Abstract
Bacterial aggregation and patchiness play an important role in a variety of ecological processes such as competition, adaptation, epidemics, and succession. Here, we demonstrate that hydrodynamics of their environment can lead to their aggregation. This is specially important since microbial habitats are rarely at rest (e.g., ocean, blood stream, flow in porous media, and flow through membrane filtration processes). In order to study the dynamics of bacterial collection in a vortical flow, we utilize a microfluidic system to mimic some of the important microbial conditions at ecologically relevant spatiotemporal scales. We experimentally demonstrate the formation of “ring”-shaped bacterial collection patterns and subsequently the formation of biofilm streamers in a microfluidic system. Acoustic streaming of a microbubble is used to generate a vortical flow in a microchannel. Due to bacteria's finite-size, the microorganisms are directed to closed streamlines and trapped in the vortical flow. The collection of bacteria in the vortices occurs in a matter of seconds, and unexpectedly, triggers the formation of biofilm streamers within minutes. Swimming bacteria have a competitive advantage to respond to their environmental conditions. In order to investigate the role of bacterial motility on the rate of collection, two strains of Escherichia coli bacteria with different motilities are used. We show that the bacterial collection in a vortical flow is strongly pronounced for high motile bacteria.
INTRODUCTION
Motility, or self-propulsion, is an inevitable part of most microorganisms' life cycles. It is developed as a survival skill to help them cope with heterogeneous environmental circumstances, such as temporal and spatial changes in stimuli.1 Another indication of the importance of motility is a variety of swimming strategies established to escape predators. It has been shown that motility plays a significant ecological role in marine life, especially in forming microbial patches in optimal positions. Patchiness, or spatial heterogeneity, has shown a remarkable influence on adaptation, epidemics, succession, maintenance of species diversity, and many other ecological processes.2 Surrounded by fluids in microorganisms' habitats, these processes are inseparable from fluid flow. However, the effect of the fluid flow on the swimming, its interaction with swimmers, and aggregation patterns of microorganisms is still ambiguous. Studies concerning the influence of fluid flow on dynamics of motile microorganisms and their aggregation are mainly numerical.3, 4, 5 However, the effects of the flow on microorganisms' swimming orientation have been experimentally reported.6 The limited number of experimental work can be contributed to technological shortcomings in creating and controlling flow environments similar to microbial habitats. Over the last few years, microfluidics has proven to be a promising tool for microscale studies due to its flexible geometry, fast fabrication, and ease of use. Also, by utilizing its small dimensions, one can mimic some of the important microbial environmental conditions.
Here, we use microfluidics as a means of creating fast and tunable vortical flow, utilizing oscillations of a microbubble. When exposed to ultrasound waves, bubbles go through translational and radial oscillations which rectify their surrounding fluid to a steady vortical flow, known as “cavitation” or “acoustic” streaming.7, 8 For an oscillating bubble adjacent to a boundary, the induced vortical flow consists of one pair of vortices. We show that bacteria collect in these vortices due to their finite-size. More quantification reveals that collection is faster and more intense for bacteria with higher motility. Although there are other promising techniques to create microvortices in order to trap or collect particles in microfluidic systems,9, 10, 11, 12, 13, 14, 15 cavitation microstreaming was used here due to its simple formation, ease of control, and the fact that collection is independent of the particle charge. Unexpectedly, within a matter of minutes after bacterial collection in the vortical flow, microbial colonies similar to biofilm streamers are observed which, to the best of our knowledge, have not been reported before. The typical size of an observed biofilm streamer, when completely stretched, and 2–3 min after it is appeared in the vortex, is approximately 40 μm, which is about 20 times the length of a single cell body.
