Abstract
The national adoption of electronic health records (EHR) promises to make an unprecedented amount of data available for clinical research, but the data are complex, inaccurate, and frequently missing, and the record reflects complex processes aside from the patient's physiological state. We believe that the path forward requires studying the EHR as an object of interest in itself, and that new models, learning from data, and collaboration will lead to efficient use of the valuable information currently locked in health records.
Introduction
The national push for electronic health records (EHR)1 will make an unprecedented amount of clinical information available for research; approximately one billion patient visits may be documented per year in the USA. These data may lead to discoveries that improve understanding of biology, aid the diagnosis and treatment of disease, and permit the inclusion of more diverse populations and rare diseases. EHR can be used in much the same way that paper records have been used, with manual extraction and interpretation of clinical information. The big promise, however, lies in large-scale use, automatically feeding clinical research, quality improvement, public health, etc. Such uses require high-quality data, which are often lacking in EHR. In this paper, we investigate a path forward for exploiting EHR data.
Challenges
Unfortunately, the EHR carries many challenges.2
Completeness
The data are largely missing in several ways. Data are occasionally missing by mistake, in the sense that data that would normally be expected to be recorded are lacking. Data are often missing in the sense that patients move among institutions for their care so that individual institutional databases contain only part of their care, and health information exchange is insufficiently pervasive to address the issue; the result is data fragmentation for research and discontinuity of clinical care. Data are also missing in the sense that they are only recorded during healthcare episodes, which usually correspond to illness. In addition, much information is implicit, under the assumption that the human reader will infer the missing information (eg, pertinent negative findings). The result is a time series that is very far from the rigorous data collection normally employed in formal experiments. Referring to the statistical taxonomy of missingness,3 health record data are certainly not missing at random, and might facetiously even be referred to as ‘almost completely missing’.
Accuracy
The data are frequently inaccurate,4 resulting in a loss of predictive power.5 Errors can occur anywhere in the process from observing the patient, to conceptualizing the results, to recording them in the record, and recording is influenced by billing requirements and avoidance of liability. Whereas some errors may be treated as random, many errors—such as influence from billing—are systematic. In addition, there is often mismatch between the nominal definition of a concept and the intent of the author. For example, PERRLA is an acronym commonly used in the eye examination that stands for ‘pupils equal, round, and reactive to light and accommodation’. It is unclear, however, how often clinicians actually test for each of the properties. In the CUMC database, 2% of patients who were missing one eye were documented in a narrative note as being PERRLA—an impossibility because two eyes are required to have equal pupils—and another 8% of those patients were documented as being PERRLA on the left or on the right, which is a misuse of the term. A researcher looking for subjects whose pupils were equal or accommodated normally could not rely on a notation of PERRLA in the chart.
Complexity
Healthcare is highly complex. It includes a mixture of many continuous variables and a large number of discrete concepts. For example, at CUMC, there are 136 035 different concepts that may be stored in the database. There is an enormous amount of work being done to create knowledge structures to define the data, including formal definitions, classification hierarchies, and inter-concept relationships (eg, clinical element model).6 Maintaining such a structure will remain a challenge, however. There may also be local variation both in structure7 and in definition and use,8 and even within an institution definitions vary over time. Much of the most important data in the record—such as symptoms and thought processes—are stored as narrative notes, which require natural language processing9 to generate a computable form. Temporal attributes are highly complex, with time scales from seconds to years and with different levels of uncertainty.10
Bias
The above challenges, including systematic errors, can result in significant bias when health record data are used naively for clinical research. For example, in one EHR study of community-acquired pneumonia,11 patients who came to the emergency department and died quickly did not have many symptoms entered into the EHR. As a result, an attempt to repeat Fine's pneumonia study12 using EHR data showed that the apparently healthiest patients died at a higher rate than sicker patients. Ultimately, healthcare data reflect a complex set of processes13 (figure 1), with many feedback loops. For example, physicians request tests relevant to the patient's current condition, and testing guides the diagnosis, which determines the treatment and future testing. Such feedback loops produce non-linear recording effects that do not reflect the underlying physiology that researchers may be attempting to study. Put another way, EHR data are not merely research data with noise and missing values. The extent and bias of the noise and missingness are sufficient to require fundamentally different methods to analyse the data.
Figure 1.
