Abstract
Background and Aims
Spontaneous male sterility is an advantageous trait for both constructing efficient pollination control systems and for understanding the developmental process of the male reproductive unit in many crops. A triallelic genetic male-sterile locus (BnMs5) has been identified in Brassica napus; however, its complicated genome structure has greatly hampered the isolation of this locus. The aim of this study was to physically map BnMs5 through an integrated map-based cloning strategy and analyse the local chromosomal evolution around BnMs5.
Methods
A large F2 population was used to integrate the existing genetic maps around BnMs5. A map-based cloning strategy in combination with comparative mapping among B. napus, Arabidopsis, Brassica rapa and Brassica oleracea was employed to facilitate the identification of a target bacterial artificial chromosome (BAC) clone covering the BnMs5 locus. The genomic sequences from the Brassica species were analysed to reveal the regional chromosomal evolution around BnMs5.
Key Results
BnMs5 was finally delimited to a 0·3-cM genetic fragment from an integrated local genetic map, and was anchored on the B. napus A8 chromosome. Screening of a B. napus BAC clone library and identification of the positive clones validated that JBnB034L06 was the target BAC clone. The closest flanking markers restrict BnMs5 to a 21-kb region on JBnB034L06 containing six predicted functional genes. Good collinearity relationship around BnMs5 between several Brassica species was observed, while violent chromosomal evolutionary events including insertions/deletions, duplications and single nucleotide mutations were also found to have extensively occurred during their divergence.
Conclusions
This work represents major progress towards the molecular cloning of BnMs5, as well as presenting a powerful, integrative method to mapping loci in plants with complex genomic architecture, such as the amphidiploid B. napus.
Keywords: Brassica napus, Brassica rapa, Brassica oleracea, genetic male sterility, triallelic genic male-sterile locus BnMs5, comparative mapping, map-based cloning
INTRODUCTION
Male sterility has been used as one of the most important pollination control systems in rapeseed. The application potential of a male sterility system, however, mainly relies on its reliability and effectiveness. In principle, only those mutants with stable and complete sterility, the availability of 100 % sterile population and no restriction of restorers could be potentially applied in a commercial production. A genetic male sterility (GMS) line Rs1046AB could qualify these requirements, thereby regarded as a possible alternative to the extensively used Polima cytoplasmic male sterility (Pol CMS) in China. The male sterility in Rs1046AB was transferred from a spontaneous mutation in Yi3A (Li et al., 1985). It was observed that approximately half of the plants in Rs1046AB remain male-sterile while the rest are completely fertile (Lu et al., 2004; Hong et al., 2006). Cytological observations showed that the development of microspore mother cells (MMCs) in Rs1046A is arrested at least during the early stage of meiosis I (Wu and Yang, 2008), leading to a relatively stable and complete male sterility phenotype. Systematic genetic analysis and molecular marker assays have shown that the male sterility of the GMS line Rs1046AB inherits according to a monogenically multiallelic locus with three different alleles (Song et al., 2005, 2006a, b; Liu et al., 2008). A similar hereditary mode has also been reported to be involved with different male sterility loci in Brassica rapa (Feng et al., 2009) and Brassica napus (Dong et al., 2012). These cases reflect the fact that the multiple-allele hereditary mode is a common phenomenon in Brassica species.
To differentiate this male-sterile locus from those of other spontaneous GMS lines in B. napus, such as BnMs1 and BnMs2 in S45AB (Pan et al., 1988), BnMs3 and BnMs4 in 9012AB (Chen et al., 1998; possibly identical to the MSL system and the Syngenta-patented NMS system discussed by Li et al., 2012), this locus has been uniformly addressed as BnMs5. Accordingly, the three alleles of this locus are individually assigned as BnMs5a (restorer type, corresponding to the previous Rf or Mf), BnMs5b (male-sterile type, corresponding to the previous Ms) and BnMs5c (normal male-fertile type, corresponding to the previous ms), with the dominance relationship of BnMs5a > BnMs5b > BnMs5c (Song et al., 2006a). Thus, plants carrying the genotype of BnMs5bBnMs5b or BnMs5bBnMs5c would be sterile, while other plants homozygous for BnMs5c or carrying the BnMs5a allele will be fertile. Consistently, genotypes of the homozygous restorer lines, male sterility lines and maintainer lines are BnMs5aBnMs5a, BnMs5bBnMs5b and BnMs5cBnMs5c, respectively. The cross between a homozygous male sterility line and a maintainer line will yield a complete male-sterile population (BnMs5bBnMs5c). It then can be used as the female line, and gives birth to fertile F1 hybrids (BnMs5aBnMs5c or BnMs5aBnMs5b) when mating with the male line (restorers).
Although the amphidiploid B. napus represents a complicated genome derived from the natural hybridization between ancestors of the diploid B. rapa (A genome donor) and B. oleracea (C genome donor), map-based cloning is still an effective way for the isolation of genes responsible for obvious phenotypic variation v (Yi et al., 2010; Dun et al., 2011; Li et al., 2012). The first step in map-based cloning is the construction of a high-resolution genetic map around the target gene. Towards this goal, some attempts have been made to enhance the molecular marker density flanking BnMs5. For example, Lu et al. (2004) firstly targeted the BnMs5b allele to a genetic interval of 9·6 cM via a backcross population. Hong et al. (2006) developed several BnMs5a-linked sequence-characterized amplified region (SCAR) markers by comparison of two near isogenic lines (NILs). Using another similar GMS line 609AB, Song et al. (2006b) co-localized BnMs5a and BnMs5b, providing the first molecular marker proof for allelism between the previously designated Ms and Mf genes. Later, a high-density genetic map based on a new F2 population was established, in which the BnMs5 locus co-segregating with one marker is bracketed by the closest flanking markers with respective distance of 0·1 and 1·2 cM (Liu et al., 2008). In conclusion, there seem to be substantial markers around BnMs5; however, it is difficult to estimate their arrangement order with the target gene based solely on their respective genetic distances from BnMs5 in the maps mentioned above. Moreover, it remains unclear whether these markers are informative enough for constructing a fine physical map of BnMs5. The second step in map-based cloning is the physical identification of the genomic region covering the gene of interest. As the genome sequence of B. napus is not yet available, the physical delimitation of the gene depends mainly on the recovery of large insert DNA clones, such as bacterial artificial chromosomes (BACs). However, due to the existence of multiple copies of a conserved block inherited from the ancestral karyotype (Parkin et al., 2005; Schranz et al., 2006), validation of target B. napus BAC clones is always complicated. Fortunately, with the release of the B. rapa reference genome sequence (Wang et al., 2011) and the availability of the Brassica oleracea Genome Database (Bolbase), this difficulty can be overcome to a great extent by comparative mapping between B. napus and B. rapa or B. oleracea.
Here we report on the physical delimitation of BnMs5 to a 21-kb interval on a B. napus BAC clone by a map-based cloning strategy, and the comparative sequence analysis around the target locus among closely related Brassica species.