Biofilms are communities of bacteria surrounded by self-secreted extracellular polymeric substances (EPS). They are present in nature in a variety of architectures; one of which is known as biofilm streamers.16, 17, 18 While attached to a surface from one side, streamers have a free tail moving with the flow and have been reported to form under the condition of laminar19 and turbulent flows.20 Upon formation of biofilms, microbial resistance to antibiotics raises up to 1000 times compared to that of planktonic bacteria.21 Combined with the fact that over 80% of microbial infections in the human body are caused by biofilms,22 it is clear that biofilms are to be avoided in health-related issues such as infections, wound healing or medical implants. Therefore, detection of bacteria and biofilm is important in medical diagnosis as well as in microbiological analysis of food, water, and environmental samples.23, 24
Biofilms cost the U.S. alone billions of dollars every year due to human and animal infections,25, 26 industrial equipment damage,27 product contamination, energy losses,28 and biofouling of membranes29 (e.g., it is estimated that up to 30% of the total operating costs of some Water Factories are related to controlling biofouling30). Biofilms, however, can also be beneficial; they are used for waste treatment and remediation,31 cleaning oil spills, filtering wastewater, and forming bio-barriers to prevent groundwater contamination.28, 32 Microbial biofilms in porous media are investigated as a means of preventing “leakage” of , which can occur due to the permeability of the cap-rock, at subsurface geological carbon sequestration sites.33 Vortical structures and secondary flows are present in porous media even at small Reynolds numbers. More recently, Rusconi et al.19 have shown that secondary vortical motion at the corners of curved microchannels leads to the formation of biofilm streamers. Despite its widespread implications, the underlying hydrodynamics of bacterial aggregation and the formation of biofilm streamers in the presence of vortical flows is currently poorly understood. To understand the formation and growth of biofilm streamers, the dynamics of bacterial aggregation at ecologically relevant spatiotemporal scales in the presence of flow must be studied. In this paper, we use microfluidics as a promising method to generate a vortical flow and investigate the behavior of bacteria in a heterogeneous flow field. Our experimental results show that vortical flow of an oscillating bubble leads to the collection of bacteria and subsequently the formation of filamentous biofilm streamers and bacterial active swimming, i.e., motility, can increase the bacterial collection rate.
MATERIAL AND METHODS
Microfluidic experimental setup
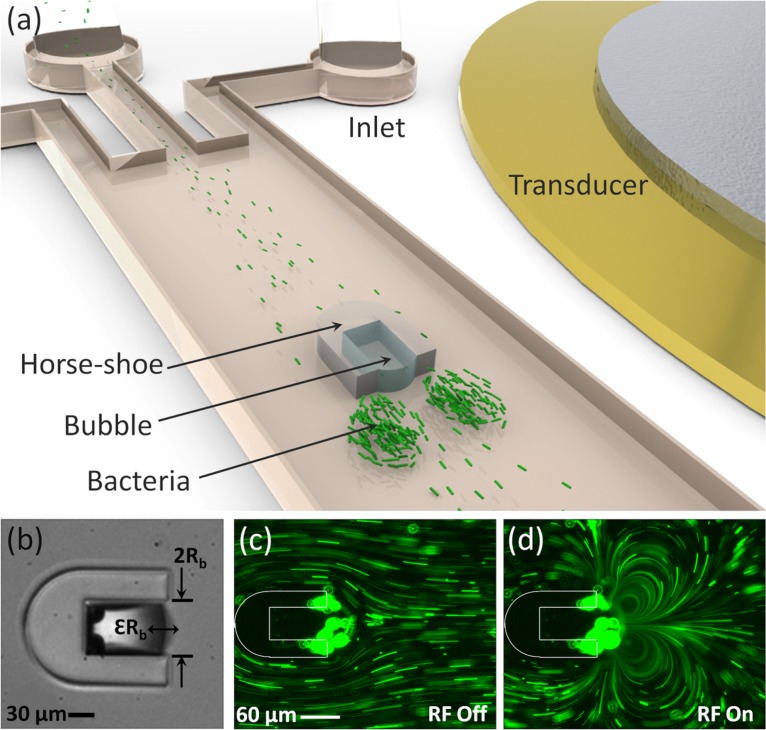
To study the interaction of bacterial suspension with a vortical flow field, we use a microfluidic device as a convenient tool to generate a vortical flow. Our device consists of three inlets, a horseshoe structure downstream of the inlets within the main channel, and one outlet. The design of horseshoe structure was first introduced by Ahmed et al.34 for rapid mixing in microfluidics systems. A schematic of the microfluidic device is shown in Fig. 1a. The main channel has a rectangular cross section 570 μm wide × 90 μm deep. All of the inlets have an equal width of 80 μm and the same depth as the main channel. The microfluidic channel is fabricated out of polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning, USA) using standard soft-lithography and bonded to a cover glass using oxygen plasma.35Escherichia coli (E. coli) suspension is injected from the inlets into the main channel via a syringe pump (KDS210, Scientific). As a result, the microchannel is filled with the fluid, however, the horseshoe structure reliably retains an air bubble of a semi-cylindrical shape. Upon applying a radio frequency (RF) signal to an adjacent piezoelectric transducer (Model No. 273-073, RadioShack®), the air-fluid interface begins to oscillate. This oscillation generates pressure and velocity fluctuations in the adjacent fluid and creates a pair of vortices next to the oscillating bubble. This phenomenon is called “cavitation microstreaming” and is more pronounced when the bubble is oscillating at its resonance frequency.7, 8 It should be noted that no vortices are generated in the absence of a bubble even in the presence of the piezoelectric excitation. Fig. 1b shows the amplitude of oscillation at the interface of a microbubble trapped in the horseshoe structure exposed to acoustic waves at 30 kHz frequency and voltage . Trajectories of 1.9 μm fluorescent spherical polystyrene beads in water are used to demonstrate the flow field adjacent to the horseshoe structure before and after applying the acoustic wave in Figs. 1c, 1d, respectively. The motion of particles and bacteria is observed through an inverted microscope (Nikon Ti2000) with a 10× objective lens and recorded via a CCD camera (CoolSNAP, PHOTOMETRICS©). It will be shown later that collection in the vortices is not observed with these tracer particles at the applied voltage of , but only with the larger bacterial cells.
