Abstract
Agricultural productivity to meet growing demands of human population is a matter of great concern for all countries. Use of green compounds to achieve the sustainable agriculture is the present necessity. This review highlights the enormous use of harsh surfactants in agricultural soil and agrochemical industries. Biosurfactants which are reported to be produced by bacteria, yeasts, and fungi can serve as green surfactants. Biosurfactants are considered to be less toxic and eco-friendly and thus several types of biosurfactants have the potential to be commercially produced for extensive applications in pharmaceutical, cosmetics, and food industries. The biosurfactants synthesized by environmental isolates also has promising role in the agricultural industry. Many rhizosphere and plant associated microbes produce biosurfactant; these biomolecules play vital role in motility, signaling, and biofilm formation, indicating that biosurfactant governs plant–microbe interaction. In agriculture, biosurfactants can be used for plant pathogen elimination and for increasing the bioavailability of nutrient for beneficial plant associated microbes. Biosurfactants can widely be applied for improving the agricultural soil quality by soil remediation. These biomolecules can replace the harsh surfactant presently being used in million dollar pesticide industries. Thus, exploring biosurfactants from environmental isolates for investigating their potential role in plant growth promotion and other related agricultural applications warrants details research. Conventional methods are followed for screening the microbial population for production of biosurfactant. However, molecular methods are fewer in reaching biosurfactants from diverse microbial population and there is need to explore novel biosurfactant from uncultured microbes in soil biosphere by using advanced methodologies like functional metagenomics.
Keywords: Biosurfactants, Sustainable agriculture, Pesticide, Plant pathogen, Remediation, Metagenomics
Introduction
Increase in agricultural productivity to meet ever growing food demands of human population is a matter of great concern for all countries. United Nation’s Economic and Social Commission for Asia and the Pacific (ESCAP) conducted a theme study in April 2009 entitled “Sustainable Agriculture and Food Security in Asia and the Pacific” in which the importance of revitalization of native soil systems for improved crop yield was emphasized. Such revitalization processes can be carried out in an eco-friendly manner using various biological amendments. Many microorganisms found in rhizosphere (the soil under the influence of plant roots) share a mutualistic relationship with plants conferring marked beneficial effects on plants. Several mechanisms are reported by which rhizobacteria help in plant growth promotion (Gamalero and Glick 2011; Zahir et al. 2004; Glick et al. 2007). Hence, rhizosphere biology is considered to be the most intensive area of research in agriculture.
Surfactants have several functional properties, well known, and exploited in many commercial sectors. There are numerous areas of agriculture which also requires surfactants. A review by Deleu and Paquot (2004) enlists the major area where surfactants are employed. It is reported in year 2004 that approximately ± 0.2 million tons of surfactants are used in crop protection and agrochemical formulations. Several reports have highlighted the advantages of green surfactants (biosurfactant derived from microbes) over the synthetic surfactant. Since there are fewer reports stating the application of biosurfactants in agriculture, the review emphasizes on the significance of biosurfactants and biosurfactant producing microbes from soil especially rhizosphere in agriculture sector.
Biosurfactants are low molecular weight surface-active compounds widely produced by bacteria, yeast and fungi. These amphiphilic biomolecules reduce the surface tension at the air/water interfaces and the interfacial tension at oil/water interfaces (Satpute et al. 2010a; Banat et al. 2010). There are different types of biosurfactant stated based on their physico-chemical properties as glycolipids, lipopeptides, neutral lipids, phospholipids, fatty acids, and polymeric (Pacwa-Plociniczak et al. 2011; Cameotra et al. 2010; Amaral et al. 2010). Biosurfactants are derived from renewable resources and are low or nontoxic, biodegradable, demonstrate excellent surface activity, possess high specificity, show effectiveness under extreme conditions, and can be reused through regeneration too as compared to synthetic surfactants (Koglin et al. 2010; Xu et al. 2011a; Lima et al. 2011b), hence commercially exploited. Biosurfactants have the potential to be applied in pharmaceutical, cosmetics, petroleum, and food industries (Rodrigues et al. 2006; Nitschke and Costa 2007; Lourith and Kanlayavattanakul 2009; Banat et al. 2010; Das et al. 2010). A potent lipopeptide biosurfactant, “surfactin” is reviewed as a versatile bioactive molecule having ability to inhibit fibrin clot formation and is also applied for enhanced oil recovery. In addition, it has demonstrated antifungal, antiviral, antitumor, insecticidal, and antimycoplasma activities. Surfactin can also be used as bioremediation agent for treatment of soil and water (Mulligan 2005). These properties of surfactin reflect its potential commercial applications (Sen 2010). Recently, biosurfactant and biosurfactant producing microbes are used for production of nanoparticles. The area of the biosurfactant mediated process of nanoparticle synthesis is emerging as part of green chemistry (Kiran et al. 2011). Biosurfactants are used for increasing the stability of microbubble technology which further has wide scope in molecular imaging, disease diagnosis, in delivery systems for drugs and genes, cost-effective water purification, and sewage treatment (Zhang et al. 2011b; Xu et al. 2011b). In conclusion, these diverse biosurfactants are useful tool for biotechnology and advantageous for the mankind. Several patents related to biosurfactant producing microbes mainly for Pseudomonas spp., Acinetobacter spp., Bacillus spp., and Candida spp., types of biosurfactant, the process for production, and industrial application of biosurfactant have been issued (Shete et al. 2006), indicating the potential of these microbe derived biomolecules.
Agriculture related applications of biosurfactants
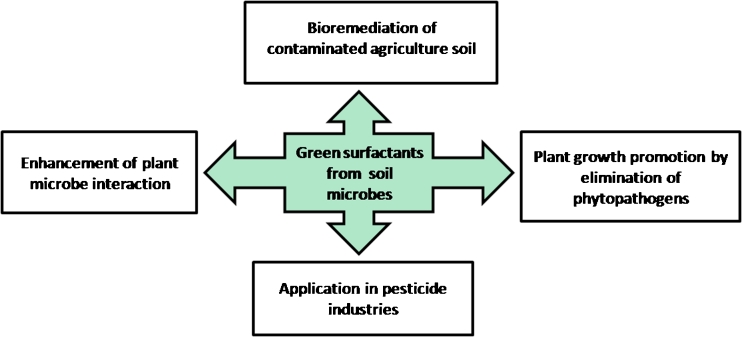
Taking into account the dual hydrophobic/hydrophilic nature of biosurfactant from microbial sources (Singh et al. 2007), these green surfactants have more advantages over the chemically synthesized surfactants. These biosurfactants can be widely exploited in areas related to agriculture for enhancement of biodegradation of pollutants to improve the quality of agriculture soil, for indirect plant growth promotion as these biosurfactants have antimicrobial activity and to increase the plant microbe interaction beneficial for plant. These biosurfactants can replace the harsh surfactant presently used in pesticide industries as these natural surfactants are found to be utilized as carbon source by soil inhabiting microbes (Scott and Jones 2000; Takenaka et al. 2007; Lima et al. 2011a) and this accounts for the biological removal of biosurfactants from the agricultural soil. The following part of the review highlights on the reports on role of biosurfactants and biosurfactant producing microbes in the most important commercial sector viz agriculture (Fig. 1).
Fig. 1.
Multifunctional prospective of biosurfactants in agriculture
Improvement of soil quality
The productivity of agriculture land is affected by presence of organic and inorganic pollutants that impart abiotic stress on the cultivated crop plant. To increase the quality of such soil contaminated by hydrocarbon and heavy metals, process of bioremediation is required. Microorganisms producing biosurfactant and and/or biosurfactants can be effectively used for removal of hydrocarbons as well as heavy metals (Sun et al. 2006). As biosurfactants are known to enhance bioavailability and carry out biodegradation of hydrophobic compounds, different technologies such as soil washing technology and clean up combined technology employ biosurfactant for effective removal of hydrocarbon and metal, respectively (Pacwa-Plociniczak et al. 2011; Liu et al. 2010; Partovinia et al. 2010; Gottfried et al. 2010; Coppotelli et al. 2010; Kang et al. 2010; Pei et al. 2009; Camilios Neto et al. 2009; Zhao and Wong 2009; Robles-González 2008; Sheng et al. 2008; Santos et al. 2008; Aşçi et al. 2008; Juwarkar et al. 2007; Hickey et al. 2007; Shin et al. 2006; Mulligan 2005; Kildisas et al. 2003; Maier et al. 2001). A very important phenomenon of desorption of hydrophobic pollutants tightly bound to soil particles is accelerated by biosurfactants. This is very crucial for bioremediation process. Biosurfactants can also enhance the degradation of certain chemical insecticides which are accumulated in the agricultural soil (Zhang et al. 2011a; Singh et al. 2009; Sharma et al. 2009; Wattanaphon et al. 2008; White et al. 2006; Neilson et al. 2003). There are several reports which suggest role of biosurfactants in improving the health of agriculture soil by the process of soil remediation. There are reports on pesticide biodegradation supported by surfactin (Mata-Sandoval et al. 2001) and degradation of chlorinated hydrocarbon supported by glycolipids (Awashti et al. 1999). Biosurfactant from Lactobacillus pentosus has demonstrated reduction by 58.6 % to 62.8 % of octane hydrocarbon from soil (Moldes et al. 2011) thus exhibiting the biodegradation accelerator property of biosurfactant. It has been observed that a biosurfactant producing species of Burkholderia isolated from oil-contaminated soil may be a potential candidate for bioremediation of variety of pesticide contamination (Wattanaphon et al. 2008). Many researchers have observed that the efficiency of biosurfactant in removal of organic insoluble pollutants from soil is more as compared to synthetic surfactants (Cameotra and Bollag 2003; Urum et al. 2003; Vipulanandan and Ren 2000; Schippers et al. 2000; Dean et al. 2001; Straube et al. 2003). Literature also states the requirement of iron for increased production of biosurfactant by Pseudomonas sp. and further enhancement of poly aromatic hydrocarbons (PAH’s) bioavailability (Santos et al. 2008). Rhamnolipids are found to be useful in removal of poly aromatic hydrocarbons (Poggi-Varaldo and Rinderknecht-Seijas 2003) and pentachlorophenol (Mulligan and Eftekhari 2003) from soil. Thus biosurfactant can be applied in agriculture soil to enhance soil quality. However, high cost for production of biosurfactant yet restricts the application of these green surfactants for bioremediation of soil contaminated by crude oil and/or petroleum (Moldes et al. 2011). Use of agro-industrial waste for production of green surfactants which can further be used for biodegradation of hydrocarbons from soil (Benincasa 2007) needs imperative examination.