MATERIALS AND METHODS
Plant materials and population construction
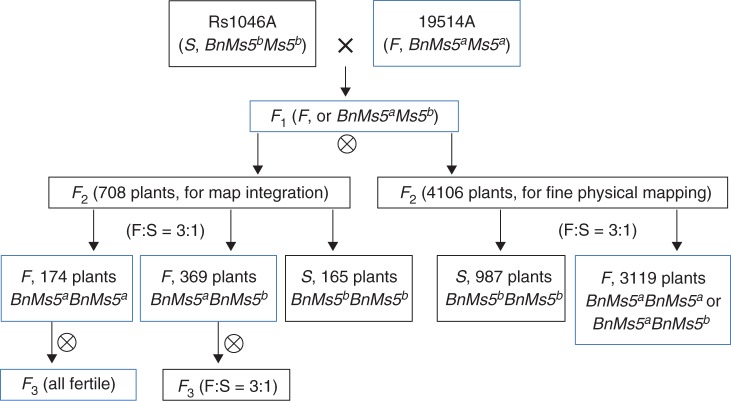
The homozygous two-type B. napus GMS line Rs1046AB (A and B individually represent the male-sterile and male-fertile plants) has been described previously (Lu et al., 2004). Rs1046AB can be maintained by sibmating its sterile plants (BnMs5bBnMs5b) with fertile plants (BnMs5aBnMs5b), always resulting in 1 : 1 fertility segregation progenies (Hong et al., 2006). The Pol CMS restorer line 7-5 (BnMs5cBnMs5c) bred in our group is a temporary maintainer of Rs1046A, while another double haploid (DH) line 19514A (a temperature-sensitive Pol CMS line, genotyped as BnMs5aBnMs5a) is a restorer of Rs1046A (Liu et al., 2008). Using the 190 F2 plants derived from the cross between Rs1046A and 19514A, Liu et al. (2008) achieved a high-density genetic map of BnMs5. For the sake of improving the resolution of this map, we enlarged this F2 population to a total number of 4814 (including the above 190 plants, outlined in Fig. 1). From this collection, 708 plants were randomly selected to construct an integrated genetic map of previously published markers, with all the fertile plants selfed to get their F3 progenies. For the remaining 4106 F2 plants, only sterile plants were chosen for the identification of recombination events between tightly associated markers and BnMs5. Individual fertility of all the F2 plants and F3 progenies were visually examined at the flowering stage.
Fig. 1.
A schematic diagram of the F2 population developed for map integration and fine mapping of the BnMs5 locus (F, fertile; S, sterile). ⊗ indicates selfing of the selected plants.
DNA extraction, RNA isolation and polymerase chain reaction (PCR)
Genomic DNA was extracted individually from young leaves at seedling stage according to a modified CTAB method (Doyle and Doyle, 1987). Every DNA sample was adjusted to a working concentration of 25 ng μL−1. Total RNA was purified using Trizol® (Invitrogen, Carlsbad, CA, USA) mainly from buds over four different developmental stages as described by Wan et al. (2010), such as premeiosis stage (bud length ≤1 mm), meiosis stage (1 mm ≤ bud length ≤ 2 mm), tetrad to vacuolated microspore stage (2 mm ≤ bud length ≤ 3·5 mm) and bicellular to late pollen grain stage (3·5 mm ≤ bud length ≤ 5 mm). First-strand cDNA was synthesized using RevertaidTM First Strand cDNA Synthesis Kit (Fermentas, Lithuania) according to the manufacturer's instruction.
The amplified fragment length polymorphism (AFLP) procedure was performed following the protocol described by Lu et al. (2004). PCR amplification with SCAR as well as insertion/deletion (In/Del) primers was prepared according to the protocol described by Li et al. (2012). Semi-quantitative reverse transcriptase PCR (RT-PCR) analysis was performed with specific primers designed from the candidate genes. Expression of the β-actin gene was used as a positive internal control. The semi-RT PCR programmes were as follows: denaturation at 94 °C for 2 min and then 32 cycles of amplification (94 °C for 30 s, variable annealing temperature depending on specific primer sets for 30 s, and 72 °C for 60 s) followed by a final extension at 72 °C for 10 min. All the PCR products were resolved on 1·5 % agarose gel or 6 % denaturing polyacrylamide gel (PAGE).
Integration of genetic maps around BnMs5
All the previously published BnMs5-associated markers were first evaluated among the parental lines Rs1046A and 19514A, as well as the homozygous fertile and sterile bulk made up from the F2 plants (Liu et al., 2008). In cases where known markers could not display a polymorphism, new primers were designed based on the markers' flanking sequences. These polymorphic markers were then analysed in all the 708 F2 plants. Data including the individual phenotype and the marker genotype were combined for linkage analysis using the software package MAPMAKER/EXP 3·0 (Lander et al., 1987; Lincoln et al., 1992), with the same parameter setting as described by Liu et al. (2008). Genetic distances were converted into centiMorgans (cM) using the Kosambi mapping function (Kosambi, 1944). A local genetic linkage map of BnMs5 was drawn using Mapdraw v2·5 (Liu and Meng, 2003). Some BnMs5-associated SCAR markers have also been anchored to a B. napus universal genetic map based on the TNDH population (Qiu et al., 2006; Shi et al., 2009), to determine the chromosome localization of BnMs5 indirectly.
Comparative mapping and development of markers from the collinear region
Sequences of the markers located on the integrated linkage map were recruited as queries to search homologous Arabidopsis genes (TAIR, http://www.arabidopsis.org) by using the BLASTN program with the parameters suggested by Parkin et al. (2005). The homologous B. rapa and B. oleracea genes for each marker were also identified from the web-based database BRAD (http://brassicadb.org/brad/index.php, chromosome v1·1; Cheng et al., 2011), Bolbase (http://119.97.203.210/bolbase/index.html) and NCBI (http://www.ncbi.nlm.nih.gov/) through BLASTN, except for the cutoff of E-value ≤1e-10. In Arabidopsis, only the orthologues with the highest E-value was analysed, while all the top-three orthologues in B. rapa and B. oleracea were considered and further assigned to the different subgenomes (Wang et al., 2011) of B. rapa and B. oleracea by referring to their annotations online (BRAD and Bolbase). The linear arrangement relationship of these molecular markers in the B. napus genome was compared with that of their homologues in the Arabidopsis, B. rapa and B. oleracea genomes, to decide whether there is a collinear region around BnMs5 in these related species.