Figure 1.
(a) Schematic of the microfluidic device with a built-in horseshoe structure. Bacterial suspension is injected from the inlets and collects in a pair of vortices induced by the oscillating microbubble trapped in the horseshoe. (b) The microbubble trapped in a horseshoe structure undergoes weakly linear oscillations. is the amplitude of oscillation of a bubble with diameter. A pair of vortices is generated upon applying the radio frequency signal. 1.9 μm polystyrene microbeads are used as flow tracers to demonstrate the flow field when the transducer is (c) off and (d) on. Structure of the horseshoe is shown with white lines for visualization.
Bacterial strains and growth conditions
Escherichia coli strains RP437 with high motility and (derived from K-12) with low motility36, 37 are used in the experiments. These two strains are transformed with plasmid pQE8-GFP-DD, which has an IPTG-inducible T5 promoter, to drive the expression of green fluorescent protein (GFP) in E. coli. To form single colonies, RP437-GFP and -GFP are grown on LB-Agar plates (1.0% tryptone + 0.5% yeast extract + 1.0% NaCl + 1.5% Agar) with 50 μg/ml ampicillin at a temperature of overnight. Colonies of these two strains are cultured in 5 ml TB (1.0% tryptone + 0.8% NaCl) liquid medium with 50 μg/ml ampicillin in a shaking incubator ( and 220 rpm) overnight. From these cultures, 50 μl is added into 5 ml TB liquid medium with 50 μg/ml ampicillin (1:100 dilution) and grown in the shaking incubator (VWR©, Model 1570) at and 220 rpm for approximately 4 h before 5 μl of 1 M IPTG solution is added. Growth is measured by the optical density at 600 nm (OD600) and it was recorded at 0.25 for both strains at the beginning of the experiments. After the OD600 was measured in the culture, the bacteria were not further diluted and were injected in the channel with OD600 of 0.25.
RESULTS AND DISCUSSION
Bacterial collection in the vortex
Once the vortical flow is generated, bacteria start to collect within the vortices as demonstrated in Fig. 2. Fluorescent light intensity (FLI), which is an indication of the bacterial concentration, significantly increases with time in the vortex pair (Figs. 2a, 2b, 2c, 2d). To better demonstrate the position of collected bacteria with respect to the vortices, the contours of FLI obtained from the experiments are overlaid on streamlines from the numerical simulation results (Figs. 2e, 2f, 2g, 2h). The numerical results are obtained using ANSYS two-dimensional Navier-Stokes solver on the geometry described in Sec. 2A. The volumetric flow rate of injected liquid into the channel is 1 μl/min in both experiments and numerical simulations. Inlet and outlet boundary conditions have been used for right and left boundaries of the domain, respectively. No-slip boundary condition is imposed on the top and bottom boundaries and walls of the horseshoe structure. The bubble interface oscillates with time as , where ε is the bubble's dimensionless amplitude of oscillation, is the radius of the bubble, f is the frequency of oscillation of the bubble, and t is time. No-slip boundary condition is used for the bubble interface rather than zero shear stress since surfactant present in the medium can effectively rigidify the surface of the microbubble.38 The simulations are done for a pure fluid to visualize the flow field. Since the volume fraction of bacteria and particles is low, the flow field is not affected by their presence. However, when bacteria is collected in the vortices, they can modify the flow field, however, this is not captured in our numerical simulation. If the RF signal is maintained for a relatively long time (more than 30 min), the bubble starts to enlarge, which is successively followed by a size change in vortices, eventually affecting and diminishing the collection. This enlargement of the bubble is mostly due to the translational oscillation of the bubble due to its excitation at resonance frequency and can be reversed, i.e., shrunken back to its original size, by shortly increasing the pressure in the microchannel by injecting fluid from both inlet and outlet for a short time. It is also worth noting that the experimentally observed asymmetry in bacterial collection in the two vortices can be caused by asymmetric flow due to different modes of bubble oscillation, microchannel geometry deficiencies, and asymmetry in the inlet flow.
