Abstract
Background
Ascertainment of potential subjects has been a longstanding problem in clinical research. Various methods have been proposed, including using data in electronic health records. However, these methods typically suffer from scaling effects—some methods work well for large cohorts; others work for small cohorts only.
Objective
We propose a method that provides a simple identification of pre-research cohorts and relies on data available in most states in the USA: merged public health data sources.
Materials and methods
The Utah Population Database Limited query tool allows users to build complex queries that may span several types of health records, such as cancer registries, inpatient hospital discharges, and death certificates; in addition, these can be combined with family history information. The architectural approach incorporates several coding systems for medical information. It provides a front-end graphical user interface and enables researchers to build and run queries and view aggregate results. Multiple strategies have been incorporated to maintain confidentiality.
Results
This tool was rapidly adopted; since its release, 241 users representing a wide range of disciplines from 17 institutions have signed the user agreement and used the query tool. Three examples are discussed: pregnancy complications co-occurring with cardiovascular disease; spondyloarthritis; and breast cancer.
Discussion and conclusions
This query tool was designed to provide results as pre-research so that institutional review board approval would not be required. This architecture uses well-described technologies that should be within the reach of most institutions.
Keywords: medical informatics applications, clinical trials, patient selection, public health informatics, clinical research informatics
Background and significance
Finding and retaining a cohort of research participants in clinical investigations, also called ascertainment, is a well-known problem for biomedical researchers. Several excellent reviews have catalogued the problems complicating recruitment.1–3 With the growing interest in translational research, finding participants to join clinical investigations promises to become ever more critical.4 In this article we describe an ascertainment solution that relies on public health data sources available in most states in the USA. We do not limit ourselves to clinical trials; rather we focus on the broader issue of ascertainment for any clinical investigation. We introduce here a general approach which could be used across a spectrum of studies, from small, investigator-initiated studies to full-blown randomized controlled trials, and studies in health services or public health research.
Recruitment and participation failure
Research studies fail for many reasons. Estimates of the fraction of randomized controlled trials that fail, or that require extension because of problems with enrollment, range as high as 60%.5 To provide an initial assessment of recruitment failure in the broader context of all clinical investigations, we performed a study with our institutional review board (IRB) at the University of Utah. We reviewed the enrollment figures for all clinical investigations between 2007 and 2011 that were closed for any reason, including a successfully completed study. For the 726 studies for which we had adequate enrollment data, 182 (25.1%) met or exceeded their stated recruiting goals. The remaining 74.9% failed to meet their original recruitment expectations. Recruitment failure does not necessarily mean that the study failed. It is possible that the initial sample size was overestimated. However, in that case one would expect a positive result that led to early termination and a publication, which was not the case for the vast majority of these studies.
Using electronic health data to enrich the ascertainment pool
Researchers increasingly are using automated methods to enrich an ascertainment sample with potential recruits who already meet key inclusion/exclusion criteria. The use of electronic health records (EHRs) for recruitment will probably increase in the USA as EHR technology becomes even more widespread, especially after the recently enacted Health Information Technology for Economic and Clinical Health (HITECH) Act and its EHR incentive program.6 Some of the very first uses of mining data in EHRs for recruitment came about as a result of the HIV epidemic; papers in 1993–5 by Tu et al and Carlson et al are representative of these early efforts.7 8 With the introduction of the Informatics for Integrating Biology and the Bedside (i2b2) platform,9 many major medical centers adopted a common data architecture and display interface for organizing and querying clinical data. The i2b2 interface introduced a straightforward, intuitive way to raise queries that is accessible to clinicians and researchers. It evolved from early work by Murphy et al.10 The i2b2 query interface supports structured data, such as laboratory data and ICD9-CM diagnoses.
Others have mined the natural language of clinical notes to identify cohorts.11 12 I2b2 was extended by Weber et al into the Shared Health Research Information Network, which supported distributed, aggregating queries across multiple institutional repositories.10 13 A conceptually similar system, the HMO Research Network, similarly permits cross-institutional queries with a population-health focus.14 The electronic MEdical REcords and GEnomics (eMERGE) Network, dedicated to advancing translational research through multi-institution data and biospecimen sharing, has also been shown to be useful in cohort ascertainment.15
However, even a large university-based medical center, or a small collection of them, may have a patient population insufficient to provide a recruitment pool for many studies. The need to maintain an unbiased sample of willing participants (both cases and controls), and not to overburden them with multiple trials requests, often outstrips a single site's population base.2 One approach to overcoming that problem is to use the Web as a recruitment tool, as in ResearchMatch or ClinicalTrials.gov.16 17 An increasingly common technique for aggregating data across multiple clinical centers is to build a virtual repository—that is, one with no persistent aggregation of data.18 19
Objective
In this paper we describe a new perspective on patient ascertainment, taking its inspiration from early work in probabilistic linking of public health data files.20–23 In the USA, each state funds a Department of Health that collect vital statistics on births, deaths, marriages, and divorces. Most states maintain repositories containing inpatient hospital admissions, ambulatory surgeries, emergency department visits, and a cancer registry. Some state health departments have developed Web-based data query systems to provide controlled access to these data.24 We describe here how it is feasible to query a large repository of linked health data sources (the Utah Population Database) to identify potential research cohorts.