Construction of a physical map and analysis of candidate genes
The JBnB BAC library (Rana et al., 2004), constructed from genomic DNA of a winter-type European cultivar ‘Tapidor’ (genotyped as BnMs5cBnMs5c, data not shown), was employed for construction of the physical map around BnMs5. A Southern blotting method (Rana et al., 2004) was applied to screen the candidate BAC clones but at a high stringency condition for hybridization (65 °C) and washing (0·1× SSC and 0·1 % SDS at 65 °C). Probes were amplified from the genomic DNA of ‘Tapidor’ and sequenced to confirm the authenticity. Plasmid DNA of the resulting positive BAC clones was prepared using the QIAGEN Large-Construct Kit (Qiagen, Valencia, CA, USA). The positive BAC clones were effectively grouped based on their DNA fingerprints as revealed by a PCR-based strategy. These PCR primers were designed from the B. rapa and B. oleracea genes orthologous to the Arabidopsis genes from At1g10180 to At1g10400. For each locus, three reverse primers were individually developed to specifically match the predicted genes of B. rapa, B. oleracea and the conservation region between them, while the only forward primer always anchors to the conservation region. The BAC clone carrying the target locus was then determined by a comprehensive evaluation, including PCR analysis with a BnMs5-associated co-dominant marker, sequence alignment of PCR products and comparative mapping with the two diploid genomes. The positive BAC clones were shotgun sequenced or partially sequenced by a long-fragment PCR method using Phusion® High-Fidelity DNA Polymerase (Finnzymes Oy, Espoo, Finland). The website-based software FGENSH (http://www.softberry.com) was employed to predict the putative open reading frames (ORFs) located in the target region. The Plant Repeat Databases (http://plantrepeats.plantbiology.msu.edu/search.html) was searched to reveal the potential transposon elements from the candidate genomic region. The coding sequences of predicted genes were analysed through BLASTN or BLASTX in TAIR for gene function annotation.
RESULTS
Fertility segregation in the F2 population
From the 708 F2 plants, 543 were male-fertile while 165 were male-sterile. Fertility survey in the F3 progenies showed that 174 lines were all fertile while the other 369 lines showed a fertile-sterile segregation (Fig. 1). The distribution of three genotypes (BnMs5aBnMs5a, BnMs5aBnMs5b, BnMs5bBnMs5b) in the F2 population was statistically consistent with the ratio of 1 : 2 : 1 (χ2 = 1·500, P = 0·472). Similarly, 987 male-sterile plants were picked up from the other 4106 F2 plants (Fig. 1), indicating that only one locus is responsible for the fertility segregation in this population (χ21:3 = 2·027, P = 0·155).
Integration of different genetic maps around BnMs5
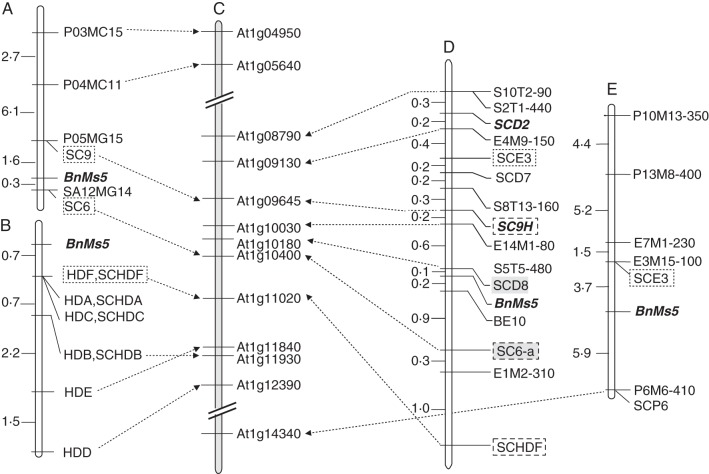
In total, 190 of the 708 F2 plants were used to map the BnMs5 locus previously, and 20 AFLP markers with relatively even arrangement on both sides have been identified (Liu et al., 2008). This suggests that high DNA sequence variations around BnMs5 exist between the two parental lines. Hence, we attempted to concentrate and compare the previously published BnMs5-linked markers based on this F2 population, except for some markers which showed a relatively large genetic distance from the target gene, such as P03MC15 and P04MC11 in Fig. 2A and three distal markers outside of SCE3 in Fig. 2E. PCR analysis showed that the closest flanking SCAR markers (SC6 and SC9, Fig. 2A) developed by Song et al. (2006b) displayed a monomorphic band between the parental lines Rs1046A and 19514A, but new primers designed from their flanking sequences (Supplementary Data Table S1) could re-amplify the potential DNA variations at other loci. SC9-a and SC9-b (Table 1) relative to SC9 can individually mark the presence of the BnMs5a and BnMs5b allele in the F2 population, or they can be used jointly as a co-dominant marker (SC9H) when performing a multiplex PCR. SC6-a (Table 1) relative to SC6 displays a BnMs5a-specific dominant marker. The SCAR markers from the maps constructed by Hong et al. (2006) and Lu et al. (2004) were also evaluated. The results showed that the original polymorphism could be detected directly by all of the SCAR markers from Fig. 2B and Fig. 2E (SCE3 and SCP6) in the F2 population. Rough mapping of these integrated SCAR markers in the 190 F2 plants showed that SCE3 and SC9H are distributed on the same side of BnMs5 as S10T2-90, while the other six SCAR markers are located on the opposite side. Given the loose linkage relationship of some makers with BnMs5, we evaluated only those markers located between S10T2-90 and SCHDF in all the 708 F2 plants. This assay led to a refined genetic map, on which SCD8 (a SCAR marker converted from the AFLP marker S5T5-480, Table 1) and SC6-a delimit BnMs5 to a 1·1-cM genetic region (Fig. 2D).
Fig. 2.
Genetic map integration of BnMs5 and its comparison with Arabidopsis. (A) The genetic map published by Song (2006b); (B) the genetic map published by Hong et al. (2006); (C) the schematic distribution of Arabidopsis genes on chromosome 1 orthologous to the BnMs5-linked markers; (D) the integrated and refined genetic map; (E) the genetic map published by Lu et al. (2004). Dotted lines with a single arrow indicate the corresponding relationship between the polymorphic markers and their orthologues. Markers in (D) indicated by grey bars were used as probes for BAC library screening. Three markers integrated into D are shown by dashed-line boxes. The bold and italic markers were mapped on the A8 chromosome of the TNDH genetic map. Genetic distances are shown in cM.
Table 1.
