Introduction
Rapid advances over the past several decades in neuroimaging and cyberinfrastructure technologies have brought explosive growth in the web-based warehousing, availability, and accessibility of imaging data on a broad array of neurodegenerative and neuropsychiatric disorders and conditions.[1-4] This growth has been driven largely by the demand for multi-scale data in the investigation of fundamental disease processes; the need for interdisciplinary cooperation to integrate, query, and interpret the data; and the movement of science in general toward freely available and openly accessible information. In response to this substantial need for capacity to store and exchange data online in meaningful ways in support of data analysis, hypothesis testing, and future reanalysis or even repurposing, the electronic collection, organization, annotation, storage, and distribution of clinical, genetic, and imaging data are by now essential activities in the contemporary biomedical and translational discovery process. The result has been the prolific development and emergence of complex computational infrastructures that serve as repositories of databases and provide critical functionalities such as sophisticated image analysis algorithm pipelines and powerful three-dimensional visualization and statistical tools.[5-9] The statistical and operational advantages of collaborative, distributed team science in the form of multi-site consortia continue to push this approach in a diverse range of population-based investigations.
New era of collaborative, interdisciplinary team science
The ongoing convergence and integration of neuroscientific infrastructures worldwide is heading ultimately to the creation of a global virtual imaging laboratory. Through ordinary web browsers, large-scale image data sets and related clinical data and biospecimens, algorithm pipelines, computational resources, and visualization and statistical toolkits are easily accessible to users regardless of their physical location or disciplinary orientation. The promise of this investigatory environment-without-walls, and its incipient marshalling of scientific talent and facilitation of collaboration across multiple disciplines, is accelerating various translational initiatives with high societal impact, such as early or pre-symptomatic diagnosis and prevention of Alzheimer’s disease.
Neuroimaging is now a major focus for multi-institutional research on progressive changes in brain architecture, biomarkers of treatment response, and the differential effects of disease on patterns of cognitive activation and connectivity.[10] Prominent research consortia and multi-site clinical trials have focused on Alzheimer’s disease, pediatric brain cancer, and fetal alcohol syndrome, in addition to multi-institutional collaborative programs for mapping the normal brain.[11, 12] Current leading-edge mapping consortia are focusing on the human brain as a complex network of connectivity and aim for a comprehensive structural description of the brain’s network architecture.[13] This collaborative effort, the human connectome (http://www.humanconnectomeproject.org/), is exploring and generating new insights on the organization of the brain’s structural connections and their role in shaping functional dynamics and brain plasticity. Such large-scale efforts necessitate close coordination of image data collection protocols, ontology development, computational requirements and sharing.
Key areas of e-science impact in neuroscience and neurology
Multi-site neuroimaging studies are dramatically accelerating the pace and volume of discoveries regarding major brain disease and the contrasts between normal and abnormal brain structure and function.[14] The large-scale, purpose-driven data sets generated by these consortia can then be used by the broader community to model and predict clinical outcomes as well as guide clinicians in selecting treatment options for various neurological diseases. Multisite trials are an important element in the study of a disease or the process of evaluating an intervention. Linking together multiple sites facilitates the recruitment of large samples that yield high statistical power for both main analyses as well as secondary analyses of subgroups. Generalizability of results to the level of the population also is maximized. Because data come from multiple sites, investigators can explore how a treatment’s effects vary across geographically diverse sites and how such variation relates to site characteristics, and to cultural and socioeconomic characteristics of the patients who participated in the studies. Such information can directly inform clinical decision-making at the level of the patient and guide the selection of treatment options. These research efforts are imperative for guiding treatment recommendations for neurological disorders domestically and internationally as well as at the level of the individual patient. Multicenter collaborations strengthen understanding of brain diseases that affect all walks of life, all ages, and all cultures, thus enabling accelerated translation of neuroimaging trial outcomes directly into clinical applications.
Large archives of neuroimaging data are also creating innumerable opportunities for re-analysis and mining that can lead to new findings of use in basic research or in the characterization of clinical syndromes. Access to databases of neuroanatomical morphology has led to the development of content-driven approaches for exploration of brains that are anatomically similar, revealing patterns embedded within entire (sub)sets of neuroimaging data.[4]
Provenance, or the description of the history of a set of data, has grown more important with the proliferation of research consortia-related efforts in neuroimaging.[5, 15] Knowledge about the origin and history of an image is crucial for establishing data and results quality; detailed information about how it was processed, including the specific software routines and operating systems that were used, is necessary for proper interpretation, high-fidelity replication, and re-use and re-purposing of the data. New mechanisms have emerged for describing provenance in a simple and easy-to-use environment, alleviating the burden of documentation from the user while still providing a rich description of an image’s source history. This combination of ease of use and highly descriptive metadata is greatly facilitating the collection of provenance and subsequent sharing of large data sets.
