Abstract
HPV account for most of the incidence of cervical cancer. Approximately 90% of anal cancers and a smaller subset (<50%) of other cancers (oropharyngeal, penile, vaginal, vulvar) are also attributed to HPV. The L1 protein comprising HPV vaccine formulations elicits high-titre neutralizing antibodies and confers type restricted protection. The L2 protein is a promising candidate for a broadly protective HPV vaccine. In our previous study, we found the most prevalent high-risk HPV infectious serotypes were HPV-16 and HPV-58 among women of Southwest China. To explore gene polymorphisms and intratypic variations of HPV-16 and HPV-58 L1/L2 genes originating in Southwest China, HPV-16 (L1: n = 31, L2: n = 28) and HPV-58 (L1: n = 21, L2: n = 21) L1/L2 genes were sequenced and compared to others described and submitted to GenBank. Phylogenetic trees were then constructed by Neighbor-Joining and the Kimura 2-parameters methods (MEGA software), followed by an analysis of the diversity of secondary structure. Then selection pressures acting on the L1/L2 genes were estimated by PAML software. Twenty-nine single nucleotide changes were observed in HPV-16 L1 sequences with 16/29 non-synonymous mutations and 13/29 synonymous mutations (six in alpha helix and two in beta turns). Seventeen single nucleotide changes were observed in HPV-16 L2 sequences with 8/17 non-synonymous mutations (one in beta turn) and 9/17 synonymous mutations. Twenty-four single nucleotide changes were observed in HPV-58 L1 sequences with 10/24 non-synonymous mutations and 14/24 synonymous mutations (eight in alpha helix and four in beta turn). Seven single nucleotide changes were observed in HPV-58 L2 sequences with 4/7 non-synonymous mutations and 3/7 synonymous mutations. The result of selective pressure analysis showed that most of these mutations were of positive selection. This study may help understand the intrinsic geographical relatedness and biological differences of HPV-16/HPV-58 and contributes further to research on their infectivity, pathogenicity, and vaccine strategy.
Introduction
Human Papillomavirus (HPV) virions are one of the most important pathogenic agents for cervical cancer, which accounts for a worldwide cancer burden in women second only to breast cancer [1], [2]. Approximately 90% of anal cancers and a smaller subset (<50%) of other cancers (oropharyngeal, penile, vaginal, and vulvar) are also attributed to HPV. In total, HPV accounts for 5.2% of the worldwide cancer burden. HPVs 16 and 18 are responsible for 70% of cervical cancer cases and, especially HPV-16, for a large proportion of other cancers [3]. On the basis of their oncogenic potential, HPV types that infect the genital tract are classified as low risk (LR) and high risk (HR) [4]. Low-risk HPVs (including HPV-6, 11, 42, 43, and 44) are mainly associated with benign genital warts, while high-risk HPVs (including HPV-16, 18, 31, 33, 39, 45, 51, 52, 56, 58, 59, and 68) are the etiological agents of cervical cancer, a disease that affects approximately 500,000 women worldwide [5]. In our previous study, we found the most prevalent high-risk HPV infectious serotypes were HPV-16 and HPV-58 among women of Southwest China.
The HPV genome is packaged within a non-enveloped, icosahedral capsid composed of 72 pentamers of the major capsid late protein (L1) and an unknown number of the minor capsid proteins L2 [6], [7]. The pentamers of L1 expressed in heterologous systems that assemble into virus-like particles (VLPs) [8] are the components used in the design of prophylactic vaccines. The L1 protein comprising HPV vaccine formulations elicits high-titre neutralizing antibodies and confers type-specific and long-lasting protection against persistent infection and associated cervical neoplasia attributable to HPV vaccine types [8], [9]. However, there has been no vaccine designed that can prevent all HPV infections owing to the lack of cross-reactivity between L1 proteins of different HPV types.
The inner conical hollow of L1 pentamers can be occluded with a monomer of L2 [6]. An N-terminal “external loop” of L2 contains cross-neutralizing epitopes, which can be the target of neutralizing and cross-neutralizing antibodies as well [10]–[12]. Analysis of FUTURE I/II and PATRICIA data suggested cross-protective vaccine efficacy against infections and lesions associated with HPV 31, 33, and 45 [13]. Therefore, targeting L2 may be an acceptable approach for a candidate vaccine. However, its low abundance in natural capsids (only 12–72 molecules per 360 copies of L1) limits its immunogenicity [14]. Currently, we have no efficient vaccines against L2 to prevent infection with these high-risk HPV types.
Due to the high prevalence of HPV not only among asymptomatic women but also in samples of different neoplasias worldwide, the association between intratypical variants of HPV-16 L1 has been described in several papers. Nevertheless, data concerning molecular variants of HPV-16 and HPV-58 L2 are still limited, necessitating further studies that would be essential to expand knowledge of the different variants. Furthermore, there is little data regarding the intratypical variants of HPV-16 L1 in Southwest China.