Primers used for genetic mapping of the BnMs5 locus and probe preparation
| Marker designation | Primer sequence (5′ → 3′) | Annealing temperature (°C) | Polymorphism | Fragment size (bp) |
|---|---|---|---|---|
| SC9-a | F: CACATAGAAGACAGAGAAGACC | 60 | Linked to BnMs5a | 453 |
| R: GCGGCGCGTTGTATTCTTCTC | ||||
| SC9-b | F: TCCGAGCTTGAGCCCTTGTCT | 60 | Linked to BnMs5b | 661 |
| R: AGTCTCACACTACTTCAAGCAG | ||||
| SC6-a | F: GAAACGCCAAGATTGGAAC | 62 | Linked to BnMs5a | 1035 |
| R: GCTTCTGTCTAATGTTAAGTCG | ||||
| BE10 | F: TTGGGTATGGTGTTGGGTTT | 62 | Co-dominant | 398 and 305 |
| R: CCATGATTGCTGATGACCTG | ||||
| SCD8 | F: TCGAGTACTTGGTCCGTT | 61 | Linked to BnMs5a | 450 |
| R: GAGCTCATAAAACCTTTAACTA | ||||
| SC6B3 | F: CATTCATCTTTCACCACCAT | 62 | Not applicable | 827 |
| R: TTGATACAAAACCCACCACA |
These SCAR markers were genotyped in the TNDH population as well. SCD2 can be directly mapped to the top end of the B. napus A8 chromosome between marker IGF5276b and pW184 (Shi et al., 2009), and SC9H was added closely on the same chromosome (data not shown). In turn, we also found that the single sequence repeat (SSR) marker HAU348 neighbouring on SCD2 on the TNDH A8 chromosome is located between SC6 and SCHDF in our F2 population (data not shown). These results together confirm that the BnMs5 locus resides on the B. napus A8 chromosome.
Collinearity analysis around BnMs5 between B. napus and Arabidopsis
The sequences of all the markers (Table S1) on Fig. 2D were aligned with the Arabidopsis genome. As shown in Fig. 2C and Table 2, where it was possible to search for markers in the Arabidopsis genome, they all hit the best homologues on chromosome 1. Alignment of other markers (Fig. 2A, B) not included in Fig. 2D further extended this syntenic region to a 3·5-Mb region (from At1g04950 to At1g14340, Fig. 1C). This clearly suggests that, near BnMs5, only one putative Arabidopsis region with good collinearity can be identified. The two markers, S10T2-90 and SCHDF, covering a genetic region of about 5·0 cM on the B. napus genome, span an Arabidopsis orthologue of about 865 kb (from At1g08790 to At1g11020, Fig. 2C). This means that inside the collinear region around BnMs5 every 1 cM B. napus genetic unit approximately corresponds to a 173-kb physical region in Arabidopsis, much smaller than the average (340 kb cM−1) calculated from the whole-genome comparative mapping between B. napus and Arabidopsis (Parkin et al., 2005). Moreover, the closest flanking markers (SCD8 and SC6H) cover 1·1 cM in the genetic map, which delimit only a fragment of 80 kb comprising 21 predicted Arabidopsis genes (from At1g10180 to At1g10400, Fig. 2C). Thus, the integrated genetic map seems to be feasible for tagging the BnMs5 locus on a single B. napus BAC clone.
Table 2.
Sequence comparison of BnMs5-linked markers with Arabidopsis and the two diploid ancestors of B. napus
| Marker designation | Comparison with Arabidopsis gene orthologue | Comparison with B. rapa predicted gene |
Comparison with B. oleracea predicted gene |
||||
|---|---|---|---|---|---|---|---|
| MF1† | MF2 | LF | Subgenome I‡ (MF1) | Subgenome II (MF2) | Subgenome III (LF) | ||
| S10T2-90 | At1g08790 (4e-09)* | N | A8, Bra030741 (3e-30) | A6, Bra018600 (1e-16) | N | C8, Bol022172 (6e-22) | C5, Bol041237 (4e-26) |
| SCD2 | N | N | N | N | N | N | N |
| E4M9-150 | At1g09130 (2e-43) | N | A8, Bra030756 (1e-55) | N | N | C8, Bol022147 (7e-51) | N |
| SCE3 | N | N | N | N | N | N | N |
| SCD7 | N | N | N | N | N | N | N |
| SC9H | At1g09645 (3e-66) | N | A8, Bra030790 (1e-105) | A6, Bra020004 (1e-73) | C8, Bol006601 (3e-80 ) | C8, Bol022103 (2e-10) | C5, Bol036815 (1e-73) |
| E14M1 | At1g10030 (2e-49) | N | A5, Bra018466 (5e-49) | A6, (Bra019965 (8e-48) | N | C8, Bol022082 (5e-49) | C5, Bol036770 (7e-42) |
| SCD8 | At1g10180 (2e-53) | N | A5, Bra018457 (1e-179) | N | N | C8, Bol022073 (1e-177) | N |
| BE10 | At1g10270 (2e-71) | A9, Bra031706 (5e-67) | A5, Bra018451 (0·0) | A6, (Bra019952 (1e-61) | C8, Bol031243 (2e-57 ) | C8, Bol022062 (1e-143) | C5, Bol036755 (2e-60) |
| SC6-a | At1g10400 (e-123) | N | A5, Bra018440 (0·0) | N | N | C8, Bol022050 (0·0) | N |
| E1M2-310 | N | N | N | N | N | N | N |
| SCHDF | At1g11020 (2e-31) | A9, Bra031736 (3e-29) | A5, Bra018414 (3e-41) | N | C8, Bol031295 (2e-29 ) | C8, Bol022016 (1e-43) | N |
| HDE | At1g11840 (1e-38) | N | A8, Bra016811 (9e-47) | A6, Bra019830 (1e-30) | N | N | C5, Bol036627 (3e-28) |
| SCHDB | At1g11930 (1e-73) | A9, Bra031797 (5e-28) | A8, Bra016802 (4e-87) | N | C8, Bol031341 (7e-30) | C8, Bol029087 (5e-68) | N |
| HDD | At1g12390 (5e-69) | A9, Bra026972 (3e-72) | A8, Bra016774 (5e-89) | A6, Bra019762 (1e-52) | C8, Bol031365 (2e-76) | C8, Bol029110 (2e-88) | N |
| P6M6-410 | At1g14340 (4e-41) | N | A8, Bra016705 (1e-67) | A6, Bra019660 (6e-45) | N | C8, Bol029203 (6e-51) | C5, Bol038029 (4e-30) |
*The E-value calculated via BLASTN is presented after the orthologues in Arabidopsis, B. rapa and B. oleracea, respectively.
†The medium fractionated blocks, the most fractionated blocks (MF2) and the least fractionated blocks (LF) are respectively abbreviated as MF1, MF2 and LF as proposed by Wang et al. (2011).
‡The concepts of Subgenome I, Subgenome II and Subgenome III are individually matched to MF1, MF2 and LF, as used to describe the evolution of the B. oleracea genome (Bolbase, http://119.97.203.210/bolbase/index.html).
N indicates that for a given marker there is no orthologue detected in the Arabidopsis genome, or the B. rapa or B. oleracea subgenomes (MF1, MF2 and LF). The three columns in bold type emphasize the homologous regions with the best synteny.
