Abstract
The past decade has seen an explosion in use of the 14C bomb-pulse to do fundamental cell biology. Studies in the 1960’s used decay counting to measure tissue turnover when the atmospheric 14C/C concentration was changing rapidly. Today bulk tissue measurements are of marginal interest since most of the carbon in the tissue resides in proteins, lipids and carbohydrates that turn over rapidly. Specific cell types with specialized functions are the focus of cell turnover investigations. Tissue samples need to be fresh or frozen. Fixed or preserved samples contain petroleum-derived carbon that has not been successfully removed. Cell or nuclear surface markers are used to sort specific cell types, typically by fluorescence-activated cell sorting (FACS). Specific biomolecules need to be isolated with high purity and accelerator mass spectrometry (AMS) measurements must accommodate samples that generally contain less than 40 micrograms of carbon. Furthermore, all separations must not add carbon to the sample. Independent means such as UV absorbance must be used to confirm molecule purity. Approaches for separating specific proteins and DNA and combating contamination of undesired molecules are described.
Introduction
The near doubling of the atmospheric 14CO2 concentration from 1955–1963 due to atmospheric above ground testing of nuclear weapons and its subsequent decline is documented by a high resolution record [1–5]. This rapid rise, sharp peak, and exponential decline is known as the 14C bomb pulse. Most of the tests occurred at relatively few sites in the Northern Hemisphere and it took several years for the excess 14C to distribute homogeneously throughout the atmosphere [2]. The excess 14CO2 entered the food chain through photosynthesis and completely labeled the terrestrial biosphere. The variation in the 14C provides an isotopic chronometer of molecular biosynthesis since 1955. This paper describes the use of the bomb pulse as a tracer in cell and molecular biology.
The elevation of 14C in the biosphere and humans that occurred due to above ground nuclear weapons testing was documented almost immediately [6,7]. Throughout the 1960s and 1970s the rise and fall of 14C/C in the biosphere continued to draw attention [8–11]. Since the 14C/C level was changing rapidly in the 1960s, it was relatively straightforward to determine carbon turnover rates of many tissues [8–11]. These studies used 14C decay counting and required grams of carbon from tissue samples for analyses. Different cell types were also not separated in these tissues due to sample size requirements.
The development of AMS in the late 1970s enabled analyses of much smaller samples and examination of carbon turnover in specific biological structures. Initially researchers concentrated on forensic applications of bomb pulse. The first biological application by Druffel and Mok (1983) dated gallstones [12], containing significant amounts of organic material (mostly cholesterol and bile pigments) from patients who lived in the northern hemisphere. The next biological application of the bomb pulse involved separating human lung parenchymal elastic fibers that were determined to be as old as the individual using aspartate racemization and 14C analyses [13]. The longevity of the elastin was suspected at the time, but nobody had previously thought of using the bomb pulse to probe human tissue turnover in this manner. Bomb pulse dating was largely ignored in the biological community for the next several years, until the pathological structures associated with Alzheimer’s disease, neurofibrillary tangles (NFT) and senile plaques (SP), were separated gravimetrically and analyzed for 14C/C post mortem using healthy tissue as controls [14]. The ages of the structures were averages accumulated over the lifetimes of the donors. In half the cases examined, the average ages of the SP and NFT predated the onset of clinical symptoms of Alzheimer’s disease while the other half possessed average SP and NFT ages corresponding to the onset of symptoms or younger [14]. The lens of the eye also possesses long-lived proteins. Lynnerup et al (2008) showed that the proteins of the lens had very limited turnover or repair [15]. Recent studies of arterial plaques show that the deposits that clog arteries accumulate over many years [16,17].
A leap occurred in biological analysis with the 14C bomb-pulse when the analysis of 14C/C in DNA to ascertain cell birth date was first performed. Since DNA only acquires significant new carbon at cell division, the 14C/C of genomic DNA is a time stamp of cell birth date [18]. Genomic DNA dating has been used to confirm that neurogenesis does not occur in the human cerebellum [18] or cortex [19] after birth. The lack of bomb pulse carbon in neuronal DNA of subjects born before 1955 indicated that DNA repair provides an insignificant amount of new carbon after cell division [18]. It has also been used to determine that adipocytes turnover approximately every 10 years [20] and cardiomyocytes turnover at a low rate [21]. Using bromodeoxyuridine (BrdU) and 14C/C analyses of DNA from pancreatic β-cells, it was determined insulin producing β-cells turnover at a 1–2% annual rate through early adulthood and then cease to turnover after age 30 [22].