Feedback loops in the electronic health record. The state of the patient varies, and it determines not only the value of the measurements in the record, but also the type and timing of the measurements.
State of the art
Fortunately, it appears that EHR do contain sufficient information: clinicians generally use health records effectively. They learn to navigate the complexity of the record and to fill in implicit information. Reusing the information for research should be possible, but having a clinician interpret the record for every case is infeasible for large studies.
To address the challenges, the task is generally broken into two steps. The first step, which can be called phenotyping or feature extraction, transforms the raw EHR data into clinically relevant features. The second step uses these features for traditional research tasks—such as measuring associations for discovery or assessing eligibility for a trial—as if a research coordinator had manually entered and verified the features. For the most part, the EHR challenges are addressed in the first step so that large EHR databases can become large research databases that can then undergo traditional analysis.
Studies employing large-scale EHR data have begun to appear,14–19 and most of them employ this two-step approach. The state of the art in feature extraction is to use a heuristic, iterative approach to generate queries that run across the entire EHR database. For example, clinical experts may read each record for a subset of subjects and create a curated dataset. A knowledge engineer generates a heuristic rule that maps record data to each variable in the study (eg, physician notes, billing codes, and medications may all be used to infer the presence of a disease). The rule is tested on the curated subset, and the rule is modified iteratively until sensitivity and specificity reach some threshold. The rule is then applied to the entire cohort.
While this avoids most case-by-case review, it still requires feature-by-feature authoring of queries. These methods are themselves time consuming;20 furthermore, there is much potentially useful information that is not used, the queries may be time consuming to maintain, and knowledge engineers and clinical experts bring their own biases. To draw an analogy with computational biology, imagine attempting high throughput research in which each investigator had to spend months verifying each of thousands of variables before collecting data. As we move to large-scale mining of the EHR, defining the queries has become a bottleneck. Efforts like eMERGE21 are showing significant progress in generating and sharing queries across institutions,22 23 but local variations remain, and defining even a small number of phenotypes can take a group of institutions years. Despite advances in ontologies and language processing, the process remains largely unchanged since the earliest days,24 using detective work and alchemy to get golden phenotypes from base data.
Next-generation phenotyping
There are several ways to improve on the current state. One approach improves on the current phenotyping process, either by making it more accurate or by reducing the knowledge engineering effort. We refer to the latter as ‘high-throughput phenotyping’. The term could be applied to the current state of the art because even a manually generated query can be run on a large database, but we suggest reserving the term for truly high-throughput approaches that do not require years to generate a handful of phenotypes. A high-throughput approach should generate thousands of phenotypes with minimal human intervention such that they could be maintained over time.
To improve phenotyping substantially, we believe that there needs to be a radical shift in approach and that the answer lies in a familiar place for informatics: a combination of top-down knowledge engineering and bottom-up learning from the data. In particular, we believe that we need a better understanding of the EHR. The EHR is not a direct reflection of the patient and physiology, but a reflection of the recording process inherent in healthcare with noise and feedback loops. We must study the EHR as an object in itself, as if it were a natural system. This better understanding will then naturally support both broad-based outcome-oriented research and physiological research.