Figure 2.
Contours of FLI, which indicate bacterial concentration, are plotted for RP437 E. coli at different time snapshots. (e)-(h) Bacteria collect in the vortex pair as shown by FLI contours overlaid on the flow streamlines (solid blue lines).
The underlying collection mechanism is based on the finite-size of cells. However, bacterial motility increases the rate and amount of collection. Bacteria enter the microchannel with a mean upstream velocity as the Poiseuille flow is dominant far away from the bubble in the microchannel. Near the oscillating bubble, cavitation microstreaming is dominant and the maximum streaming velocity can be approximated as , where ϵ is the ratio of bubble's amplitude of oscillation to its radius.39 It should be noted that within the range of applied voltage in our experiments, amplitude of oscillation is a linear function of applied voltage. A voltage within the range of is applied to the piezoelectric which leads to a streaming velocity of near the bubble interface. Upon the generation of cavitation microstreaming, the inlet flow must pass through a gap between the bubble and closed streamlines adjacent to the bubble. If the effective radius of the bacteria is larger than the gap width, they cannot penetrate the bubble and are forced to enter the closed streamlines, i.e., vortices. The same mechanism was recently used by Wang et al.39 as a size-sensitive sorting concept in a microfluidic system. They utilized an oscillating microbubble to trap polystyrene particles of radius 5 μm.40 More details on the working mechanism can be found in Ref. 39. The gap width can be estimated as , where H is the width of the channel. To confirm the working mechanism, we used spherical polystyrene particles (Bangs Lab, Inc.) with a mean diameter of 1.9 μm. As predicted, no collection was observed in the applied range of voltage. By increasing the voltage to , gap width decreases to about 1 μm, which is small enough to accumulate the microparticles in the vortices. When a rod-shaped object, such as E. coli, enters the gap, its orientation dictates whether it can pass through the gap or be pushed into the vortex. Within the range of streaming velocity near the bubble used in our experiments, rod-shaped bacteria are not necessarily aligned with the streamlines.6E. coli has a rod-shaped body approximately 2 μm long and 1 μm wide.41, 42 Considering 5 μm long bundled flagella, the overall length of the bacteria exceeds the gap width and the bacteria injected into the inlet are trapped in the vortex and their concentration increases with time. Another mechanism that contributes to trapping and collection near an oscillating bubble is due to the radiation force of an acoustic wave, acting on floating objects with different density compared to the background fluid.43 For a spherical particle with radius , radiation force is calculated from , where , and r are the density of the medium, radial surface velocity amplitude of the bubble, and radial distance, respectively. B is given by , where is the density of the spherical object.43 At f = 30 kHz and voltage range used in this work, the magnitude of the radiation force at 1 μm and 60 μm from the bubble's surface is about 0.8 μN and 0.001 pN, respectively. Since the radiation force has a very short range, , its role on the initial bacterial collection is negligible, even though it may affect initial formation of the biofilm streamer around the bubble. Shear-induced migration, which is responsible for the cross-streamline migration towards the low shear rate region, i.e., center of the vortex, is another mechanism that can lead to particle trapping within the vortices.10 However, shear-induced migration is not the driving mechanism in these experiments due to low volume fraction of bacteria. Shear-induced migration velocity scales as , where is the bacteria volume fraction. Also, to evaluate any effect of bacterial swimming towards the oxygen (bubble), i.e., oxytaxis, we conducted experiments with bacteria in a substrate with bubbles with no background flow and acoustic waves and we observed that the concentration of bacteria increases around the bubble after about 6–7 h. Therefore, given the short time of our experiments, we can safely rule out any possibility of bacterial swimming towards the bubble due to oxytaxis.