Methods
Researchers must obtain approval from local Human Subject Ethics Boards before embarking on a bomb-pulse study. Each country has its own specific laws governing human subject research based on the Helsinki Declaration [23] that is an update and expansion of principles from the Nuremberg Code [24]. Human subjects in bomb-pulse biology studies are often deceased as tissues are most often obtained from autopsies or tissue banks. Permission to harvest tissues for research must be obtained from surviving family members when obtaining tissue at autopsy. Tissue banks have releases in place for providing de-identified tissue. Tissues may also be obtained from surgery (e.g., liposuction or explanted organ). Living subjects must give written informed consent to use their tissue for research. Tissues must always be de-identified, but information such as date of birth, date of death, sex, and cause of death are available.
It is easier to interpret data if one avoids subjects born while the atmospheric 14C/C was changing most rapidly. During the rise and initial fall of the 14CO2 bomb-pulse from 1955–1965 the pulse possessed significant geographical variation and the 14C/C of local food supplies were more variable than either before the pulse or a couple years after the peak [2]. For subjects born after 1965, the change in atmospheric 14C/C has been decreasing only, simplifying interpretation when carbon turnover exists. For subjects born before the pulse, any elevation in 14C/C from prebomb level is clear evidence that new carbon is entering the sample. Likewise, if a subject born before the bomb-pulse has no bomb carbon in a specific structure, neuronal DNA for example, it is clear evidence that no new cells have been generated since 1955 [18]. For some types of cells, turnover changes or stops at a particular age. Perl et al (2010) showed that pancreatic beta cell turnover occurs until early adulthood and then promptly stops at age 30 [22].
The laboratories used for the biological sample preparation for measuring bomb pulse 14C obviously need to be free of radiocarbon contamination. It can be a challenge to find a lab free of legacy 14C tracer contamination at many institutions. Procedures for checking the 14C cleanliness of surfaces and air supplies are in the literature [25,26]. All surfaces readily touched need to be swiped to check for contamination. Shared instruments such as centrifuges, shakers, fume hoods, and biosafety cabinets are often contaminated with 14C. Strictly controlling access to AMS preparation laboratories significantly reduces the chance of casual tracking of contamination into the lab [25,26].
Tissues currently need to be fresh or frozen for bomb-pulse dating. Tissues that are fixed are always contaminated with fossil carbon. Fixing tissues chemically modifies molecules with carbon containing compounds to resist degradation. Formalin, paraffin or polymers used to embed tissues for microtome slices resist decontamination and have not been successfully removed for dating samples. Traditional histology slide preparation replaces water with paraffin. The experience of our lab with collaborators around the world is that fossil carbon from fixed samples can never be completely removed, probably due to formaldehyde crosslinks. Thinly sectioned histology samples have not been used for bomb-pulse dating due to fossil carbon contamination from fixing the tissue and the small amount of carbon in biomolecules of interest in slices only a few μm thick.
Tissue selection
Whole tissue turnover studies conducted before the AMS era measured the turnover of the dominant carbon sources in organs; predominantly proteins, carbohydrates, and lipids. These classes of biomolecules generally turnover frequently, even if a cell does not divide. Today cell biology examines specific cell types or structures, so techniques to specifically isolate and purify cells and molecules of interest need to be used. Often a marker must be developed or verified for isolation of a specific cell type since a population of cells must be used in order to obtain enough carbon for analysis. One can use cell surface markers, nuclear surface markers, specific enzyme expression, cell size, or structure density to separate a target cell type from other cells. Cell size separations are probably the easiest to perform but are less specific. Agglomerations of small cells can sort as one large cell or adhere to a large cell and influence the purity of the sort. Additional analyses need to be completed to quantify the sort purity. Density separations typically need a high-speed centrifuge and construction of a density gradient with a solute [14]. Solvent extraction techniques can be used to separate lipophilic from hydrophilic molecules. Aqueous-organic interfaces, either liquid-liquid or liquid-solid, separate hydrophilic from lipophilic molecules nicely in most cases. However, removing all the carbon from the organic solvent in a lipophilic molecule sample can be a challenge. In most cases, an organic solvent is petroleum-derived and free of 14C, but volatile organic solvents can be used [27]. Preventing physical entrapment of organic phase solvent in a lipophilic sample during drying generally requires some method development aided by use of controls of known isotope ratio. Animal tissue from slaughterhouses known to be from animals 2–3 years old is helpful when establishing protocols. Researchers can check for retention of solvent that can skew the 14C/C ratio.