Overview of the Utah Population Database (UPDB)
The UPDB is a research resource at the University of Utah. It includes genealogies of the founders of Utah and their Utah descendants that were obtained from the Utah Family History Library. UPDB has been extended by including linked datasets from public health sources that provide information not available in any single dataset. State-wide data include information from driver license records, the Utah and Idaho cancer registries, Utah hospital inpatient records and ambulatory surgery events, Utah vital records (births, fetal deaths, marriages, divorces, and deaths), the social security death index, and voter registration records.
The number of records for each data source is shown in table 1; it also indicates which of these sources were made available to the tool described in this paper. Over 18.9 million source records have been linked for over 6.5 million distinct individuals. Most Utah families are represented; some having as many as 11 generations of pedigree data. In addition, EHR data covering patients in the state's two largest healthcare networks (Intermountain Healthcare and the University of Utah Health Care system) have been linked to individuals within the UPDB. A master subject index has been created between these sources and the UPDB. This linking process, and the quality of probabilistic record linking, have been previously described.25 26
Table 1.
Sources of data and number of records comprising the Utah Population Database (UPDB)
| Sources of data | Number of records |
|---|---|
| Genealogies (Family History Library)* | 1619006* |
| Utah Cancer Registry* | 280046* |
| Cancer Data Registry of Idaho | 135816 |
| Vital Records (UDOH) | 4213781 |
| Births 1915–77 970774 Births 1978–2010* 1465125* Marriages and divorces 987779 Deaths 1904–67 333144 Deaths 1968–2010* 447908* Fetal deaths 9051 |
|
| Inpatient hospital discharge claims (UDOH)* | 3835954* |
| Ambulatory surgery (UDOH)† | 3399967 |
| Driver licenses (Utah Department of Public Safety) | 3309006 |
| Social security death* | 581368* |
| Utah voter registration | 1586961 |
| 18961905 |
*Indicates items made available to the tool described in this paper.
†New dataset in UPDB and not yet available to the tool described here.
UDOH, Utah Department of Health.
Because investigators do not have direct access to the UPDB, we have designed and implemented a system called Utah Population Database Limited (UPDBL) to allow Web access to aggregated, deidentified UPDB data. The overall strategy was to create a rapid query system to determine the availability of specific research cohorts. UPDBL's design requirements were to (1) allow users to select cohorts using information from multiple, integrated UPDB datasets; (2) allow users access to some family or pedigree information; (3) provide results as pre-research so that IRB approval would not be required; and (4) incorporate a query tool methodology that returns deidentified, summary data. When researchers discover that a cohort exists that meets basic research eligibility criteria, they can move forward with a grant or IRB application.
Materials and methods
Implementation of the UPDBL system
The UPDBL system consists of two parts: the UPDBL database and the UPDBL query tool. The UPDBL database is maintained as an independent, limited version of the UPDB. It contains de-normalized person, relationship, and record detail tables, and tables related to managing queries, users, and logging use of the query tool. The UPDBL query tool provides a Web-enabled interface, where researchers generate queries and view aggregate results from the UPDBL.
Architecture of the UPDBL system
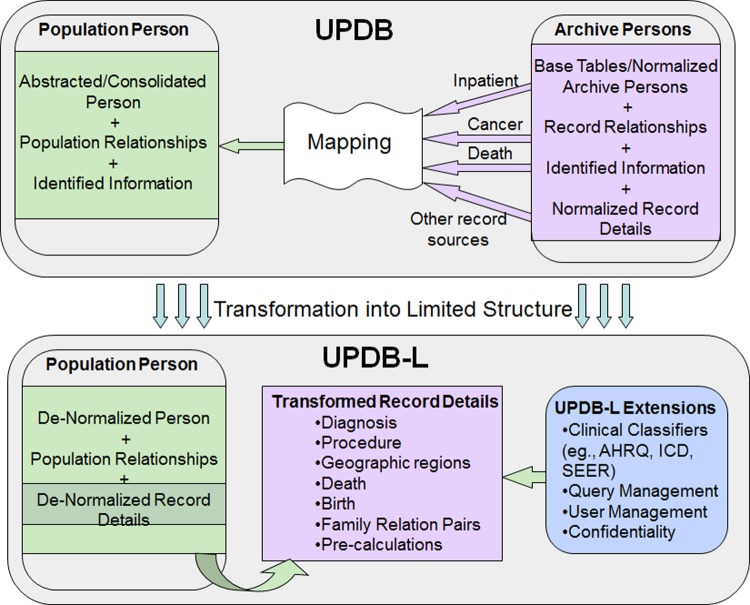
The design of the UPDBL is a relational database containing demographic information on unique individuals loaded into the PopulationPerson table (see figure 1), the base table that connects or links to all the other health data tables. There are over 152 million relative pairs which include all first-, second-, and third- (first cousin) degree relationships. UPDBL also includes clinical details from multiple sources. These incorporate the International Classification of Diseases (ICD) coding system, the International Classification of Diseases for Oncology (ICDO), and Surveillance, Epidemiology, and End Results (SEER) ‘recodes’ (for cancer site specification). UPDBL does not include identifying information such as name, address, or social security number. Only a subset of UPDB data is available for interrogation in UPDBL. However, information from restricted datasets such as driver license and voter registration is used to derive current residence information at the state health-district level for UPDBL. There are also tables in UPDBL that track users and queries, providing information for security auditing.