To investigate the effect of bacterial motility, experiments were repeated with two strains of E. coli exhibiting different motilities. strain with measured motility around 1 μm/s and RP437 strain with motility of 8 μm/s were used as low and high motile bacteria, respectively. Motility for these two strains is measured using image analysis and cell tracking. The FLI profile of the experimental images was exported via ImageJ from a 155 μm line along one of the vortices for both strains (Fig. 3 inset). Results of these image analyses are shown in Figs. 3a, 3b for low () and high motile (RP437) E. coli, respectively. Although in both cases, bacterial concentration increases with time, low motile E. coli bacteria collect at a considerably lower rate in the vortex pair. Faster and more intense collection of high motile bacteria, can be explained by their higher cross stream velocity. Bacterial motility via flagella is episodic and punctuated by frequent tumbles and causes E. coli to actively change swimming direction, known as “run-and-tumble.”44 A motile swimmer has the ability to migrate cross streamlines utilizing its motility. Although, for a motile swimmer within the vortex, the probability of its swimming towards a streamline with higher vorticity (inner streamline) is almost the same as that of its swimming towards an outer streamlines, when passing through the gap between the bubble and closed streamlines, due to the fact that the swimmer cannot penetrate the bubble, it must swim towards the vortex. Therefore, the overall flux of bacteria due to motility is into the vortex. Hence, active swimming increases the rate of collection of bacteria.
Figure 3.
FLI, indicating bacterial concentration, is plotted along the red-line across the vortex, as shown in the inset, for (a) low motile () and (b) high motile (RP437) E. coli versus time. Applied voltage is (enhanced online).
Further experiments with two other E. coli strains, BW25113 and JW2039, were conducted to test our hypothesis that motility is the main factor in enhancement of rate and amount of bacterial collection for RP437. These two strains, BW25113 and JW2039, have identical genotypes except that in JW2039, the colanic acid (or CA which is the main component of EPS in E. coli) synthesis gene wcaF is knocked out, and its CA production is significantly inhibited45 but they have the same motility. The experimental results with these strains confirmed that their amount and rate of collection within the vortices are the same. Therefore, our suggested mechanism is consistent with the observations for these two strains as well. These results also suggest that CA production is not important in the rate and amount of collection of bacteria in the vortical flow.
Riedel et al.46 have previously shown that flagellar intertwining and hydrodynamic interactions cause sperm to swim as vortices. In that work, vortex array is generated due to the swimming motion of the sperms and thus the velocity field is of the same order as sperm's swimming speed, whereas here the vortex pair is initiated by the cavitation streaming and the velocity field in the vortical structure is much larger than bacteria swimming velocity. In addition the ratio of cell length scale to vortex length scale and initial cell density is much smaller than the ones in Riedel et al.46 and we expect that flagellar intertwining not to be a driving mechanism in the cell collection reported here.
Bacterial aggregation and biofilm formation
After running the experiments for about 5–15 min (in some cases even as short as 2–3 min), aggregates of bacteria, very similar to biofilm streamers, were observed in the microfluidic channel (Fig. 4a). It should be noted that no bacterial aggregation or biofilm streamer is observed adjacent to the bubble in the absence of the vortical flow. The fast formation of the biofilm streamer in the presence of the cavitation streaming vortices suggests that high-Peclet convection due to the vortices is important. Filamentous biofilms, called streamers, are found ubiquitously in nature. They are observed in turbulent flows20 and also in curved channels under conditions of laminar flow where it is shown that exopolysaccharides (EPS) are correlated to their formation.19 Figure 4b shows the formation of these biofilm streamers at a higher initial bacterial concentration (OD600 = 2) and after 30 min that are formed next to an oscillating bubble with the shape of two vortices. Tails of the streamers in the vortical flow of several oscillating microbubbles are connected to each other via filamentous biofilms (Fig. 4b). Movies of Figure 4 show more details on the initial steps of formation of biofilm streamers. Extracellular structures (capsular and O-antigen polysaccharides, and potentially pili), which can cause bacteria to adhere to one another, might drive the streamer formation. When concentration of planktonic bacteria reaches a threshold, the signals produced by bacteria accumulate in relatively high concentration.47 Thus, intercellular signaling referred to as quorum sensing is activated.48, 49, 50 Bacteria floating in suspension also produce signals, but at low bacterial concentration the signal is diffused through the surrounding medium.47 Quorum sensing can regulate cooperative production of exopolysaccharides. When a phenotypic transformation to the biofilm state occurs, cells create an extracellular matrix composed of polysaccharides, proteins, DNA, detritus from lysed cells, and abiotic material like precipitates.28 However, the experimental time frame here is quite short and the convective movement of fluid through streamer structures is sufficiently high that quorum sensing molecules may not remain within these structures.
Figure 4.
(a) A biofilm streamer formed after three minutes of applying the voltage (OD600 = 0.25, ). Rapid E.coli biofilm formation in (b) a microfluidic channel after 30 min (OD600 = 2). Aggregate of E. coli formed the shape of two vortices in cavitation streaming of an oscillating bubble (). The bacteria remained aggregated even after the removal of the RF signal. (c) A microfluidic channel with oscillating bubbles () on the side walls (OD600 = 2) (enhanced online).
