Abstract
One of the most exciting recent advances in the epigenetic field is the discovery that 5-methylcytosine (5mC) in DNA can be iteratively oxidized by a family of proteins known as Tet proteins to generate 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). These 5mC derivatives can be further processed by thymine-DNA glycosylase (TDG) followed by base excision repair or by replication-dependent dilution leading to DNA demethylation. Given the similarity between 5mC and its oxidation derivatives, many of the conventional techniques used for 5mC analysis cannot distinguish between 5mC and 5hmC/5fC/5caC. Here, we describe 2D-TLC and mass spectrometry methods that we have successfully used in differentiating 5mC from its oxidative derivatives as well as in characterizing the enzymatic activity of Tet proteins both in vitro and in vivo.
1. INTRODUCTION
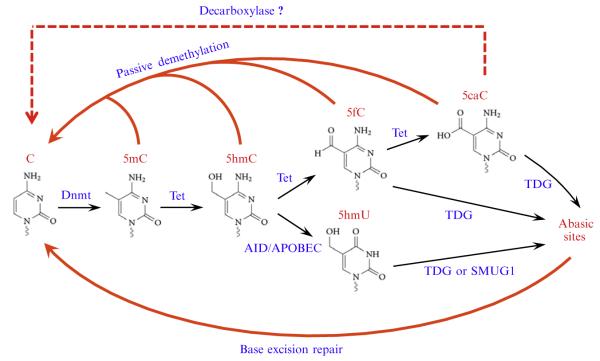
One of the recent advances in the epigenetic filed is the discovery that Tet family proteins are capable of catalyzing the oxidation of 5-methylcytosine (5mC), a well-characterized epigenetic mark, into 5-hydroxymethylcytosine (5hmC) in mammalian DNA (Ito et al., 2010; Tahiliani et al., 2009). Remarkably, more recent studies have shown that 5hmC can be further oxidized by Tet proteins to generate 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (He et al., 2011; Ito et al., 2011), which have also been detected in the mouse genome (Ito et al., 2011; Pfaffeneder et al., 2011). These new findings suggest that Tet protein-catalyzed iterative oxidation of 5mC could be the initial steps in DNA demethylation pathways (Fig. 5.1). Indeed, immunostaining of the zygotic DNA have shown that loss of 5mC in the male pronucleus correlates with the appearance of 5hmC, 5fC, and 5caC (Gu et al., 2011; Inoue, Shen, Dai, He, & Zhang, 2011; Inoue & Zhang, 2011; Iqbal, Jin, Pfeifer, & Szabo, 2011; Wossidlo et al., 2011), which are gradually diluted in a replication-dependent manner during mouse preimplantation development (Inoue et al., 2011; Inoue & Zhang, 2011). In addition to this passive demethylation process, both 5fC and 5caC, but not 5mC and 5hmC, can be actively removed from the genome by thymine-DNA glycosylase (TDG) through a base excision repair pathway (He et al., 2011; Maiti & Drohat, 2011). Thus, Tet-mediated iterative oxidation of 5mC plays an important role in regulating DNA methylation dynamics.
Figure 5.1.
Proposed models of Tet-initiated DNA demethylation pathways. DNA methylation (5mC) is established and maintained by DNA methyltransferases (Dnmt). In mammals, 5mC can be oxidized by the Tet proteins to generate 5hmC, 5fC, and 5caC. These 5mC derivatives, specifically 5fC and 5caC, can be actively removed through base excision repair initiated by the glycosylase TDG or can be passively removed through replication-dependent dilution during preimplantation development. 5hmC may also be deaminated by AID/APOBEC into 5hmU, which can be removed by TDG or SMUG1. In addition, a putative decarboxylase may directly convert 5caC to C.
However, conventional approaches for DNA methylation studies, including bisulfite genomic sequencing and methylation-sensitive restriction enzyme digestion, cannot discriminate 5mC from other 5mC oxidation products such as 5hmC (Huang et al., 2010; Jin, Kadam, & Pfeifer, 2010). It was also reported that 5fC and 5caC are interpreted as unmodified C in bisulfite genomic sequencing (Booth et al., 2012; He et al., 2011). To overcome these technical difficulties and to study Tet-mediated iterative oxidation, several techniques have been developed which allow quantification and genome-wide mapping of the cytosine derivatives. These techniques include (1) thin-layer chromatography (TLC) analysis of modified nucleotides (Ito et al., 2010; Kriaucionis & Heintz, 2009; Tahiliani et al., 2009), (2) liquid chromatography and mass spectrometry (LC-MS) analysis (Globisch et al., 2010; Ito et al., 2011; Munzel et al., 2010), (3) cytosine modification-specific antibodies (Ficz et al., 2011; Williams et al., 2011; Wu et al., 2011), (4) glucosylation of 5hmC (Kinney et al., 2011; Szwagierczak, Bultmann, Schmidt, Spada, & Leonhardt, 2010), (5) chemical/enzymatic labeling or conversion of modified cytosine (Booth et al., 2012; Pastor et al., 2011; Song, Szulwach, et al., 2011; Yu et al., 2012), and (6) single-molecule, real-time sequencing (Flusberg et al., 2010; Song, Clark, et al., 2011).