Figure 1.
Overview of the transformation process that converts the Utah Population Database (UPDB) to Utah Population Database Limited (UPDBL). The details have been abstracted, showing the general flow from identifiable data sources (top figure) to a de-identified, flat data structure (bottom figure) accessible to the UPDBL query tool (lower right). AHRQ, Agency for Healthcare Research and Quality; ICD, International Classification of Diseases; SEER, Surveillance, Epidemiology, and End Results. This figure is only reproduced in colour in the online version.
Transformation scheme
Because data come from disparate original datasets that vary by era-of-creation, structural transformations convert the original UPDB database to the UPDBL version. This facilitates consistency when building queries and efficient performance. An overview of the transformation process is shown in figure 1 and includes:
De-normalization of person, diagnosis, procedure, and birth records, which flatten the data structures to simplify and facilitate query generation.
Pre-calculation of age and date ranges at the time of data loading for improved performance.
Computation of combined ICD code, ICD revision, and ICDO data fields to improve structured query language generation for complex logical queries involving many different codes and code versions.
Generation of first-degree relationships linking individuals, stored in a bidirectional manner (eg, father–child and child–father) from the original unidirectional UPDB relationship table.
Reformatting birth records which vary greatly over time in information and structure. As part of the UPDBL transformation, older records are restructured to fit the newer schema.
Dictionaries, based on the Agency for Healthcare Research and Quality Clinical Classification system and the SEER recode system, have been created to provide a powerful and intuitive user interface for finding appropriate diagnosis codes when building queries.
The UPDBL query tool
The UPDBL query tool is based on a conceptual architecture previously developed by the research informatics group at the Huntsman Cancer Institute, University of Utah. It is made available as a Web-based Java application that provides a rich front-end graphical user interface. It enables researchers with minimal training to design, build, store, and run queries, and to view aggregate results for their cohort.
Query generation
The UPDBL user interface assists users in building potentially complex queries that may span several types of health records and historical coding revisions. The query-building interface is oriented towards creating the logical structure of the query. Users choose individual criteria involving demographic, diagnosis, procedure, and birth data elements. Multiple levels of query criteria are placed within a visual logical tree structure as the user builds the query (for an example, see figure 5). Users can choose the type of logical condition that will be applied between the criteria (AND/OR). The UPDBL design allows users to create queries with any degree of logical complexity. If users select multiple criteria they are considered to be AND'd together within the criteria. Diagnosis and procedure criteria can also be NOT'd. Users can also create additional levels of logic to find people with a given condition who have a relative who matches another specific condition.
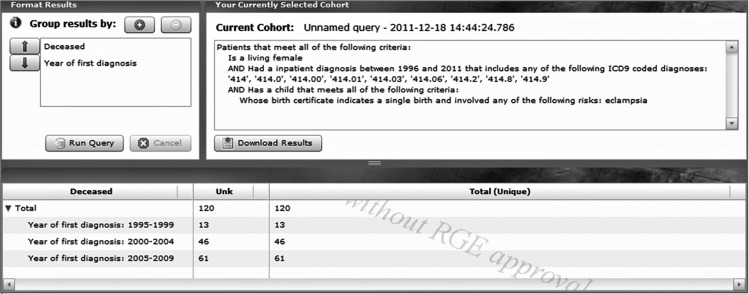
Figure 5.
A sample screen shot showing the results of a moderately complex query specifying living women who had an inpatient diagnosis of ischemic heart disease and who also had an eclampsia-related disorder during a pregnancy with a single infant. The ‘Current cohort’ pane (upper right) summarizes in words the user's selections using previous selection screens. Results of the query are displayed in the bottom pane. The column headed ‘Unk’ indicates that there are no known death certificates for the women in this cohort.
Aggregation of results
After building a query, users run their query to produce a set of counts aggregated by a combination of user-selected data fields, such as demographic, birth, diagnosis, procedure, and dates/age ranges. The query tool generates an n-dimensional aggregation, with the first level being presented as columns for each value and each subsequent level being represented by a nested tree structure of rows. Results are given as totals (and a total number of distinct persons) for each row and column. Users can download the results of their queries to Excel. Examples of these steps are shown in the figures mentioned in the ‘Results’ section.