Multimodal classification of images has advanced the utility of atlases of neuropathology through standardized 3D coordinate systems that integrate data across patients, techniques, and acquisitions.[11, 16, 17] Atlases with a well-defined coordinate space, together with algorithms to align data with them, have enabled the pooling of brain mapping data from multiple subjects and sources, including large patient populations, and facilitated reconstruction of the trajectories of neurodegenerative diseases like Alzheimer’s as they progress in the living brain.[18-22] Automated algorithms can then utilize atlas descriptions of anatomical variance to guide image segmentation, tissue classification, functional analysis, and pathology detection.[7, 23, 24] Statistical representations of anatomy resulting from the application of atlasing strategies to specific subgroups of diseased individuals have revealed charactersitics of structural brain differences in a number of diseases, including Alzheimer’s disease, HIV/AIDS, unipolar depression, Tourette syndrome, and autism.[25]
Atlas-based descriptions of variance offer statistics on degenerative rates and can elucidate clinically relevant features at the systems level. Atlases have identified differences in atrophic patterns between Alzheimer’s disease and Lewy body dementia, and differences between atrophy rates across clinically defined subtypes of psychosis. Atlases have also revealed the association between genes and brain structure.[26] Based on well-characterized patient groups, population-based atlases contain composite maps and visualizations of structural variability, asymmetry, and group-specific differences. Pathological change can be tracked over time, and generic features resolved, enabling these atlases to offer biomarkers for a variety of pathological conditions as well as morphometric measures for genetic studies or drug trials.
Brain atlases can now accommodate observations from multiple modalities and from populations of subjects collected at different laboratories around the world. These probabilistic systems show promise for identifying patterns of structural, functional, and molecular variation in large imaging databases, for pathology detection in individuals and groups, and for determining the effects of age, gender, handedness, and other demographic or genetic factors on brain structures in space and time. Integrating these observations to enable statistical comparison has already provided a deeper understanding of the relationship between brain structure and function.
This chapter considers and assesses the clinical implications of enabling large numbers of scientists to work in tandem with the same large data sets in the context of one such effort, the Alzheimer’s Diseases Neuroimaging Initiative (ADNI). Two facets in particular of this project exemplify the clinical value of large-scale neuroimaging databases in research and in patient care: (1) disease diagnosis and progression tracking, including the diagnostic value of image databases in demarcating abnormal and normal ranges of biospecimens; and (2) role of neuroimages in statistical powering, subject stratification, and incisive endpoints and outcomes of clinical trials.
Alzheimer’s Diseases Neuroimaging Initiative (ADNI)
The Alzheimer’s Diseases Neuroimaging Initiative (ADNI) exemplifies a remarkably successful, open, shared, and efficient database.[27-30] ADNI brings together geographically distributed investigators with diverse scientific capabilities for the intensive study of biomarkers that signify and track the progression of Alzheimer’s disease. The quantity of imaging, clinical, cognitive, biochemical, and genetic data acquired and generated throughout the project have required powerful informatics systems and mechanisms for processing, integrating, and disseminating these data not only to support the research needs of the investigators who make up the ADNI cores but also to provide widespread data access to the greater scientific community. At the junction of this collaborative endeavor, the UCLA Laboratory of Neuro Imaging (LONI) has provided an infrastructure to facilitate data integration, access, and sharing across a diverse and growing community of multidisciplinary scientists.
ADNI is composed of eight cores responsible for conducting the study along with external investigators authorized to use ADNI data. The various information systems employed by the cores result in an intricate flow of data into, out of, and among information systems and institutions. Ultimately, the data flow into the ADNI data repository (http://adni.loni.ucla.edu/), where they are made available to the community. This well-curated scientific data repository enables data to be accessed by researchers across the globe and to be preserved over time. To date, over 1,300 investigators have been granted access to ADNI data, resulting in extensive download activity that exceeds one million downloads of imaging, clinical, biomarker, and genetic data.
Image Data Workflow
The ADNI Informatics Core provides a user-friendly, web-based environment for storing, searching, and sharing data acquired and generated by the ADNI community. In the process, the LONI Image and Data Archive (IDA) has grown to meet the evolving needs of the ADNI community through continuing development of an increasingly interactive environment for data discovery and visualization. The automated systems developed to date include components for de-identification and secure archiving of imaging data from the 57 ADNI sites, management of the image workflow whereby raw images transition from quarantine status to general availability and then proceed through preprocessing and post-processing stages; integration of non-imaging data from other cores, management of data access and data sharing activities and provision of a central, secure repository for disseminating data and related information to the ADNI community.
The imaging cores perform quality control and preprocessing of the MR and PET images; the ADNI image analysts perform post-processing and analysis of the preprocessed images and related data; the biochemical samples are processed and the results compiled; and investigators download and analyze data as best fits their individual research needs.