Cell surface markers can be utilized using magnetic activated cell sorting (MACS) or fluorescence activated cell sorting (FACS). MACS is attractive since it can be done in parallel with inexpensive equipment. Antibodies to specific cell surface markers are attached to small magnetic beads. The beads are mixed in a cell suspension and then pulled out to the side of the tube with a magnetic rack. The beads can be returned to suspension by removing the magnet, rinsed to remove any unwanted cells that were inadvertently trapped, and separated again with a magnetic rack. The isolated cells can then be stripped from the magnetic beads and harvested for analysis. Generally in FACS, a fluorescent label is attached to an antibody that recognizes a target protein, either on the cell membrane or a different cellular structure. FACS is a form of flow cytometry that sorts cells or nuclei one-by-one into bins based upon the presence of fluorescence from the antibody. Nuclear cell markers are generally more specific than cell surface markers [18,19,21], the cell nucleus is more specific to cell type or function than its surface. Also, nuclei are essentially little spheres and are much easier to sort than irregularly shaped neurons [18,19] or large cardiomyocytes [21]. Both MACS and target protein FACS use an antibody to identify a target cell type and perform a binary sort whose purity depends upon the specificity of the antibody. MACS can process larger samples in parallel and is faster than FACS. FACS images provide excellent visualization of a sort and are often preferred for figures in publications. A subset of MACS isolations should be checked for purity with FACS (see Fig. 1). The choice of MACS or FACS depends on preference of the researcher and whether there is a need to keep the cells viable after sorting to confirm function [28–30].
Figure 1.
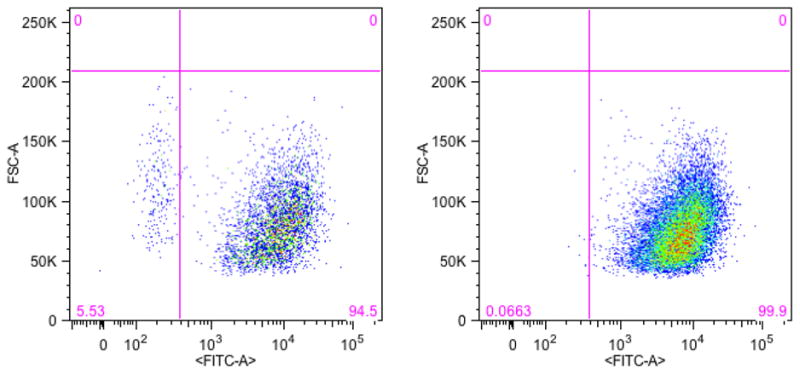
FACS scatter plots of CD44+ surface protein in T24 human bladder cancer cells. The lower right quadrant of each plot contains the CD44+ cell population. Panel A shows the distribution of CD44+ (lower right quadrant) and CD44− (lower left quadrant). Panel B shows the same distribution after a MACS sort for CD44+ cells. Here MACS concentrated the CD44+ population through removal of the CD44− population. FACS confirmed the effectiveness of MACS.