Regulatory considerations
A number of regulatory activities were required to enable the release of the UPDBL. The project was approved by the University of Utah IRB. The University of Utah Resource for Genetic and Epidemiologic Research (RGE) provides oversight for UPDB and reviewed each version before release. The RGE review committee includes a representative for each data contributor and is described in detail elsewhere.27
Confidentiality
To minimize the potential for re-identification of an individual through repeated queries we implemented a data constraint strategy. The final query constraints were worked out in conjunction with a consultant from the Inter-University Consortium for Political and Social Research, University of Michigan; this organization assists data producers in making data ready for public release. Two key issues emerged from the consultation. The first was whether ‘sensitive’ medical diagnoses should be restricted. We decided that this restriction would be inherently subjective and arbitrary, concluding that as long as the geographic location was large, we could include all diagnoses with negligible risk. As a result, location is available at the level of the entire state and the 12 Utah health districts, and no data are provided by city, county, or zip code. This is a close approximation of the geographical identifier rule in the Health Insurance Portability and Accountability Act (HIPAA) Safe Harbor standard. The second decision was determining the level of familial information that would be released. We agreed that deidentified groupings of first-degree (parent–child, siblings), second-degree (aunt/uncle–niece/nephew, grandparent–grandchild), and third-degree relatives (only first cousins) would be acceptable.
We also decided to use a two-level confidentiality approach. Microlevel confidentiality focuses on individuals within the database while the macrolevel deals with issues associated with the authorized users. Microlevel confidentiality followed the methodology discussed by Rudolph et al28 to deal with the disclosure risk associated with a user running multiple queries on small result sets. In returning aggregated results, the UPDBL implements the following: (1) cells of less than 5 are denoted by an asterisk (*) and the number is removed from the column/row total; (2) ages are grouped in 5-year intervals with age 90+ as one category; (3) year of birth, diagnosis, and death are grouped in 5-year intervals; and (4) geographic locations are grouped into 12 Utah health districts.
At the macrolevel, since the UPDBL query tool is a resource available to individuals associated with institutions of higher learning or non-profit health organizations who intend to conduct research, we set a strict authorization process. This included (1) a Web-enabled process for the registration of users; (2) completion of a data user agreement that was reviewed by the University of Utah general counsel and University of Utah privacy officer; (3) a confidentiality reminder each time a user logs in; and (4) on-going logging of all registered users and all submitted queries. The information provided at the time of registration (such as full name, institutional affiliation, email, and phone number) is manually authenticated for users who are outside specific domains at the University of Utah. In the data-use agreement, the user agrees that ‘I will not link the query system data with any other data that, after linkage, might allow identification of an individual represented in the query system data.’ The data use agreement can be viewed online.29 Last, the results of a UPDBL query may be used internally to provide preparatory information for research projects; however, they may not be published in abstracts, posters, manuscripts, etc, without permission from the RGE.
Results
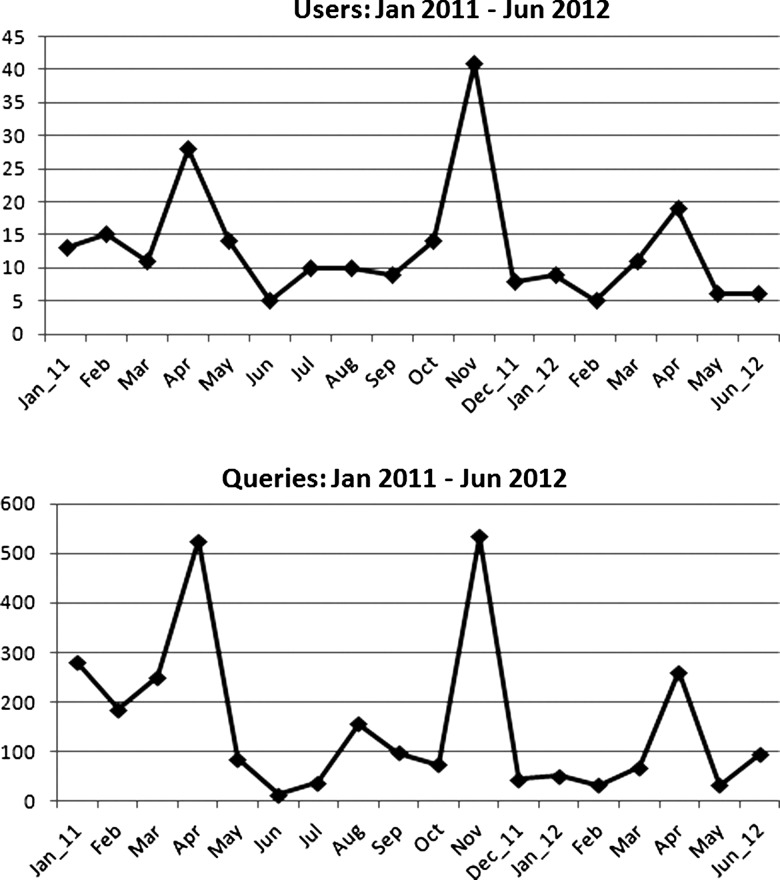
After the initial design of the UPDBL query tool, input was solicited from focus groups of investigators with previous research experience using UPDB. The first release of the UPDBL query tool was in 2009, and a major update was released in 2011. At each release, user focus groups helped to guide design. The query tool provides an extensive help section and context-sensitive tips, as well as five training videos, so that researchers can quickly understand how to navigate the query process. Since its release, 241 users from 17 institutions have signed the user agreement and used the query tool. These users represent a wide range of disciplines such as biomedical informatics, cardiology, family and preventive medicine, internal medicine, neurology, obstetrics and gynecology, pediatrics, pharmacotherapy, psychiatry, and surgery. To provide a sense of current research usage, we selected data for an 18-month period after completion of the development; thus usage by the development staff is not included. For the period from January 2011 through June 2012, the number of users each month is presented in the top panel of figure 2 with the number of queries each month in the bottom panel. The three peaks are the result of regular training sessions for UPDBL, which are open to university faculty and staff. Note that the shapes of both time series are similar, suggesting that a modest number of users (from 5 to 40 per month) undertake fairly deep explorations (mean of 14.6 queries per user).