In order to explore the effect of EPS in the fast biofilm streamer formation, we used E. coli JW2039 which is deficient in producing EPS and its parent strain BW25113. The CA, an EPS of E. coliK-12, synthesis gene wcaF was knocked out in JW2039, resulting in its significantly inhibited production of CA.45, 51 The preliminary experiments did not reveal a significant difference in the collection rate and amount between the results of two strains. However, no biofilm streamer was observed for these two strains which may be due to fact that BW25113 has been reported as a poor biofilm producer in the literature36 and EPS production of JW2039 is inhibited.
CONCLUSIONS
We have shown that a vortical flow of an oscillating bubble can play an important role in bacterial collection and eventually triggers the formation of biofilms. The fast formation of biofilm streamers in a vortical flow can be a tipping point in developing techniques to study the formation and microbial features of biofilms. In conventional techniques, bacteria are incubated on nutrient media in order to generate large enough concentrations of microorganisms required for detection procedures which could take days. Therefore, portable point-of-care clinical and environmental diagnostic chips for rapid microbiological tests have yet to be developed. Our experimental results can pave the way for developing disposable and portable microfluidic devices to be used in microbiological tests. Furthermore, our experimental results have shown that the rate of bacterial collection in a vortical flow is pronounced for high motile bacteria.
ACKNOWLEDGMENTS
The authors are very thankful to Dr. Tony Huang for valuable help on the earlier stages of the experiments. We are also grateful to Sixing Li and Nydia Morales Soto for their help and guidance on culturing the bacteria strains. We would like to thank Dr. Wayne Curtis for valuable discussions. This work is supported by NSF Grant No. CBET-1150348-CAREER.
References
- Fenchel T., “ Microbial behavior in a heterogeneous world,” Science 296(5570), 1068 (2002). 10.1126/science.1070118 [DOI] [PubMed] [Google Scholar]
- Legendre P. and Fortin M. J., “ Spatial pattern and ecological analysis,” Plant Ecol. 80(2), 107–138 (1989). 10.1007/BF00048036 [DOI] [Google Scholar]
- Ardekani A. M. and Gore E., “ Emergence of a limit cycle for swimming microorganisms in a vortical flow of a viscoelastic fluid,” Phys. Rev. E 85, 056309 (2012). 10.1103/PhysRevE.85.056309 [DOI] [PubMed] [Google Scholar]
- Durham W. M., Climent E., and Stocker R., “ Gyrotaxis in a steady vortical flow,” Phys. Rev. Lett. 106(23), 238102 (2011). 10.1103/PhysRevLett.106.238102 [DOI] [PubMed] [Google Scholar]
- Luchsinger R. H., Bergersen B., and Mitchell J. G., “ Bacterial swimming strategies and turbulence,” Biophys. J. 77(5), 2377–2386 (1999). 10.1016/S0006-3495(99)77075-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos and Stocker R., “ Microorganisms in vortices: A microfluidic setup,” Limnol. Oceanogr. 4, 392–398 (2006). 10.4319/lom.2006.4.392 [DOI] [Google Scholar]
- Nyborg W. L., Acoustic Streaming (Academic, New York, 1965), Vol. 2. [Google Scholar]
- Longuet-Higgins M. S., “ Viscous streaming from an oscillating spherical bubble,” Proc. R. Soc. London, Ser. A 454(1970), 725 (1998). 10.1098/rspa.1998.0183 [DOI] [Google Scholar]
- Kim T., Doh I., and Cho Y. H., “ On-chip three-dimensional tumor spheroid formation and pump-less perfusion culture using gravity-driven cell aggregation and balanced droplet dispensing,” Biomicrofluidics 6, 034107 (2012). 10.1063/1.4739460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo L. Y., Hou D., Maheshswari S., and Chang H. C., “ Electrohydrodynamic surface microvortices for mixing and particle trapping,” Appl. Phys. Lett. 88(23), 233512 (2006). 10.1063/1.2212275 [DOI] [Google Scholar]
- Lim D. S. W., Shelby J. P., Kuo J. S., and Chiu D. T., “ Dynamic formation of ring-shaped patterns of colloidal particles in microfluidic systems,” Appl. Phys. Lett. 83(6), 1145–1147 (2003). 10.1063/1.1600532 [DOI] [Google Scholar]