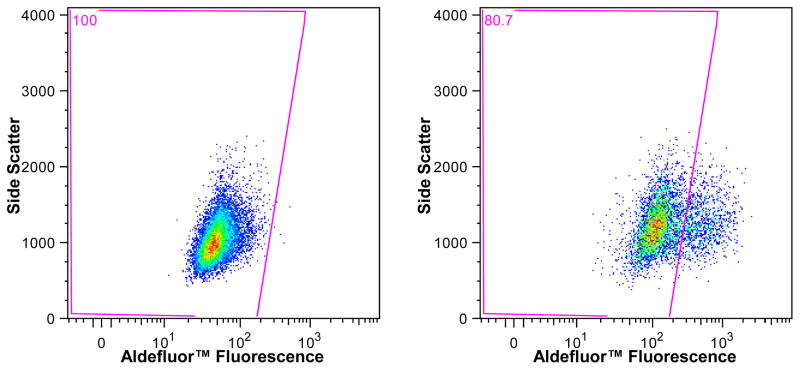
FACS can also be used to separate cells based on increased expression of specific proteins or enzymes. The fluorescent label recognizes a specific enzyme and produces a visualization of a concentration gradient during a sort. Instead of sorting into neat binary bins, gates to distinguish the cut-off between cell types are set with specific controls (Fig. 2) [31–33]. Surface markers touted as specific to cancer stem cells have been found to be common to many tumor cells [31–33]. Separating cells by enzyme concentration is sorting by cell function, which generally is the motivation for determining cell turnover.
Figure 2.
Aldefluor™ sort of the T24 bladder cancer cell line. The gate in the left plot is set to include all cells incubated with a specific inhibitor of the enzyme aldehyde dehydrogenase. This gate is then applied to the test sample, cells without the inhibitor, seen in the plot on the right, to isolate the AldefluorHigh cells that comprise 19.3% of the population.
Molecule separation
Additional separation techniques are usually desired to isolate specific biomolecules from the sorted cell for analysis. Typically DNA, a specific protein, or protein class is the target molecule. It is easiest to separate molecules in the liquid phase, or by selectively precipitating either your target or everything else out of the liquid phase.
The need to minimize carbon addition during sample generation wherever possible is obvious. If carbon must be added (e.g. ethanol precipitation), try to pick a molecule that is soluble in water and can be removed by rinsing. It is best to avoid molecular biology kits since kits are proprietary and manufacturers usually will not reveal contents, making it difficult to determine the sources of carbon they may contain. Our experience over nearly 15 years of using DNA columns is that petroleum-derived carbon from the column contaminates the DNA (Table 1). The column contamination isn’t a problem if the experiment is separating DNA-adducts of a 14C-labeled compound, the 5–20 μg of dead carbon is just part of the carrier carbon (0.5–1.0 mg C) added to the sample. But when doing bomb-pulse dating, the variability of the carbon contamination and the necessity of measuring small, neat samples adversely influences precision. Size exclusion columns for protein separation or buffer exchange behave similarly, shedding fragments of column into the eluent that are not readily removed. We have found size exclusion spin filters work well to remove small molecule debris. Spin filters are manufactured with glycerin on the filter to maintain the integrity of the filter. We find several rinses with distilled, deionized water removes the glycerine. Columns for separating DNA or protein often deposit significant petroleum derived carbon into samples that resists removal.
Table 1.
Depression of F14C of DNA after separation using DNA columns. The carbon mass was not measured directly for samples prepared in February and March 1998 using sealed tube combustion and reduction by Vogel’s method [49].
| Sample | Year Analyzed | Contemporary F14C at analysis | Carbon Mass (μg) | Measured F14C |
|---|---|---|---|---|
| Rat liver DNA | June 1998 | 1.107 | 116 | 0.963±0.019 |
| Rat liver DNA | June 1998 | 1.107 | 92 | 0.988±0.015 |
| Rat liver DNA | June 1998 | 1.107 | 72 | 0.867±0.016 |
| Human Blood DNA | February 1998 | 1.111 | ~500 | 1.101±0.012 |
| Human Blood DNA | February 1998 | 1.111 | ~500 | 1.112±0.013 |
| Human Blood DNA | February 1998 | 1.111 | ~500 | 1.097±0.013 |
| Rat liver DNA | March 1998 | 1.111 | ~300 | 1.070±0.013 |
| Rat liver DNA | March 1998 | 1.111 | ~300 | 1.090±0.023 |
| Rat liver DNA | March 1998 | 1.111 | ~300 | 1.097±0.013 |
| Human Blood DNA | June 2005 | 1.064 | 51 | 0.991±0.007 |
Procedures for achieving DNA preparations suitable for neat AMS measurement are in the literature [18–22]. Although carbon containing solvents are used, they are rinsed extensively and pure samples are produced. Care must be taken to avoid the formation of hard pellets that resist resuspension. Generally multiple precipitations and resuspensions are required to achieve a pure separation and hard pellets can resist resuspension. Also, nontarget molecules can sometimes be physically trapped in precipitate. Serial precipitations and resuspensions can eliminate this source of impurity. When preparing DNA, protein contamination is a significant concern. Proteinase and RNAase enzymes are used to digest protein and RNA. An advantage of sorting nuclei rather than whole cells is the reduction in protein load during DNA purification [18,19,21]. It is helpful to use a contemporary control tissue to establish procedural purity. Residual petroleum-derived carbon is easily detected. The purity of the DNA extraction is checked using ultraviolet/ visible absorbance and checking the 260/280 nm ratio [18–22]. A 260/280 nm absorbance ratio greater than 1.8 isconsidered pure.