Figure 2.
Utah Population Database Limited (UPDBL) usage patterns from the most recent 18 months available, showing number of distinct users and number of queries, January 2011 to June 2012 (UPDBL was released in August 2009). Note that the shapes of both time series are similar, suggesting that a modest number of users (from 5 to 40 per month) undertake fairly deep explorations (mean of 14.6 queries per user).
To demonstrate the capability of the UPDBL query tool, we provide three case examples; queries were made entirely through the user interface and required no additional data processing or programing.
Pregnancy complications co-occurring with cardiovascular disease
In a recent publication Mongraw-Chaffin et al30 investigated the contribution of pregnancy complications, specifically pre-eclampsia, to the risk of subsequent deaths from cardiovascular disease. We modified this research question to identify a potential cohort of women who had eclampsia or pre-eclampsia (identified from birth certificates) who also experienced an inpatient hospital event for ischemic heart disease.
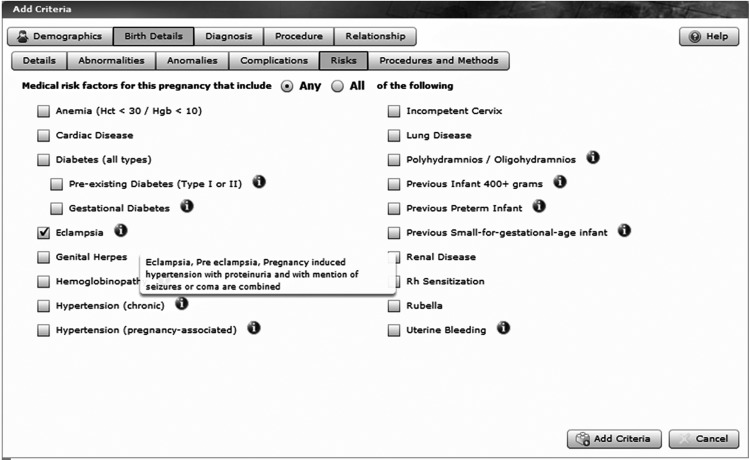
The first step of building the cohort identifies women who had a birth from 1978 through 2009, with the stipulation that this was a single birth and that the women experienced eclampsia, see figure 3. This category includes eclampsia, pre-eclampsia, and pregnancy-induced hypertension with proteinuria and with a mention of seizures or coma (note the ‘infobutton’ information box explaining the semantics of this selection). We add criteria to the query to include those who had an inpatient visit between 1996 and 2009 with one of the ICD9 codes for ischemic heart disease as seen in figure 4. We have grouped the cohort by whether they are alive or dead, and by the year of diagnosis. This query requires access to a number of linked datasets, including birth certificates, hospital inpatient discharge records, and death certificates. As shown in figure 5, the results provide a potential study cohort of 120 living women who meet all the criteria.
Figure 3.
A sample screen shot showing the selection of birth details, specifically eclampsia, pre-eclampsia, and pregnancy-induced hypertension with proteinuria and with a mention of seizures or coma, which are all combined under ‘eclampsia’ as shown to the user in the pop-up help balloon.
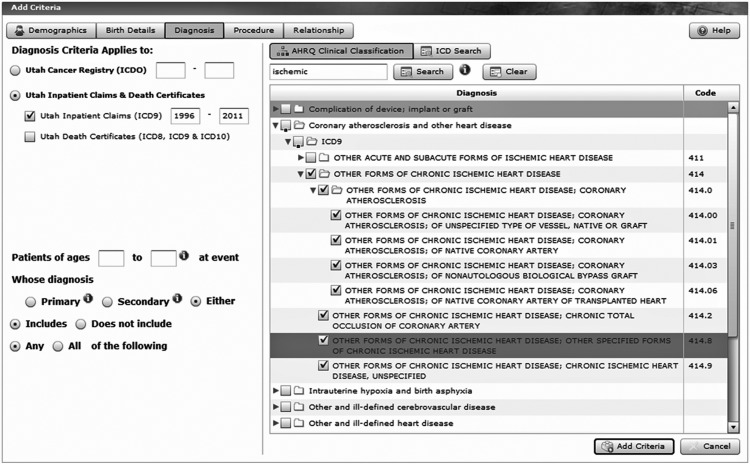
Figure 4.
A sample screen shot showing how diagnoses are selected in Utah Population Database Limited. Ischemic heart disease was the target, as defined in the Agency for Healthcare Research and Quality Clinical Classification taxonomy, and the query is to draw from the Utah state-wide inpatient claims and death certificate data sources (upper left). The key word ‘ischemic’ was typed in the search window (upper center) to start the taxonomy traversal.