Here, we describe two methods that we have been using in studying the Tet enzymatic activity. The 2D-TLC method is easy to follow and does not require additional instruments, while the mass spectrometry method is more accurate and sensitive and can be used to quantify the levels of cytosine derivatives in genomic DNA.
2. EXPRESSION AND PURIFICATION OF TET PROTEINS
For production of recombinant proteins, cDNAs encoding the catalytic domains of mouse Tet1 (aa1367–2039), Tet2 (aa916–1921), Tet3 (aa697–1668), and their corresponding catalytic mutants (Ito et al., 2010) are cloned into a modified pFastBac-HTb (Invitrogen) insect cell expression vector inframe with a FLAG tag at the N-terminus. Constructs are transformed into DH10Bac Escherichia coli (Invitrogen) following manufacturer’s instructions to generate bacmid DNA. Since recombinant bacmid DNA is ≥ 135 kbp, most silica-gel-membrane-based plasmid miniprep kits are not suitable for bacmid DNA isolation, anion-exchange-resin-based kits should be used instead (e.g., PureLink HiPure Plasmid Miniprep Kit (Invitrogen)). Alternatively, we found that crude purification of bacmid DNA using conventional alkaline lysis followed by ethanol precipitation could also give satisfying results in the subsequent Sf9 cell transfections. To verify the successful transposition of Tet cDNAs to bacmid DNA, polymerase chain reaction (PCR) analysis is performed with pUC/M13 forward (5′-CCCAGTCACGACGTTGTAAAACG-3′) and reverse (5′-AGCGGATAACAATTTCACACAGG-3′) primers. A PCR band of ~2400 bp+ the size of the insert indicates successful transposition, otherwise, an ~350 bp band will show up. Once correct bacmid DNA is generated, baculovirus can be produced in Sf9 cells.
Sf9 cells are maintained at a density of 0.5–6×106 cells/ml in Sf-900 II SFM medium (Invitrogen) containing 10% fetal bovine serum in a spinner flask at 27 °C. Before transfection, cells are seeded in a six-well plate with 9×105 cells/well and allowed to attach for 1 h at 27 °C. Dilute 1 μg of bacmid DNA (or the amount from 100 μl of bacteria if crude bacmid DNA is used) and 8 μl of Cellfectin II (Invitrogen) separately with 100 μl of Grace’s Insect Medium (unsupplemented). Combine the diluted bacmid with diluted Cellfectin II reagent and incubate at room temperature for 30 min. While the transfection complexes are forming, remove the medium from the plate and wash cells once with 2 ml of Grace’s Insect Medium (unsupplemented). Remove the wash medium, add 800 μl of Grace’s Insect Medium (unsupplemented) to the transfection mixture and gently add the mixture onto the cells immediately. Incubate cells for 5 h at 27 °C before replacing the transfection mixture with 2 ml of complete growth medium (Sf-900 II SFM medium containing 10% fetal bovine serum). After 96 h, collect the medium and centrifuge at 500×g for 5 min to remove floating cells. Transfer the supernatant containing baculovirus to a new tube and store the P1 viral stock at 4 °C protected from light. To verify the successful production of virus, expression of recombinant Tet proteins in the transfected cells can be examined by Western blot, although the expression may be weak at this stage. To amplify the P1 viral stock, seed 2×106 cells/well in a six-well plate, add 50 μl of P1 viral stock, and incubate for 72 h. Collect the P2 viral stock as per the P1 stock. We usually further amplify the P2 viral stock to generate P3 viral stock of higher titer. Seed 9×106 cells/dish in a 10 cm dish, add 250 μl of P2 viral stock, and incubate for 72 h. The titer of the P3 viral stock is usually ~2×108 pfu/ml.