- Ota H., Kodama T., and Miki N., “ Rapid formation of size-controlled three dimensional hetero-cell aggregates using micro-rotation flow for spheroid study,” Biomicrofluidics 5(3), 034105 (2011). 10.1063/1.3609969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp K. V., Yazdi S. H., and Davison S. M., “ Localized flow control in microchannels using induced-charge electroosmosis near conductive obstacles,” Microfluidics Nanofluidics 10(6), 1257–1267 (2011). 10.1007/s10404-010-0756-3 [DOI] [Google Scholar]
- Pethig R., “ Review article—dielectrophoresis: Status of the theory, technology, and applications,” Biomicrofluidics 4, 022811 (2010). 10.1063/1.3456626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S. C., Mach A. J., and Di Carlo D., “ High-throughput size-based rare cell enrichment using microscale vortices,” Biomicrofluidics 5, 022206 (2011). 10.1063/1.3576780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley P., Sauer K., Davies D. G., and Costerton J. W., “ Biofilms as complex differentiated communities,” Annu. Rev. Microbiol. 56(1), 187–209 (2002). 10.1146/annurev.micro.56.012302.160705 [DOI] [PubMed] [Google Scholar]
- O'Toole G., Kaplan H. B., and Kolter R., “ Biofilm formation as microbial development,” Annu. Rev. Microbiol. 54(1), 49–79 (2000). 10.1146/annurev.micro.54.1.49 [DOI] [PubMed] [Google Scholar]
- Valiei A., Kumar A., Mukherjee P. P., Liu Y., and Thundat T., “ A web of streamers: Biofilm formation in a porous microfluidic device,” Lab Chip 12, 5133–5137 (2012). 10.1039/c2lc40815e [DOI] [PubMed] [Google Scholar]
- Rusconi R., Lecuyer S., Guglielmini L., and Stone H. A., “ Laminar flow around corners triggers the formation of biofilm streamers,” J. R. Soc. Interface 7(50), 1293 (2010). 10.1098/rsif.2010.0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley P., Lewandowski Z., Boyle J. D., and Lappin-Scott H. M., “ Oscillation characteristics of biofilm streamers in turbulent flowing water as related to drag and pressure drop,” Biotechnol. Bioeng. 57, 536–544 (1998). [DOI] [PubMed] [Google Scholar]
- Mah T. F., Pitts B., Pellock B., Walker G. C., Stewart P. S., and O'Toole G. A., “ A genetic basis for pseudomonas aeruginosa biofilm antibiotic resistance,” Nature 426, 306–310 (2003). 10.1038/nature02122 [DOI] [PubMed] [Google Scholar]
- Lewis K., “ Persister cells, dormancy and infectious disease,” Nat. Rev. Microbiol. 5(1), 48–56 (2007). 10.1038/nrmicro1557 [DOI] [PubMed] [Google Scholar]
- Hou D., Maheshwari S., and Chang H. C., “ Rapid bioparticle concentration and detection by combining a discharge driven vortex with surface enhanced Raman scattering,” Biomicrofluidics 1, 014106 (2007). 10.1063/1.2710191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K., Kaprelyants A. S., Salina E. G., Schuler M., and Markx G. H., “ Construction by dielectrophoresis of microbial aggregates for the study of bacterial cell dormancy,” Biomicrofluidics 4(2), 022810 (2010). 10.1063/1.3435336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., and Greenberg E. P., “ Bacterial biofilms: A common cause of persistent infections,” Science 284, 1318–1322 (1999). 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- Turakhia M. H., Cooksey K. E., and Characklis W. G., “ Influence of a calcium-specific chelant on biofilm removal,” Appl. Environ. Microbiol. 46(5), 1236–1238 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech I. B. and Sunner J., “ Biocorrosion: Towards understanding interactions between biofilms and metals,” Curr. Opin. Biotechnol. 15, 181–186 (2004). 10.1016/j.copbio.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Klapper I. and Dockery J., “ Mathematical description of microbial biofilms,” SIAM Rev. 52, 221–265 (2010). 10.1137/080739720 [DOI] [Google Scholar]
- Vrouwenvelder J. S., Graf von der Schulenburg D. A., Kruithof J. C., Johns M. L., and van Loosdrecht M. C. M., “ Biofouling of spiral-wound nanofiltration and reverse osmosis membranes: A feed spacer problem,” Water Res. 43, 583–594 (2009). 10.1016/j.watres.2008.11.019 [DOI] [PubMed] [Google Scholar]
- Flemming H. C., “ Reverse osmosis membrane biofouling,” Exp. Therm. Fluid Sci. 14, 382–391 (1997). 10.1016/S0894-1777(96)00140-9 [DOI] [Google Scholar]