Heavy metals are present as pollutant in agriculture soil. The heavy metal pollution originates from excessive use of metal salt-based fungicides, sewage, and sludge amendments applied on the agricultural fields. These heavy metals serve as essential micronutrients and are required for various important physiological processes in plant metabolism. However, it can be detrimental to plant growth at higher concentrations causing damage to plant in form of root tissue necrosis and purpling of foliage. A recent review summarizes the role of biosurfactants and biosurfactant producing microorganisms in bioremediation of heavy metals and hydrocarbon pollutants. There are several reports on potential properties of biosurfactants produced by Pseudomonas sp, Bacillus sp., and Acinetobacter sp. for removal of heavy metals from contaminated soil and even acceleration of biodegradation of pesticides (Pacwa-Plociniczak et al. 2011; Kassab and Roane 2006). Rhamnolipid biosurfactant produced by species of pseudomonads are reported to remove toxic metals from soil (Herman et al. 1995). Further, biosurfactant such as rhamnolipid and surfactin are known to remove heavy metals such as Ni, Cd, Mg, Mn, Ca, Ba, Li, Cu, and Zn (ions) from soil with a new method of foaming-surfactant technology (Neilson et al. 2003; Mulligan and Wang 2004; Mulligan et al. 2001).
Augmentation of soil health requires removal of less water soluble pollutants by use of surfactants. The synthetic surfactants are also used for removal of nonpolar organic contaminants (NOC) from soil. However, these surfactants are required at higher concentration and also affect the microbial biodegradation (Colores et al. 2000). Thus, rather than use of harmful synthetic surfactants overproducer of biosurfactants can be the most useful for bioremediation (Rosenberg and Ron 1998). Uses of certain supplements such as cyclodextrin also appear in literature which can enhance the degradation of hydrocarbons by soil microbial population (Bardi et al. 2000).
Plant pathogen elimination
Several biosurfactants from microbes have antimicrobial activity against plant pathogens and therefore they are considered to a promising biocontrol molecule for achieving sustainable agriculture. Biosurfactants produced by rhizobacteria are known to have antagonist properties (Nihorimbere et al. 2011). An agricultural application of chemical surfactants and biosurfactants also facilitates biocontrol mechanism of plant growth promoting microbes such as parasitism, antibiosis, competition, induced systemic resistance, and hypovirulence (Singh et al. 2007). In bulk, the surfactants are used in agriculture to enhance the antagonistic activities of microbes and microbial products (Jazzar and Hammad 2003; Kim et al. 2004). Several in vitro and in situ studies have demonstrated the role of surfactants in improving the insecticidal activities of other systems (Jazzar and Hammad 2003; Gronwald et al. 2002; Krishnayya and Grewal 2002). In addition, these surfactants are used in combination with fungus (Myrothecium verrucaria) to eradicate weed species which affect the land productivity and also the spread of such weed species have adverse effect on biodiversity (Boyette et al. 2002) and surfactants are also reported to inhibit the aflatoxin production by Aspergillus sp. which infects crops such as cottonseed, peanuts and corn during storage as well as at agricultural field (Rodriguez and Mahoney 1994). Thus the surfactants (synthetic and biological) play diverse role in plant pathogen elimination directly or indirectly and at different processes related to agriculture.
The following section states few examples of biosurfactants as an essential component for inhibition of phytopathogens. Biosurfactant producing rhizospheric isolates of Pseudomonas and Bacillus have exhibited biocontrol of soft rot causing Pectobacterium and Dickeya spp. (Krzyzanowska et al. 2012). Rhamnolipids have demonstrated inhibition of zoospore forming plant pathogens that have acquired resistance to commercial chemical pesticides (Sha et al. 2011; Kim et al. 2011; Hultberg et al. 2008a) and another investigation has shown that rhamnolipid can stimulate plant immunity which is considered as an alternative strategy to reduce the infection by plant pathogens (Vatsa et al. 2010). Recent investigation has also established rhamnolipid as an insecticidal compound. Kim and co-workers (2011) have isolated biosurfactant from a strain of Pseudomonas, which has demonstrated insecticidal activity against green peach aphid (Myzus persicae). Plant growth-promoting Pseudomonas putida produces biosurfactants that can cause lysis of zoospores of the oomycete pathogen Phytophthora capsici; causative agent of damping-off of cucumber (Kruijt et al. 2009). The lipopeptide biosurfactant produced by strains of Bacillus exhibits growth inhibition of phytopathogenic fungi like Fusarium spp., Aspergillus spp., and Biopolaris sorokiniana. Such biosurfactant can be used as biocontrol agent (Velho et al. 2011). Brevibacillus brevis strain HOB1 produces surfactin isoform and this lipopeptide biosurfactant has demonstrated strong antibacterial and antifungal property which can be exploited for control of phytopathogens (Haddad 2008). Antifungal properties of biosurfactant produced by strains of Pseudomonas fluorescens is well documented in literature (Nielsen and Sørensen 2003). Hultberg et al. (2008b)) have reported that fluorescent pseudomonad’s with the biosurfactant producing ability can inhibit the growth of fungal pathogens such as Pythium ultimum (causative agent of damping off and root rot of plants), Fusarium oxysporum (causes wilting in crop plants), and Phytophthora cryptogea (causes rotting of fruits and flowers). Pseudomonas sp. are reported as biocontrol agents against Verticillium microsclerotia; a causative agent of Verticillium wilt mainly in potatoes. The biosurfactant produced by this Pseudomonas sp. is considered to play major role in inhibition of in vitro viability of Verticillium sp. (Debode et al. 2007). Strains of Pseudomonas sp. terminate the growth of pathogenic fungi Rhizoctonia solani (causes several plant diseases) and Phythium ultimum (causes damping off and root rot of plants) by production of dual functioning compounds tensin, viscosin and viscosinamid. The dual function includes biosurfactant and antifungal activity (Andersen et al. 2003). Colletotrichum gloeosporioides, causative agent for anthracnose on papaya leaves is reported to be controlled by biosurfactant producing Bacillus subtilis isolated from soil (Kim et al. 2010). A possible plant pathogen Pseudomonas aeruginosa was reported to be inhibited by biosurfactant produced by Staphylococcus sp., isolated from crude oil-contaminated soil (Eddouaouda et al. 2012). The above examples prove that the green surfactants are well documented in literature for plant growth promotion by their detrimental effect on pathogens. Hence, these biosurfactants and/or biosurfactant producing microbes are potential substitutes for the harsh chemical pesticides and insecticides being currently used in agriculture. Apart from these anti-phytopathogenic properties, addition of biosurfactant is also known to accelerate compositing process by providing favorable conditions for microbial growth (Zhang et al. 2011a, b) and thus offers an additional advantage of use of these green surfactants., The biosurfactants which have antagonist properties against phytopathogens may also affect the other flora of the system. Thus, to construct a potent green surfactant with specificity against the phytopathogens, the chemical composition of the biosurfactant may be varied by altering the production strategies.
Asset for beneficial plant microbe interaction
To provide beneficial effect to the plants by rhizobacteria, it is very important for these microbes to interact with the plant surfaces such as roots (Nihorimbere et al. 2011). Microbial factors such as motility, ability to form biofilm on root surface and release of quorum sensing molecules are required to establish association with the plant. It is reviewed that quorum sensing molecules such as acyl homoserine lactone (AHL) are required for synthesis of antifungal compounds by the rhizobacteria. Studies also indicate that the concentration of these molecules is high in rhizosphere as compared to that to bulk soil (soil away from plant roots) suggesting the role of AHL and AHL-like molecules in rhizosphere competence (ability of beneficial microorganism to colonize the root surface). These AHL are also reported to contribute in regulation of exopolysaccharide essential for biofilm formation (Newton and Fray 2004; Loh et al. 2002). Dusane et al. (2010) have recently reported that the biosurfactant (rhamnolipid) produced by Pseudomonas spp. regulates the process of quorum sensing (cell to cell communication). It is also reported that biosurfactants affect the motility of microorganisms, participate in signaling and differentiation as well as in biofilm formation (Ron and Rosenberg 2011; Berti et al. 2007; Van Hamme et al. 2006; Kearns and Losick 2003). Hence, these green surfactants are important parameters for microbes to achieve a beneficial association with the plant roots and improve the growth of the plant. Further these biosurfactants produced by rhizobacteria increase the bioavailability of hydrophobic molecules which may serve as nutrients. Biosurfactants produced by soil microbes provide wettability to soil and support proper distribution of chemical fertilizers in soil thus assisting plant growth promotion. Reviewing the functions of biosurfactant indicates the essential role of these green compounds for sustainable agriculture.