To express the Tet proteins, prepare 1 l of Sf9 cells in mid log growth phase (1.5–2×106 cells/ml) in a spinner flask, infect the cells with 20 ml P3 viral stock (the multiplicity of infection (MOI) is ~2) for 72 h. Cells are collected and washed once with ice-cold phosphate buffered saline. Cell pellet is then resuspended in 40 ml of LysisBuffer F (50 mM HEPES, 500 mM NaCl,2 mM MgCl2, 2 mM DTT, 0.2% NP-40, 20% glycerol, 1× protease inhibitors without EDTA (Roche), pH 8.0) and transferred to a Dounce homogenizer with a type A pestle. Homogenize slowly for 30 times over a 30-min period on ice and centrifuge the cell lysate at 14,000 ×g for 20 min at 4 °C. While centrifuging the cell lysate, wash 1 ml of 50% slurry of FLAG M2 beads (Sigma) with 20 ml Lysis Buffer F in a 50 ml conical tube, spin at 500×g for 5 min, remove the supernatant. Transfer the supernatant of the cell lysate to the tube and rotate at 4 °C for 3 h. After the incubation, collect beads by spinning at 500×g for 5 min at 4 °C, wash beads with 30 ml Wash Buffer (50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 20% glycerol) for three times, and elute the protein with 500 μl Wash Buffer containing 200 μg/ml 3×FLAG peptide (Sigma) by rotating at 4 °C for 30 min. Aliquot the eluted protein and store at −80 °C. The purity of the eluted protein can be determined by Coomassie staining of an SDS-PAGE.
3. TET ACTIVITY ASSAYS
3.1. Preparation of the substrates
To set up the in vitro enzymatic assays, proper DNA substrates are required. If the reaction products are to be analyzed by TLC assay, the modified cytosine should be placed in the context of a restriction site that can be digested regardless of the modification. Since TaqI (with a recognition site of TCGA) is insensitive to all cytosine modifications (Ito et al., 2011), we have been using double-stranded 20-mer DNA containing a TaqI site as our substrate with the following sequences:
Taq20-F: 5′-GTTCAGCTTXGATCACGCTC-3′
Taq20-R: 5′-GAGCGTGATXGAAGCTGAAC-3′
(X represents a modified cytosine)
However, if mass spectrometry is to be used as the detection method, the modified cytosine can be placed anywhere in the strand. Besides the above 20-mer DNA, we have also been using a double-stranded 38-mer DNA containing nine modified cytosines in each strand as in natural situation where the modified cytosines might be clustered together:
38mer-F: 5′-AGCCXGXGCXGXGCXGGTXGAGXGGCXGCT CCXGCAGC-3′
38mer-R: 5′-GCTGCXGGAGCXGCCXCTCXACCXGCXCXGC XCXGGCT-3′
Oligonucleotides containing 5mC and 5hmC can be directly purchased from Integrated DNA Technologies, while those containing 5fC and 5caC are synthesized as described (Dai & He, 2011). However, the phosphoramidite forms of both 5fC and 5caC have become commercially available recently from Glen Research. Therefore, oligonucleotides containing 5fC and 5caC can now be synthesized in most oligonucleotide synthesis facilities.
After synthesis of the oligonucleotides containing the desired modification, the oligonucleotides are annealed in 1×NEB Buffer 2 by boiling the mixture for 5 min and slowly cooling down to room temperature. High-quality double-stranded DNA can be further purified by polyacrylamide gel electrophoresis if necessary.
3.2. In vitro enzymatic assays
For in vitro enzymatic activity assays, 0.5 μg of various double-stranded DNA substrates are incubated with 1.2 μg of wild-type or catalytic mutant Tet proteins (~1:6 enzyme/substrate ratio if the 20-mer substrate is used; 50 μl reaction) in the presence of 50 mM HEPES (pH 7.9), 100 mM NaCl, 75 μM Fe(NH4)2(SO4)2, 2 mM ascorbate, 1 mM α-ketoglutarate (2-oxoglutarate), 1 mM DTT and 1 mM ATP at 37 °C for 40 min. Since Tet proteins are quickly inactivated under the reaction condition, it is necessary to shorten the incubation time to 2.5 min if kinetic studies are to be performed (Ito et al., 2011). The reactions are stopped by the addition of 10 volumes of ice-cold Buffer PN (QIAGEN), and the oligonucleotides are then purified using QIAquick Nucleotide Removal Kit (QIAGEN) following manufacturer’s instructions. Now, the oligonucleotides are ready to be analyzed by either 2D-TLC or mass spectrometry.
