Abstract
Background
RNA interference (RNAi) has the potential to be more selective than small molecule inhibitors and can be used to target proteins, such as RAS, that are currently undruggable. The purpose of our study was to determine the optimal co-targeting strategy of the commonly mutated PI3K/AKT/mTOR and RAS pathways by a selective RNAi approach in colorectal cancer (CRC) cell lines possessing co-existent PIK3CA and KRAS mutations.
Methods
Human CRC cell lines HCT116 and DLD-1 were treated with a panel of siRNAs directed against the PI3K/AKT/mTOR and RAS pathways; proliferation, apoptosis, and protein expression were assessed. Combined treatment with siRNA and 5-fluorouracil (5-FU) was then evaluated.
Results
PIK3CA and KRAS siRNAs were most effective as single treatments; combined treatments with PIK3CA + KRAS siRNA resulted in a more pronounced inhibition of CRC proliferation. Either KRAS siRNA alone or combined PIK3CA + KRAS siRNA treatments increased apoptosis in HCT116 cells but not in the DLD-1 cell line. Inhibition of 4E-BP1 phosphorylation correlated with increased apoptosis. Additionally, siRNA treatment combined with 5-FU further inhibited CRC cell proliferation.
Conclusions
Combined PIK3CA + KRAS siRNA treatments offer an effective therapy against CRCs with co-existing mutations in both pathways. Decreased 4E-BP1 phosphorylation correlates with increased apoptosis and may provide a biomarker indicative of treatment success. Furthermore, siRNA directed to PIK3CA and KRAS may be used to enhance the effects of current chemotherapy.
Keywords: small interfering RNA (siRNA), RNA interference (RNAi), colorectal cancer, PI3K, KRAS, chemotherapy
INTRODUCTION
Colorectal cancer (CRC) is the second leading cause of cancer related deaths in the United States 1. The 5-year survival for localized disease is 90%; however, once distant organs are involved, survival drops to approximately 12% 1. Although progress has been made in survival for earlier stage disease, only minimal improvement has been noted in patients with systemic metastases 2. The poor prognosis associated with advanced disease indicates a need for more effective and targeted therapies in the treatment of metastatic CRC.
Phosphatidylinositol 3-kinase (PI3K), a ubiquitous lipid kinase composed of an 85 kDa regulatory subunit and a 110 kDa catalytic subunit, exerts many of its effects through its downstream effectors AKT and mTOR 3. The PI3K pathway has been associated with the growth and progression of many cancer types, including CRC, with mutations seen in approximately 40% of CRCs 4. The most commonly occurring mutations in this pathway are in the PIK3CA gene, which encodes the p110α catalytic subunit of PI3K 5. Increased expression of PI3K/AKT/mTOR pathway components has been demonstrated in CRC and surrounding stroma and correlates with tumor stage 6–8. Furthermore, these components play an important role in promoting CRC metastasis 6, 9, 10.
Mutations of the RAS pathway also occur frequently in CRCs. KRAS and BRAF mutations are present in up to 50% of CRCs and co-exist with PIK3CA mutations in approximately 30% 11, 12. These pathways can cooperate to drive the growth of a number of cancer types, including CRC 13. Moreover, inhibition of either of these pathways alone often results in only minimal anti-tumor effects in cancers harboring mutations in both pathways 14. This appears to be due to activation of shared downstream targets of the PI3K/AKT/mTOR and RAS pathways, such as 4E-BP1 14. These findings argue for the necessity of a co-targeting strategy for effective treatment of cancers possessing simultaneous mutations in these pathways. Although data supports the benefit of combined inhibition of these pathways, the use of small molecule inhibitors has certain drawbacks, including the potential for increased toxicity.
Small interfering RNA (siRNA) are 21–23 nucleotide RNA sequences capable of binding to and destroying complementary RNA strands thereby silencing gene expression 15. The highly selective and specific nature of siRNA offers the potential for more effective and less toxic treatments as compared to more traditional therapies 16. Furthermore, RNA interference (RNAi) can be used to knock down targets, such as RAS, that are currently undruggable 17. Although there are numerous benefits to RNAi, complications associated with in vivo treatments, such as rapid renal clearance, phagocytosis, aggregation with serum proteins, and degradation by endogenous nucleases, have limited its application 15, 16. Many of these issues have been overcome through the chemical modification of the siRNA structure 15. Furthermore, advances in nanotechnology have significantly improved systemic delivery of siRNA. An important example of this is a recently reported study demonstrating the first human trial showing specific gene inhibition in solid cancers by systemically delivered siRNA 18. As RNAi becomes an increasingly viable therapeutic option, it is important to establish the most effective targeted gene silencing for the treatment of specific cancer types. The purpose of our study was to determine the optimal RNAi therapy for CRCs possessing co-existent PIK3CA and KRAS mutations through the use of a novel siRNA co-targeting strategy.