Potential of biosurfactants in pesticide industries
Surfactants are must as adjuvant with fungicides, insecticides, and herbicides. The synthetic surfactant presently used in pesticides industries act as emulsifying, dispersing, spreading and wetting agent and enhance the efficiency of pesticides. In addition, these surfactants are used in insecticides in modern agriculture as these have defensive properties (Rostas and Blassmann 2009). Different types of surfactants such as anionic, cationic, amphoteric, and nonionic are presently being used at several pesticide manufacturing industries (Mulqueen 2003). Thus surfactants are widely used in formulation of pesticides. However, it is important to note that the surfactant present in pesticides formulations in access becomes accumulated in soil and affects the texture, color and growth of the plant. These harmful pesticides also become leached from soil to the ground water (Blackwell 2000). Pesticide residues are known to persist for years in soil and spreads in air and water too. These even remain on surface of fruit and vegetables (Street 1969). In addition, the synthetic surfactants are considered as potent organic pollutants in soil (Petrovic and Barcelo 2004). Considering the adverse effect of pesticides and surfactants associated with the pesticides, there is need to use the environmentally safe biosurfactants to replace these harmful surfactants in the million dollar pesticide industries thus preventing the pollutions (Hopkinson et al. 1997). Research related to exploration of soil bacteria which can utilize the chemical surfactant in agriculture soil as carbon source can be another alternative to this environmental problem. There is a report on bacteria belonging to Pseudomonas sp. and Burkholderia sp. from paddy field to degrade surfactants (Nishio et al. 2002). Agriculture important products like pesticides formed with the assistance of biosurfactant can be widely used on agricultural fields. The need for agrochemical industries is to develop effective formulation technology and to achieve this goal; many companies can employ mixture of biosurfactants in different combinations with the polymers to make excellent formulations for agricultural applications.
Microbes from soil environment as producers of biosurfactants
Several researchers indicate that variety of environmental niches such as soil, water, and leaf surface are explored for biosurfactant producing bacteria (Burch et al. 2011). Many rhizosphere and plant associated microbes are known to produce biosurfactant indicating the potential role of biosurfactant in plant-microbe interaction and further application of biosurfactant in agriculture (Amani et al. 2010; Singh et al. 2009; Kruijt et al. 2009; Nielsen and Sørensen 2003; Bodour et al. 2003; Nielsen et al. 2002; Takeyama et al. 2002). The table 1 summarizes the genera that appear in literature as biosurfactant producers from rhizosphere as well as contaminated soil. The literature affirms the prevalence of biosurfactant producing bacteria in environment and that biosurfactants also have applications in agriculture and related industries as they play vital role in soil remediation, plant pathogen elimination and by increasing the bioavailability of nutrient for beneficial plant associated microbes.
Table 1.
Recent reports on agri-potential biosurfactant producing microbes from contaminated soil and rhizosphere
| Microorganism | Source | Reference/s |
|---|---|---|
| Pseudomonas aeruginosa | Indigenous flora of apple; petroleum-contaminated soil | Abbasi et al. (2012); Kumar et al. (2012) |
| Bacillus cereus Bacillus megaterium B. thuringiensis | Land farming soil | Cerqueira et al. (2012) |
| Stenotrophomonas maltophilia | Land farming soil | Cerqueira et al. (2012) |
| Psuedomonas nitroreducens | Petroleum-contaminated soil | Onwosi and Odibo (2012) |
| Acinetobacter sp. | Petroleum-contaminated soil | Chen et al. (2012) |
| Staphylococcus sp. | Crude oil-contaminated soil | Eddouaouda et al. (2012) |
| Pseudomonas sp. | Crude oil-contaminated soil | Hua and Wang (2012) |
| Pseudomonas aeruginosa Bacillus cereus | Petrochemical waste-contaminated soil | Cerqueira et al. (2011) |
| Bacillus subtilis | Endosulfan sprayed cashew plantation soil containing hydrophobic substances | Sekhon et al. (2011) |
| Serratia marcescens | Hydrocarbon-contaminated soil | Roldán-Carrillo et al. (2011) |
| Enterobacter cloacae Pseudomonas sp. | Heavy crude oil-contaminated soil | Darvishi et al. (2011) |
| Streptomyces rochei | Heavy crude oil-contaminated soil | Chaudhary et al. (2011) |
| Pseudomonas fluorescens | Rhizosphere of fique | Sastoque-Cala et al. (2010) |
| Pseudomonas aeruginosa | Petroleum-contaminated soil | Nie et al. (2010) |
| Rhodococcus fascaians | Antarctic soil | Gesheva et al. (2010) |
| Bacillus subtilis | Soil | Kim et al. (2010) |
| Bacillus mojavensis | Endophytic bacteria from maize | Snook et al. (2009) |
| Sphingomonas paucimobilis | Phenanthrene-contaminated soil microcosm | Coppotelli et al. (2010) |
| Pseudomonas sp. | Agriculture soil | Singh et al. (2009) |
| Pseudomonas sp. | Oil-contaminated soil | Cameotra and Singh (2009) |
| Pseudomonas putida | Rhizosphere of black pepper | Kruijt et al. 2009 |
| Pseudomonas aeruginosa | Oil-contaminated soil | de Lima et al. (2009) |
| Burkholderia cenocepacia | Fuel oil-contaminated soil | Wattanaphon et al. (2008) |
| Rhodococcus wratislaviensis | Soil | Tuleva et al. (2008) |
| Nocardia otitidiscaviarium | Contaminated soil | Zeinali et al. (2007) |
| Pseudomonas aeuroginosa | Diesel-contaminated soil | Chen et al. (2007) |
| Pantoea sp. | Ornithogenic soil of Antarctica | Vasileva-Tonkova and Gesheva (2007) |
| Pseudomonas aeruginosa Bacillus subtilis | Petroleum oil-contaminated soil | Das and Mukherjee (2007) |
| Pseudomonas sp. | Rhizosphere of white and red cocoyam plants | Perneel et al. (2007) |
| Pseudomonas chlororaphis | Soil | Gunther et al. (2005) |
| Acinetobacter junii | Long Beach soil | Menezes Bento et al. (2005) |
| Pseudomonas fluorescens | Sugar beet rhizosphere | Nielsen and Sørensen (2003) |
| Flavobacterium sp. | Hydrocarbon/metal-contaminated soil | Bodour et al. (2003) |
| Bacillus sp. | Soil | Takeyama et al. (2002) |
| Pseudomonas fluorescens | Petroleum-contaminated soil | Barathi and Vasudevan (2001) |
Molecular methods for profiling of biosurfactant producing community from agriculture soil
The conventional methods used for screening microbes for biosurfactant production are well complied (Satpute et al. 2010b; Walter et al. 2010). Ecological niches contaminated with hydrocarbon are the most recommended sites for the isolation of biosurfactant producing microbes. Techniques for purification of biosurfactant includes thin layer chromatography, high pressure liquid chromatography and phase separation technology (Baker and Chen 2010; Heyd et al. 2008) followed is the characterization of the biomolecule by infra red, gas chromatography mass spectrometry, nuclear magnetic resonance and fast atom bombardment mass spectrometry (Petrovic and Barcelo 2004; Satpute et al. 2010b). High throughput methods are also developed by automation and miniaturization for screening of biosurfactant producers (Walter et al. 2010). Recently, MALDI-TOF mass spectrometry is reported for detection and separation of biosurfactants (Kurtzman et al. 2010). Along with the traditional methods, molecular techniques are being implemented to detect presence of biosurfactant producing bacteria. Techniques such as PCR, cloning, sequencing, homology analysis, and transposon mutagenesis appear in the literature. PCR based techniques targeting genes involved either in synthesis of biosurfactant (for, e.g., srfA3, sfp, coma, licA3, rhlA, rhlB, rhlC, swrW) or regulation of biosurfactant production (for ,e.g., rhlR, rhlI, dnaK) have been employed (Simpson et al. 2011; Neilson et al. 2010; Hommais et al. 2008; Tanikawa et al. 2006; Dubern et al. 2005; Hsieh et al. 2004) mainly for Bacillus spp., Pseudomonas spp., and Serratia spp. Bioinformatics approach such as “mine” the genome of are used in few studies which has led to identification of a nonribosomal peptide biosynthetic gene cluster that codes for proteins involved in the production of structurally related linear lipopeptides (Berti et al. 2007). The molecular approach is concentrated for very few bacterial strains and there is need to explore novel biosurfactant from uncultured microbes in soil biosphere by using advanced methodologies like functional metagenomics. This approach will also append tremendous knowledge of genes pool related to biosurfactant production; still undiscovered. The data generated from such high throughput studies will accelerate application of biosurfactant in agriculture as well as other fields. The following are the steps that can be employed for molecular characterization of biosurfactant production in bacteria from selected habitat especially hydrocarbon/crude oil/ heavy metal-contaminated agricultural soil:
Total DNA directly can be extracted from soil samples and subsequently analyzed either by characterizing particular sequences targeted and amplified by PCR.
PCR products can be analyzed by cloning or genetic fingerprint. Genetic fingerprint consists in a rapid and simple electrophoretic analysis of the PCR products enabling the analysis of the genetic structure of the community.
Characterization of cloned sequences enables assessment of the genetic diversity of a community and can reveal the phylogenetic affiliation of the community members.
Similarly, the sequencing of bands of fingerprint profiles can lead to identification of particular populations and/or type of biosurfactant dominant in the selected niche.
Functional community can be analyzed by FISH, SIP, DNA microarray technology and can also help to assess the genetic structure of biosurfactant producing communities.
Apart from the above methodologies, real time PCR can also be employed to understand the biosurfactant producing population present in a particular niche in comparison with the total bacterial community profile.