Collagen, keratin, and hemoglobin are proteins routinely dated using radiocarbon. Collagen is most widely used because bones can be well preserved and the mineral component of bone protects the collagen in some circumstances. Bone turnover is well known and a variety of circulating markers are used to assess bone health and disease. Collagen is found in many tissues besides bone but has not been analyzed in bomb-pulse applications. More than 20 years ago, Nelson (1991) showed that the CO2 released from the reaction of ninhydrin with free amino acids could be used for carbon isotopic studies of proteins, specifically bone collagen [34]. In this volume Hodgins (2012) describes an approach to use ninhydrin to date collagen-based emulsions in photographic prints [35]. The technique liberates only the carboxyl carbon in amino acids however, utilizing up to 40% of the carbon available in a protein sample.
Although high performance liquid chromatography (HPLC) has not yet been used to separate intact proteins for bomb pulse dating, HPLC is effective in separating proteins and keeps them in the liquid phase. HPLC has been used for separations of mixtures for compound specific radiocarbon analysis [36,37]. HPLC has also been utilized to separate individual amino acids for radiocarbon dating [38,39], so any protein could potentially be analyzed for radiocarbon content provided sufficient mass is available. There are several devices commercially available that use isoelectric focusing to separate proteins by pI while keeping them in the liquid phase. They have been used to separate labeled protein in 14C tracer applications [40] but not unlabeled proteins for dating. The proprietary ampholytes used to establish the pH gradients during isoelectric focusing contain high levels of carbon designed to get low solubility proteins into the liquid phase and can be difficult to remove. Two-dimensional (2D) gels used to separate proteins by molecular weight and isoelectric point always contaminate a protein for protein dating. The protein and gel must be separated with extensive processing to hope for sufficient purity for dating. Our experience is that the gel is never fully removed from the protein with physical means. Performing a protease digestion with HPLC amino acid analysis or ninhydrin reaction may work, but it has not been reported. Protein mass spectrometry is also useful for identifying proteins in a separation but mass yields would be very small and not practical for radiocarbon analyses.
AMS Analysis
AMS analysis is often challenging in bomb-pulse dating due to small sample size. This volume contains papers that describe specialized approaches to measure samples containing less than 10 μg C [41–44]. We find drying liquid samples in a lyophilizer using an oilless pump works well to minimize sample loss and keep fossil and contemporary backgrounds low. We minimize time in the lyophilizer, typically achieving dryness for sets of 20 samples containing 1 mL of water each in about 8 hours. Fossil carbon backgrounds are monitored by co-lyophilization of small standard and background samples. We make dilute solutions of NIST Oxalic Acid II and IAEA C-6 sucrose and pipette 20–80 μg C samples of each into sample tubes for co-lyophilizaton with unknowns. The depression in the isotope ratio of these standards is used to determine the amount of fossil carbon background. Contemporary carbon backgrounds are monitored using ~100 μg pieces of Queets A wood. The elevation of 14C/C in the isotope ratio of the ancient wood is used to determine the amount of contemporary carbon background.