3.3. In vivo enzymatic assays in HEK293T cells
HEK293T cells contain low levels of endogenous 5hmC, 5fC, and 5caC. Since high transfection efficiency (>90%) and high Tet protein expression can be achieved in HEK293T cells, it provides an ideal system for analyzing enzymatic activity of Tet proteins in vivo. To overexpress Tet proteins in HEK293T cells, cDNAs encoding the full-length or catalytic domains of mouse Tet proteins and their corresponding catalytic mutants are cloned into a modified pcDNA3 (Invitrogen) mammalian cell expression vector in-frame with a FLAG tag at the N-terminus. 8×105 cells/well are seeded into a six-well plate on the day before transfection. The cells are transfected with 2 μg of Tet expression constructs with FuGENE HD (Roche; 1:3 plasmid/FuGENE HD ratio) following manufacturer’s instructions. 48 h after transfection, the transfected cells are harvested and genomic DNA is extracted using a DNeasy Blood & Tissue Kit (QIAGEN; include the optional RNase A treatment step during genomic DNA extraction and elute with Buffer EB). The genomic DNA is now ready to be analyzed by either 2D-TLC or mass spectrometry.
4. ANALYSIS OF THE CYTOSINE DERIVATIVES BY 2D-TLC
TLC is a classic method that separates different nucleotides based on their differential migration rates on TLC plates. TLC assays have been used successfully for analyzing 5hmC in previous studies (Koh et al., 2011; Kriaucionis & Heintz, 2009; Tahiliani et al., 2009). However, under previous TLC conditions, 5hmC and 5fC have almost identical migration patterns, and 5caC fails to migrate (Ito et al., 2011). To overcome this technical problem, we developed a modified 2D-TLC assay using a more acidic buffer as the second developing condition. For 2D-TLC analysis, the in vitro enzymatic reaction products (or 10 μg of genomic DNA) are digested with 40 U of TaqI (use 100 U for genomic DNA) overnight, and then 20 U of calf intestinal alkaline phosphatase (CIAP) are added, followed by two more hours of incubation. The digested DNA is purified with QIAquick Nucleotide Removal kit. The DNA is then endlabeled with 40 U of T4 polynucleotide kinase and 15 μCi of [γ-32P]-ATP (25 Ci/mmol) for 1 h, ethanol-precipitated, and redissolved in 40 μl of water. Then, the labeled DNA is heat-denatured and digested into nucleotides with 200 U of nuclease S1 (Sigma) in the presence of 0.5 mM ZnSO4, 14 mM sodium acetate (pH 5.2) at 37 °C for 2 h (total volume is 50 μl) before being analyzed by 2D-TLC.
For 2D-TLC assay, 1.5 μl of the digestion product is spotted on one corner of a 20×20 cm Polyethyleneimine cellulose F TLC plate (Merck), and the TLC plate is developed using TLC Buffer 1 (isobutyric acid: NH4OH: H2O=66:2:20). After complete drying, the TLC plate is developed in the second direction using TLC Buffer 2 (isopropanol: HCl: H2O 70:15:15). After drying, the TLC plate is exposed to an X-ray film for autoradiography or a storage phosphor screen for better quantitation. A representative 2D-TLC assay result is shown in Fig. 5.2. To confirm the migration patterns of the cytosine derivatives, standard nucleosides (Berry & Associates) can be labeled with deoxycytidine kinase (Proteinkinase.de) in the presence of [γ-32P]-ATP and analyzed in parallel with the samples using the same 2D-TLC procedure.
Figure 5.2.

A representative autoradiograph of 2D-TLC analysis of the enzymatic reaction product derived from 5mC-containing 20-mer oligo DNA incubated with Tet2 protein.