Raw Image Data
In keeping with the objectives of the ADNI project to make data available to the scientific community, without embargo, while meeting the needs of the core investigators, the IDA developed the image data workflow shown in Figure 1. Initially, each acquisition site uploads image data to the repository via the IDA, a web-based application that incorporates a number of data validation and data de-identification operations, including validation of the subject identifier, validation of the dataset as human or phantom, validation of the file format, image file de-identification, encrypted data transmission, database population, secure storage of the image files and metadata, and tracking of data accesses. The image archiving portion of the system is both robust and simple, with new users requiring little, if any, training. Key system components supporting the process of archiving raw data are:
The subject identifier is validated against a set of acceptable, site-specific IDs.
Potentially patient-identifying information is removed or replaced. Raw image data are encoded in the DICOM, ECAT and HRRT file formats, from different scanner manufacturers and models (e.g., SIEMENS Symphony, GE SIGNA Excite, PHILIPS Intera, etc.). The Java applet de-identification engine is customized for each of the image file formats deemed acceptable by the ADNI imaging cores and any files not of an acceptable format are bypassed. Since the applet is sent to the upload site, all de-identification takes place at the acquisition site and no identifying information is transmitted.
Images are checked to see that they “look” appropriate for the type of upload. Phantom images uploaded under a patient identifier are flagged and removed from the upload set. This check is accomplished using a classifier that has been trained to identify human and phantom images.
Image files are transferred encrypted (HTTPS) to the repository in compliance with patient-privacy regulations.
Metadata elements are extracted from the image files and inserted into the database to support optimal storage and access. Customized database mappings were constructed for the various image file formats to provide consistency across scanners and image file formats.
Newly received images are placed into quarantine status, and the images are queued for those charged with performing MR and PET quality assessment.
Quality assessment results are imported from an external database and applied to the quarantined images. Images passing quality assessment are made available and images not passing quality control tagged as failing quality control.
Once raw data undergo quality assessment and are released from quarantine, they become immediately available to authorized users.
Figure 1.
Clinical and imaging data flows from the acquisition sites into separate clinical and imaging databases. Quality assessments and preprocessed images are generated by the imaging cores and returned to the central archive where image analysts obtain, process, and return to the archive further derived image data and results.
Processed Image Data
Preprocessed images are the recommended common set for analysis. The goals of preprocessing are to produce data standardized across site and scanner and with certain image artifacts corrected. Usability of processed data for further analysis requires an understanding of the data provenance, or information about the origin and subsequent processing applied to a set of data. To provide almost immediate access to preprocessed data in a manner that preserved the relationship between the raw and preprocessed images and that captured processing provenance, we utilize an Extended Markup Language (XML) schema that defines required metadata elements as well as standardized taxonomies. The system supports uploading large batches of preprocessed images in a single session with minimal interaction required by the person performing the upload. A key aspect of this process is agreement on the definitions of provenance metadata descriptors. Using standardized terms to describe processing minimized variability and aids investigators in gaining an unambiguous interpretation of the data.
Preprocessed images are uploaded by the quality control sites on a fairly continuous basis. In order to minimize duplicate analyses, an automated data collection component was implemented whereby newly uploaded preprocessed scans are placed into predefined, shared data collections. These shared collections, organized by patient diagnostic group (normal control, MCI, AD) and visit (baseline, 6 month, etc.), together with a redesigned user interface (Figure 2) that clearly indicates which images have not previously been downloaded, greatly reduces the time and effort needed to obtain new data. The same process may be used for post-processed data allowing analysts to share processing protocols via descriptive information contained in the XML metadata files.
Figure 2.
The ADNI Data Collections interface provides access to shared data collections and is organized to meet the workflow needs of the analysts. The ability to easily select only images not previously downloaded by the user saves time and effort.
Data integration
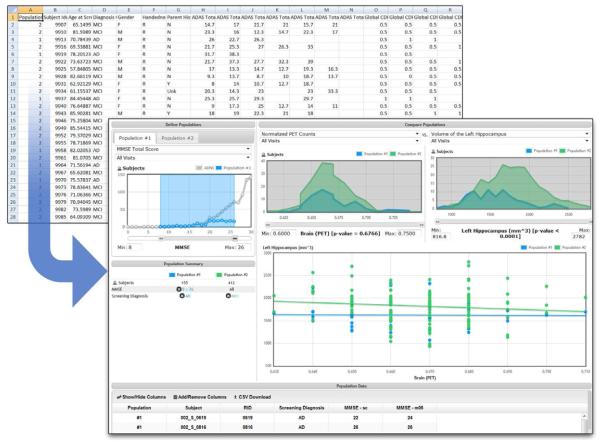
A subset of data from the clinical database is also integrated into the IDA to support richer multimodal queries across the combined set. (Figure 3) The selection of the initial set of clinical data elements was based on user surveys in which participants identified the elements they thought would be most useful in supporting their investigations. Because the clinical data originate in an external database, automated methods for obtaining and integrating the external data also validate and synchronize the data from the two sources and ensure that data from the same subject visit are combined.
Figure 3.
This interface enables creation of groups from complex queries and comparison in a variety of graphical formats.