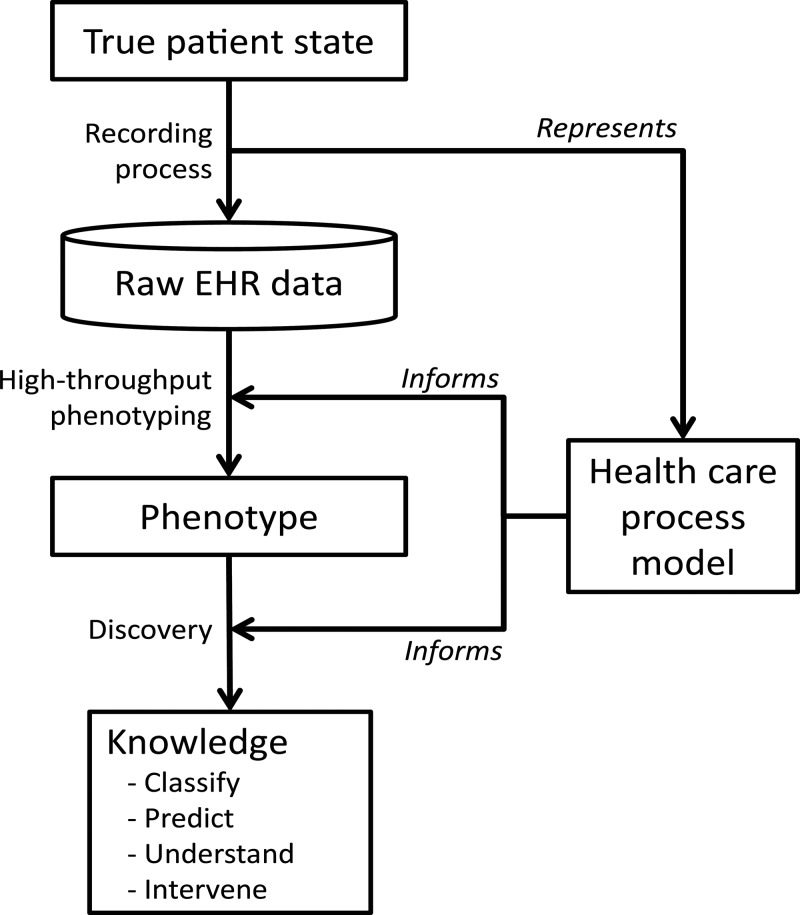
One component is a healthcare process model that represents how processes occur and how data are recorded (figure 2). Some aspects of the healthcare process model are being defined, for example, through research related to SNOMED, Health Level 7, and the Clinical Element Model,25–27 but they do not directly address the recording process, so additional modeling efforts are likely to be needed. Such efforts might group variables into types and might include temporal patterns of data capture. Given the complexity of healthcare and the number of human and organizational influences, a top-down model is unlikely to be sufficient. Therefore, a second component is also needed: we must mine the EHR data to learn the idiosyncrasies of the healthcare process and their effects on the recording process. That is, we believe that the interactions and dependencies are too complex to model and predict at a detailed level (eg, intention vs definition, team interactions), so empirical measurement of the relationships among data elements will be essential.
Figure 2.
Phenotyping and discovery. The raw electronic health record (EHR) data are an indirect reflection of the true patient state due to the recording process. Attempts to create phenotypes and discover knowledge must account for the recording. The healthcare process model represents the salient features of the recording process and informs the phenotyping and discovery.
A rigorous model populated with characteristics learned from the data could improve phenotyping in several ways. For example, it may be possible to map raw data, such as a time series of diagnosis codes, to a probability of disease.28 If biases can be quantified—for example, the degree to which a given variable tends to over or underestimate a feature—then one could avoid sources that are most biased, or one could combine sources that have bias in opposite directions. The process of generating a phenotype query would then become less heuristic and more data driven.
A full review of the data mining methods appropriate to phenotyping is beyond the scope of a perspective, but the following are particularly relevant. First is simply characterizing the raw data with frequencies, co-occurrences, and—when possible—predictive value with respect to desired phenotypes (eg, how accurate are International Classification of Disease, version 9 codes). Dimension reduction using algorithms like principal component analysis (empirical orthogonal functions)29 30 addresses the many disparate variables that comprise an EHR. Instead of top-down defined phenotypes, it may be appropriate to define latent variables that have high predictive value using techniques such as latent Dirichlet allocation31 32 or other methods.33 The ability to find similar cases is often useful to define cohorts for machine learning, and has been done with symbolic and computational techniques.34 35 Clinical databases can be stratified into more regular subsets, producing more stable results.36 Natural language processing37 is of course essential to phenotype EHR data due to the narrative content.
While time has long been a research topic in informatics,38 39 further work may be needed. This includes temporal modeling and abstraction40 41 (including temporal treatment of narrative data),42 as well as purely numeric approaches, including non-linear time series analysis drawn from the physics literature. The latter includes aggregation of short time series,43 particularly as applied to health record data and modified to accommodate non-equally spaced time series.44 Researchers have noted that missingness itself is a useful feature in producing phenotypes.45
We can also improve the use of EHR data at the second step, the discovery stage, which may include classification (eg, clinical trial eligibility), prediction (eg, readmission rate), understanding (eg, physiology), and intervention. Sensitivity to EHR bias may depend on the goal: prediction may be accurate even if important confounders are not measured in the EHR, but unmeasured confounders could mislead our understanding of physiology.