MATERIALS AND METHODS
Cell lines, siRNA, reagents, and antibodies
HCT116 and DLD-1 cell lines were obtained from American Type Culture Collection (Manassas, VA). ON-TARGETplus SMARTpool siRNAs directed against PIK3R1 (L-003020), PIK3CA (L-003018), AKT1 (L-003000), AKT2 (L-003001), RICTOR (L-016984), RAPTOR (L- 004107), KRAS (L-005069), BRAF (L-003460), MEK1 (L-003571), MEK2 (L-003573), ERK1 (L-003592), ERK2 (L-003555) or a non-targeting control pool (D-001810-10) were purchased from Dharmacon (Lafayette, CO). Lipofectamine RNAiMAX transfection reagent was obtained from Invitrogen (Grand Island, NY). Cell Death Detection ELISAplus was acquired from Roche (Indianapolis, IN). For quantitative real time PCR (qRT-PCR), an RNeasy collection kit was obtained from Qaigen (Valencia, CA) and a high capacity cDNA reverse transcription kit as well as a TaqMan Gene Expression Master Mix and TaqMan probes for human PIK3CA (Hs00907957), KRAS (Hs00364284), and GAPDH (#4333764) from Applied Biosystems (Austin, TX). Monoclonal antibodies against p110α, pAKT, total AKT, AKT2, pERK 1/2, total ERK 1/2, RICTOR, RAPTOR, p4E-BP1, and total 4E-BP1 were purchased from Cell Signaling (Danvers, MA); AKT1, BRAF, MEK1, and MEK2 from Santa Cruz Biotechnology (Santa Cruz, CA); KRAS from Abcam (Cambridge, MA); and p85α from Millipore (Billerica, MA).
siRNA transfection
Cells were transfected using Lipofectamine RNAiMAX reagent according to the manufacturer’s protocol. The initial panel of single siRNA treatments was tested using 50 nM of non-targeting control (NTC), PIK3R1, PIK3CA, AKT1, AKT2, RICTOR, RAPTOR, KRAS, BRAF, MEK1, MEK2, ERK1, or ERK2 siRNA. Combination siRNA treatments were performed using either 100 nM NTC siRNA, 50 nM PIK3CA siRNA + 50 nM NTC siRNA, 50 nM KRAS siRNA + 50 nM NTC siRNA, or 50 nM PIK3CA siRNA + 50 nM KRAS siRNA in order to maintain an equivalent total concentration of siRNA in each treatment group.
Cell proliferation
Cells were plated at a density of 25,000 cells/well in 24 well plates. Cells were transfected 12 h later as described above. Seventy-two h after transfection, cells were trypsinized and counted using a Beckman-Coulter Vi Cell XR cell viability analyzer (Fullerton, CA). For applicable experiments, media was exchanged 4 h after transfection and replaced with media containing 5-fluorouracil (5-FU) at the described concentrations. Media was again exchanged with fresh drug media at 36 h and cells counted at 72h as described above.
DNA Fragmentation ELISA
Cells were plated at a density of 50,000 cells/well in 24 well plates. Cells were transfected 12 h later as described above. The media was exchanged for fresh growth media 4 h later. Growth media was removed 24 h following transfection, and cells were serum starved for an additional 24 h. Apoptosis was measured by DNA fragmentation using the Cell Death Detection ELISAplus as previously described 19.
qRT-PCR
Cells were plated in 6 well plates at a density of 120,000 cells/well. siRNA transfections were performed as described above. RNA was isolated at 48 h using an RNeasy mini kit. cDNA was synthesized using a high capacity cDNA reverse transcription kit. qRT-PCR was performed with a TaqMan gene expression master mix and TaqMan probes for PIK3CA, KRAS, or GAPDH. Relative quantification of PIK3CA and KRAS expression was performed based on threshold cycle (CT) normalized to GAPDH using the 2−ΔΔCT method.
Western blot analysis
Cells were plated in 6 well plates at a density of 120,000 cells/well. siRNA transfections were performed as described above. Whole cell lysates were collected 72 h following transfection. Western blot analysis was performed as previously reported 19 for relevant siRNA targeted proteins and the downstream effectors pAKT, total AKT, pERK, total ERK, p4E-BP1 and total 4E-BP1.
Statistical analysis
Data were analyzed using two-sample t-test. p < 0.05 was considered statistically significant.