5. ANALYSIS OF THE CYTOSINE DERIVATIVES BY MASS SPECTROMETRY
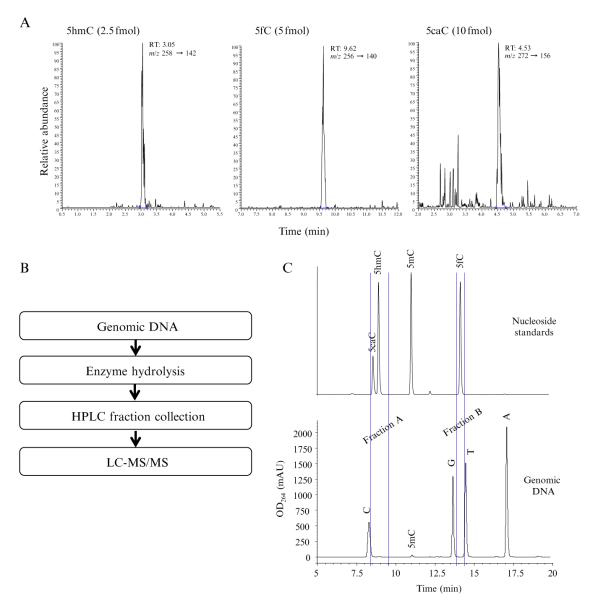
For quantitative analysis of the cytosine derivatives, we developed a sensitive and specific method using liquid chromatography-tandem mass spectrometry (LC-MS/MS) with multiple reactions monitoring (MRM) to simultaneously detect and quantitate cytosine derivatives (Ito et al., 2011). The instrument we have been using is an ultra-performance liquid chromatography system (Waters) coupled to a TSQ-Quantum Ultra triple-quadrupole mass analyzer (ThermoFinnigan) with a heat-assisted electrospray ionization source, which can detect and quantitate as low as 10 fmol of cytosine derivatives (Fig. 5.3A).
Figure 5.3.
Analysis of the cytosine derivatives using mass spectrometry. (A) Representative LC-MS/MS chromatograms at the limits of detection for 5hmC (2.5 fmol), 5fC (5 fmol), and 5caC (10 fmol), showing excellent signal-to-noise ratio. (B) Schematic representation of the procedures used to quantify endogenous cytosine derivatives in genomic DNA. (C) HPLC analysis of nucleoside standards (top panel) and hydrolyzed genomic DNA. The fractions corresponding to the retention time of 5hmC/5caC (fraction A) and 5fC (fraction B) are collected for further LC-MS/MS analysis.
To prepare the samples, in vitro enzymatic reaction products or 2.5 μg of genomic DNA are heat-denatured, hydrolyzed with 90 U of nuclease S1 in Buffer 1 (0.5 mM ZnSO4, 14 mM sodium acetate, pH 5.2) at 37 °C for at least 1 h (total volume is 44.5 μl), followed by the addition of 5 μl 10×Buffer 2 (560 mM Tris–HCl, 30 mM NaCl, 10 mM MgCl2, pH 8.3), 0.5 μg of phosphodiesterase I and 2 U of CIAP for an additional 1 h (final volume is 50 μl). Then, the digested DNA is filtered with Nanosep3K to remove enzymes and undigested DNA (if any). For the in vitro enzymatic reaction products, dilute the samples to 200 μl, and subject 10 μl of the diluted samples to LC-MS/MS analysis. For genomic DNA, directly subject 10 μl of the filtered samples to LC-MS/MS analysis.
Separation prior to mass spectrometry is achieved using a 2.1×100 mm HSS T3 1.8 μm column (Waters) with gradient elution at a flow rate of 0.2 ml/min using 0.02% acetic acid in water as mobile phase A and methanol as mobile phase B. The gradient is 0→3.5 min, 3% B, 3.5→12.5 min, 3%→16.2% B, 12.5→13 min, 16.2%→30% B, 13→15 min, 30% B, 15→16 min, 30%→3% B, 16→20 min, 3% B. The mass spectrometer is set to positive mode, precursor to product ion transitions for cytosine and its derivatives are monitored using the parameters in Tables 5.1 and 5.2. External standard calibration is used for the quantitation. To correct for matrix effects, the calibration curves for nucleoside standards (Berry & Associates) are generated by mixing the nucleosides with the enzymes in the digestion buffer and then filtered using the same procedures used for the samples. Calibration curves are constructed for each batch of analysis. It is also important to include one standard sample every 10 samples to check the signal stability of the instrument.
Table 5.1.
MRM transition parameters for monitoring cytosine and its derivatives
| Nucleoside | Transition (m/z) | Collision energy (eV) |
|---|---|---|
|
| ||
| C | 228→112 | 15 |
|
| ||
| 5mC | 242→126 | 15 |
|
| ||
| 5hmC | 258→142 | 15 |
|
| ||
| 5fC | 256→140 | 15 |
|
| ||
| 5caC | 272→156 | 15 |
Table 5.2.