Infrastructure mechanics
A robust and reliable infrastructure is a necessity for supporting a resource intended to serve a global scientific community. The hardware infrastructure of the Informatics Core provides high performance, security, and reliability at each level. The fault-tolerant network infrastructure has no single points of failure. A firewall appliance protects and segments the network traffic, permitting only authorized ingress and egress. Multiple redundant database, application, and web servers ensure service continuity in the event of a single system failure and also provide improved performance through load balancing of requests across the multiple machines. To augment the network-based security practices and to ensure compliance with privacy requirements, the servers utilize SSL encryption for all data transfers. Post-transfer redundancy checking on the files is performed to guarantee the integrity of the data. Backup systems are designed to ensure data integrity and to protect data in the event of catastrophic failure.
ADNI policy requires participating sites to upload new data within 24 hours of acquisition. To prevent performance degradation, the application servers are divided by upload/download functionality. In order to prevent a single downloader from dominating a web server with multiple requests, the activity of each downloader is monitored and his/her download rate is throttled accordingly. These measures help to ensure ADNI data and resources are equitably shared with maximal efficiency.
Data Access and Security
Access to ADNI data is restricted to those who are site participants and those who have applied for access and received approval from the project’s Data Sharing and Publication Committee. An online application and review feature is integrated into the IDA so that applicant information and committee decisions are recorded in the database and the e-mail communications acknowledging application receipt, approval, or disapproval are automatically generated. Different levels of user access control the system features available to an individual. All data uploads, changes, and deletions are logged.
More than 100,000 image data sets (more than 5 million files) and related clinical, imaging, biomarker, and genetic data sets are available to approved investigators. More than one million downloads of raw, pre- and post-processed scans have been provided. Clinical, biomarker, image analysis results, and genetic data have been downloaded more than 5,000 times. Data download activity has increased annually since the data first became available in 2007. There are users from across the globe accessing the archive around the clock.
Data Management
There are Data User Management tools for reviewing data use applications, managing manuscript submissions, and sending notifications to investigators whose annual ADNI update is due. There is also a set of Project Summary tools that support interactive views of upload and download activities by site, user, time period, and provide exports of the same. Other information, documents, and resources geared toward apprising investigators about the status of the study and data available in the archive are provided through the ADNI web site (http://adni.loni.ucla.edu/).
The Informatics Core provides a mechanism to distribute and share data, results, and information not only among the project participants but also the scientific community at large. This informatics model enables a far more extensive array of analytic strategies and approaches to interpreting these data. These dissemination aspects of ADNI are among the most important to the project’s success. The databasing, querying, examination, and processing of data sets from multiple subjects necessitate efficient and intuitive interfaces, responsive answers to searches, coupled analyses workflows, and comprehensive provenance with a view toward promoting independent re-analysis and study replication. As such, a new interface enabling interactive data mining and analyses has been developed.
Lessons Learned by the ADNI Informatics Core
Successful informatics solutions must build a trust with the communities they seek to serve and provide dependable services and open policies to researchers. Several factors contribute to database utility, including whether it actually contains viable data and these are accompanied by a detailed description of their acquisition (e.g., meta-data); whether the database is well organized and the user interface is easy to navigate; whether the data are derived versions of raw data or the raw data themselves; the manner in which the database addresses the sociological and bureaucratic issues that can be associated with data sharing; whether it has a policy in place to ensure that requesting authors give proper attribution to the original collectors of the data; and the efficiency of secure data transactions. These systems must provide flexible methods for data description and relationships among various meta-data characteristics. Moreover, those that have been specifically designed to serve a large and diverse audience with a variety of needs and that possess the qualities described above, represent the types of databases that can have the greatest benefit to scientists looking to study a disease, assess new methods, examine previously published data, or explore novel ideas using the data.
Lessons learned to date by the Informatics Core include the following principles: (1) The data archive information must be open and unrestricted. (2) The database must be transparent in terms of activity and content. (3) The duties and responsibilities of stakeholder individuals and institutions must be clearly and precisely specified. (4) Technical and semantic interoperability between the database and other online resources (data and analyses) is the optimal approach. (5) Clear curation systems governing quality control, data validation, authentication, and authorization must be in place. (6) The systems must be operationally efficient and flexible. (7) Clear policies of respect for intellectual property and other ethical and legal requirements must be in place in advance of access. (8) Management accountability and authority are obligatory. (9) A solid technological architecture and supporting expertise are essential. (10) Systems for user support must be reliable. (11) HIPAA compliance must be thorough and ever-adaptable to changes in regulatory and statutory fashion.