Constructing models
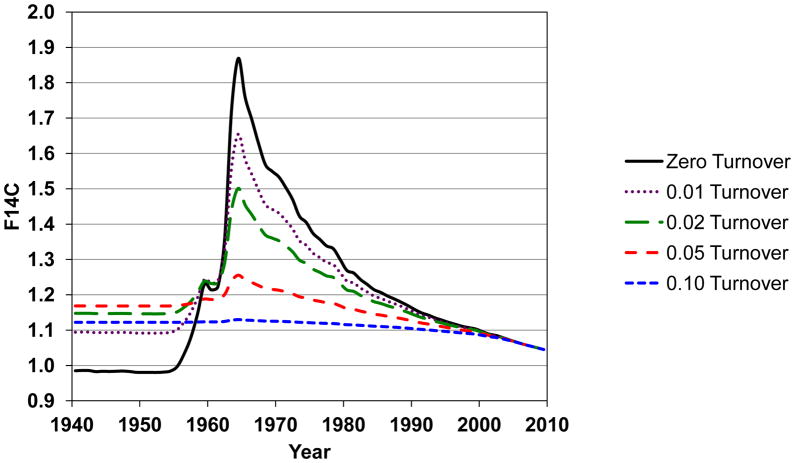
When turnover occurs, a model must be constructed to account for the difference between the 14C/C at the time of birth and its value at measurement [45]. More than one model can be constructed from the same set of data, and multiple scenarios should be evaluated. When determining the age of a deposit or pathological structure, elevation of 14C/C compared to the contemporary level at harvest gives an average age of the structure. By using subjects of different ages, a model of cell turnover can be constructed. Carbon turnover flattens the bomb-pulse curve. Figure 3 depicts the 14C/C of a generic biological molecule that undergoes carbon turnover. The curves in Fig. 3 depict the 14C/C in 2011 of biological carbon molecules initially synthesized at a given date that undergo different annual carbon turnover rates. In this model we assumed there was no physiological preference in turnover, any carbon atom in a population was equally likely to turnover each year. For example, if a tissue has 10% annual carbon turnover, 90% of the carbon is unchanged and 10% of the carbon gets the next year’s 14C/C ratio from the published annual growing season average (zero turnover). The new total 14C/C is calculated and the process is repeated annually to produce the curves with different turnover rates in Fig. 3. Subjects born before 1955 always possess an increased 14C/C ratio compared to their birth level if turnover occurs. Subjects born on the rise of the pulse can exhibit an increased or decreased 14C/C depending on the rate of turnover and where they fall on the curve. Subjects born after the pulse always exhibit a decreased 14C/C if turnover occurs because the newer carbon always has a lower 14C/C. If a small population of cells undergoes turnover in a primarily static pool, the deviation from zero turnover can be small.
Figure 3.
Effect of carbon turnover on the 14C/C concentration of biomolecules in the upper latitude Northern Hemisphere. Turnover flattens the 14C/C bomb-pulse curve if all carbon atoms are equally likely to turnover in any given year. Average annual turnover rates of 0, 1, 5 and 10% are depicted.
The diets of a study population need to be considered when interpreting individual data and when constructing models [46]. Significant marine components to the diet have an influence on δ13C and 14C/C. Geographical differences in δ13C of specific biomolecules due to food sources are reported [47,48]. In practice, subject de-identification and the fact that many samples are acquired at autopsy prevent detailed knowledge of diet or specific stresses that may have influenced carbon resorption or scavenging mechanisms. As differences in cell turnover between healthy and disease states start to be studied, human subject recruitment protocols will include criteria that constrain some of these uncertainties.
Conclusion
Bomb-pulse dating in cell and molecular biology is challenging. Obtaining clean separations of specific biomolcules has been the most difficult task. Every system is different, so sorting parameters change among cell types. We have found that markers touted as specific in the literature can be generic, common to many cell types. Samples tend to be small and more challenging to analyze by AMS than typical samples. Small samples are more sensitive to contamination so relevant backgrounds and controls need to be included that can detect retention of solvents. Independent analyses for molecular purity should also be performed since AMS does not distinguish carbon sources. Despite the challenges, bomb-pulse dating possesses tremendous power. No other technique can measure molecular or cellular turnover at such high precision in humans over decades.
