) [Internet]. Vienna: Technische Universität Wien, 2003. http://www.ci.tuwien.ac.at/Conferences/DSC-2003/ (accessed 14 Apr 2012). [Google Scholar]
- 25. McMurry A. System Use Cases - SHRINE [Internet]. Open.med. 2011. https://open.med.harvard.edu/display/SHRINE/System+Use+Cases (accessed 20 May 2012). [Google Scholar]
- 26. Simard JF, Neovius M, Hagelberg S, et al. Juvenile idiopathic arthritis and risk of cancer: a nationwide cohort study. Arthritis Rheum 2010;62:3776–82 [DOI] [PubMed] [Google Scholar]
- 27. CARRA Policies [Internet] Childhood Arthritis & Rheumatology Research Alliance. http://www.carragroup.org/content_dsp.do?pc=Policies (accessed 8 Apr 2012).
- 28. Wallace CA, Ilowite NT. Project Information: 1RC1AR058605–01. NIH RePORTER—NIH Research Portfolio Online Reporting Tools Expenditures and Results [Internet]. National Institutes of Health. http://projectreporter.nih.gov/project_info_description.cfm?aid=7832335 (accessed 14 Apr 2012).
- 29. Mina R, Scheven von E, Ardoin SP, et al. Consensus treatment plans for induction therapy of newly diagnosed proliferative lupus nephritis in juvenile systemic lupus erythematosus. Arthritis Care Res (Hoboken) 2012;64:375–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huber AM, Robinson AB, Reed AM, et al. Consensus treatments for moderate juvenile dermatomyositis: beyond the first two months. Results of the second Childhood Arthritis and Rheumatology Research Alliance consensus conference. Arthritis Care Res (Hoboken) 2012;64:546–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huber AM, Giannini EH, Bowyer SL, et al. Protocols for the initial treatment of moderately severe juvenile dermatomyositis: results of a Children's Arthritis and Rheumatology Research Alliance Consensus Conference. Arthritis Care Res (Hoboken) 2010;62:219–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DeWitt EM, Kimura Y, Beukelman T, et al. Consensus treatment plans for new-onset systemic juvenile idiopathic arthritis. Arthritis Care Res (Hoboken). Published Online First: 30 January 2012.10.1002/acr.21625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beukelman T, Patkar NM, Saag KG, et al. 2011 American College of Rheumatology recommendations for the treatment of juvenile idiopathic arthritis: initiation and safety monitoring of therapeutic agents for the treatment of arthritis and systemic features. Arthritis Care Res (Hoboken) 2011;63:465–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li SC, Feldman BM, Higgins GC, et al. Treatment of pediatric localized scleroderma: results of a survey of North American pediatric rheumatologists. J Rheumatol 2010;37:175–81 [DOI] [PubMed] [Google Scholar]
- 35. Saltz J, Oster S, Hastings S, et al. caGrid: design and implementation of the core architecture of the cancer biomedical informatics grid. Bioinformatics 2006;22:1910–16 [DOI] [PubMed] [Google Scholar]
- 36. Curtis LH, Weiner MG, Boudreau DM, et al. Design considerations, architecture, and use of the Mini-Sentinel distributed data system. Pharmacoepidemiol Drug Saf 2012;21(Suppl 1):23–31 [DOI] [PubMed] [Google Scholar]
- 37. Platt R, Carnahan RM, Brown JS, et al. The US Food and Drug Administration's Mini-Sentinel program: status and direction. Pharmacoepidemiol Drug Saf 2012;21(Suppl 1):1–8 [DOI] [PubMed] [Google Scholar]
- 38. National Database for Autism Research [Internet] National Institutes of Health (US). http://ndar.nih.gov (accessed 13 Mar 2012).
- 39. Olive M, Rahmouni H, Solomonides T, et al. SHARE road map for HealthGrids: methodology. Int J Med Inform 2009;78(Suppl 1):S3–12 [DOI] [PubMed] [Google Scholar]
- 40. CONNECT Community Portal [Internet] Office of the National Coordinator for Health Information Technology. US Dept of Health and Human Services. http://connectopensource.org (accessed 14 Apr 2012).
- 41. Query Health Initiative [Internet] Standards & Interoperability (S&I) Framework. http://queryhealth.org (accessed 13 Mar 2012).
- 42. IOM (Institute of Medicine) Digital Infrastructure for the Learning Health System: The Foundation for Continuous Improvement in Health and Health Care: Workshop Series Summary. Washington (DC): National Academies Press (US), 2011 [PubMed] [Google Scholar]
- 43. IOM (Institute of Medicine) Learning What Works: Infrastructure Required for Comparative Effectiveness Research: Workshop Summary. Washington (DC): National Academies Press (US), 2011 [PubMed] [Google Scholar]
- 44. Green ED, Guyer MS; National Human Genome Research Institute Charting a course for genomic medicine from base pairs to bedside. Nature 2011;470:204–13 [DOI] [PubMed] [Google Scholar]
- 45. Mandl KD, Kohane IS. No small change for the health information economy. N Engl J Med 2009;360:1278–81 [DOI] [PubMed] [Google Scholar]
- 46. McMurry AJ, Gilbert CA, Reis BY, et al. A self-scaling, distributed information architecture for public health, research, and clinical care. J Am Med Inform Assoc 2007;14:527–33 [DOI] [PMC free article] [PubMed] [Google Scholar]