Determination of the BnMs5-originated subgenome in diploid ancestors
The draft genome sequence of B. rapa accession Chiifu-401-42 has been published recently (Wang et al., 2011), and the draft genome sequence of B. oleracea is also available for sequence alignment and comparative genomics analysis. On the basis of these sequences, triplication of the two-diploid genome relative to Arabidopsis has been clearly revealed, and the concept of the least fractionated blocks (LF), the medium fractionated blocks (MF1) and the most fractionated blocks (MF2) was raised to describe the three subgenomes of the mesopolyploid B. rapa (Wang et al., 2011) and B. oleracea (Bolbase). To determine which of the six subgenomes provided the genomic region around BnMs5 in B. napus, we performed comparative mapping between B. napus and the two diploid ancestors. As shown in Table 2, all the markers which can hit a homologous Arabidopsis gene were able to identify at least one orthologue in B. rapa as well as B. oleracea. As expected, the genomic region showing synteny with BnMs5 can be identified in all six subgenomes, and the arrangement of orthologues in each subgenome is completely in line with the order of markers in B. napus genome except for the subgenome MF2 in B. rapa which includes two discontinuous chromosome segments (A5 and A8, Table 2). The similarities between markers and their orthologues vary between different subgenomes. In a global view, it is unambiguous that the subgenomes with the best collinearity are MF2 in both B. rapa and B. oleracea. In combination with the localization on B. napus A8, the MF2 subgenome of B. rapa should contain the ancestor of BnMs5. In B. rapa MF2, a no less than 230-kb A5 fragment (from Bra018466 to Bra018414) covering both closest flanking markers of BnMs5 split the A8 syntenic region. Consequently, differentiation of the MF2 subgenome bracketed by Bra018440 (homologous to SC6-a) to Bra018457 (homologous to SCD8) from other subgenomes would contribute to identification of target BAC clones from the homologous ones.
Screening and characterization of BAC clones
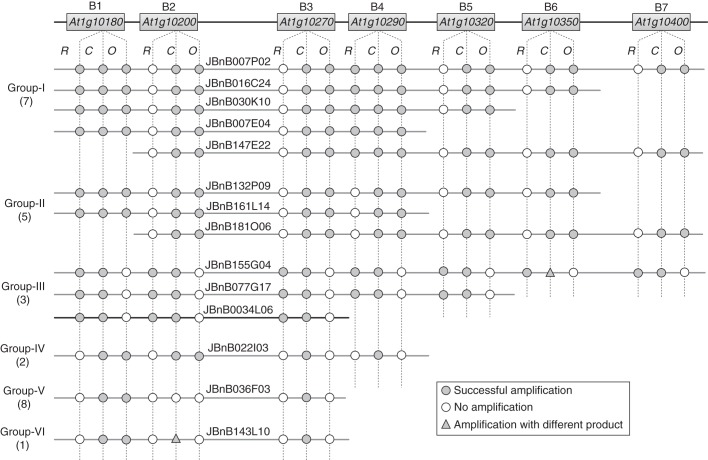
The physical distance between Bra018440 and Bra018457 is about 60 kb in B. rapa, far less than the average insertion length (145 kb, Rana et al., 2004) of the JBnB BAC library. This suggest, per se, that the BnMs5 candidate region can probably be delimited to a single B. napus BAC by using SCD8 and SC6-a as hybridization probes. The specific 450-bp and 827-bp bands were then individually amplified by SCD8 and SC6B3 (designed from the flanking sequence of marker SC6, Table 1). Using these as probes identified a total of 28 positive BAC clones. To eventually identify the target BAC clone carrying the BnMs5 locus, we tried to group all BAC clones first. Based on the B. rapa and B. oleracea predicted genes in the MF2 subgenomes homologous to seven Arabidopsis genes between At1g10180 and At1g10400 (Table S2), 21 pairs of primers were designed (Table S3). According to the genotypes revealed by these primers, 26 BAC clones can be effectively divided into at least six groups (Fig. 3). The remaining two clones were not considered as only one primer pair can generate a PCR product from them. As shown in Fig. 3, the primers relative to the conservation regions between B. rapa and B. oleracea MF2 subgenomes can amplify products with expected size in most cases, whereas obvious differences can be found between the specific primers. All primers specific to the B. oleracea MF2 subgenome can yield expected products in the clones from Group I and II but fail in the clones from Group III. In contrast, the seven primers specific to the B. rapa MF2 subgenomes can generate anticipated products from Group III clones, but only one and two of them amplified products in Group I and II, respectively. Consequently, we assumed that the target BAC clone might be included by the highly homologous groups from Group I to III, whereas the other three groups seem to be from genomic regions with lower similarity due to fewer amplicons by these primers. Therefore, clone JBnB007P02 from Group I covering both probes was first shotgun-sequenced to obtain more sequence information about these groups. An insertion of 196 kb was recovered from JBnB007P02, on which SCD8 and SC6-a delimit an interval of 105 kb.
Fig. 3.
Grouping of the BAC clones by PCR analysis. Arabidopsis genes under the primers are orthologous to the B. rapa and B. oleracea MF2 genes used for primer design. In each group, only those BAC clones with representative PCR patterns are listed (all the clones are indicated in Fig. 4), with the total number of members given in parentheses following group designation. R, O and C indicate the primer combinations specific to B. rapa, B. oleracea and the conservation region between them (Table S3), respectively. The clone highlighted in black is the target clone carrying BnMs5.
To further narrow down the physical region containing BnMs5, numerous primers were designed according to the JBnB007P02 sequence. As it was not clear whether JBnB007P02 is a target BAC clone or just one of the multiple paralogous copies, primers were exclusively identical to the B. rapa MF2 sequence when variations existed between B. rapa and JBnB007P02. Polymorphism examination showed that only the co-dominant marker (BE10, Table 1) is associated with the BnMs5 locus. Genotyping of all the 708 F2 plants detected two recombination events between BE10 and BnMs5. Linkage mapping indicated that BE10 is 0·2 cM away from BnMs5 on the side of SC6H (Fig. 2). This result was further evidenced by a survey of the 987 F2 sterile plants (Table S4).
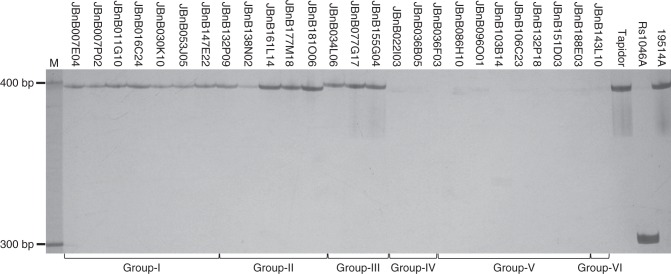
Given the tight linkage between BnMs5 and BE10, we further analysed all the 26 positive BAC clones with this marker. As shown in Fig. 4, BE10 could generate specific bands from all the clones from Group I to Group III but no amplification from Group IV to Group VI clones, although sequence variation of two dispersed nucleotides exists between the 3′-end of BE10 left primer and the Group I clone JBnB007P02. Sequence analysis showed that the products amplified from the Group I and Group II clones such as JBnB007P02 have an identical size of 395 bp, 3 bp smaller than those bands (398 bp) from the three Group III clones (JBnB034L06, JBnB077G17 and JBnB155G04). More importantly, we found that the products from Group III BAC clones have the same sequence with the BnMs5-associated polymorphic bands from both mapping parental lines and ‘Tapidor’, except for a 93-bp deletion in Rs1046A; by contrast, the BE10 band from JBnB007P02 shares only 91 % identity with the parental line 19514A (Table S1). Together with the above analysis, it can be concluded that the BAC clones of Group III should most likely carry the BnMs5 locus.