RESULTS
siRNA directed to PIK3CA and KRAS is most effective in the inhibition of CRC proliferation
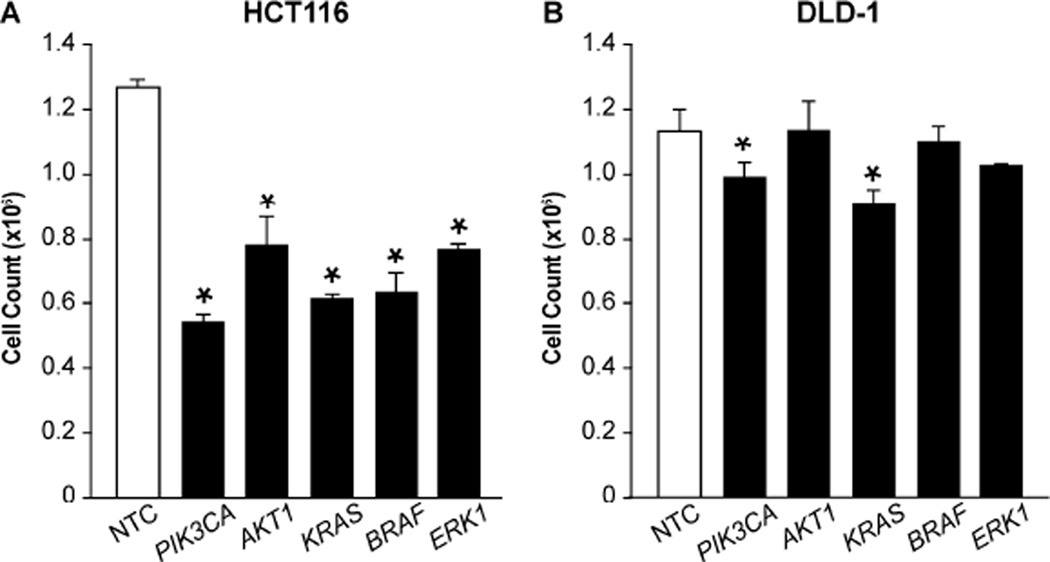
We first determined which siRNA was most effective in the treatment of CRC (Fig. 1). The HCT116 and DLD-1 cell lines were selected for testing due to the co-existent PIK3CA and KRAS mutations present in both cell lines. Based on previous studies, a panel of siRNA directed toward components of the PI3K/AKT/mTOR (PIK3R1, PIK3CA, AKT1, AKT2, RICTOR, or RAPTOR) or RAS (KRAS, BRAF, MEK1, MEK2, ERK1, or ERK2) pathways was selected for comparison. Cell viability was analyzed at 72 h to determine relative responsiveness to siRNA treatment. Proliferation in the HCT116 cell line was significantly decreased using siRNA directed to PIK3R1, PIK3CA, AKT1, AKT2, KRAS, BRAF, MEK1, MEK2, ERK1, or ERK2. The DLD-1 cell line, on the other hand, proved more resistant to treatments, with PIK3CA and KRAS siRNA resulting in the only significant reductions in proliferation. Collectively, the greatest effects for PI3K/AKT/mTOR and RAS pathway components were noted with PIK3CA and KRAS siRNA treatments, respectively.
Figure 1. PIK3CA and KRAS siRNAs most effectively inhibit CRC proliferation.
(A) HCT116 and (B) DLD-1 human colon cancer cells were plated in 24 well plates at a density of 25,000 cells/well. Cells were transfected 12 h later with a panel of single siRNAs directed to the PI3K/AKT/mTOR (PIK3R1, PIK3CA, AKT1, AKT2, RICTOR, RAPTOR) or RAS (KRAS, BRAF, MEK1, MEK2, ERK1, ERK2) pathways. Proliferation was assessed by cell counting at 72 h. Results for selected siRNA treatments are shown (* p < 0.05 vs. control).
Combination of PIK3CA + KRAS siRNA treatments enhance anti-proliferative effects compared with single siRNA treatments
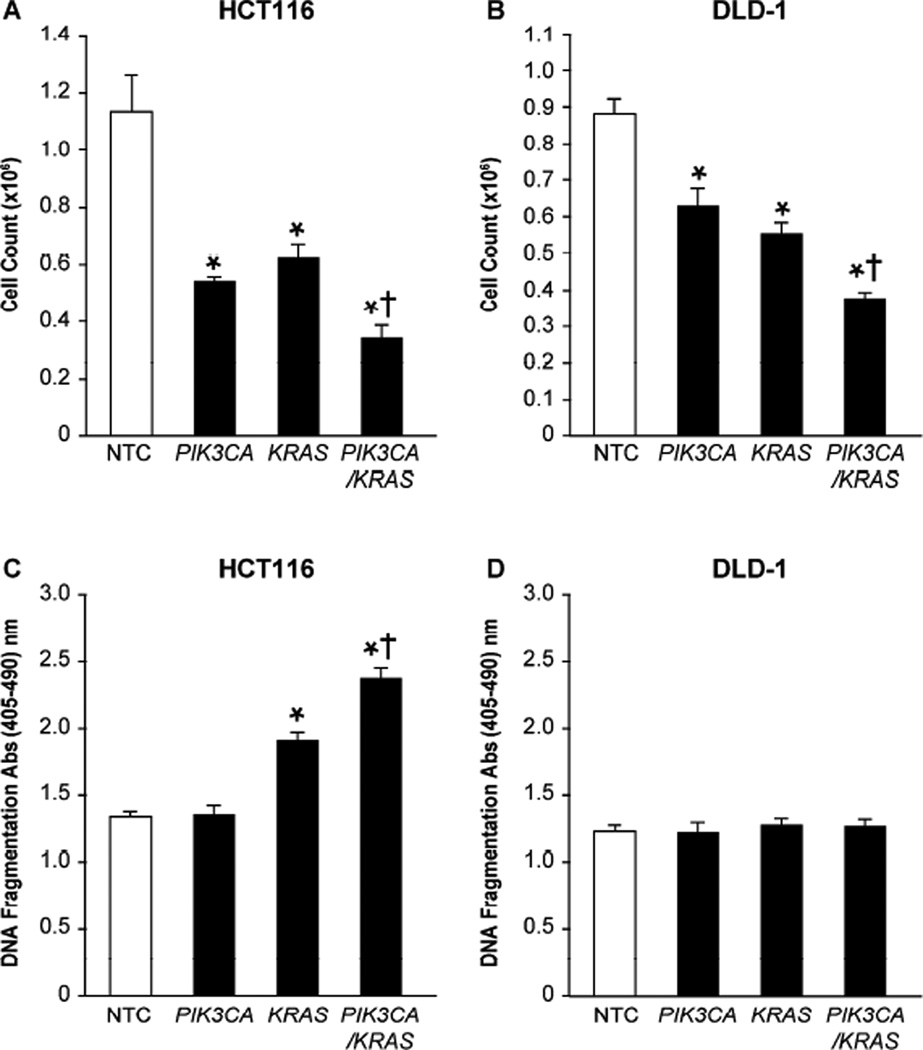
After confirming PIK3CA and KRAS as the most effective siRNAs in their respective pathways, we hypothesized that a combination strategy would result in greater effects relative to single agents in cell lines possessing activating mutations in both pathways. This was determined by treating HCT116 and DLD-1 cells with non-targeting control siRNA, siRNA directed to PIK3CA or KRAS, or combination PIK3CA + KRAS siRNAs and evaluating the effects on cellular proliferation and apoptosis. In both HCT116 and DLD-1 cells, individual PIK3CA or KRAS siRNA treatments significantly decreased proliferation relative to control (Fig. 2A and 2B). Furthermore, combination treatments significantly reduced proliferation relative to either PIK3CA or KRAS treatment alone.