Acknowledgments
Support was provided by grants from the National Center for Research Resources (5P41RR013461-14) and the National Institute of General Medical Sciences (8 P41 GM103483-14) from the National Institutes of Health, NEI 5R21EY018722 and LLNL LDRD (10-LW-033). This work performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stuiver M, Reimer PJ, Baziunas TF. Radiocarbon. 1998;40:1127. [Google Scholar]
- 2.Hua Q, Barbetti M. Radiocarbon. 2004;46:1273. [Google Scholar]
- 3.Levin I, Kromer B. Radiocarbon. 2004;46:1261. [Google Scholar]
- 4.Graven HD, Guilderson TP, Keeling RF. J Geophys Res-Atmos. 2012;117:D02302. [Google Scholar]
- 5.Levin I, Naegler T, Kromer B, Diehl M, Francey RJ, Gomez-Pelaez AJ, Steele LP, Wagenbach D, Weller R, Worthy DE. Tellus. 2010;62B:26. [Google Scholar]
- 6.Rafter TA, Fergusson GJ. Science. 1957;126:557. doi: 10.1126/science.126.3273.557. [DOI] [PubMed] [Google Scholar]
- 7.Broecker WS, Schubert A, Olson EA. Science. 1959;130:331. doi: 10.1126/science.130.3371.331-a. [DOI] [PubMed] [Google Scholar]
- 8.Libby WF, Berger R, Ross JF, Alexander GV, Mead JF. Science. 1964;146:1171. doi: 10.1126/science.146.3648.1170. [DOI] [PubMed] [Google Scholar]
- 9.Nydal R, Lovesth K, Syrstad O. Nature. 1971;232:418. doi: 10.1038/232418a0. [DOI] [PubMed] [Google Scholar]
- 10.Harkness DD, Walton A. Nature. 1972;240:302. doi: 10.1038/240302a0. [DOI] [PubMed] [Google Scholar]
- 11.Stenhouse MJ, Baxter MS. Nature. 1977;267:828. doi: 10.1038/267828a0. [DOI] [PubMed] [Google Scholar]
- 12.Druffel EM, Mok HYI. Radiocarbon. 1983;25:629. [Google Scholar]
- 13.Shapiro SD, Endicott SK, Province MA, Pierce JA, Campbell EJ. J Clin Invest. 1991;87:1828. doi: 10.1172/JCI115204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovell MA, Robertson JD, Buchholz BA, Xie CS, Markesbury WR. Neurbiology Aging. 2002;23:179. doi: 10.1016/s0197-4580(01)00281-0. [DOI] [PubMed] [Google Scholar]
- 15.Lynnerup N, Kjeldsen H, Heegaard S, Jacobsen C, Heinemeier J. PLoS ONE. 2008;3:e1529. doi: 10.1371/journal.pone.0001529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonçalves I, Stenström K, Skog G, Mattsson S, Nitulescu M, Nilsson J. Circ Res. 2010;106:1174. doi: 10.1161/CIRCRESAHA.109.211201. [DOI] [PubMed] [Google Scholar]
- 17.Hägg S, Salehpour M, Noori P, Lundström J, Possnert G, Takolander R, Konrad P, Rosfors S, Ruusalepp A, Skogsberg J, Tegnér J, Björkegren J. PLoS ONE. 2011;6:e18248. doi: 10.1371/journal.pone.0018248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisén J. Cell. 2005;122:133. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 19.Bhardwaj RD, Curtis MA, Spalding KL, Buchholz BA, Fink D, Björk-Eriksson T, Nordborg C, Druid H, Eriksson PS, Frisén J. PNAS. 2006;103:12564. doi: 10.1073/pnas.0605177103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergman OI, Blomqvist L, Hoffstedt J, Näslund E, Britton T, Concha H, Hassan M, Rydén M, Frisén J, Arner P. Nature. 2008;453:783. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 21.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisén J. Science. 2009;324:98. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perl S, Kushner JA, Buchholz BA, Meeker AK, Stein GM, Hsieh M, Kirby M, Pechhold S, Liu EH, Harlan DM, Tisdale JF. J Clin Endocrinol Metab. 2010;95:E234. doi: 10.1210/jc.2010-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helsinki Declaration, World Medical Association. 59th WMA General Assembly; Seoul, Korea. October 2008; [accessed 23 August 2012]. http://www.wma.net/en/30publications/10policies/b3/index.html. [Google Scholar]