ADNI Scientific Advances and Future Vision
With a current enrollment and ongoing longitudinal follow-up of over 800 subjects with mild cognitive impairment (MCI) and mild Alzheimer’s disease (AD), as well as cognitively normal older individuals, across 57 project implementation sites in North America, the ADNI databases encompass a substantial convergence of data from medical evaluations; clinical, cognitive, functional, and behavioral assessments; biochemical analytics; structural and functional neuroimages; and genetic assessments. New and important insights into the neurobiology of the AD spectrum have emerged from the collective analytic effort to date,[27, 31] particularly, as intended, in the realm of biomarkers as indirect measures or diagnostics of disease severity and as dynamic trackers of disease progression, with attendant implications for treatment trial design with respect to sample size and statistical powering as well as subject stratification.[28, 32, 33]
ADNI investigators have used multiple imaging biomarkers to quantify disease progression and measure various aspects of AD pathology. These include PET for amyloid, fluoro-deoxyglucose PET for metabolic decline, and MRI for brain atrophy, as well as risk factors that influence these measures (e.g., ApoE, cardiovascular risks).[2, 34-38] Patients with MCI and AD experience progressive brain atrophy and this has proved an especially fertile area for investigators to illuminate predictability in regional and temporal patterns of damage through neuroimaging as patients are followed longitudinally through disease progression and the trajectory of neurodegeneration is documented through imaging. In vivo measures and quantification of subregional atrophy, such as changes in cortical thickness or structure volume, hold great promise of improved diagnosis as well as assessment of the neuroprotective effects of newly developed therapies undergoing early and late phase trials [39-41]. Through 3D mapping of gray matter atrophy, ADNI researchers have found that cortical areas affected earlier in the disease process are more severely damaged than those that are affected late, suggesting that structural MRI is reliable not only as an in vivo disease-tracking technique but also may prove useful in evaluating disease-modifying therapies.[42, 43] Indeed, ADNI MRI metrics indicate that degree of neurodegeneration of medial temporal structures is a reliable antecedent imaging marker of imminent conversion from MCI to AD, with decreased hippocampal volume as the most robust marker. Validation of imaging biomarkers is thus critical as they can render clinical trials of disease modifying agents more efficacious by identifying individuals who are at highest risk for progression to AD.[44]
ADNI data has been used in the elucidation of associations between various imaging, CSF, genetic and clinical measures in different cohorts. ADNI investigators have examined brain morphometric changes alone that occur during disease progression using cross sectional and longitudinal MRI data.
Morphometry
Specific morphometric measures such as regional cortical thickness provide excellent sensitivity to group differences. In a cross-sectional study of normal controls, MCI and AD subjects, differences were found mostly in hippocampus and entorhinal cortex, which had the largest effect sizes, along with other temporal regions, the temporal horn of the lateral ventricle, rostral posterior cingulate and some parietal and frontal regions. [45] Additional atrophy was seen in AD patients relative to controls in the inferior parietal, banks of the superior temporal sulcus, retrosplenial and some frontal regions. Similar results were shown by Karow et al. [46]. The trajectories of change over the course of the disease varies though. [45] Mesial and temporal regions, exhibited a linear rate of atrophy through both MCI stages to AD, whereas the lateral temporal middle gyrus, retrosplenial, inferior parietal and rostral mid-frontal cortices, exhibited accelerated atrophy later in the disease.
Leung et al. [47] also found higher rates of hippocampal atrophy in MCI converters to AD than non-converters. McDonald et al. [48] examined regional rates of neocortical atrophy in two groups of MCI patients with different degrees of impairment. As the disease progressed, atrophy migrated from the medial and inferior lateral temporal, inferior parietal and posterior cingulate, to the superior parietal, prefrontal and lateral occipital cortex and then to the anterior cingulate cortex. The least impaired MCI patients showed the greatest rates of atrophy in the medial temporal cortex. Using a variety of approaches these findings are quite consistent. Hua et al. [49] and Leow et al. [20] both used tensor based morphology (TBM) producing annualized 3D maps of structural changes. Schuff et al. [50] focused on changes in hippocampal volume and McEvoy et al. [51] calculated an atrophy score based on regions of interest most associated with AD atrophy. Collectively, these studies showed atrophy spreading from the medial temporal lobe to the parietal, occipital and frontal lobes over the course of the disease, with MCI patients in general having a more anatomically restricted AD-like pattern of change. MCI subjects who converted to AD within the time frame of the study had a more AD-like pattern of atrophy.
Shape
Beyond simple volumetric analysis, is the assessment of changes regions of interest shape. Qiu et al. [52] used diffeomorphic metric mapping to reveal that the anterior segment of the hippocampus and the basolateral complex of the amygdala had the most surface inward deformation in MCI and AD patients, coupled with a complementary outward deformation in the lateral ventricles. Similarly Apostolova et al. [53] found enlargement of the lateral ventricles with disease progression.