Spondyloarthritis
This example is based on a pre-research query from a clinical researcher who ran a series of queries to identify patients with spondyloarthritis (SpA). SpA is the name given to a family of inflammatory rheumatic diseases affecting the spine and other joints, as well as ligaments and tendons. It predominantly affects teens and young adults. This researcher wanted to determine if there was a sufficient pool of people to examine rates of cardiovascular disease and selected risk factors in patients with SpA. Using inpatient records and death certificates, 1441 subjects were identified with SpA, including those with ankylosing spondylitis, psoriatic arthritis, or inflammatory bowel disease-related arthritis. For these patients, the researcher queried diagnosis of myocardial infarction, ischemic cerebrovascular disease, or peripheral vascular disease and cardiovascular risk factors of hypertension, hyperlipidemia, diabetes mellitus, and obesity. The results are shown in table 2.
Table 2.
Results of UPDBL queries on patients with spondyloarthritis and with cardiovascular complications (overall n=1441)
| Spondyloarthritis diagnosis% (n) | |
|---|---|
| Cardiovascular diagnosis | |
| Ischemic cerebrovascular disease | 0.76 (11) |
| Myocardial infarction | 7.84 (113) |
| Peripheral vascular disease | 2.64 (38) |
| Cardiovascular risk factors | |
| Hypertension | 47.19 (680) |
| Hyperlipidemia | 14.37 (207) |
| Diabetes mellitus | 18.46 (266) |
| Obesity | 13.32 (192) |
UPDBL, Utah Population Database Limited.
Breast cancer
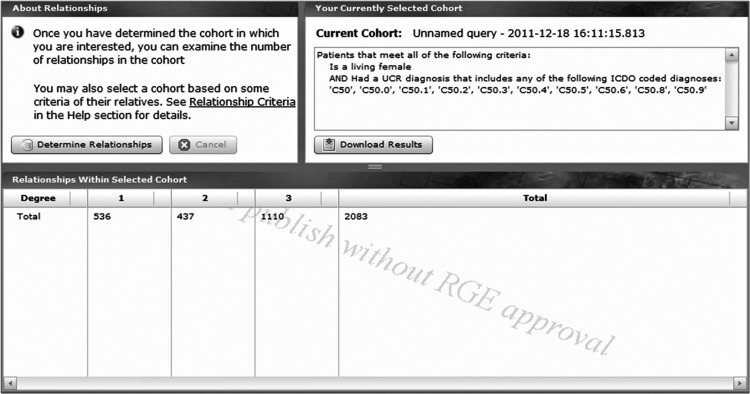
This case example uses the pedigree data in the UPDBL; these data consist of family history data that have been extended by vital records, particularly birth certificates. Consider a hypothetical researcher into breast cancer who wants to conduct a familial study that includes patients with breast cancer who are related as siblings, as mother–daughter pairs, as aunt–niece pairs, or as first cousins. That researcher can greatly reduce the recruitment effort if he/she can identify these related individuals at the time of ascertainment. A query was submitted requesting information on all women diagnosed with breast cancer in the Utah Cancer Registry and who are alive. The result set was18 281. Next we wanted to know how many of these patients with breast cancer were related. The query tool includes a tab to ‘Determine relationships’ and is shown in figure 6. There were 536 sets of living first-degree relatives (siblings, mother–daughter) who both had breast cancer, 437 sets of living second-degree relatives (aunt–niece, grandmother–granddaughter) and 1100 sets of living third-degree relatives (first cousins).
Figure 6.
A sample screen shot showing how many first- (n=536), second- (n=437), and third- (n=1110) degree relatives are known to exist in the Utah Population Database for the individuals from a Utah Population Database Limited query of the number of living women with breast cancer.
Discussion
We built the UPDBL query tool to provide researchers with a mechanism for obtaining a realistic estimate of potential research cohorts. The tool provides results in the aggregate, and thus it does not require pre-approval by Utah IRB nor does it require the involvement of information technology staff.
The pre-eclampsia example illustrates how a researcher could read an informative research article and immediately explore UPDBL for the possibility of finding similar data. As a side note, if a woman had multiple admissions for ischemic heart disease, only one will be returned by the query tool. While this constrains multiple counts of the same diagnosis for the same person, it also limits the ability of the UPDBL query tool to be used to study readmissions.
The researcher who used UPDBL to assess a possible cohort with spondyloarthritis and cardiovascular disease now has a study approved by the University of Utah IRB and the RGE. The staff that manages the UPDB has created an individual-level dataset from the UPDB (the original source of data for the UPDBL) to use in her project.
Although hypothetical, the breast cancer example could provide a cancer researcher with information about a cohort of breast cancer survivors who also have a living close relative with breast cancer. This is a salient topic in Utah; the UPDB was used in the original study that identified the first two breast cancer genes, BRCA1 and BRCA2.31 These women and the unaffected women in their families would be appropriate for a screening study to determine whether they should receive tailored recommendations for the frequency of screening mammograms. The RGE has developed a set of policies and procedures for contacting potential subjects to participate in such research which has been described by Wylie and Mineau.27
As discussed above, the UPDB has been linked to the EHR of healthcare networks in the state. Once cohorts of interest are identified and researchers develop IRB-approved protocols, these two resources can provide the basis for subject recruitment into clinical research studies.