Fig. 4.
Amplification pattern of the marker BE10 among all the BAC clones from six groups. M, GeneRulerTM 100-bp DNA ladder (Fermentas).
A physical map around BnMs5 and prediction of a candidate gene
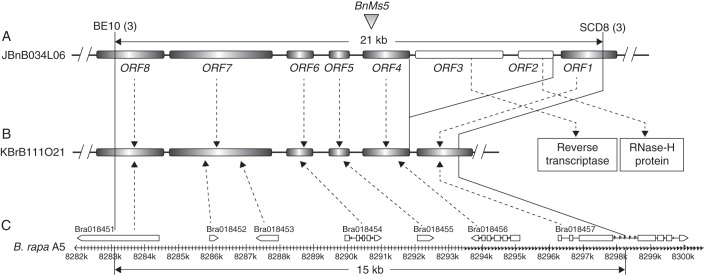
With JBnB007P02 as a query, a B. rapa sequenced BAC clone KBrB111O21 (GenBank code: CU695277) was searched. KBrB111O21 is derived from a BAC library constructed from genomic DNA of ‘Chiifu’, the same cultivar used for B. rapa genome sequencing. KBrB111O21 has an identical sequence to the Group III BAC clones at the BE10 locus, and also covers the fragment of SCD8. Moreover, KBrB111O21 can be wholly mapped to the same B. rapa A5 chromosomal segment as the BnMs5-linked markers. Given that BE10 is just about 15 kb away from SCD8 on KBrB111O21, we adopted an overlapping long-PCR method to isolate the candidate region from the GroupbIII BAC clone JBnB034L06, with the sequence of KBrB111O21 as a reference. This strategy recovered the 21-kb sequence between SCD8 and BE10 on JBnB034L06. BLASTN analysis showed that JBnB034L06 shares 99 % identity with KBrB111O21, and the greater size in JBnB034L06 resulted from a 5·7-kb fragment insertion. Comparatively, JBnB034L06 has only a 76 % similarity with JBnB007P02; however, they share better conservation in the regions containing putative functional genes and differ mainly in the transposon-related interval as described below. The almost identical sequence between JBnB034L06 and KBrB111O21 further demonstrated that the former is the target BAC clone carrying the BnMs5 locus.
Gene prediction of the 21-kb sequence in JBnB034L06 showed that there are six putative ORFs and two incomplete ones where SCD8 and BE10 individually reside (Fig. 5A). This prediction is essentially consistent with the gene annotation in the B. rapa MF2 subgenome, except that ORF7 corresponds to two neighbouring B. rapa genes (Bra018452 and Bra018453). ORF1, ORF6, ORF8 and partial coding sequence (CDS) of ORF7 are highly conserved with the Arabidopsis genes At1g10180, At1g10200, At1g10270 and At1g10220, respectively. ORF2 and ORF3 are located in the insertion fragment relative to KBrB111O21, annotated as a reverse transcriptase and a RNase-H protein, respectively. Both of them have no orthologue in the collinear Arabidopsis region, and belong to the typical retrotranscription transposon class (Wicker et al., 2007). ORF4 and ORF5 are unlike any Arabidopsis genes at the DNA level. Preliminary RT-PCR analysis showed that all the predicted functional genes are expressed in anthers from class 1 to class 4 of the temporary maintainer 7-5 (Fig. S1).
Fig. 5.
A high-resolution physical map around BnMs5 and its comparison with the B. rapa orthologue. The physical map of the candidate region restricted by markers BE10 and SCD8 in B. napus BAC clone JBnB034L06 is schematically shown in (A), and their orthologues in B. rapa BAC clone KBrB111O21 and B. rapa A5 chromosome are shown in (B) and (C), respectively. Dotted lines with a single arrow indicate the corresponding relationship between the predicted genes from B. napus and B. rapa. Vertical solid lines related to BE10 and SCD2 indicate their (or their orthologues') positions on the respective region. The ORFs as indicted by black bars in (A) and (B) were predicted by FGENSH, while the gene annotation information in (C) was obtained from BRAD. ORF2 and ORF3 emphasized with white bars were two transposon-related genes, which were predicted in the B. napus genome but missed in the B. rapa genome (A). The numbers of recombinants for BE10 and SCD8 are shown individually in parentheses following the markers. The physical map distance is shown in kb.
DISCUSSION
In this study, we described the genetic and physical mapping of a triallelic male sterility locus in B. napus. The BnMs5 locus was finally delimited to a 21-kb DNA fragment of a BAC clone on the B. napus A8 chromosome, which covers six predicted functional genes and two transposon-related elements.
Identification of the target BAC clones
A total of 28 positive BAC clones were extracted from a B. napus BAC clone library, highlighting the need to select the target clones from multiple homologues. In fact, it is always a challenge to anchor the target locus to a specific BAC (or contig) in polyploid crops when applying a Southern blotting method in library screening. Here, we employed a combinational strategy to achieve this. We used hybridization probes with a low copy number for screening of the B. napus BAC library. Because substantial gene losses following polyploid formation have occurred extensively in B. rapa (Wang et al., 2011) as well as in B. oleracea, the orthologues of different Arabidopsis genes differ in copy number in the amphidiploid B. napus genome. Here, each of the probe fragments has only one orthologue in both B. rapa and B. oleracea. Theoretically then, there are only two copies for each probe in the B. napus genome. In this case, the number of resulting BAC clones would be greatly reduced, and accordingly the task of grouping these clones would be relieved. We next attempted to identify the B. rapa and B. oleracea subgenomes that have the best collinearity with the B. napus genomic region covering the BnMs5 locus by a comparative genomics approach. In most cases, one marker hit multiple orthologues in the B. rapa and B. oleracea genomes, while comparison of all these orthologues enabled us to anchor those with the highest similarity to the same chromosome segment relative to the MF2 subgenomes. In combination with the localization of BnMs5 on the B. napus A8 chromosome, it can be concluded that BnMs5 should originate from the B. rapa MF2. The candidate BAC clones can thus be chosen roughly by judging whether they have the identical gene composition with the identified B. rapa subgenome through PCR analysis. As expected, this method enabled us to group all the BAC clones into at least six groups, and the target BAC clone should reside in Group I to III clones. Finally, we further differentiated these highly homologous BAC clones according to their genotypes of a BnMs5-associated marker. The BE10-specific band from the Group III BAC clones shows 100 % nucleotide identity with the polymorphic fragments from the mapping parents, highlighting that the target clones covering the BnMs5 locus should be included in this group. Similar cases have also been reported in isolation of BnMs1 (Yi et al., 2010) and BnMs3 (Li et al., 2012). This result is consistent with the observation that the B. rapa-specific primers have all the expected amplicons in Group III clones, as further evidenced by sequence comparison between JBnB034L06 and KBrB111O21. The display of three genotypes at the BE10 locus among BAC clones is possibly involved with the multi-copy status of the BE10 fragment in the B. napus genome, as BE10 is homologous to At1g10270, which has three orthologues in both B. rapa and B. oleracea genomes. The reason why only one band can be amplified from ‘Tapidor’ but bands varying in size from BAC clones results from the DNA variation at the 3′-end of the BE10 left primer. When the genomic DNA of ‘Tapidor’ was used as template, the left primer could only competitively bind to the target region (as with JBnB034L06); however, it can also bind to the highly identical sequences (as with JBnB007P02) in the absence of a perfect matching site.