Even if EHR bias or noise cannot be measured, it may be possible to factor it out. In one study, patient data were normalized to reduce interpatient variance, improving the estimation of the correlation among variables.46 In another, a derived property (mutual information) was used in place of traditional parameters such as glucose because they had too much variation between patients.47 In other cases, when the biases and noise cannot be eliminated, perhaps they can be understood. For example, it may be useful to characterize discovered associations as being due to the healthcare process (eg, physician's intention) versus due to physiology.46 Although it is challenging, a number of techniques may be used to infer causation, including dynamic Bayesian networks,48 Granger causality,49 and logic-based paradigms.50 Recent work demonstrated the control of the confounding effects of covariates51 with a demonstration of drugs' effects on electrocardiogram QT intervals.
Discussion
We believe that the full challenge of phenotyping is not broadly recognized. For example, one review of mining EHRs52 discusses interoperability and privacy as key challenges, but otherwise focuses on the promise of the data rather than the data challenges, which are arguably more difficult to solve.
We believe that the phenotyping process needs to become more data driven and that we need to learn more about the recording process. We have sometimes used the phrase, ‘the physics of the medical record’, to point out the likely direction forward. It will require study of the EHR as if it were a natural object worthy of study in itself, and it may be helpful to employ the general paradigm of physics, which involves modeling and aggregation. It will be helpful to pull in expertise and algorithms from many fields, including non-linear time series analysis from physics,53 new directions in causality from philosophy,50 psychology, economics, of course our usual collaborators in computer science and statistics, and even new models of research that engage the public.
Our hope is that by exploiting our ample data, we can surpass human performance and produce even more reliable phenotypes and accurate associations. To draw an analogy, a CT scanner uses data that are feasible to collect—namely external x-ray images—and deconvolves them to produce an image that reflects clinically relevant but hidden internal anatomical features. Similarly, we need to use data that are feasible to collect from EHR and deconvolve them to produce clinically relevant phenotypes that are only implicit in the raw data. Furthermore, the advanced use of EHR data, which are becoming both deep in content and broad in coverage of the nation's population, may open new ways to look at clinical research, studying detailed physiology (including fine laboratory measurements) over large populations, in what might be called population physiology.47 To draw one more analogy from physics, we can move from studying weather—individual phenotypes—to studying climate—properties of phenotypes over populations and time.
Systematic changes in the adoption and use of EHR, such as those promoted by the HITECH incentive program (meaningful use),1 will probably have large effects on how EHR data get used in research. For example, structured data entry for meaningful use, quality measurement, or value-based purchasing should improve the volume and quality of data available to research. Variables that have been notoriously difficult to collect, such as smoking history, may become more broadly available. On the other hand, forced data entry can introduce biases that are difficult to detect or correct. Health information exchange, which pulls together not only multiple EHR but also new data sources such as pharmacy fill data, should reduce data fragmentation, although researchers will need to contend with heterogeneous data definitions and data entry cultures. Therefore, even in a new era of the increased use of EHR, a deep understanding of EHR data will be critical.
Furthermore, this must not be a one-way street; improved understanding of the EHR must be fed back to improve the EHR. For example, better understanding of missing data, inaccuracies, and biases could lead to improved user interfaces, data definitions, and even workflows. The long-term vision of an EHR platform that supports clinical care, research, and public health will only be achieved with better understanding and true innovation.
Footnotes
Contributors: The authors are responsible for: the conception and design, acquisition of data, and analysis and interpretation of data; drafting the article and revising it; and final approval of the version to be published.
Funding: This work was funded by a grant from the National Library of Medicine, ‘Discovering and applying knowledge in clinical databases’ (R01 LM006910).
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Correction notice: This article has been corrected since it was published Online First. It is now unlocked.