Electrospray parameters for the mass spectrometer
| ESI mode | Positive |
|---|---|
|
| |
| Spray voltage | 3000 V |
|
| |
| Vaporizer temperature | 250 °C |
|
| |
| Capillary temperature | 285 °C |
|
| |
| Sheath gas flow rate | 35 arb |
|
| |
| AUX gas flow rate | 25 arb |
6. ANALYSIS OF THE ENDOGENOUS LEVEL OF CYTOSINE DERIVATIVES BY MASS SPECTROMETRY
For endogenous 5fC and 5caC in genomic DNA, they are first enriched by HPLC fractionation before being analyzed by mass spectrometry (Fig. 5.3B and C) (Ito et al., 2011). Before analyzing genomic DNA, the retention time for each of the cytosine derivatives needs to be determined using standard nucleosides (Fig. 5.3C). We have noticed that the retention time of 5caC is sensitive to pH of the digestion buffer and the mobile phase, thus the standard nucleosides should be prepared in the sample digestion buffer before HPLC analysis, and the pH of mobile phase A should also be accurately adjusted. To avoid carry-over of the standard nucleosides, it is necessary to make several blank injections before sample analysis. We also include the fractions from the last blank injection in the subsequent LC-MS/MS analysis to make sure there is no carry-over.
To enrich 5fC and 5caC, 20–100 μg of genomic DNA is heat-denatured, hydrolyzed with nuclease S1 (20 U/μg of DNA) in Buffer 1 (0.5 mM ZnSO4, 14 mM sodium acetate, pH 5.2) at 37 °C for at least 1 h (total volume is 250 μl), followed by the addition of 30 μl 10× Buffer 2 (560 mM Tris–HCl, 30 mM NaCl, 10 mM MgCl2, pH 8.3), 5 μg of phosphodiesterase I and 20 U of CIAP for an additional 1 h (final volume is 300 μl), and filtered with Nanosep3K to remove enzymes and undigested DNA (if any). Then, 275 μl of the filtered samples are subjected to HPLC (Agilent 1200 with a 4.6×150 mm, 3 μm, Atlantis T3 column) with gradient elution at a flow rate of 0.8 ml/min using 5 mM ammonium formate in water (pH 4.0) as mobile phase A and methanol as mobile phase B. The gradient is 0→15 min, 3%→30% B, 15→17 min, 30% B, 17→18 min, 30%→3% B, 18→25 min, 3% B. During the HPLC separation, fractions corresponding to the retention time of 5hmC, 5fC, and 5caC are collected for LC-MS/MS quantitation, while C and 5mC are directly quantitated with external standard curves using the UV detector signals (Fig. 5.3C). The collected fractions are then dried with a Speed-Vac and redissolved with 20 μl of water. Of which, 15 μl is subjected to LC-MS/MS analysis as described above. External standard calibration is used for the quantitation. We suggest make new calibration curve for each batch of analysis and include one standard for every 10 samples to confirm the signal stability of the instrument.
Considering the sample loss during sample enrichment, the recovery rates of 5hmC, 5fC, and 5caC need to be determined and used to correct the percentages of the cytosine derivatives in total cytosines. To determine the recovery rates, standard 5hmC, 5fC, and 5caC nucleosides are mixed at five different levels with 15 nmol (if 20 μg of genomic DNA is analyzed) each of C, G, T, A in the digestion buffer, then the standard mixtures are analyzed by the same procedures as that used for genomic DNA samples. Finally, measured percentages (cytosine derivative/total cytosine) in the standard mixtures are plotted against the actual percentages. The recovery rates determined are usually ~50% for all the cytosine derivatives and can be used to correct the measured results for genomic DNA samples.
ACKNOWLEDGMENTS
We thank Dr. James A. Swenberg and Leonard B. Collins (UNC) for their help in the development of the mass spectrometry method. Y. Z. is a HHMI Investigator. This work was supported by NIH (GM68804 and U01DK089565) and the HHMI.
REFERENCSE
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Dai Q, He C. Syntheses of 5-formyl- and 5-carboxyl-dC containing DNA oligos as potential oxidation products of 5-hydroxymethylcytosine in DNA. Organic Letters. 2011;13:3446–3449. doi: 10.1021/ol201189n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nature Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLos One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PloS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Research. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Research. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney SM, Chin HG, Vaisvila R, Bitinaite J, Zheng Y, Esteve PO, et al. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. The Journal of Biological Chemistry. 2011;286:24685–24693. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. The Journal of Biological Chemistry. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, et al. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angewandte Chemie (International Ed. in English) 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angewandte Chemie (International Ed. in English) 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- Song CX, Clark TA, Lu XY, Kislyuk A, Dai Q, Turner SW, et al. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nature Methods. 2011;9:75–77. doi: 10.1038/nmeth.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature Biotechnology. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Research. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the Mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nature Communications. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes & Development. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]