Relation to Cognition
It has been well established that structures within the temporal lobe decline in AD. Due to their critical role in the formation of memories, it is one of the first functions to be measurably affected in AD. Within the temporal lobe, hippocampal atrophy is a common structural biomarker since it is among the earliest to degenerate in AD. Leow et al. [20] found a strong association between several cognitive scores and temporal lobe atrophy in MCI patients. Similarly, Morra et al. [54] found that bilateral hippocampal atrophy at baseline was strongly correlated with the Mini Mental State Exam. Using TBM, Hua et al. [19] found that baseline temporal lobe atrophy was associated with both baseline and change in the CDR-SB in MCI and AD patients, but with change in the MMSE only in the AD group, providing further evidence for the acceleration of atrophic change with disease progression.
Genetics
Morra et al. [55], Wolz et al. [56], Hua et al. [49] and Risacher et al. [57] all found that carriers of the APOE4 allele had higher rates of hippocampal atrophy than non-carriers. Vemuri et al. [58] also found that the APOE4 genotype contributed to MRI atrophy. Hua et al. [49] found that the APOE4 allele had a dose-dependent detrimental risk with greater atrophy in the hippocampus and temporal lobe in homozygotes than heterozygotes in MCI and AD groups. The recently identified AD risk allele, GRIN2b was associated with higher rates of temporal lobe atrophy in the pooled group, but more weakly than APOE4 [59]. Other thus far unidentified genetic risk factors likely contribute to AD, with epidemiological studies suggesting, maternal history of the disease increases the risk of developing AD.
Other interesting Associations
Using the related method of TBM, Ho et al. [60] created regional maps of changes in brain tissue and used the resulting Jacobian values to represent brain tissue excess or deficit relative to a template. They found that lower brain volume in the frontal, parietal, occipital and temporal lobes was associated with higher Body Mass Index (BMI) in MCI and AD patients and that ventricular expansion correlated with higher BMI in AD but not MCI patients.
PET
FDG-PET has been used by several groups to investigate relationships between cerebral glucose hypometabolism and other factors including cognitive measures and CSF biomarkers in MCI and AD cohorts. Wu et al. [61] showed hypometabolic rates in posterior cingulate/precuneus and parietotemporal regions. Chen et al. [62] investigated declines in CMRgl in statistically predefined ROIs associated with AD over 12 months in the ADNI cohort and found significant changes in MCI and AD groups compared to controls bilaterally in the posterior cingulate, medial and lateral parietal, medial and lateral temporal, frontal and occipital cortex. These, and many other, papers support the use of glucose metabolism as a sensitive measure of cognition in AD.
Summary
Given the vagaries of disease modification and of what eventually will prove a valid, reliable and compelling empirical basis for distinguishing between true disease modification and symptomatic treatment effects, ADNI investigators are bringing multimodal neuroimaging techniques to the cutting edge of utility as dynamic biomarkers, sensitive to disease state and progression across different stages.[18, 33, 63] From here, the next step is neuroimages as reliable treatment trial endpoints and clinical intervention outcomes with respect to longitudinal changes in brain volume, enhancing both the feasibility and the cost efficiencies of trials through improved signal detection of novel treatments.[64] Inasmuch as AD is a devastating neurodegenerative disease against which no effective treatment is now known, ADNI and similar discovery initiatives worldwide give solid hope for the future in patient care.
Acknowledgments
The original work was primarily funded by the ADNI (Principal Investigator: Michael Weiner; NIH grant number U01 AG024904). ADNI is funded by the National Institute of Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and the Foundation for the National Institutes of Health, through generous contributions from the following companies and organizations: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, the Alzheimer’s Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging (ISOA), with participation from the U.S. Food and Drug Administration. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. This study was also supported by grant P41 RR013642 from the National Center for Research Resources (NCRR), National Institutes of Health (NIH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Evans AC. Large-scale morphometric analysis of neuroanatomy and neuropathology. Anatomy and embryology. 2005;210(5-6):439–46. doi: 10.1007/s00429-005-0045-1. [DOI] [PubMed] [Google Scholar]
- 2.Frisoni GB, et al. Virtual imaging laboratories for marker discovery in neurodegenerative diseases. Nature reviews. Neurology. 2011 doi: 10.1038/nrneurol.2011.99. [DOI] [PubMed] [Google Scholar]
- 3.Toga AW. Guest Editorial: A vision of computational anatomy. Biomed Comput Rev. 2009;5(3):1. [Google Scholar]