Screening unculturable microbes for biosurfactant production: A metagenomic approach
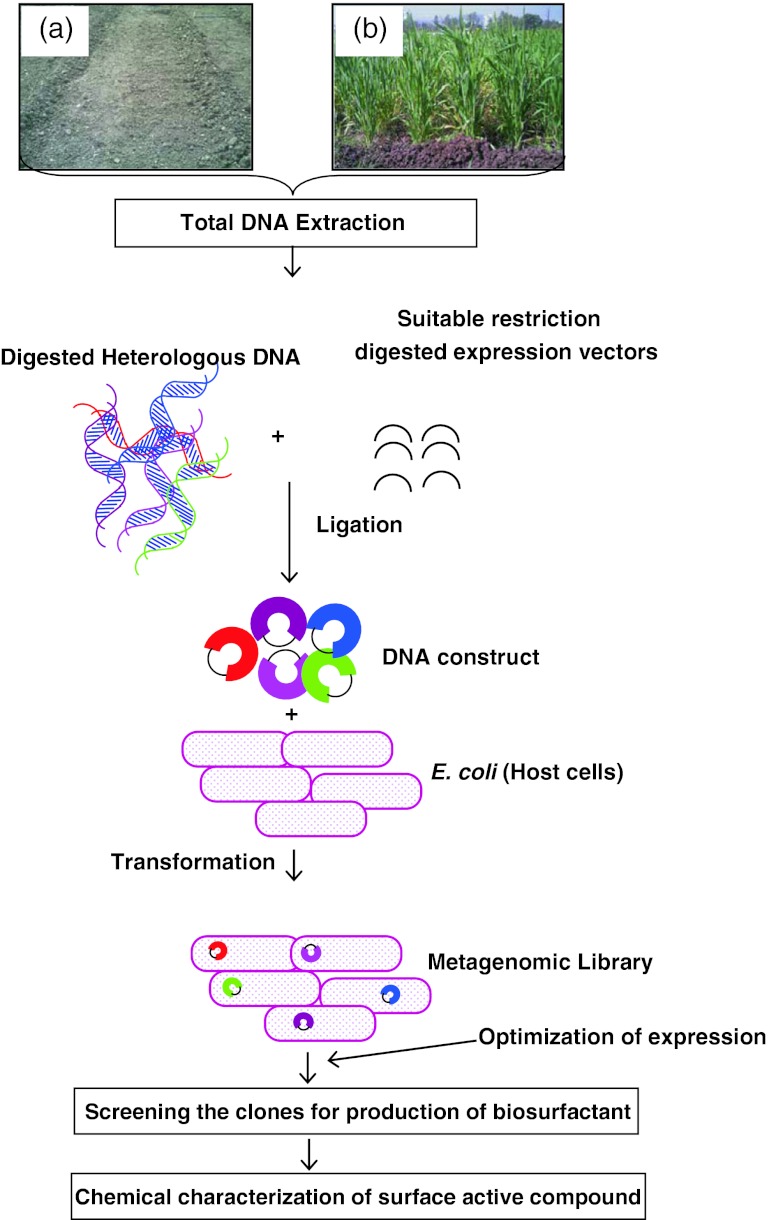
Metagenomics is the culture-independent genomic analysis of microbial communities. The term was derived from the statistical concept of meta-analysis (the process of statistically combining separate analysis) and genomics (the comprehensive analysis of an organism’s genetic material) (Schloss and Handelsman 2003; Rondon et al. 2000). Thus, this technique is a powerful tool for exploring novel compounds from uncultured bacteria associated with natural ecosystems. No PCR is involved in the metagenomics and hence PCR biases can be ruled out. Other advantage is that whole soil DNA is cloned and sequenced, thus metagenomics offers the opportunity to capture operons or genes encoding pathways that may direct the synthesis of complex molecules such as biosurfactants. It is observed that the genes that encode for proteins/enzymes involved in the pathway of biosurfactant synthesis are usually clustered in a region of chromosome. The gene related to biosynthesis of bacterial surfactants lie on gene cluster of approximately 3,000–7,000 bp. Hence it is possible to imply the metagenomic approach to obtain novel biosurfactant from uncultured bacteria associated with contaminated agricultural soil and rhizosphere. Most of the reports on commercially significant biosurfactant are from pathogenic bacterial strains and thus metagenomic approach is must for production of biosurfactants and in supplement there is greater possibility to search for novel biosurfactant by this technique. Fig 2 summaries the steps for functional metagenomic to mine the novel biosurfactants from uncultured bacteria. The location for extraction of total metagenomic DNA should be selected which may include hydrocarbon, pesticides or heavy metal-contaminated agricultural soil and/or rhizosphere. The total DNA extracted should be fragmented by restriction enzymes and inserted in a suitable expression vector. The DNA construct should be transformed in host like E.coli and all the metagenomic clones should be screened for production of biosurfactant by known conventional and/or high through put techniques. The novel surface active compound should be further chemically characterized. The sequence of the clone positive for biosurfactant production should be sequenced and analyzed. The functional metagenomic approach seems a promising technique for mining novel green surfactants which can replace the harsh chemical surfactants widely employed in agriculture as well as other sectors.
Fig. 2.
Schematic representation of construction and screening of metagenomic libraries from contaminated agriculture soil (a) and rhizosphere (b) for novel biosurfactant from uncultured bacteria
Conclusion
Surfactants have several applications in agriculture and agrochemical industries. However, there is rare use of biosurfactants which are more environmental friendly. Exact role of surfactant in facilitating other systems as biocontrol agents is yet not much understood and warrants investigations. Such studies will help in replacing the harsh chemical surfactants with green ones. There is need to work on the production cost of green surfactants to achieve net economic gain from application of biosurfactants in agriculture as well as other sectors. The use of agriculture waste for overproduction of biosurfactants also requires a more serious thought. The chemical compositions of biosurfactants reported potent biocontrol agents can be altered by changing the production scheme. This approach may lead to biosynthesis of highly target specify green surfactant/s. The high prevalence of biosurfactants and biosurfactant producing bacteria in rhizosphere is a positive indication for its potent role in sustainable agriculture. Mainly species of Pseudomonas and Bacillus appear in literature as producers of biosurfactants indicating that only limited genera have been studied till date. A modern approach such as functional metagenomics is the utmost essential which will even lead to discovery of novel green surfactants. Intense work on green surfactants is a priority to prevent the adverse effects of synthetic surfactants largely employed in many commercial sectors including agrochemical industries. Hence, it can be concluded that a cumulative input by researchers from various fields such as molecular biology, biochemistry, microbiology, computational biology, environmental science is indispensable.
References
- Abbasi H, Hamedi MM, Lotfabad TB, Zahiri HS, Sharafi H, Masoomi F, Moosavi-Movahedi AA, Ortiz A, Amanlou M, Noghabi KA. Biosurfactant-producing bacterium, Pseudomonas aeruginosa MA01 isolated from spoiled apples: physicochemical and structural characteristics of isolated biosurfactant. J Biosci Bioeng. 2012;113:211–219. doi: 10.1016/j.jbiosc.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Amani H, Mehrnia MR, Sarrafzadeh MH, Haghighi M, Soudi MR. Scale up and application of biosurfactant from Bacillus subtilis in enhanced oil recovery. Appl Biochem Biotechnol. 2010;162:510–523. doi: 10.1007/s12010-009-8889-0. [DOI] [PubMed] [Google Scholar]
- Amaral PF, Coelho MA, Marrucho IM, Coutinho JA. Biosurfactants from yeasts: characteristics, production and application. Adv Exp Med Biol. 2010;672:236–249. doi: 10.1007/978-1-4419-5979-9_18. [DOI] [PubMed] [Google Scholar]
- Andersen JB, Koch B, Nielsen TH, Sørensen D, Hansen M, Nybroe O, Christophersen C, Sørensen J, Molin S, Givskov M. Surface motility in Pseudomonas sp. DSS73 is required for efficient biological containment of the root-pathogenic microfungi Rhizoctonia solani and Pythium ultimum. Microbiol. 2003;149:37–46. doi: 10.1099/mic.0.25859-0. [DOI] [PubMed] [Google Scholar]
- Aşçi Y, Nurbaş M, Açikel YS. A comparative study for the sorption of Cd(II) by K-feldspar and sepiolite as soil components and the recovery of Cd(II) using rhamnolipid biosurfactant. J Environ Manage. 2008;88:383–392. doi: 10.1016/j.jenvman.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Awashti N, Kumar A, Makkar R, Cameotra S. Enhanced biodegradation of endosulfan, a chlorinated pesticide in presence of biosurfactant. J Environ Sci Heal B. 1999;34:793–803. [Google Scholar]
- Baker SC, Chen CY. Enrichment and purification of lipopeptide biosurfactants. Adv Exp Med Biol. 2010;672:281–288. doi: 10.1007/978-1-4419-5979-9_21. [DOI] [PubMed] [Google Scholar]
- Banat IM, Franzetti A, Gandolfi I, Bestetti G, Martinotti MG, Fracchia L, Smyth TJ, Marchant R. Microbial biosurfactants production, applications and future potential. Appl Microbiol Biotechnol. 2010;87:427–444. doi: 10.1007/s00253-010-2589-0. [DOI] [PubMed] [Google Scholar]
- Barathi S, Vasudevan N. Utilization of petroleum hydrocarbons by Pseudomonas fluorescens isolated from a petroleum-contaminated soil. Environ Int. 2001;26:413–416. doi: 10.1016/s0160-4120(01)00021-6. [DOI] [PubMed] [Google Scholar]
- Bardi L, Mattei A, Steffan S, Marzona M. Hydrocarbon degradation by a soil microbial population with beta-cyclodextrin as surfactant to enchance bioavailability. Enzyme Microb Technol. 2000;27:709–713. doi: 10.1016/s0141-0229(00)00275-1. [DOI] [PubMed] [Google Scholar]
- Benincasa M. Rhamnolipid produced from agroindustrial wastes enhances hydrocarbon biodegradation in contaminated soil. Curr Microbiol. 2007;54:445–449. doi: 10.1007/s00284-006-0610-8. [DOI] [PubMed] [Google Scholar]
- Berti AD, Greve NJ, Christensen QH, Thomas MG. Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J Bacteriol. 2007;189:6312–6323. doi: 10.1128/JB.00725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell PS. Management of water repellency in Australia and risks associated with preferential flow, pesticide concentration and leaching. J Hydrology. 2000;231–232:384–395. [Google Scholar]