- Tchobanoglous G., Burton F. L., and Stensel H. D., Wastewater Engineering: Treatment and Reuse, 4th ed. (Metcalf and Eddy Inc., McGraw Hill, New York, 2003). [Google Scholar]
- Harvey R. W., Smith R. L., and George L., “ Effect of organic contamination upon microbial distributions and heterotrophic uptake in a Cape Cod, Mass., aquifer,” Appl. Environ. Microbiol. 48, 1197–1202 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. C., Phillips A. J., Hiebert R., Gerlach R., Spangler L. H., and Cunningham A. B., “ Biofilm enhanced geologic sequestration of supercritical CO2,” Int. J. Greenhouse Gas Control 3, 90–99 (2009). 10.1016/j.ijggc.2008.05.002 [DOI] [Google Scholar]
- Ahmed D., Mao X., Shi J., Juluri B. K., and Huang T. J., “ A millisecond micromixer via single-bubble-based acoustic streaming,” Lab Chip 9(18), 2738–2741 (2009). 10.1039/b903687c [DOI] [PubMed] [Google Scholar]
- Xia Y. and Whitesides G. M., “ Soft lithography,” Annu. Rev. Mater. Sci. 28(1), 153–184 (1998). 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- Wood T. K., González Barrios A. F., Herzberg M., and Lee J., “ Motility influences biofilm architecture in Escherichia coli,” Appl. Microbiol. Biotechnol. 72(2), 361–367 (2006). 10.1007/s00253-005-0263-8 [DOI] [PubMed] [Google Scholar]
- Zhuang W., Yuan D., Li J. R., Luo Z., Zhou H. C., Bashir S., and Liu J., “ Highly potent bactericidal activity of porous metal-organic frameworks,” Adv. Healthcare Mater. 1(2), 225–238 (2012). 10.1002/adhm.201100043 [DOI] [PubMed] [Google Scholar]
- Elder S. A., “ Cavitation microstreaming,” J. Acoust. Soc. Am. 31, 54 (1959). 10.1121/1.1907611 [DOI] [Google Scholar]
- Wang C., Jalikop S. V., and Hilgenfeldt S., “ Size-sensitive sorting of microparticles through control of flow geometry,” Appl. Phys. Lett. 99, 034101 (2011). 10.1063/1.3610940 [DOI] [Google Scholar]
- Wang C., Jalikop S. V., and Hilgenfeldt S., “ Efficient manipulation of microparticles in bubble streaming flows,” Biomicrofluidics 6(1), 012801 (2012). 10.1063/1.3654949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H. C., E. coli in Motion (Springer Verlag, 2004). [Google Scholar]
- Watari N. and Larson R. G., “ The hydrodynamics of a run-and-tumble bacterium propelled by polymorphic helical flagella,” Biophys. J. 98(1), 12–17 (2010). 10.1016/j.bpj.2009.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. L., “ Particle gathering and microstreaming near ultrasonically activated gas-filled micropores,” J. Acoust. Soc. Am. 84(4), 1378–1387 (1988). 10.1121/1.396636 [DOI] [PubMed] [Google Scholar]
- Stocker R., “ Reverse and flick: Hybrid locomotion in bacteria,” Proc. Natl. Acad. Sci. U.S.A. 108(7), 2635–2636 (2011). 10.1073/pnas.1019199108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y. and Zhang T., “ Probing roles of lipopolysaccharide, fimbria and colanic acid in the attachment of Escherichia coli strains on inert surfaces.” Langmuir 27(18), 11545–11553 (2011). 10.1021/la202534p [DOI] [PubMed] [Google Scholar]
- Riedel I. H., Kruse K., and Howard J., “ A self-organized vortex array of hydrodynamically entrained sperm cells,” Science 309, 300–303 (2005). 10.1126/science.1110329 [DOI] [PubMed] [Google Scholar]
- National Research Council, Opportunities for Environmental Applications of Marine Biotechnology: Proceedings of the October 5-6, 1999, Workshop (National Academy, 2000). [PubMed]
- Fuqua W. C., Winans S. C., and Greenberg E. P., “ Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators,” J. Bacteriol. 176, 269–275 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S. and Williams P., “ Quorum sensing and social networking in the microbial world,” J. R. Soc. Interface 6(40), 959–978 (2009). 10.1098/rsif.2009.0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joint I., “ Bacterial conversations: talking, listening and eavesdropping. a NERC discussion meeting held at the Royal Society on 7 December 2005,” J. R. Soc. Interface 3(8), 459–463 (2006). 10.1098/rsif.2006.0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese P. N., Pratt L. A., and Kolter R., “ Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture,” J. Bacteriol. 182(12), 3593–3596 (2000). 10.1128/JB.182.12.3593-3596.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]