Figure 2. Combined PIK3CA + KRAS siRNA treatments enhance anti-proliferative effects.
(A) HCT116 and (B) DLD-1 cells were plated in 24 well plates at a density of 25,000 cells/well. Cells were transfected 12 h later with 100 nM NTC siRNA, 50 nM PIK3CA siRNA + 50 nM NTC siRNA, 50 nM KRAS siRNA + 50 nM NTC siRNA, or 50 nM PIK3CA siRNA + 50 nM KRAS siRNA. Proliferation was assessed by cell counting 72 h following transfection. (C) HCT116 and (D) DLD-1 cells were plated in 24 well plates at a density of 50,000 cells/well. siRNA transfections were performed using the same treatment groups described above. Media was exchanged for fresh growth media 4 h later. Serum starved conditions were initiated 24 h after transfection and continued for an additional 24 h. Forty-eight h post transfection, apoptosis was measured by DNA fragmentation using Cell Death Detection ELISAplus (* p < 0.05 vs. control; † p < 0.05 vs. PIK3CA siRNA or KRAS siRNA alone).
More selected responses were noted with apoptosis as measured by DNA fragmentation. For the HCT116 cell line, KRAS siRNA significantly increased apoptosis relative to control; however, PIK3CA siRNA alone resulted in no discernable effect. Despite negligible apoptotic effects with isolated PIK3CA treatment, when PIK3CA siRNA was combined with KRAS siRNA, apoptosis was significantly increased relative to both control and KRAS siRNA treatments alone (Fig. 2C). Although increased apoptosis was noted in the HCT116 cell line, no significant increase was observed for DLD-1 (Fig. 2D).
Phosphorylation of the downstream effector 4E-BP1 correlates with apoptosis
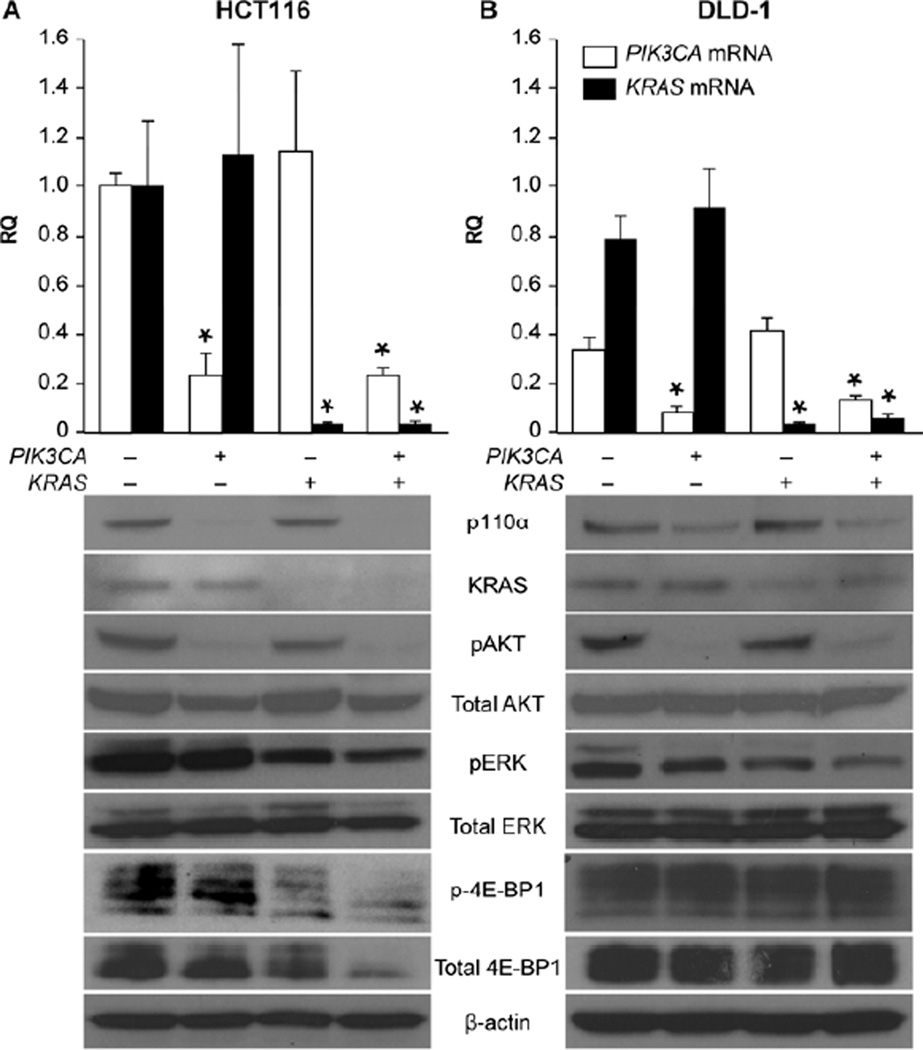
4E-BP1 has recently been identified as a downstream effector of both the PI3K/AKT/mTOR and RAS pathways and integrates the function of these pathways 14. In this present study, treatments that caused the greatest increase in apoptosis (KRAS and combination PIK3CA + KRAS siRNA treatments in the HCT116 cell line) correlated with reduced phosphorylation of 4E-BP1; similarly, only a minimal decrease in phosphorylation was noted with PIK3CA siRNA in the HCT116 cell line and no detectable difference was observed for any treatment in the DLD-1 cell line, again correlating with effects on apoptosis (Fig. 3A and 3B). Knockdown of siRNA targets