Open Access: This is an Open Access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 3.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/3.0/
References
- 1.Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. N Engl J Med 2010;363:501–4 [DOI] [PubMed] [Google Scholar]
- 2.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc 2013;20:144–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heitjan DF, Basu S. Distinguishing “missing at random” and “missing completely at random”. Am Statistician 1996;50:207–13 [Google Scholar]
- 4.Hogan WR, Wagner MM. Accuracy of data in computer-based patient records. J Am Med Inform Assoc 1997;4:342–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sagreiya H, Altman RB. The utility of general purpose versus specialty clinical databases for research: warfarin dose estimation from extracted clinical variables. J Biomed Inform 2010;43:747–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao C, Parker CG, Oniki TA, et al. An OWL meta-ontology for representing the clinical element model. AMIA Annu Symp Proc 2011;1372–81 [PMC free article] [PubMed] [Google Scholar]
- 7.Pryor TA, Hripcsak G. Sharing MLM's: an experiment between Columbia-Presbyterian and LDS Hospital. Proc Annu Symp Comput Appl Med Care 1993:399–403 [PMC free article] [PubMed] [Google Scholar]
- 8.Hripcsak G, Kuperman GJ, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inf Med 1998;37:1–7 [PubMed] [Google Scholar]
- 9.Friedman C, Hripcsak G. Natural language processing and its future in medicine. Acad Med 1999;74:890–5 [DOI] [PubMed] [Google Scholar]
- 10.Hripcsak G, Zhou L, Parsons S, et al. Modeling electronic discharge summaries as a simple temporal constraint satisfaction problem. J Am Med Inform Assoc 2005;12:55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hripcsak G, Knirsch C, Zhou L, et al. Bias associated with mining electronic health records. J Biomed Discov Collab 2011;6:48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fine MJ, Auble TE, Yealy DM, et al. A prediction rule to identify low-risk patients with community-acquired pneumonia. N Engl J Med 1997;336:243–50 [DOI] [PubMed] [Google Scholar]
- 13.Boustani MA, Munger S, Gulati R, et al. Selecting a change and evaluating its impact on the performance of a complex adaptive health care delivery system. Clin Interv Aging 2010;5:141–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurreeman F, Liao K, Chibnik L, et al. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet 2011;88:57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brownstein JS, Murphy SN, Goldfine AB, et al. Rapid identification of myocardial infarction risk associated with diabetes medications using electronic medical records. Diabetes Care 2010;33:526–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation 2010;122:2016–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen DP, Morgan AA, Butte AJ. Validating pathophysiological models of aging using clinical electronic medical records. J Biomed Inform 2010;43:358–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kullo IJ, Fan J, Pathak J, et al. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc 2010;17:568–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hripcsak G, Austin JHM, Alderson PO, et al. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology 2002;224:157–63 [DOI] [PubMed] [Google Scholar]
- 20.Wilcox AB, Hripcsak G. The role of domain knowledge in automating medical text report classification. J Am Med Inform Assoc 2003;10:330–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarty CA, Chisholm RL, Chute CG, et al. ; eMERGE Team The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Annu Symp Proc 2011;2011:274–83 [PMC free article] [PubMed] [Google Scholar]
- 23.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med 2011; 3:79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warner HR. Knowledge sectors for logical processing of patient data in the HELP system. Proceedings of the International Conference on Interactive Techniques in Computer-Aided Design; Bologna, Italy New York: IEEE, 1978:401–4 [Google Scholar]
- 25.Scott P, Worden R. Semantic mapping to simplify deployment of HL7 v3 Clinical Document Architecture. J Biomed Inform 2012;45:697–702. [DOI] [PubMed] [Google Scholar]
- 26.Heymans S, McKennirey M, Phillips J. Semantic validation of the use of SNOMED CT in HL7 clinical documents. J Biomed Semantics 2011;2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao C, Parker CG, Oniki TA, et al. An OWL meta-ontology for representing the clinical element model. AMIA Annu Symp Proc 2011;2011:1372–81 [PMC free article] [PubMed] [Google Scholar]