- 24.Trials of War Criminals before the Nuremberg Military Tribunals under Control Council Law No 10. Vol. 2. Washington, D.C: U.S. Government Printing Office; 1949. Nuremberg Code; pp. 181–182. [Google Scholar]
- 25.Buchholz BA, Freeman SPHT, Haack KW, Vogel JS. Nucl Instr & Meth, B. 2000;172:404. [Google Scholar]
- 26.Zermeño P, Kurdyla DK, Buchholz BA, Heller SJ, Kashgarian M, Frantz BR. Nucl Instr & Meth, B. 2004;223–24:293. [Google Scholar]
- 27.Arner P, Bernard S, Salepour M, Possnert G, Leibl J, Steier P, Buchholz BA, Eriksson M, Arner E, Hauner H, Skurk T, Ryden M, Frayn KN, Spalding KL. Nature. 2011;478:110. doi: 10.1038/nature10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaines P, Wojchowski DM. Biotechniques. 1999;26:683. doi: 10.2144/99264st04. [DOI] [PubMed] [Google Scholar]
- 29.Fong CY, Peh GSL, Gauthaman K, Bongso A. Stem Cell Rev and Rep. 2009;5:72. doi: 10.1007/s12015-009-9054-4. [DOI] [PubMed] [Google Scholar]
- 30.Gerashchenko BI. Cytometry Part A. 2011;79A:179. doi: 10.1002/cyto.a.21021. [DOI] [PubMed] [Google Scholar]
- 31.Falso MJS, Buchholz BA, deVere White RW. Anticancer Research. 2012;32:733. [PMC free article] [PubMed] [Google Scholar]
- 32.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CHM, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer Res. 2006;66:9339. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 33.Visvader JE, Lindeman GJ. Nature Rev Cancer. 2008;8:755. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 34.Nelson DE. Science. 1991;251:552. doi: 10.1126/science.1990430. [DOI] [PubMed] [Google Scholar]
- 35.Hodgins G. Dating photographs using the bomb-spike. Nucl Instr & Meth, B this volume. 2012:XXX. [Google Scholar]
- 36.Pearson A, McNichol AP, Benitez-Nelson BC, Hayes JM, Eglinton TI. Geochimica et Cosmochimica Acta. 2001;65:3123. [Google Scholar]
- 37.Smittenberg RH, Hopmans EC, Schouten S, Sinninghe Damsté JS. J Chromatogr A. 2002;978:129. doi: 10.1016/s0021-9673(02)01427-9. [DOI] [PubMed] [Google Scholar]
- 38.Tripp JA, McCullagh JSO, Hedges REM. J Sep Sci. 2006;29:41. doi: 10.1002/jssc.200500247. [DOI] [PubMed] [Google Scholar]
- 39.McCullagh JSO, Marom A, Hedges REM. Radiocarbon. 2010;52:620. [Google Scholar]
- 40.Buchholz BA, Haack KW, Sporty JL, Buckpitt AR, Morin D. Nucl Instr & Meth, B. 2010;268:1324. doi: 10.1016/j.nimb.2009.10.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liebl J. 14C bomb peak dating of human DNA samples at the microgram level. Nucl Instr & Meth, B this volume. 2012:XXX. [Google Scholar]
- 42.Wacker L. A versatile gas interface for routine radiocarbon analyses with a gas ion source. Nucl Instr & Meth, B this volume. 2012:XXX. [Google Scholar]
- 43.Ognibene TJ, Salazar GA. Instillation of hybrid ion source on the 1-MV LLNL BioAMS spectrometer. Nucl Instr & Meth, B this volume. 2012:XXX. doi: 10.1016/j.nimb.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salehpour M, Hakansson K, Possnert G. Nucl Instr & Meth, B this volume. 2012:XXX. [Google Scholar]
- 45.Bernard S, Frisén J, Spalding KL. Nucl Instr & Meth, B. 2010;268:1295. [Google Scholar]
- 46.Georgiadou E, Stenström K. Radiocarbon. 2010;52:800. [Google Scholar]
- 47.Slatkin DN, Friedman L, Irsa AP, Micca PL. Science. 1985;228:1004. doi: 10.1126/science.4001927. [DOI] [PubMed] [Google Scholar]
- 48.Alkass K, Buchholz BA, Druid H, Spalding KL. Forensic Sci Inter. 2011;209:34. doi: 10.1016/j.forsciint.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogel JS. Radiocarbon. 1992;34:344. [Google Scholar]