Confidentiality
There are a number of approaches available to maintain the confidentiality of individuals within large databases. It was essential to provide practices that would mitigate threats while providing investigators with tools to determine the feasibility of research cohorts. In addition to the constraints such as scrubbing small cells, etc, the macrolevel constraints provide an additional level of confidentiality by placing restrictions on the query tool users who must register and complete a user agreement. As described by Malin et al, the ability to join deidentified data with an identified resource can result in the reidentification of individuals.32 The user agreement restricts the user from joining results to auxiliary data sources and includes legal consequences.
Limitations
The portability of the query tool has not been used as part of an open-source initiative. Requests for collaborative development and identification of additional funding sources could provide this opportunity in the future. However, the conceptual approach described here is portable to nearly all states in the USA. Also, we do not routinely audit queries submitted by a user. As noted by Dwork, query auditing is computationally problematic.33 However, if misuse of the tool is discovered we can identify the user based on audit logs and pursue a number of punitive options.
Future development
Other state-wide health datasets of interest include the Emergency Department Database, which is an administrative database available in many states, and the All Payer Claims Database that is being developed for a number of states including Utah. The All Payer Database compiles medical and pharmacy claims data across healthcare insurances providers (payers) and would provide valuable information for both inpatient and outpatient encounters. As more data sources are added and the query tool becomes more complex, we envisage conducting a user survey to evaluate their satisfaction over time, similar to the method suggested by Rajput et al.34
Conclusions
To deal with the reasons that research studies fail, investigators need tools that provide solid pre-research data for use in funding applications. The UPDBL query tool provides access to query-integrated, linked health data sources. The tool's user interface guides a researcher through the creation of a query and does so in a way that does not require any special technical proficiency on the part of the user.
Public health data sources are available in most states in the USA. While UPDB does include access to detailed pedigree data that may not be available in other places, we have demonstrated using several examples that a query tool built on integrated public health sources alone is quite powerful. In contrast to EHR-based ascertainment approaches, which are usually limited to a single healthcare network, the use of public health sources promises the broadest possible survey of potential cohorts.
Acknowledgments
We are indebted to Dr Melissa Reily for permission to use her spondyloarthritis data table. We wish to thank Dr Felicia LeClere for her evaluation of the Utah Population Database Limited query tool.
Footnotes
Contributors: JFH and GPM conceived the article, framed the overall study, oversaw the data collection and use of case examples, and wrote the ‘Background-Significance’ and ‘Discussion/Conclusion’ sections. SJC, SCH, AH, CS, and AMF built and tested the Utah Population Database Limited system and wrote/edited the technical descriptions. All the authors contributed to the editing and final approval of the manuscript. JFH is the guarantor of the paper.
Funding: This work was supported by the National Center for Research Resources grants R01-RR021746 and UL1-RR025764. This work was partially supported (Utah Population Database and the Utah Cancer Registry) by the National Cancer Institute grants P30CA042014 and HHSN 261201000026C; by the Huntsman Cancer Foundation; and by the Utah Department of Health and University of Utah.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Campbell MK, Snowdon C, Francis Det al. Recruitment to randomised trials: strategies for trial enrollment and participation study. The STEPS study. Health Technol Assess 2007;11:iii, ix–105 [DOI] [PubMed] [Google Scholar]
- 2.Sung N, Crowley W, Jr, Genel Met al. Central challenges facing the national clinical research enterprise. JAMA 2003;289:1278. [DOI] [PubMed] [Google Scholar]
- 3.Watson JM, Torgerson DJ. Increasing recruitment to randomised trials: a review of randomised controlled trials. BMC Med Res Methodol 2006;6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Embi PJ, Payne PR. Clinical research informatics: challenges, opportunities and definition for an emerging domain. J Am Med Inform Assoc 2009;16:316–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puffer S, Torgerson D. Recruitment difficulties in randomised controlled trials. Controlled Clin Trials 2003;24:S214–15 [Google Scholar]
- 6.The HITECH Act: Medicare and Medicaid Electronic Health Records (EHR) Incentive Programs. 2011. http://www.cms.gov/ehrincentiveprograms (accessed 3 June 2011).