was also demonstrated by Western blot analysis. This was again confirmed by qRT-PCR, which showed targeted mRNA knockdown in the PIK3CA, KRAS, and combination PIK3CA + KRAS siRNA treatment groups in both cell lines. Additionally, as expected, decreased phosphorylation of AKT and ERK was observed in cells treated with PIK3CA or KRAS siRNA, respectively.
Figure 3. Decreased phosphorylation of 4E-BP1 correlates with increased apoptosis.
(A) HCT116 and (B) DLD-1 cells were plated in 6 well plates at a density of 1.2×105 cells/well. Cells were transfected 12 h later with 100 nM NTC siRNA, 50 nM PIK3CA siRNA + 50 nM NTC siRNA, 50 nM KRAS siRNA + 50 nM NTC siRNA, or 50 nM PIK3CA siRNA + 50 nM KRAS siRNA. RNA was isolated 48 h post transfection and PIK3CA and KRAS mRNA levels measured using qRT-PCR (* p < 0.05 vs. control; RQ = relative quantity). HCT116 and DLD-1 cells were again plated and transfected under the same conditions to evaluate corresponding protein expression. Whole cell lysates were collected at 72 h and analyzed by Western blot (20 to 60 ug total protein per lane). β-actin was used as a loading control.
PIK3CA and KRAS mutational status alters the response of CRC to siRNA treatment
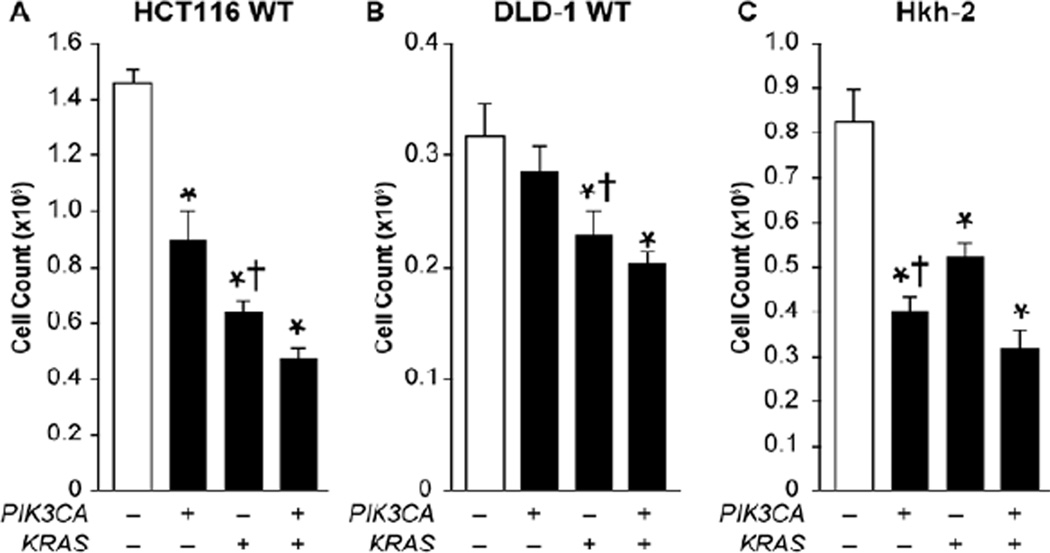
While siRNAs directed to PIK3CA and KRAS produced the greatest effects on proliferation in both cell lines, we hypothesized that oncogenic activation of the PI3K/AKT/mTOR and RAS pathways, resulting from PIK3CA and KRAS mutations, respectively, would influence response to these treatments. In order to determine the role that PIK3CA mutation plays in treatment sensitivity, cell viability was tested in HCT116 and DLD-1 isogenic cells in which the mutant PIK3CA allele is deleted 20. These isogenic cells, therefore, offer the unique ability to evaluate a change specifically in the gene of interest while maintaining an otherwise identical genetic background. We found that PIK3CA siRNA was less effective as a single treatment, particularly in the DLD-1 isogenic cells in which PIK3CA siRNA treatment did not achieve significance relative to control (Fig. 4A and 4B). Furthermore, in both HCT116 and DLD-1 isogenic cells, KRAS siRNA was significantly more effective as an individual treatment compared to individual PIK3CA siRNA treatment.