Local chromosomal evolution around BnMs5
The high-resolution genetic linkage map and partial genomic sequences around BnMs5 in B. napus together with the available genome sequences of closely related species provide an opportunity to investigate local chromosome evolution processes around this male-sterility locus. The collinear fragment around BnMs5 spans at least 3·5 Mb on Arabidopsis chromosome 1, which is actually a part of the long conserved Block A covering the Arabidopsis sequences from At1g01560 and At1g19330 (Schranz et al., 2006). This result coincides with the localization of BnMs5 in the B. napus A8 chromosome, one end of which shows good collinearity with Block A (Parkin et al., 2005). Because B. napus A8 was evolved from the ancestral B. rapa A8, it can be reasonably concluded that the B. rapa ancestor of BnMs5 should reside on A8. However, the B. rapa MF2 fragment covering both closest flanking markers of BnMs5 is located on A5 according to the BRAD annotation, although relatively distal markers still correspond to A8. This observation may be interpreted as an accidental translocation of a portion of A5 carrying BnMs5 with A8 occurring in our mapping population, but we prefer to explain this via an accidental experiment error when assembling the next-generation sequencing data as a result of the following. First, no Arabidopsis Block A fragment was found on B. napus A5 in previous comparative mapping (Parkin et al., 2005) and map integration (Wang et al., 2011). Secondly, the B. rapa BAC clone KBrB111O21, which originates from the A5 target region on BRAD, was actually reported to be mapped on the B. rapa A8 chromosome previously (NCBI annotation CU695277). Lastly, the good collinearity relationship around BnMs5 between B. napus A8 and Block A was inherited from B. rapa A8, so it is not reasonable to suggest that this relationship is maintained in the offspring but disrupted in the ancestor, and a small fragment from A5 can be integrated into A8 but retain perfect synteny with Arabidopsis chromosome 1.
The BnMs5 locus originates from the B. rapa MF2 subgenome, indicating that a considerable amount of Arabidopsis orthologues around BnMs5 might have been discarded during genome evolution. This is matches the fact that only four of the eight candidate genes could match homologues from the Arabidopsis collinear region, while another six were completely lost. Subsequent chromosome evolution around BnMs5 in B. napus was probably driven by the transposon-related sequences, because the insertion fragment in JBnB034L06 relative to KBrB111O21 includes two transposon-related genes. Numerous dramatic genomic expansion events such as transposon movements have occurred in the B. oleracea MF2 subgenome, driving its size between SCD8 and BE10 to be more than four times as large as the B. rapa MF2 subgenome. Accordingly, we do not know whether JBnB007P02 has evolved from the B. oleracea MF2 subgenome.
The other five BAC clone groups may be individually associated with the genomic regions relative to the MF1 and LF subgenomes of B. rapa as well as the MF2, MF1 and LF subgenomes of B. oleracea. However, it remains impractical to establish the correlations between each clone group and the putative subgenome, especially after sequence analysis of JBnB007P02. As JBnB034L06 is suggested to have evolved from the MF2 subgenomes of B. rapa, we initially hypothesized that the highly homologous JBnB007P02 may correspond to the B. oleracea MF2 subgenome, due to the close similarity between JBnB007P02 and JBnB034L06 (data not shown). Nevertheless, the syntenic region restricted by SCD8 and BE10 in the B. oleracea MF2 subgenome actually spans a physical region of 81 kb covering 12 genes from Bol022062 to Bol022073 (Table S2), more than twice the size of the corresponding fragment (24 kb) in JBnB007P02. Furthermore, JBnB007P02 has much higher sequence identity with the MF2 subgenome of B. rapa than that of B. oleracea (data not shown). These facts seem to suggest that JBnB007P02 possibly originated from an extra chromosomal segment duplication event around BnMs5 in the B. napus genome given the divergence from its diploid ancestors, although the possibility cannot be excluded that JBnB007P02 was inherited from the B. oleracea MF2 subgenome but accompanying large-scale fragment loss. This speculation is also favoured by the fact that each of the probe fragments (SCD8 and SC6B3) has only one copy in the two diploid genomes but three or four copies in B. napus. Regardless, the combination of these analyses indicated that BnMs5 may reside in a hotspot region of chromosomal evolution. In the next step, these different groups of BAC clones need to be characterized further, to clarify how many paralogues of the BnMs5 locus are retained in the B. napus genome and how they diverged from each other.
Functional prediction of the BnMs5 candidate genes
In the present study, six putative functional genes were predicted from the candidate region, except for the transposon-related ORF2 and ORF3. ORF1 is an orthologue of At1g10180, a member (Exo84B) of the exocyst complex involved in cytokinesis and cell plate maturation (Fendrych et al., 2010). Mutations in some members of this complex could lead to defects such as etiolated hypocotyl elongation, pollen germination and pollen tube growth in Arabidopsis (Hála et al., 2008). ORF6 is highly conserved with At1g10200, which translates a WLIM-domain-containing protein. Arabidopsis WLIM proteins play prominent roles in the regulation of actin cytoskeleton organization and dynamics in sporophytic tissues and pollen (Papuga et al., 2010). ORF7 corresponds to two independent predicted genes in B. rapa, i.e. Bra018452 and Bra018453. Bra018452 has a very low match with a galactose oxidase/kelch repeat-containing F-box family protein (AT5G02990), while Bra018453 inherits a 150-bp sequence from At1G10220 encoding a zinc finger protein. ORF8 is homologous to At1g10270, encoding a GLUTAMINE-RICH PROTEIN23 (GRP23), which is essential for early embryogenesis (Ding et al., 2006). The predicted ORF4 is partially conserved with some hypothetical Arabidopsis proteins, while ORF5 shows low similarity with a hydroxyproline-rich glycoprotein family protein. It remains difficult to establish the possible relationship between the predicted genes and the cytological sterility phenotype in Rs1046A.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Professor Jinling Meng from Huazhong Agricultural University for kindly providing the JBnB BAC clones and genomic DNA of ‘Tapidor’, and mapping the two SCAR markers into the TNDH genetic map. We also thank Dr Laiqiang Song from the Jiangxi Academy of Agricultural Science for providing the original marker sequences of SC6 and SC9, and Ms Charlotte Kirchhelle from Technical University of Munich for critical reading of the manuscript. This work was supported by NSFC (30570999), IFS (C/4788-1) and Doctoral Fund of the Ministry of Education of China (20090146110011).