- 7.Tu SW, Kemper CA, Lane NM, et al. A methodology for determining patients’ eligibility for clinical trials. Methods Inf Med 1993;32:317–25 [PubMed] [Google Scholar]
- 8.Carlson RW, Tu SW, Lane NMet al. Computer-based screening of patients with HIV/AIDS for clinical-trial eligibility. Online J Curr Clin Trials 1995; Doc No 179:(3347 words; 32 paragraphs) [PubMed] [Google Scholar]
- 9.i2b2. The i2b2 Home Page. 2012 (accessed 27 June 2012). http://www.i2b2.org [Google Scholar]
- 10.Murphy SN, Gainer V, Chueh HC. A visual interface designed for novice users to find research patient cohorts in a large biomedical database. AMIA Annual Symposium Proceedings/AMIA Symposium AMIA Symposium (Evaluation Studies Research Support, Non-US Gov't) 2003:489–93 [PMC free article] [PubMed]
- 11.Pakhomov S, Weston SA, Jacobsen SJ, et al. Electronic medical records for clinical research: application to the identification of heart failure. Am J Manag Care (Comparative Study Research Support, N.I.H., Extramural Validation Studies) 2007;13:281–8 [PubMed] [Google Scholar]
- 12.Seyfried L, Hanauer DA, Nease D, et al. Enhanced identification of eligibility for depression research using an electronic medical record search engine. Int J Med Inform (Research Support, N.I.H., Extramural) 2009;78:e13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy SN, Weber G, Mendis Met al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc: JAMIA (Research Support, N.I.H., Extramural) 2010;17:124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieu TA, Hinrichsen VL, Moreira A, et al. Collaborations in population-based health research: the 17th Annual HMO Research Network Conference, 23–25 March 2011, Boston, MA, USA. Clin Med Res (Research Support, N.I.H., Extramural) 2011;9:137–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schildcrout JS, Basford MA, Pulley JMet al. An analytical approach to characterize morbidity profile dissimilarity between distinct cohorts using electronic medical records. J Biomed Inform 2010;43:914–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Research Match: Online Recruitment. 2011 (accessed 3 June 2011). http://www.researchmatch.org
- 17.ClinicalTrials.gov: Online Recruitment. 2011 (accessed 3 June 2011). http://www.clinicaltrials.gov
- 18.Fultz S, Skanderson M, Mole Let al. Development and verification of a ‘virtual’ cohort using the National VA Health Information System. Med Care 2006;44:S25. [DOI] [PubMed] [Google Scholar]
- 19.Livne OE, Schultz ND, Narus SP. Federated querying architecture with clinical & translational health IT application. J Med Systems 2011;2011:1–14 [DOI] [PubMed] [Google Scholar]
- 20.Howe GR. Use of computerized record linkage in cohort studies. Epidemiol Rev 1998;20:112–21 [DOI] [PubMed] [Google Scholar]
- 21.Adams MM, Wilson HG, Casto DL, et al. Constructing reproductive histories by linking vital records. Am J Epidemiol 1997;145:339–48 [DOI] [PubMed] [Google Scholar]
- 22.Howe GR, Lindsay J. A generalized iterative record linkage computer system for use in medical follow-up studies. Comput Biomed Res 1981;14:327–40 [DOI] [PubMed] [Google Scholar]
- 23.Newcombe HB, Kennedy JM, Axford SJ, et al. Automatic linkage of vital records. Science 1959;130:954–9 [DOI] [PubMed] [Google Scholar]
- 24.Braithwaite G, Haggard LM. Utah's IBIS-PH: an innovative user interface solution for Web-based data query systems. J Public Health Manag Pract 2006;12:146–54 [DOI] [PubMed] [Google Scholar]
- 25.Duvall SL, Fraser AM, Kerber RA, et al. The impact of a growing minority population on identification of duplicate records in an enterprise data warehouse. Stud Health Technol Inform 2010;160:1122–6 [PubMed] [Google Scholar]
- 26.Duvall SL, Fraser AM, Rowe K, et al. Evaluation of record linkage between a large healthcare provider and the Utah Population Database. J Am Med Inform Assoc 2012;19:e54–e59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wylie JE, Mineau GP. Biomedical databases: protecting privacy and promoting research. Trends Biotechnol 2003;21:113–16 [DOI] [PubMed] [Google Scholar]
- 28.Rudolph BA, Shah GH, Love D. Small numbers, disclosure risk, security, and reliability issues in web-based data query systems. J Public Health Manag Practice 2006;12:176. [DOI] [PubMed] [Google Scholar]
- 29.UPDBL. UPDBL Data Use Agreement. 2009 (accessed 27 June 2012). http://hci-updblapp.hci.utah.edu/updbl/Help/UPDBL.htm.
- 30.Mongraw-Chaffin ML, Cirillo PM, Cohn BA. Preeclampsia and cardiovascular disease death: prospective evidence from the child health and development studies cohort. Hypertension 2010;56:166–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldgar DE, Cannon-Albright LA, Oliphant Aet al. Chromosome 17q linkage studies of 18 Utah breast cancer kindreds. Am J Hum Genet (Research Support, U.S. Gov't, P.H.S.) 1993;52:743–8 [PMC free article] [PubMed] [Google Scholar]
- 32.Malin B, Karp D, Scheuermann RH. Technical and policy approaches to balancing patient privacy and data sharing in clinical and translational research. J Investig Med (Research Support, N.I.H., Extramural) 2010;58:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dwork C. A firm foundation for private data analysis. Commun ACM 2011;54:86–95 [Google Scholar]
- 34.Rajput ZA, Mbugua S, Amadi Det al. Evaluation of an Android-based mHealth system for population surveillance in developing countries. J Am Med Inform Assoc 2012;19:655–9 [DOI] [PMC free article] [PubMed] [Google Scholar]