Figure 4. siRNA directed to the mutated pathway of isogenic cells more effectively inhibits proliferation.
(A) HCT116 Wild Type (WT) – mutant PIK3CA allele deleted, (B) DLD-1 WT – mutant PIK3CA allele deleted, and (C) Hkh-2 – HCT116 cells with mutant KRAS allele deleted, were plated in 24 well plates at a density of 25,000 cells/well. Cells were transfected 12 h later with 100 nM NTC siRNA, 50 nM PIK3CA siRNA + 50 nM NTC siRNA, 50 nM KRAS siRNA + 50 nM NTC siRNA, or 50 nM PIK3CA siRNA + 50 nM KRAS siRNA. Proliferation was assessed by cell counting at 72 h (* p < 0.05 vs. control; † p < 0.05 vs. alternate single siRNA treatment).
To determine the effects that KRAS mutation has on responsiveness of CRC to siRNA treatments, HCT116 isogenic cells in which the KRAS mutant allele is deleted by homologous recombination were used 21. Therefore, in contrast to the HCT116 and DLD-1 PIK3CA wild type isogenic cells, the Hkh-2 cells retain their mutant PIK3CA expression but have wild type KRAS expression. While KRAS siRNA treatments still reduced proliferation relative to control, PIK3CA siRNA was significantly more effective as a single agent therapy in these cells (Fig. 4C). Overall, when either the mutant PIK3CA or KRAS allele was deleted in the respective isogenic cell lines, siRNA directed to the mutated pathway was significantly more effective than siRNA directed to the non-mutated pathway.
siRNA directed to PIK3CA and KRAS enhances inhibition of proliferation associated with 5-FU
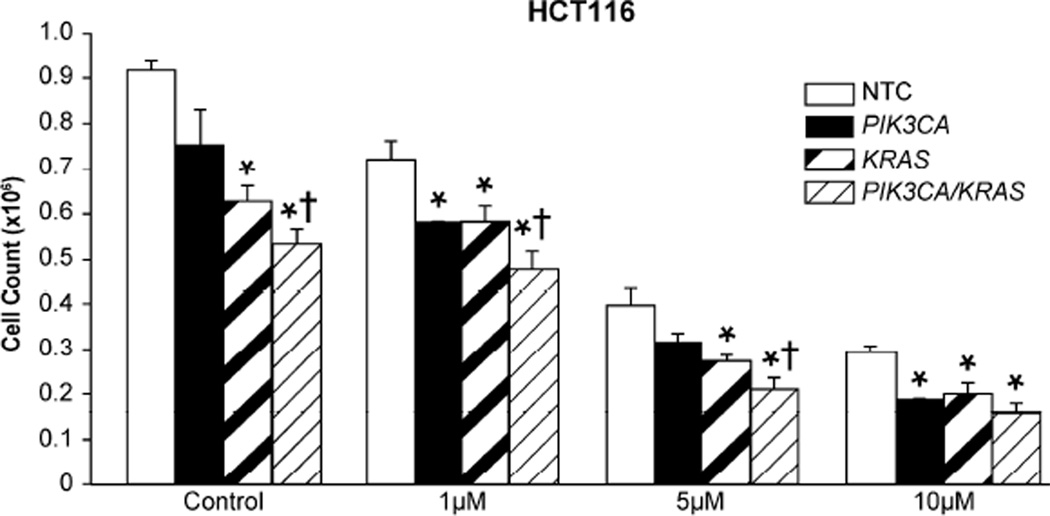
Given the beneficial effects that were noted with siRNA therapy, we next determined if siRNA treatment could enhance the effects of traditional chemotherapy. HCT116 cells were transfected with siRNA and subsequently treated with increasing doses of 5-FU. 5-FU combined with PIK3CA siRNA resulted in significantly decreased proliferation at both the 1 uM and 10 uM doses and with KRAS siRNA at all doses tested (Fig. 5). Additionally, 5-FU and combination PIK3CA + KRAS siRNA treatments resulted in the greatest overall decrease in proliferation compared with cells treated with single siRNAs and 5-FU. Furthermore, combined PIK3CA + KRAS siRNA in tandem with 5-FU (5 uM) resulted in a greater decrease in proliferation than 5-FU alone at twice the concentration.
Figure 5. siRNA treatments enhance inhibition of proliferation associated with 5-FU.
HCT116 cells were plated in 24 well plates at a density of 25,000 cells/well. Cells were transfected 12 h later with 100 nM NTC siRNA, 50 nM PIK3CA siRNA + 50 nM NTC siRNA, 50 nM KRAS siRNA + 50 nM NTC siRNA, or 50 nM PIK3CA siRNA + 50 nM KRAS siRNA. Media was exchanged 4 h later for either control media containing DMSO only, or media containing 1, 5, or 10 uM of 5-FU. Media was again exchanged for fresh drug media at 36 h post transfection. Proliferation was assessed by cell counting at 72 h (* p < 0.05 vs. control; † p < 0.05 vs. PIK3CA siRNA or KRAS siRNA alone).