- Bodour AA, Drees KP, Maier RM. Distribution of biosurfactant-producing bacteria in undisturbed and contaminated arid Southwestern soils. Appl Environ Microbiol. 2003;69:3280–3287. doi: 10.1128/AEM.69.6.3280-3287.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyette CD, Walker HL, Abbas HK. Biological control of kudzu (Pueraria lobata); with an isolate of Myrothecium verrucaria. Biocontrol Sci Technol. 2002;12:75–82. [Google Scholar]
- Burch AY, Browne PJ, Dunlap CA, Price NP, Lindow SE. Comparison of biosurfactant detection methods reveals hydrophobic surfactants and contact-regulated production. Environ Microbiol. 2011;13:2681–2691. doi: 10.1111/j.1462-2920.2011.02534.x. [DOI] [PubMed] [Google Scholar]
- Cameotra SS, Bollag JM. Biosurfactant-enhanced bioremediation of polycyclic aromatic hydrocarbons. Crit Rev Environ Sci Technol. 2003;30:111–126. [Google Scholar]
- Cameotra SS, Singh P. Synthesis of rhamnolipid biosurfactant and mode of hexadecane uptake by Pseudomonas species. Microb Cell Fact. 2009;11:8–16. doi: 10.1186/1475-2859-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameotra SS, Makkar RS, Kaur J, Mehta SK. Synthesis of biosurfactants and their advantages to microorganisms and mankind. Adv Exp Med Biol. 2010;672:261–280. doi: 10.1007/978-1-4419-5979-9_20. [DOI] [PubMed] [Google Scholar]
- Camilios Neto D, Meira JA, Tiburtius E, Zamora PP, Bugay C, Mitchell DA, Krieger N. Production of rhamnolipids in solid-state cultivation: characterization, downstream processing and application in the cleaning of contaminated soils. Biotechnol J. 2009;4:748–755. doi: 10.1002/biot.200800325. [DOI] [PubMed] [Google Scholar]
- Cerqueira VS, Hollenbach EB, Maboni F, Vainstein MH, Camargo FA, do Carmo M, Peralba R, Bento FM. Biodegradation potential of oily sludge by pure and mixed bacterial cultures. Bioresour Technol. 2011;102:11003–11010. doi: 10.1016/j.biortech.2011.09.074. [DOI] [PubMed] [Google Scholar]
- Cerqueira VS, Hollenbach EB, Maboni F, Camargo FA, Peralba Mdo C, Bento FM. Bioprospection and selection of bacteria isolated from environments contaminated with petrochemical residues for application in bioremediation. World J Microbiol Biotechnol. 2012;28:1203–1222. doi: 10.1007/s11274-011-0923-z. [DOI] [PubMed] [Google Scholar]
- Chaudhary P, Sharma R, Singh SB, Nain L. Bioremediation of PAH by Streptomyces sp. Bull Environ Contam Toxicol. 2011;86:268–271. doi: 10.1007/s00128-011-0211-5. [DOI] [PubMed] [Google Scholar]
- Chen J, Huang PT, Zhang KY, Ding FR. Isolation of biosurfactant producers, optimization and properties of biosurfactant produced by Acinetobacter sp. from petroleum-contaminated soil. J Appl Microbiol. 2012;112:660–671. doi: 10.1111/j.1365-2672.2012.05242.x. [DOI] [PubMed] [Google Scholar]
- Chen SY, Lu WB, Wei YH, Chen WM, Chang JS. Improved production of biosurfactant with newly isolated Pseudomonas aeruginosa S2. Biotechnol Prog. 2007;23:661–666. doi: 10.1021/bp0700152. [DOI] [PubMed] [Google Scholar]
- Colores GM, Macur RE, Ward DM, Inskeep WP. Molecular analysis of surfactant driven microbial population shifts in hydrocarbon contaminated soil. Appl Environ Microbiol. 2000;66:2959–2964. doi: 10.1128/aem.66.7.2959-2964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppotelli BM, Ibarrolaza A, Dias RL, Del Panno MT, Berthe-Corti L, Morelli IS. Study of the degradation activity and the strategies to promote the bioavailability of phenanthrene by Sphingomonas paucimobilis strain 20006FA. Microb Ecol. 2010;59:266–276. doi: 10.1007/s00248-009-9563-3. [DOI] [PubMed] [Google Scholar]
- Darvishi P, Ayatollahi S, Mowla D, Niazi A. Biosurfactant production under extreme environmental conditions by an efficient microbial consortium, ERCPPI-2. Colloids Surf B Biointerfaces. 2011;84:292–300. doi: 10.1016/j.colsurfb.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Das K, Mukherjee AK. Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresour Technol. 2007;98:1339–1345. doi: 10.1016/j.biortech.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Das P, Mukherjee S, Sivapathasekaran C, Sen R. Microbial surfactants of marine origin: potentials and prospects. Adv Exp Med Biol. 2010;672:88–101. doi: 10.1007/978-1-4419-5979-9_7. [DOI] [PubMed] [Google Scholar]
- de Lima CJ, Ribeiro EJ, Sérvulo EF, Resende MM, Cardoso VL. Biosurfactant production by Pseudomonas aeruginosa grown in residual soybean oil. Appl Biochem Biotechnol. 2009;152:156–168. doi: 10.1007/s12010-008-8188-1. [DOI] [PubMed] [Google Scholar]
- Dean SM, Jin Y, Cha DK, Wilson SV, Radosevich M. Phenanthrene degradation in soils co-inoculated with phenanthrene-degrading and biosurfactant-producing bacteria. J Environ Qual. 2001;30:1126–1133. doi: 10.2134/jeq2001.3041126x. [DOI] [PubMed] [Google Scholar]
- Debode J, De Maeyer K, Perneel M, Pannecoucque J, De Backer G, Höfte M. Biosurfactants are involved in the biological control of Verticillium microsclerotia by Pseudomonas spp. J Appl Microbiol. 2007;103:1184–1196. doi: 10.1111/j.1365-2672.2007.03348.x. [DOI] [PubMed] [Google Scholar]
- Deleu M, Paquot M. From renewable vegetables resources to microorganisms: new trends in surfactant. CR Chimie. 2004;7:641–646. [Google Scholar]
- Dubern JF, Lagendijk EL, Lugtenberg BJ, Bloemberg GV. The heat shock genes dnaK, dnaJ, and grpE are involved in regulation of putisolvin biosynthesis in Pseudomonas putida PCL1445. J Bacteriol. 2005;187:5967–5976. doi: 10.1128/JB.187.17.5967-5976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusane D, Rahman P, Zinjarde S, Venugopalan V, McLean R, Weber M. Quorum sensing; implication on rhamnolipid biosurfactant production. Biotech Genetic Eng Rev. 2010;27:159–184. doi: 10.1080/02648725.2010.10648149. [DOI] [PubMed] [Google Scholar]
- Eddouaouda K, Mnif S, Badis A, Younes SB, Cherif S, Ferhat S, Mhiri N, Chamkha M, Sayadi S. Characterization of a novel biosurfactant produced by Staphylococcus sp. strain 1E with potential application on hydrocarbon bioremediation. J Basic Microbiol. 2012;52:408–418. doi: 10.1002/jobm.201100268. [DOI] [PubMed] [Google Scholar]
- Gamalero E, Glick BR. Mechanisms Used by Plant Growth- promoting Bacteria Chapter 2. In: Maheshwari DK, editor. Bacteria in Agrobiology: Plant Nutrient Management. Berlin Heidelberg: Springer-Verlag; 2011. pp. 17–46. [Google Scholar]
- Gesheva V, Stackebrandt E, Vasileva-Tonkova E. Biosurfactant production by halotolerant Rhodococcus fascians from Casey Station, Wilkes Land, Antarctica. Curr Microbiol. 2010;61:112–117. doi: 10.1007/s00284-010-9584-7. [DOI] [PubMed] [Google Scholar]
- Glick BR, Todorovic B, Czarny J, Cheng Z, Duan J, McConkey B. Promotion of plant growth by bacterial ACC deaminase. Crit Rev Plant Sci. 2007;26:227–242. [Google Scholar]
- Gottfried A, Singhal N, Elliot R, Swift S. The role of salicylate and biosurfactant in inducing phenanthrene degradation in batch soil slurries. Appl Microbiol Biotechnol. 2010;86:1563–1571. doi: 10.1007/s00253-010-2453-2. [DOI] [PubMed] [Google Scholar]
- Gronwald JW, Plaisance KL, Ide DA, Wyse DL. Assessment of Pseudomonas syringae pv. tagetis as a biocontrol agent for Canada thistle. Weed Sci. 2002;50:397–404. [Google Scholar]
- Gunther NW, IV, Nuñez A, Fett W, Solaiman DKY. Production of rhamnolipids by Pseudomonas chlororaphis, a non pathogenic bacteria. Appl Environ Microbiol. 2005;71:2288–2293. doi: 10.1128/AEM.71.5.2288-2293.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad NI. Isolation and characterization of a biosurfactant producing strain, Brevibacilis brevis HOB1. J Ind Microbiol Biotechnol. 2008;35:1597–1604. doi: 10.1007/s10295-008-0403-0. [DOI] [PubMed] [Google Scholar]
- Herman DC, Artiola JF, Miller RA. Removal of cadmium, lead and zinc from soil by a rhamnolipid biosurfactant. Environ Sci Technol. 1995;29:2280–2285. doi: 10.1021/es00009a019. [DOI] [PubMed] [Google Scholar]