- 4.Joshi SH, Horn JD, Toga AW. Interactive exploration of neuroanatomical meta-spaces. Frontiers in neuroinformatics. 2009;3:38. doi: 10.3389/neuro.11.038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinov I, et al. Neuroimaging study designs, computational analyses and data provenance using the LONI pipeline. PloS one. 2010;5(9) doi: 10.1371/journal.pone.0013070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinov ID, et al. Efficient, distributed and interactive neuroimaging data analysis using the LONI Pipeline. Frontiers in neuroinformatics. 2009;3:22. doi: 10.3389/neuro.11.022.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shattuck DW, et al. Online resource for validation of brain segmentation methods. NeuroImage. 2009;45(2):431–9. doi: 10.1016/j.neuroimage.2008.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, et al. Robust surface reconstruction via Laplace-Beltrami eigen-projection and boundary deformation. IEEE transactions on medical imaging. 2010;29(12):2009–22. doi: 10.1109/TMI.2010.2057441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen JR, et al. Decoding continuous variables from neuroimaging data: basic and clinical applications. Frontiers in neuroscience. 2011;5:75. doi: 10.3389/fnins.2011.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pievani M, et al. Functional network disruption in the degenerative dementias. Lancet neurology. 2011 doi: 10.1016/S1474-4422(11)70158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toga AW, et al. Towards multimodal atlases of the human brain. Nature reviews. Neuroscience. 2006;7(12):952–66. doi: 10.1038/nrn2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Horn JD, Toga AW. Multisite neuroimaging trials. Current opinion in neurology. 2009;22(4):370–8. doi: 10.1097/WCO.0b013e32832d92de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sporns O. The human connectome: a complex network. Annals of the New York Academy of Sciences. 2011;1224(1):109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- 14.Biswal BB, et al. Toward discovery science of human brain function. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(10):4734–9. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackenzie-Graham AJ, et al. Provenance in neuroimaging. NeuroImage. 2008;42(1):178–95. doi: 10.1016/j.neuroimage.2008.04.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toga AW, Thompson PM. What is where and why it is important. NeuroImage. 2007;37(4):1045–9. doi: 10.1016/j.neuroimage.2007.02.018. discussion 1066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D, et al. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage. 2011;55(3):856–67. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewers M, et al. Staging Alzheimer’s disease progression with multimodality neuroimaging. Progress in neurobiology. 2011 doi: 10.1016/j.pneurobio.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua X, et al. 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. NeuroImage. 2008;41(1):19–34. doi: 10.1016/j.neuroimage.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leow AD, et al. Alzheimer’s disease neuroimaging initiative: a one-year follow up study using tensor-based morphometry correlating degenerative rates, biomarkers and cognition. NeuroImage. 2009;45(3):645–55. doi: 10.1016/j.neuroimage.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madsen SK, et al. 3D maps localize caudate nucleus atrophy in 400 Alzheimer’s disease, mild cognitive impairment, and healthy elderly subjects. Neurobiology of aging. 2010;31(8):1312–25. doi: 10.1016/j.neurobiolaging.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson PM, et al. Tracking Alzheimer’s disease. Annals of the New York Academy of Sciences. 2007;1097:183–214. doi: 10.1196/annals.1379.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morra JH, et al. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Human brain mapping. 2009;30(9):2766–88. doi: 10.1002/hbm.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westman E, et al. AddNeuroMed and ADNI: Similar patterns of Alzheimer’s atrophy and automated MRI classification accuracy in Europe and North America. NeuroImage. 2011 doi: 10.1016/j.neuroimage.2011.06.065. [DOI] [PubMed] [Google Scholar]
- 25.Sakoglu Ü, et al. Paradigm shift in translational neuroimaging of CNS disorders. Biochemical Pharmacology. 2011;81(12):1374–1387. doi: 10.1016/j.bcp.2010.12.029. [DOI] [PubMed] [Google Scholar]
- 26.Schumann G, et al. The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Molecular psychiatry. 2010;15(12):1128–39. doi: 10.1038/mp.2010.4. [DOI] [PubMed] [Google Scholar]
- 27.Aisen PS, et al. Clinical Core of the Alzheimer’s Disease Neuroimaging Initiative: progress and plans. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2010;6(3):239–46. doi: 10.1016/j.jalz.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beckett LA, et al. The Alzheimer’s Disease Neuroimaging Initiative: Annual change in biomarkers and clinical outcomes. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2010;6(3):257–64. doi: 10.1016/j.jalz.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen RC, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74(3):201–9. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toga AW, Crawford KL. The informatics core of the Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2010;6(3):247–56. doi: 10.1016/j.jalz.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiner MW, et al. The Alzheimer’s disease neuroimaging initiative: progress report and future plans. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2010;6(3):202–11. doi: 10.1016/j.jalz.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thal LJ, et al. The role of biomarkers in clinical trials for Alzheimer disease. Alzheimer disease and associated disorders. 2006;20(1):6–15. doi: 10.1097/01.wad.0000191420.61260.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavedo E, Frisoni GB. The dynamic marker hypothesis of Alzheimer’s disease and its implications for clinical imaging. The quarterly journal of nuclear medicine and molecular imaging : official publication of the Italian Association of Nuclear Medicine. 2011;55(3):237–49. [PubMed] [Google Scholar]