DISCUSSION
In this study, we evaluated the use of siRNA as a treatment for CRC possessing activating mutations in the PI3K/AKT/mTOR and RAS pathways. First, by examining a panel of siRNAs directed against the PI3K/AKT/mTOR and RAS pathways we determined that PIK3CA and KRAS are the optimal siRNA treatments in their respective pathways. Second, we showed that combined PIK3CA + KRAS treatments resulted in reduced proliferation over either agent alone. Furthermore, in the HCT116 cell line, there was an increase in apoptosis with KRAS siRNA treatment and an even greater effect with combined PIK3CA + KRAS siRNA treatment. Third, we demonstrated that phosphorylation of the downstream effector 4E-BP1 correlated with increased apoptosis. Fourth, we showed sensitivity to siRNA treatments is influenced by the mutational status of the cancer as evidenced by the reduced response to treatments targeting pathways in cell lines in which the mutant allele had been removed. Finally, we showed that when siRNA treatments are combined with the traditional chemotherapeutic agent 5-FU, there is an enhanced response over either agent alone.
Mutated pathways in human cancer, such as PI3K/AKT/mTOR and RAS, are able to provide a survival advantage over their normal counterparts; however, cancers are also often dependent on these pathways for survival. One approach to target these mutated pathways is the use of small molecule inhibitors. Unfortunately, depending on the specificity of the drug, adverse side effects frequently occur. Furthermore, targets of these drugs are limited to proteins with enzymatic activity, which means many targets, such as RAS, remain undruggable 4, 12. siRNA provides a unique alternative in that it is highly specific and has the ability to provide greater effect with less toxicity than small molecule inhibitors 15–17. We have demonstrated that siRNA specifically targeting mutated pathways in CRC provides an effective treatment and is capable of both inhibiting proliferation and inducing apoptosis. Although siRNA can be highly effective, the variable response noted between individual siRNAs indicate that selection of optimal targets is an important and necessary step in maximizing treatment effectiveness. Additionally, data from the isogenic cell lines shows that alterations in mutational profile results in a change in treatment response. Given the critical role these pathways play in cell growth and proliferation, inhibition of the non-mutated pathway still demonstrated some effect; however, targeting of the mutated pathway resulted in a significantly more pronounced response. These findings further support the importance of a mutation-based treatment strategy.
While optimal targeting of an individual pathway is critical, the overall mutational profile of the cancer needs to be taken into account. Mutations in the PI3K/AKT/mTOR and RAS pathways frequently occur in CRC and often co-exist 11, 12. Cancers that have aberrant signaling in only one of these pathways tend to demonstrate a dependency on the activated pathway and are usually sensitive to selective inhibition 22. However, when co-activation of both pathways is present, the sensitivity to inhibition of either pathway alone is often minimal 14, 22. Instead, combined targeting of both pathways can prevent resistance and is able to produce more pronounced effects 14. Our present data correlate with these findings. While there are modest effects when siRNA is used to inhibit either of these pathways alone, an increased response occurs when both pathways are targeted simultaneously. This indicates that a co-targeting siRNA strategy will likely be the most effective treatment for CRCs that possess this commonly occurring mutational profile.
4E-BP1 is a small, heat stable protein that plays an important role in cap-dependent translation by binding to and inhibiting eIF4E 23. High levels of phosphorylated 4E-BP1 have been associated with poor prognosis in a number of cancer types 24. Additionally, this protein has recently been identified as a key downstream effector of both the PI3K/AKT/mTOR and RAS pathways and appears to play an integral role in exerting their effects 14. In our study, we showed that inhibition of the PI3K pathway alone, through the use of siRNA directed to PIK3CA, resulted in minimal decreases in phosphorylation of 4E-BP1 in the HCT116 cell line. KRAS siRNA treatments produced a more pronounced response; however, the greatest decreases in p4E-BP1 occurred when cells were treated with combined PIK3CA + KRAS siRNAs. Interestingly, HCT116 cells demonstrated a similar pattern for apoptosis. Additionally, the DLD- 1 cell line showed neither an increase in apoptosis nor a decrease in phosphorylation of 4E-BP1 with any siRNA treatments. This variation is likely secondary to differences in their genetic profile. While HCT116 and DLD-1 cells both have PIK3CA and KRAS mutations, abnormal signaling resulting from additional mutations, such as a p53 mutation in DLD-1, may diminish the response to targeting of the PI3K/AKT/mTOR and RAS pathways. However, given the strong correlation of 4E-BP1 phosphorylation with induction of apoptosis, our data indicate that phosphorylation of 4E-BP1 may be of potential use as a biomarker indicative of treatment success.
While we have shown that siRNA treatment is effective as an individual therapy, some data exists supporting its use in combination with chemotherapeutic agents 19. Although a number of new agents have been introduced, 5-FU based therapy presently remains the backbone of CRC medical management in the clinical setting; however, 5-FU treatment is often complicated by toxicity, frequently resulting in early termination of therapy and decreased compliance with recommended treatment regimens 25. Furthermore, despite the use of presently available therapies, survival remains poor 1. Here we show that siRNA directed against appropriately selected targets in mutated pathways can be used as a co-treatment with 5-FU to improve the inhibition of CRC. These findings indicate that siRNA may augment 5-FU based therapy while potentially allowing for treatment with lower doses, thereby decreasing adverse effects and increasing the number of patients able to complete appropriate treatment regimens.