- Heyd M, Kohnert A, Tan TH, Nusser M, Kirschhöfer F, Brenner-Weiss G, Franzreb M, Berensmeier S. Development and trends of biosurfactant analysis and purification using rhamnolipids as an example. Anal Bioanal Chem. 2008;391:1579–1590. doi: 10.1007/s00216-007-1828-4. [DOI] [PubMed] [Google Scholar]
- Hickey AM, Gordon L, Dobson AD, Kelly CT, Doyle EM. Effect of surfactants on fluoranthene degradation by Pseudomonas alcaligenes PA-10. Appl Microbiol Biotechnol. 2007;74:851–856. doi: 10.1007/s00253-006-0719-5. [DOI] [PubMed] [Google Scholar]
- Hommais F, Oger-Desfeux C, Van Gijsegem F, Castang S, Ligori S, Expert D, Reverchon S, Nasser W. PecS is a global regulator of the symptomatic phase in the phytopathogenic bacterium Erwinia chrysanthemi 3937. J Bacteriol. 2008;190:7508–7522. doi: 10.1128/JB.00553-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkinson MJ, Collins HM, Goss GR. Pesticide formulations and application systems: ASTM Committee E-35 on Pesticides. ASTM International. 1997;17(1328):1–331. [Google Scholar]
- Hsieh FC, Li MC, Lin TC, Kao SS. Rapid detection and characterization of surfactin-producing Bacillus subtilis and closely related species based on PCR. Curr Microbiol. 2004;49:186–191. doi: 10.1007/s00284-004-4314-7. [DOI] [PubMed] [Google Scholar]
- Hua F, Wang H. Uptake modes of octadecane by Pseudomonas sp. DG17 and synthesis of biosurfactant. J Appl Microbiol. 2012;112:25–37. doi: 10.1111/j.1365-2672.2011.05178.x. [DOI] [PubMed] [Google Scholar]
- Hultberg M, Bergstrand KJ, Khalil S, Alsanius B. Characterization of biosurfactant-producing strains of fluorescent pseudomonads in a soilless cultivation system. Antonie Van Leeuwenhoek. 2008;94(2):329–334. doi: 10.1007/s10482-008-9250-2. [DOI] [PubMed] [Google Scholar]
- Hultberg M, Bergstrand KJ, Khalil S, Alsanius B. Production of biosurfactants and antibiotics by fluorescent pseudomonads isolated from a closed hydroponic system equipped with a slow filter. Antonie Van Leeuwenhoek. 2008;93:373–380. doi: 10.1007/s10482-007-9214-y. [DOI] [PubMed] [Google Scholar]
- Jazzar C, Hammad EA. The efficacy of enhanced aqueous extracts of melia azedarach leaves and fruits integrated with the Camptotylus reuteri releases against the sweet potato whitefly nymphs. Bull Insectol. 2003;56:269–275. [Google Scholar]
- Juwarkar AA, Nair A, Dubey KV, Singh SK, Devotta S. Biosurfactant technology for remediation of cadmium and lead contaminated soils. Chemosphere. 2007;68:1996–2002. doi: 10.1016/j.chemosphere.2007.02.027. [DOI] [PubMed] [Google Scholar]
- Kang SW, Kim YB, Shin JD, Kim EK. Enhanced biodegradation of hydrocarbons in soil by microbial biosurfactant, sophorolipid. Appl Biochem Biotechnol. 2010;160:780–790. doi: 10.1007/s12010-009-8580-5. [DOI] [PubMed] [Google Scholar]
- Kassab DM, Roane TM. Differential responses of a mine tailings Pseudomonas isolate to cadmium and lead exposures. Biodegradation. 2006;17:379–387. doi: 10.1007/s10532-005-9010-1. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- Kildisas V, Levišauskas D, Grigiškis S, Baškys E. Development of clean-up complex technology of soil contaminated by oil pollutants based on cleaner production concepts. Environ Res Eng Manage. 2003;25:87–93. [Google Scholar]
- Kim PI, Bai H, Bai D, Chae H, Chung S, Kim Y, Park R, Chi YT. Purification and characterization of a lipopeptide produced by Bacillus thuringiensis CMB26. J Appl Microbiol. 2004;97:942–949. doi: 10.1111/j.1365-2672.2004.02356.x. [DOI] [PubMed] [Google Scholar]
- Kim PI, Ryu J, Kim YH, Chi YT. Production of biosurfactant lipopeptides Iturin A, fengycin and surfactin A from Bacillus subtilis CMB32 for control of Colletotrichum gloeosporioides. J Microbiol Biotechnol. 2010;20:138–145. [PubMed] [Google Scholar]
- Kim SK, Kim YC, Lee S, Kim JC, Yun MY, Kim IS. Insecticidal activity of rhamnolipid isolated from Pseudomonas sp. EP-3 against green peach aphid (Myzus persicae) J Agric Food Chem. 2011;59:934–938. doi: 10.1021/jf104027x. [DOI] [PubMed] [Google Scholar]
- Kiran GS, Selvin J, Manilal A, Sujith S. Biosurfactants as green stabilizers for the biological synthesis of nanoparticles. Crit Rev Biotechnol. 2011;31:354–364. doi: 10.3109/07388551.2010.539971. [DOI] [PubMed] [Google Scholar]
- Koglin A, Doetsch V, Bernhard F. Molecular engineering aspects for the production of new and modified biosurfactants. Adv Exp Med Biol. 2010;672:158–169. doi: 10.1007/978-1-4419-5979-9_12. [DOI] [PubMed] [Google Scholar]
- Krishnayya PV, Grewal PS. Effect of neem and selected fungicides on viability and virulence of the entomopathogenic nematode steinernema feltiae. Biocontrol Sci Technol. 2002;12:259–266. [Google Scholar]
- Kruijt M, Tran H, Raaijmakers JM. Functional, genetic and chemical characterization of biosurfactants produced by plant growth-promoting Pseudomonas putida 267. J Appl Microbiol. 2009;107:546–556. doi: 10.1111/j.1365-2672.2009.04244.x. [DOI] [PubMed] [Google Scholar]
- Krzyzanowska DM, Potrykus M, Golanowska M, Polonis K, Gwizdek-Wisniewska A, Lojkowska E, Jafra S (2012) Rhizosphere bacteria as potential biocontrol agents against soft rot caused by various Pectobacterium and Dickeya spp. strains. J plant pathol 94
- Kumar CG, Mamidyala SK, Sujitha P, Muluka H, Akkenapally S (2012) Evaluation of critical nutritional parameters and their significance in the production of rhamnolipid biosurfactants from Pseudomonas aeruginosa BS-161R. Biotechnol Prog. doi:10.1002/btpr.1634 [DOI] [PubMed]
- Kurtzman CP, Price NP, Ray KJ, Kuo TM. Production of sophorolip biosurfactants by multiple species of the Starmerella (Candida) bombicola yeast clade. FEMS Microbiol Lett. 2010;311:140–146. doi: 10.1111/j.1574-6968.2010.02082.x. [DOI] [PubMed] [Google Scholar]
- Lima TM, Procópio LC, Brandão FD, Leão BA, Tótola MR, Borges AC. Evaluation of bacterial surfactant toxicity towards petroleum degrading microorganisms. Bioresour Technol. 2011;102:2957–2964. doi: 10.1016/j.biortech.2010.09.109. [DOI] [PubMed] [Google Scholar]
- Lima TM, Procópio LC, Brandão FD, Carvalho AM, Tótola MR, Borges AC. Biodegradability of bacterial surfactants. Biodegradation. 2011;22:585–592. doi: 10.1007/s10532-010-9431-3. [DOI] [PubMed] [Google Scholar]
- Liu WW, Yin R, Lin XG, Zhang J, Chen XM, Li XZ, Yang T. Interaction of biosurfactant-microorganism to enhance phytoremediation of aged polycyclic aromatic hydrocarbons (PAHS) contaminated soils with alfalfa (Medicago sativa L.) Huan Jing Ke Xue. 2010;31:1079–1084. [PubMed] [Google Scholar]
- Loh J, Pierson EA, Pierson LS, 3rd, Stacey G, Chatterjee A. Quorum sensing in plant-associated bacteria. Curr Opin Plant Biol. 2002;5:1–5. doi: 10.1016/s1369-5266(02)00274-1. [DOI] [PubMed] [Google Scholar]
- Lourith N, Kanlayavattanakul M. Natural surfactants used in cosmetics: glycolipids. Int J Cosmet Sci. 2009;31:255–261. doi: 10.1111/j.1468-2494.2009.00493.x. [DOI] [PubMed] [Google Scholar]
- Maier RM, Neilson JW, Artiola JF, Jordan FL, Glenn EP, Descher SM. Remediation of metal-contaminated soil and sludge using biosurfactant technology. Int J Occup Med Environ Health. 2001;14:241–248. [PubMed] [Google Scholar]
- Mata-Sandoval JC, Karns J, Torrents A. Influence of rhamnolipids and Triton X-100 on the biodegradation of three pesticides in aqueous and soil slurries. J Agri Food Chem. 2001;49:3296–3303. doi: 10.1021/jf001432w. [DOI] [PubMed] [Google Scholar]
- Menezes Bento F, de Oliveira Camargo FA, Okeke BC, Frankenberger WT., Jr Diversity of biosurfactant producing microorganisms isolated from soils contaminated with diesel oil. Microbiol Res. 2005;160:249–255. doi: 10.1016/j.micres.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Moldes AB, Paradelo R, Rubinos D, Devesa-Rey R, Cruz JM, Barral MT. Ex situ treatment of hydrocarbon-contaminated soil using biosurfactants from Lactobacillus pentosus. J Agric Food Chem. 2011;59:9443–9447. doi: 10.1021/jf201807r. [DOI] [PubMed] [Google Scholar]