LITERATURE CITED
- Chen FX, Hu BC, Li C, Li QS, Chen WS, Zhang ML. Genetic studies on GMS in Brassica napus L: I. Inheritance of recessive GMS line 9012A. Acta Agronomica Sinica. 1998;24:431–438. [Google Scholar]
- Cheng F, Liu SY, Wu J, Fang L, et al. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biology. 2011;11:136. doi: 10.1186/1471-2229-11-136. http://dx.doi.org/10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y-H, Liu N-Y, Tang Z-S, Liu J, Yang W-C. Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. The Plant Cell. 2006;18:815–830. doi: 10.1105/tpc.105.039495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong FM, Hong DF, Xie YZ, et al. Molecular validation of a multiple-allele recessive genic male sterility locus (BnRf) in Brassica napus L. Molecular Breeding. 2012;30:1193–1205. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Dun XL, Zhou ZF, Xia SQ, et al. BnaC.Tic40, a plastid inner membrane translocon originating from Brassica oleracea, is essential for tapetal function and microspore development in Brassica napus. The Plant Journal. 2011;68:532–545. doi: 10.1111/j.1365-313X.2011.04708.x. [DOI] [PubMed] [Google Scholar]
- Fendrych M, Synek L, Pecenková T, et al. The exocyst complex is involved in cytokinesis and cell plate maturation. The Plant Cell. 2010;22:3053–3065. doi: 10.1105/tpc.110.074351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Wei P, Piao Z, et al. SSR and SCAR mapping of a multiple-allele male-sterile gene in Chinese cabbage (Brassica rapa L.) Theoretical and Applied Genetics. 2009;119:333–339. doi: 10.1007/s00122-009-1042-1. [DOI] [PubMed] [Google Scholar]
- Hála M, Cole R, Synek L, et al. An exocyst complex functions in plant cell growth in arabidopsis and tobacco. The Plant Cell. 2008;20:1330–1345. doi: 10.1105/tpc.108.059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong DF, Wan LL, Liu PW, Yang GS, He QB. AFLP and SCAR markers linked to the suppressor gene (Rf) of a dominant genetic male sterility in rapeseed (Brassica napus L.) Euphytica. 2006;151:401–409. [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Annals of Human Genetics. 1944;12:172–175. [Google Scholar]
- Lander ES, Green P, Abrahamson J, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Li J, Hong DF, He JP, et al. Map-based cloning of a recessive genic male sterility locus in Brassica napus L. and development of its functional marker. Theoretical and Applied Genetics. 2012;125:223–224. doi: 10.1007/s00122-012-1827-5. [DOI] [PubMed] [Google Scholar]
- Li SL, Qian YX, Wu ZH. Inheritance and utilization of genetic male sterility in rapeseed (Brassica napus L.) Acta Agriculturae Shanghai. 1985;1:1–12. [Google Scholar]
- Lincoln S, Daly M, Lander E. Constructing genetic linkage maps with Mapmaker/exp 3·0: a tutorial and reference manual. 3rd edn. Cambridge, MA: Whitehead Institute Technical Report; 1992. [Google Scholar]
- Liu J, Hong DF, Lu W, He Q B, Yang GS. Genetic analysis and mapping of genes associated with dominant genic male sterility in rapeseed (Brassica napus L.) Genes & Genomics. 2008;30:523–532. [Google Scholar]
- Liu RH, Meng JL. Mapdraw, a Microsoft Excel macro for draw genetic linkage maps based on given genetic linkage data. Hereditas (Beijing) 2003;25:317–321. [PubMed] [Google Scholar]
- Lu GY, Yang GS, Fu T D. Molecular mapping of a dominant genic male sterility gene Ms in rapeseed (Brassica napus) Plant Breeding. 2004;123:262–265. [PubMed] [Google Scholar]
- Pan T, Zeng FY, Wu SH, Zhao Y. A study on breeding and application GMS line of low erucic acid in rapeseed (B. napus) Chinese Journal of Oil Crop Sciences. 1988;3:5–8. [Google Scholar]
- Papuga J, Hoffmann C, Dieterle M, et al. Arabidopsis LIM proteins: a family of actin bundlers with distinct expression patterns and modes of regulation. The Plant Cell. 2010;22:3034–3052. doi: 10.1105/tpc.110.075960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, et al. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171:765–781. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Morgan C, Shi J, et al. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theoretical and Applied Genetics. 2006;114:67–80. doi: 10.1007/s00122-006-0411-2. [DOI] [PubMed] [Google Scholar]
- Rana D, van den Boogaart T, O'Neill CM, et al. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. The Plant Journal. 2004;40:725–733. doi: 10.1111/j.1365-313X.2004.02244.x. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Lysak MA, Mitchell–Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends of Plant Science. 2006;11:535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Shi JQ, Li RY, Qiu D, et al. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics. 2009;182:851–861. doi: 10.1534/genetics.109.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song LQ, Fu TD, Yang GS, Tu JX, Ma CZ. Genic verification of multiple allelic gene for dominant genic male sterility in 609AB (Brassica napus L.) Acta Agronomica Sinica. 2005;31:896–902. [Google Scholar]
- Song LQ, Fu TD, Yang GS, Tu JX, Ma CZ. Allelism analysis of dominant genic male sterility gene and its restorer gene in Brassica napus. Scientia Agricultura Sinica. 2006a;39:456–462. [Google Scholar]
- Song LQ, Fu TD, Tu JX, Ma CZ, Yang GS. Molecular validation of multiple allele inheritance for dominant genic male sterility gene in Brassica napus L. Theoretical and Applied Genetics. 2006b;113:55–62. doi: 10.1007/s00122-006-0271-9. [DOI] [PubMed] [Google Scholar]
- Wan LL, Xia XY, Hong DF, Yang GS. Molecular analysis and expression of a floral organ-specific polygalacturonase gene isolated from rapeseed (Brassica napus L) Molecular Biology Reports. 2010;37:3851–3862. doi: 10.1007/s11033-010-0041-2. [DOI] [PubMed] [Google Scholar]
- Wang XW, Wang HZ, Wang J, et al. The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, et al. A unified classification system for eukaryotic transposable elements. Nature Reviews Genetics. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Wu JY, Yang GS. Meiotic abnormality in dominant genic male sterile Brassica napus. Molecular Biology. 2008;42:572–578. [PubMed] [Google Scholar]
- Yi B, Zeng FQ, Lei SL, et al. Two duplicate CYP704B1-homologous genes BnMs1 and BnMs2 are required for pollen exine formation and tapetal development in Brassica napus. The Plant Journal. 2010;63:925–938. doi: 10.1111/j.1365-313X.2010.04289.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.