In summary, our data indicate that siRNA can provide an effective, mutation-based therapy for the management of CRC. Additionally, in CRC with co-existing mutations in the commonly activated PI3K/AKT/mTOR and RAS pathways, combined targeting of these pathways is likely to produce the greatest effect. Moreover, we showed that phosphorylation of the downstream effector 4E-BP1 may serve as a biomarker indicative of treatment success and that siRNA treatment enhances the response of CRC to traditional chemotherapy. Collectively, our findings strongly support the use of siRNA as an effective therapeutic modality in the treatment of CRC.
ACKNOWLEDGEMENTS
We would like to thank Drs. Qing-Bai She, Jing Li, and Jun Song for their assistance with project design and interpretation, Drs. B. Vogelstein and V. E. Veculescu (Johns Hopkins University) for providing HCT116 and DLD-1 PIK3CA isogenic cells, and Donna Gilbreath for manuscript preparation.
Financial Support: This work is supported by grant P20CA153043 (GI SPORE) from the National Institutes of Health.
ABBREVIATIONS
- RNAi
Ribonucleic acid interference
- CRC
colorectal cancer
- 5-FU
5-fluorouracil
- PI3K
Phosphatidylinositol 3-kinase
- siRNA
Small interfering ribonucleic acid
- qRT-PCR
Quantitative real time polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have no conflicts of interest regarding publication of this manuscript.
REFERENCES
- 1.Cancer Facts & Figures 2011. Atlanta: American Cancer Society; 2011. [Google Scholar]
- 2.van der Pool AE, Damhuis RA, Ijzermans JN, de Wilt JH, Eggermont AM, Kranse R, et al. Trends in incidence, treatment and survival of patients with stage IV colorectal cancer: a population-based series. Colorectal disease : the official journal of the Association of Coloproctology of Great Britain and Ireland. 2012;14:56–61. doi: 10.1111/j.1463-1318.2010.02539.x. [DOI] [PubMed] [Google Scholar]
- 3.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 4.Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 5.Velho S, Oliveira C, Ferreira A, Ferreira AC, Suriano G, Schwartz S, Jr, et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41:1649–1654. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 6.Gulhati P, Cai Q, Li J, Liu J, Rychahou PG, Qiu S, et al. Targeted inhibition of mammalian target of rapamycin signaling inhibits tumorigenesis of colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:7207–7216. doi: 10.1158/1078-0432.CCR-09-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson SM, Gulhati P, Rampy BA, Han Y, Rychahou PG, Doan HQ, et al. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. Journal of the American College of Surgeons. 2010;210:767–776. 76–78. doi: 10.1016/j.jamcollsurg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gulhati P, Bowen KA, Liu J, Stevens PD, Rychahou PG, Chen M, et al. mTORC1 and mTORC2 Regulate EMT, Motility, and Metastasis of Colorectal Cancer via RhoA and Rac1 Signaling Pathways. Cancer Res. 2011;71:3246–3256. doi: 10.1158/0008-5472.CAN-10-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rychahou PG, Jackson LN, Silva SR, Rajaraman S, Evers BM. Targeted molecular therapy of the PI3K pathway: therapeutic significance of PI3K subunit targeting in colorectal carcinoma. Annals of surgery. 2006;243:833–842. doi: 10.1097/01.sla.0000220040.66012.a9. discussion 43-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rychahou PG, Kang J, Gulhati P, Doan HQ, Chen LA, Xiao SY, et al. Akt2 overexpression plays a critical role in the establishment of colorectal cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:20315–20320. doi: 10.1073/pnas.0810715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, et al. Highthroughput oncogene mutation profiling in human cancer. Nature genetics. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 12.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA : the journal of the American Medical Association. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 13.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 14.She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer cell. 2010;18:39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nature reviews Drug discovery. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rychahou PG, Jackson LN, Farrow BJ, Evers BM. RNA interference: mechanisms of action and therapeutic consideration. Surgery. 2006;140:719–725. doi: 10.1016/j.surg.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nature reviews Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rychahou PG, Murillo CA, Evers BM. Targeted RNA interference of PI3K pathway components sensitizes colon cancer cells to TNF-related apoptosis-inducing ligand (TRAIL) Surgery. 2005;138:391–397. doi: 10.1016/j.surg.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 22.Yu K, Toral-Barza L, Shi C, Zhang WG, Zask A. Response and determinants of cancer cell susceptibility to PI3K inhibitors: combined targeting of PI3K and Mek1 as an effective anticancer strategy. Cancer biology & therapy. 2008;7:307–315. doi: 10.4161/cbt.7.2.5334. [DOI] [PubMed] [Google Scholar]
- 23.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annual review of biochemistry. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 24.Armengol G, Rojo F, Castellvi J, Iglesias C, Cuatrecasas M, Pons B, et al. 4E-binding protein 1: a key molecular "funnel factor" in human cancer with clinical implications. Cancer Res. 2007;67:7551–7555. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 25.Kahn KL, Adams JL, Weeks JC, Chrischilles EA, Schrag D, Ayanian JZ, et al. Adjuvant chemotherapy use and adverse events among older patients with stage III colon cancer. JAMA : the journal of the American Medical Association. 2010;303:1037–1045. doi: 10.1001/jama.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]