- Mulligan CN, Wang S. Remediation of a heavy metal contaminated soil by a rhamnolipid foam. In: Thomas HR, editor. Yangt RN. Thomas Telford: Geoenvironmental engineering. Integrated management of groundwater and contaminated land. London; 2004. pp. 544–551. [Google Scholar]
- Mulligan CN, Yong RN, Gibbs BF. Heavy metal removal from sediments by biosurfactants. J Hazard Mater. 2001;85:111–125. doi: 10.1016/s0304-3894(01)00224-2. [DOI] [PubMed] [Google Scholar]
- Mulligan CN. Environmental applications for biosurfactants. Environ Pollut. 2005;133:183–198. doi: 10.1016/j.envpol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Mulligan CN, Eftekhari F. Remediation with surfactant foam of PCP- contaminated soil. Eng Geol. 2003;70:269–279. [Google Scholar]
- Mulqueen P. Recent advances in agrochemical formulations. Adv Colloid Interface Sci. 2003;106:83–107. doi: 10.1016/s0001-8686(03)00106-4. [DOI] [PubMed] [Google Scholar]
- Neilson JW, Artiola JF, Maier RM. Characterization of lead removal from contaminated soils by non-toxic soil-washing agents. J Environ Qual. 2003;32:899–908. doi: 10.2134/jeq2003.8990. [DOI] [PubMed] [Google Scholar]
- Neilson JW, Zhang L, Veres-Schalnat TA, Chandler KB, Neilson CH, Crispin JD, Pemberton JE, Maier RM. Cadmium effects on transcriptional expression of rhlB/rhlC genes and congener distribution of monorhamnolipid and dirhamnolipid in Pseudomonas aeruginosa IGB83. Appl Microbiol Biotechnol. 2010;88:953–963. doi: 10.1007/s00253-010-2808-8. [DOI] [PubMed] [Google Scholar]
- Newton JA, Fray RG. Integration of environmental and host-derived signals with quorum sensing during plant–microbe interactions. Cell Microbiol. 2004;6:213–224. doi: 10.1111/j.1462-5822.2004.00362.x. [DOI] [PubMed] [Google Scholar]
- Nie M, Yin X, Ren C, Wang Y, Xu F, Shen Q. Novel rhamnolipid biosurfactants produced by a polycyclic aromatic hydrocarbon-degrading bacterium Pseudomonas aeruginosa strain NY3. Biotechnol Adv. 2010;28:635–643. doi: 10.1016/j.biotechadv.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TH, Sørensen D, Tobiasen C, Andersen JB, Christophersen C, Givskov M, Sørensen J. Antibiotic and biosurfactant properties of cyclic lipopeptides produced by fluorescent Pseudomonas spp. from the sugar beet rhizosphere. Appl Environ Microbiol. 2002;68:3416–3423. doi: 10.1128/AEM.68.7.3416-3423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TH, Sørensen J. Production of cyclic lipopeptides by Pseudomonas fluorescens strains in bulk soil and in the sugar beet rhizosphere. Appl Environ Microbiol. 2003;69:861–868. doi: 10.1128/AEM.69.2.861-868.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihorimbere V, Marc Ongena M, Smargiassi M, Thonart P. Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnol Agron Soc Environ. 2011;15:327–337. [Google Scholar]
- Nishio E, Ichiki Y, Tamura H, Morita S, Watanabe K, Yoshikawa H. Isolation of bacterial strains that produce the endocrine disruptor, octylphenol diethoxylates, in paddy fields. Biosci Biotechnol Biochem. 2002;66:1792–1798. doi: 10.1271/bbb.66.1792. [DOI] [PubMed] [Google Scholar]
- Nitschke M, Costa S. Biosurfactants in food industry. Trends Food Sci Technol. 2007;18:252–259. [Google Scholar]
- Onwosi CO, Odibo FJ. Effects of carbon and nitrogen sources on rhamnolipid biosurfactant production by Pseudomonas nitroreducens isolated from soil. World J Microbiol Biotechnol. 2012;28:937–942. doi: 10.1007/s11274-011-0891-3. [DOI] [PubMed] [Google Scholar]
- Pacwa-Plociniczak PGA, Piotrowska-Seget Z, Cameotra SS. Environmental applications of biosurfactants: recent advances. Int J Mol Sci. 2011;12:633–654. doi: 10.3390/ijms12010633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partovinia A, Naeimpoor F, Hejazi P. Carbon content reduction in a model reluctant clayey soil: slurry phase n-hexadecane bioremediation. J Hazard Mater. 2010;181:133–139. doi: 10.1016/j.jhazmat.2010.04.106. [DOI] [PubMed] [Google Scholar]
- Pei X, Zhan X, Zhou L. Effect of biosurfactant on the sorption of phenanthrene onto original and H2O2 treated soils. J Environ Sci (China) 2009;21:1378–1385. doi: 10.1016/s1001-0742(08)62429-8. [DOI] [PubMed] [Google Scholar]
- Perneel M, Heyrman J, Adiobo A, De Maeyer K, Raaijmakers JM, De Vos P, Höfte M. Characterization of CMR5c and CMR12a, novel fluorescent Pseudomonas strains from the cocoyam rhizosphere with the biocontrol activity. J Appl Microbiol. 2007;103:1007–1020. doi: 10.1111/j.1365-2672.2007.03345.x. [DOI] [PubMed] [Google Scholar]
- Petrovic M, Barcelo D. Analysis and fate of surfactants in sludge and sludge-amended soil. Trends Analyt Chem. 2004;23:10–11. [Google Scholar]
- Poggi-Varaldo HM, Rinderknecht-Seijas N. A differential availability enhancement factor for the evaluation of pollutant availability in soil treatments. Acta Biotechnol. 2003;23:271–280. [Google Scholar]
- Robles-González IV. A review on slurry bioreactors for bioremediation of soils and sediments. Microb Cell Fact. 2008;7:5. doi: 10.1186/1475-2859-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues L, Banat IM, Teixeira J, Oliveira R. Biosurfactants: potential applications in medicine. J Antimicrob Chemother. 2006;57:609–618. doi: 10.1093/jac/dkl024. [DOI] [PubMed] [Google Scholar]
- Rodriguez SB, Mahoney NE. Inhibition of aflatoxin production by surfactants. Appl Environ Microbiol. 1994;60:106–110. doi: 10.1128/aem.60.1.106-110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán-Carrillo T, Martínez-García X, Zapata-Peñasco I, Castorena-Cortés G, Reyes-Avila J, Mayol-Castillo M, Olguín-Lora P. Evaluation of the effect of nutrient ratios on biosurfactant production by Serratia marcescens using a Box-Behnken design. Colloids Surf B Biointerfaces. 2011;86:384–389. doi: 10.1016/j.colsurfb.2011.04.026. [DOI] [PubMed] [Google Scholar]
- Ron EZ, Rosenberg E. Natural roles in biosurfactants. Environ Microbiol. 2011;3:229–236. doi: 10.1046/j.1462-2920.2001.00190.x. [DOI] [PubMed] [Google Scholar]
- Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E, Ron EZ. Surface active polymers from the genus Acinetobacter. In: Kaplan D L., editor. Biopolymers from renewable resources. Berlin: Springer; 1998. pp. 281–291. [Google Scholar]
- Rostas M, Blassmann K. Insects had it first: surfactants as a defense against predators. Proc R Soc B. 2009;276:633–638. doi: 10.1098/rspb.2008.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos EC, Jacques RJ, Bento FM, Peralba Mdo C, Selbach PA, Sá EL, Camargo FA. Anthracene biodegradation and surface activity by an iron-stimulated Pseudomonas sp. Bioresour Technol. 2008;99:2644–2649. doi: 10.1016/j.biortech.2007.04.050. [DOI] [PubMed] [Google Scholar]
- Sastoque-Cala L, Cotes-Prado AM, Rodríguez-Vázquez R, Pedroza-Rodríguez AM. Effect of nutrients and conditions of fermentation on the production of biosurfactants using rhizobacteria isolated from fique. Univ Sci. 2010;15:251–264. [Google Scholar]
- Satpute SK, Banpurkar AG, Dhakephalkar PK, Banat IM, Chopade BA. Methods for investigating biosurfactants and bioemulsifiers: a review. Crit Rev Biotechnol. 2010;30(2):127–144. doi: 10.3109/07388550903427280. [DOI] [PubMed] [Google Scholar]
- Satpute SK, Bhuyan SS, Pardesi KR, Mujumdar SS, Dhakephalkar PK, Shete AM, Chopade BA. Molecular genetics of biosurfactant synthesis in microorganisms. Adv Exp Med Biol. 2010;672:14–41. doi: 10.1007/978-1-4419-5979-9_2. [DOI] [PubMed] [Google Scholar]
- Schippers C, Gessner K, Müller T, Scheper T. Microbial degradation of phenanthrene by addition of a sophorolipid mixture. J Biotechnol. 2000;83:89–198. doi: 10.1016/s0168-1656(00)00304-7. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Biotechnological prospects from metagenomics. Curr Opin Biotechnol. 2003;14:303–310. doi: 10.1016/s0958-1669(03)00067-3. [DOI] [PubMed] [Google Scholar]
- Scott MJ, Jones MN. The biodegradation of surfactants in the environment. Biochim Biophys Acta. 2000;1508:235–251. doi: 10.1016/s0304-4157(00)00013-7. [DOI] [PubMed] [Google Scholar]