- 34.Jagust WJ, et al. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2010;6(3):221–9. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohannim O, et al. Boosting power for clinical trials using classifiers based on multiple biomarkers. Neurobiology of aging. 2010;31(8):1429–42. doi: 10.1016/j.neurobiolaging.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen RC. Alzheimer’s disease: progress in prediction. Lancet neurology. 2010;9(1):4–5. doi: 10.1016/S1474-4422(09)70330-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jack CR, Jr., et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet neurology. 2010;9(1):119–28. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jack CR, Jr., et al. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer’s disease. Brain : a journal of neurology. 2010;133(11):3336–48. doi: 10.1093/brain/awq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hampel H, et al. Biological markers of amyloid beta-related mechanisms in Alzheimer’s disease. Experimental neurology. 2010;223(2):334–46. doi: 10.1016/j.expneurol.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hampel H, et al. Biomarkers for Alzheimer’s disease therapeutic trials. Progress in neurobiology. 2010 doi: 10.1016/j.pneurobio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Holland D, et al. Subregional neuroanatomical change as a biomarker for Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(49):20954–9. doi: 10.1073/pnas.0906053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apostolova LG, et al. Three-dimensional gray matter atrophy mapping in mild cognitive impairment and mild Alzheimer disease. Archives of neurology. 2007;64(10):1489–95. doi: 10.1001/archneur.64.10.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Apostolova LG, et al. 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. Human brain mapping. 2010;31(5):786–97. doi: 10.1002/hbm.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Risacher SL, et al. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Current Alzheimer research. 2009;6(4):347–61. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fennema-Notestine C, et al. Structural MRI biomarkers for preclinical and mild Alzheimer’s disease. Hum Brain Mapp. 2009;30(10):3238–53. doi: 10.1002/hbm.20744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karow DS, et al. Relative capability of MR imaging and FDG PET to depict changes associated with prodromal and early Alzheimer disease. Radiology. 2010;256(3):932–42. doi: 10.1148/radiol.10091402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leung KK, et al. Automated cross-sectional and longitudinal hippocampal volume measurement in mild cognitive impairment and Alzheimer’s disease. Neuroimage. 2010;51(4):1345–59. doi: 10.1016/j.neuroimage.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonald CR, et al. Regional rates of neocortical atrophy from normal aging to early Alzheimer disease. Neurology. 2009;73(6):457–65. doi: 10.1212/WNL.0b013e3181b16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hua X, et al. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008;43(3):458–69. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuff N, et al. Nonlinear time course of brain volume loss in cognitively normal and impaired elders. Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.07.012. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McEvoy LK, et al. Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009;251(1):195–205. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu A, et al. Regional shape abnormalities in mild cognitive impairment and Alzheimer’s disease. Neuroimage. 2009;45(3):656–61. doi: 10.1016/j.neuroimage.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Apostolova LG, et al. Automated 3D mapping of baseline and 12-month associations between three verbal memory measures and hippocampal atrophy in 490 ADNI subjects. Neuroimage. 2010;51(1):488–99. doi: 10.1016/j.neuroimage.2009.12.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morra JH, et al. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer’s disease mild cognitive impairment, and elderly controls. Neuroimage. 2008;43(1):59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morra JH, et al. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Neuroimage. 2009;45(1 Suppl):S3–15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolz R, et al. Measurement of hippocampal atrophy using 4D graph-cut segmentation: Application to ADNI. Neuroimage. 2010;52(1):109–18. doi: 10.1016/j.neuroimage.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 57.Risacher SL, et al. Longitudinal MRI atrophy biomarkers: Relationship to conversion in the ADNI cohort. Neurobiol Aging. 2010;31(8):1401–1418. doi: 10.1016/j.neurobiolaging.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vemuri P, et al. Effect of apolipoprotein E on biomarkers of amyloid load and neuronal pathology in Alzheimer disease. Ann Neurol. 2010;67(3):308–16. doi: 10.1002/ana.21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hua X, et al. Sex and age differences in atrophic rates: an ADNI study with n=1368 MRI scans. Neurobiol Aging. 2010;31(8):1463–1480. doi: 10.1016/j.neurobiolaging.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ho AJ, et al. Obesity is linked with lower brain volume in 700 AD and MCI patients. Neurobiol Aging. 2010;31(8):1326–39. doi: 10.1016/j.neurobiolaging.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu X, et al. Assessing the reliability to detect cerebral hypometabolism in probable Alzheimer’s disease and amnestic mild cognitive impairment. J Neurosci Methods. 2010;192(2):277–85. doi: 10.1016/j.jneumeth.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen K, et al. Twelve-month metabolic declines in probable Alzheimer’s disease and amnestic mild cognitive impairment assessed using an empirically pre-defined statistical region-of-interest: findings from the Alzheimer’s Disease Neuroimaging Initiative. Neuroimage. 2010;51(2):654–64. doi: 10.1016/j.neuroimage.2010.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ewers M, et al. Neuroimaging markers for the prediction and early diagnosis of Alzheimer’s disease dementia. Trends in neurosciences. 2011 doi: 10.1016/j.tins.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valenzuela M, et al. Neuroimaging as endpoints in clinical trials: Are we there yet? Perspective from the first Provence workshop. Molecular psychiatry. 2011 doi: 10.1038/mp.2011.62. [DOI] [PubMed] [Google Scholar]