- 28.Perotte A, Hripcsak G. Using density estimates to aggregate patients and summarize disease evolution (poster) In: AMIA Summit on Translational Bioinformatics. San Francisco, CA: AMIA, 2011;138 [Google Scholar]
- 29.Pearson K. On lines and planes of closest fit to systems of points in space. Phil Mag 1901;2:559–72 [Google Scholar]
- 30.Weare BC, Navato AR, Newell RE. Empirical orthogonal analysis of Pacific sea surface temperatures. J Phys Oceanography 1976;6:671–9 [Google Scholar]
- 31.Blei DM, Ng AY, Jordan MI. Latent dirichlet allocation. J Mach Learn Res 2003;3:993–1022 [Google Scholar]
- 32.Perotte A, Bartlett N, Elhadad N, et al. Hierarchically supervised Latent Dirichlet Allocation. Twenty-Fifth Annual Conference onNeural Information Processing Systems; 12–15 December, 2011, Granada, Spain, 2011:2609–17 [Google Scholar]
- 33.Tatonetti NP, Denny JC, Murphy SN, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther 2011;90:133–42.10.1038/clpt.2011.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melton GB, Parsons S, Morrison FP, et al. Inter-patient distance metrics using SNOMED CT defining relationships. J Biomed Inform 2006;39:697–705 [DOI] [PubMed] [Google Scholar]
- 35.Hripcsak G, Knirsch C, Zhou L, et al. Using discordance to improve classification in narrative clinical databases: an application to community-acquired pneumonia. Comput Biol Med 2007;37:296–304 [DOI] [PubMed] [Google Scholar]
- 36.Altiparmak F, Ferhatosmanoglu H, Erdal S, et al. Information mining over heterogeneous and high-dimensional time-series data in clinical trials databases. IEEE Trans Inf Technol Biomed 2006;10:254–63 [DOI] [PubMed] [Google Scholar]
- 37.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc 2011;18:544–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shahar Y, Tu SW, Musen MA. Temporal-abstraction mechanisms in management of clinical protocols. Proc Annu Symp Comput Appl Med Care 1991;629–33 [PMC free article] [PubMed] [Google Scholar]
- 39.Kahn MG, Fagan LM, Tu S. Extensions to the time-oriented database model to support temporal reasoning in medical expert systems. Methods Inf Med 1991;30:4–14 [PubMed] [Google Scholar]
- 40.Sacchi L, Larizza C, Combi C, et al. Data mining with temporal sbstractions: learning rules from time series. Data Mining and Knowledge Discov 2007;15:217–47 [Google Scholar]
- 41.Moskovitch R, Shahar Y. Medical temporal-knowledge discovery via temporal abstraction. AMIA Annu Symp Proc 2009:452–6 [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou L, Hripcsak G. Temporal reasoning with medical data – a review with emphasis on medical natural language processing. J Biomed Inform 2007;40:183–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komalapriya C, Thiel M, Ramano MC, et al. Reconstruction of a system's dynamics from short trajectories. Phys Rev E 2008;78:066217. [DOI] [PubMed] [Google Scholar]
- 44.Albers DJ, Hripcsak G. A statistical dynamics approach to the study of human health data: resolving population scale diurnal variation in laboratory data. Physics Letters A 2010;374:1159–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin JH, Haug PJ. Exploiting missing clinical data in Bayesian network modeling for predicting medical problems. J Biomed Inform 2008;41:1–14 [DOI] [PubMed] [Google Scholar]
- 46.Hripcsak G, Albers DJ, Perotte A. Exploiting time in electronic health record correlations. J Am Med Inform Assoc 2011;18:i109–i115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albers DJ, Schmidt M, Hripcsak G. Population physiology: conjoining EHR dynamics with physiological modeling (abstract). In: AMIA Summit on Translational Bioinformatics. San Francisco, CA: AMIA, 2011:1 [Google Scholar]
- 48.van Gerven M, Taal B, Lucas P. Dynamic Bayesian networks as prognostic models for clinical patient management. J Biomed Inform 2008;41:515–29 [DOI] [PubMed] [Google Scholar]
- 49.Granger CW. Investigating causal relations by econometric models and cross-spectral methods. Econometrica 1969;37:424–38 [Google Scholar]
- 50.Kleinberg S, Mishra B. The temporal logic of causal structures. In: Proceedings of the 25th Conference on Uncertainty in Artificial Intelligence (UAI). 18-21 June 2009; Montreal, QC, Canada. Corvallis, Oregon: AUAI Press, 2009:303–12 [Google Scholar]
- 51.Tatonetti NP, Ye PP, Daneshjou R, et al. Data-driven prediction of drug effects and interactions. Sci Transl Med 2012;4:125ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012.10.1038/nrg3208 [DOI] [PubMed] [Google Scholar]
- 53.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012. Published Online First 2 May 2012. doi: 10.1038/nrg3208 [DOI] [PubMed] [Google Scholar]