- Sekhon KK, Khanna S, Cameotra SS. Enhanced biosurfactant production through cloning of three genes and role of esterase in biosurfactant release. Microb cell Fact 27. 2011;10:49. doi: 10.1186/1475-2859-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R. Surfactin: biosynthesis, genetics and potential applications. Adv Exp Med Biol. 2010;672:316–323. doi: 10.1007/978-1-4419-5979-9_24. [DOI] [PubMed] [Google Scholar]
- Sha R, Jiang L, Meng Q, Zhang G, Song Z. Producing cell-free culture broth of rhamnolipids as a cost-effective fungicide against plant pathogens. J Basic Microbiol. 2011;52:458–466. doi: 10.1002/jobm.201100295. [DOI] [PubMed] [Google Scholar]
- Sharma S, Singh P, Raj M, Chadha BS, Saini HS. Aqueous phase partitioning of hexachlorocyclohexane (HCH) isomers by biosurfactant produced by Pseudomonas aeruginosa WH-2. J Hazard Mater. 2009;171:1178–1182. doi: 10.1016/j.jhazmat.2009.06.116. [DOI] [PubMed] [Google Scholar]
- Sheng X, He L, Wang Q, Ye H, Jiang C. Effects of inoculation of biosurfactant-producing Bacillus sp. J119 on plant growth and cadmium uptake in a cadmium-amended soil. J Hazard Mater. 2008;155:17–22. doi: 10.1016/j.jhazmat.2007.10.107. [DOI] [PubMed] [Google Scholar]
- Shete AM, Wadhawa G, Banat IM, Chopade BA (2006) Mapping of patents on bioemulsifier and biosurfactant: a review. J Sci Indust Res 65:91–115
- Shin KH, Kim KW, Ahn Y. Use of biosurfactant to remediate phenanthrene-contaminated soil by the combined solubilization-biodegradation process. J Hazard Mate. 2006;137:1831–1837. doi: 10.1016/j.jhazmat.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Simpson DR, Natraj NR, McInerney MJ, Duncan KE. Biosurfactant-producing Bacillus are present in produced brines from Oklahoma oil reservoirs with a wide range of salinities. Appl Microbiol Biotechnol. 2011;91:1083–1093. doi: 10.1007/s00253-011-3326-z. [DOI] [PubMed] [Google Scholar]
- Singh A, Van Hamme JD, Ward OP. Surfactants in microbiology and biotechnology: part 2: application aspects. Biotechnol Adv. 2007;25:99–121. doi: 10.1016/j.biotechadv.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Singh PB, Sharma S, Saini HS, Chadha BS. Biosurfactant production by Pseudomonas sp. and its role in aqueous phase partitioning and biodegradation of chlorpyrifos. Lett Appl Microbiol. 2009;49:378–383. doi: 10.1111/j.1472-765X.2009.02672.x. [DOI] [PubMed] [Google Scholar]
- Snook ME, Mitchell T, Hinton DM, Bacon CW. Isolation and characterization of leu7-surfactin from the endophytic bacterium Bacillus mojavensis RRC 101, a biocontrol agent for Fusarium verticillioides. J Agric Food Chem. 2009;57:4287–4292. doi: 10.1021/jf900164h. [DOI] [PubMed] [Google Scholar]
- Straube WL, Nestler CC, Hansen LD, Ringleberg D, Pritchard PH, Jones-Meehan J. Remediation of polyaromatic hydrocarbons (PAHs) through landfarming with biostimulation and bioaugmentation. Acta Biotechnol. 2003;23:179–196. [Google Scholar]
- Street JC. Methods of removal of pesticides residues. Canad Med Ass J. 1969;100:154–160. [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wu L, Luo Y. Application of organic agents in remediation of heavy metals- contaminated soil. Ying Yong Sheng Tai Xue Bao. 2006;17:1123–1128. [PubMed] [Google Scholar]
- Takenaka S, Tonoki T, Taira K, Murakami S, Aoki K. Adaptation of Pseudomonas sp. strain 7–6 to quaternary ammonium compounds and their degradation via dual pathways. Appl Environ Micro. 2007;73:1797–1802. doi: 10.1128/AEM.02426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeyama H, Wada M, Matsunaga T. Screening of soil bacteria for production of biocleaner. Appl Biochem Biotechnol. 2002;98–100:319–326. doi: 10.1385/abab:98-100:1-9:319. [DOI] [PubMed] [Google Scholar]
- Tanikawa T, Nakagawa Y, Matsuyama T. Transcriptional downregulator hexS controlling prodigiosin and serrawettin W1 biosynthesis in Serratia marcescens. Microbiol Immunol. 2006;50:587–596. doi: 10.1111/j.1348-0421.2006.tb03833.x. [DOI] [PubMed] [Google Scholar]
- Tuleva B, Christova N, Cohen R, Stoev G, Stoineva I. Production and structural elucidation of trehalose tetraesters (biosurfactants) from a novel alkanothrophic Rhodococcus wratislaviensis strain. J Appl Microbiol. 2008;104:1703–1710. doi: 10.1111/j.1365-2672.2007.03680.x. [DOI] [PubMed] [Google Scholar]
- Urum K, Pekdemir T, Gopur M. Optimum conditions for washing of crude oil-contaminated soil with biosurfactant solutions. Trans Inst Chem Eng. 2003;81B:203–209. [Google Scholar]
- Van Hamme JD, Singh A, Ward OP. Physiological aspects. part 1 in a series of papers devoted to surfactants in microbiology and biotechnology. Biotechnol Adv. 2006;24:604–620. doi: 10.1016/j.biotechadv.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Vasileva-Tonkova E, Gesheva V. Biosurfactant production by antarctic facultative anaerobe Pantoea sp. during growth on hydrocarbons. Curr Microbiol. 2007;54:136–1341. doi: 10.1007/s00284-006-0345-6. [DOI] [PubMed] [Google Scholar]
- Vatsa P, Sanchez L, Clement C, Baillieul F, Dorey S. Rhamnolipid biosurfactants as new players in animal and plant defense against microbes. Int J Mol Sci. 2010;11:5095–5108. doi: 10.3390/ijms11125095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velho RV, Medina LF, Segalin J, Brandelli A. Production of lipopeptides among Bacillus strains showing growth inhibition of phytopathogenic fungi. Folia Microbiol (Praha) 2011;56:297–303. doi: 10.1007/s12223-011-0056-7. [DOI] [PubMed] [Google Scholar]
- Vipulanandan C, Ren X. Enhanced solubility and biodegradation of naphthalene with biosurfactant. J Environ Eng. 2000;126:629–634. [Google Scholar]
- Walter V, Syldatk C, Hausmann R. Screening concepts for the isolation of biosurfactant producing microorganisms. Adv Exp Med Biol. 2010;672:1–13. doi: 10.1007/978-1-4419-5979-9_1. [DOI] [PubMed] [Google Scholar]
- Wattanaphon HT, Kerdsin A, Thammacharoen C, Sangvanich P, Vangnai AS. A biosurfactant from Burkholderia cenocepacia BSP3 and its enhancement of pesticide solubilization. J Appl Microbiol. 2008;105:416–423. doi: 10.1111/j.1365-2672.2008.03755.x. [DOI] [PubMed] [Google Scholar]
- White JC, Parrish ZD, Gent MP, Iannucci-Berger W, Eitzer BD, Isleyen M, Mattina MI. Soil amendments, plant age, and intercropping impact p, p’-DDE bioavailability to Cucurbita pepo. J Environ Qual. 2006;35:992–1000. doi: 10.2134/jeq2005.0271. [DOI] [PubMed] [Google Scholar]
- Xu Q, Nakajima M, Liu Z, Shiina T. Biosurfactants for microbubble preparation and application. Int J Mol Sci. 2011;12:462–475. doi: 10.3390/ijms12010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Nakajima M, Liu Z, Shiina T. Biosurfactant from microbubble preparation and application. International J of Molecul Sci. 2011;12:462–475. doi: 10.3390/ijms12010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahir ZA, Arshad M, William T, Frankenberger WT., Jr Plant growth promoting rhizobacteria: applications and perspectives in agriculture. Adv Agron. 2004;81:97–168. [Google Scholar]
- Zeinali M, Vossoughi M, Ardestani SK. Characterization of a moderate thermophilic Nocardia species able to grow on polycyclic aromatic hydrocarbons. Lett Appl Microbiol. 2007;45:622–628. doi: 10.1111/j.1472-765X.2007.02241.x. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wang S, Yan Y. Isomerization and biodegradation of beta-cypermethrin by Pseudomonas aeruginosa CH7 with biosurfactant production. Bioresour Technol. 2011;102:7139–7146. doi: 10.1016/j.biortech.2011.03.086. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gu W, Xu P, Tang S, Xie K, Huang X, Huang Q. Effects of alkyl polyglycoside (APG) on composting of agricultural wastes. Waste Manag. 2011;31:1333–1338. doi: 10.1016/j.wasman.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Wong JW. Biosurfactants from Acinetobacter calcoaceticus BU03 enhance the solubility and biodegradation of phenanthrene. Environ Technol. 2009;30:291–299. doi: 10.1080/09593330802630801. [DOI] [PubMed] [Google Scholar]