The aim of this study is to detect the nucleotide variability, gene polymorphism and phylogeny in the L1 and L2 genes of the High-Risk HPV-16 (L1: n = 31, L2: n = 28) and HPV-58 (L1: n = 21, L2: n = 21) samples obtained from Southwest China. The most variable sequences were chosen for an analysis of the diversity of their secondary structure. Nucleotide and amino acid sequence alignments were used to evaluate variant clusters. Amino acid changes of L1 and L2 genes might affect immune responses to HPV-16 and HPV-58 capsid proteins and advance HPV vaccine strategies. The genomic characterization of HPV variants is pivotal for a deeper understanding of the intrinsic geographical relatedness and biological differences of these viruses and contributes further to research on their infectivity and pathogenicity.
Materials and Methods
Ethics Statement
Written consent was obtained from each participant. The study protocol was approved by the institutional ethics committee (Institute of Medical Biology, Chinese Academy of Medical Sciences, and Peking Union Medical College) and was in accordance with the Declaration of Helsinki for Human Research of 1974 (last modified in 2000).
Clinical Specimens
Samples examined in this study were obtained from cervical scrapings of 3000 volunteer outpatients from women in Southwest China from 2009 to 2011. After routine cytology and HC2 testing, a cell suspension from each sample was placed in a 1.5 mL Eppendorf tube and transferred to a laboratory at the Institute of Medical Biology for HPV DNA amplification. HPV typing of these specimens was performed using a nested multiplex PCR assay as described previously [15], [16]. HPV-16 (L1: n = 31; L2: n = 28), HPV-58 (L1: n = 21; L2: n = 21) sequences thereby obtained were used for molecular characterization by sequence analysis of the L1 and L2 gene.
Nucleic acid extraction and sequencing amplification
Molecular characterization was performed by sequence analysis of L1 and L2 gene amplicons. The entire region of L1 and L2 genes of HPV-16 and HPV-58 were amplified using degenerate primer pairs. Two partially overlapping fragments for each virus were amplified. All Primers were designed and synthesized by Sangon Biotech (Shanghai). The GeneBank reference sequences used for primer design were HPV-16 (NC001526 [17]) and HPV-58 (HQ537759). The primer sequences and their relative positions are listed in Table 1. The amplification of the fragments was performed in 50 ul reaction volumes containing 5 ul extracted DNA (template), 2×power taq PCR MasterMix (TaKaRa), 25 pmol of each primer (Sangon Biotech) and deionized water. The cycling conditions were as follows: 94° for 5 min followed by 30 cycles at 94° for 45 sec, 50° for 45 sec, 72° for 1 min and a final 72° extension for 7 min. Amplicons were visualized on 2% agarose gels stained with GoldViewTM Nucleic Acid Stain. Then PCR products were purified and sequenced by Sangon Biotech.
Table 1. Primers used for the molecular characterization of HPV-16 and HPV-58 L1 and L2.
| Primer name | Sequence 5′ to 3′ | Amplicon size (bp) |
| HPV16 L1 1F | ATAGTTCCAGGGTCTCCA | 743 |
| HPV16 L1 1R | AGTCCATAGCACCAAAGC | |
| HPV16 L1 2F | GAACACTGGGGCAAAGGATC | 1236 |
| HPV16 L1 2R | TACAATGAATAACCACAACA | |
| HPV16 L2 1F | TTACTTAACAATGCGACACA | 801 |
| HPV16 L2 1R | TTATCCACATCTATACCTTCA | |
| HPV16 L2 2F | CCCTGCTTTTGTAACCACTC | 776 |
| HPV16 L2 2R | CGTGCAACATATTCATCCGT | |
| HPV58 L1 1F | GTGTCATTGGAACCTGGTCCA | 985 |
| HPV58 L1 1R | GCCAAGTTTTCCAGCCCTATT | |
| HPV58 L1 2F | GCCAGTGAACCTTATGGGGAT | 771 |
| HPV58 L1 2R | TTTGCGTTTGGTGGATGGT | |
| HPV58 L2 1F | ATGGTGGTATGGTATTGT | 810 |
| HPV58 L2 1R | CTTAACTTGTTGGGTGGT | |
| HPV58 L2 2F | TCCTTTACTGAGCCATCC | 1018 |
| HPV58 L2 2R | ATAAATGCTTGTGCGTGA |
Molecular characterization and phylogenetic analysis
All sequences from a given sample were combined and used to construct alignments. ClustalX (version 1.83) multiple sequence alignment analysis was carried out to calculate the percentage of sequence similarity between the L1/L2 amplicons and the representative sequences of the HPV-16 and HPV-58 variants. Phylogenetic trees of respective HPV-16 and HPV-58 L1 and L2 sequences were constructed by the Neighbor-Joining method [18]–[20] and the Kimura 2-Parameter model by MEGA package, version4.1. A bootstrap re-sampling analysis was performed (2,000 replicates) to test the robustness of the major phylogenetic groups [17], [19], [21]. To estimate selection pressure acting on the HPV-16 and HPV-58 L1 and L2 gene sequences, synonymous and nonsynonymous nucleotide divergence for coding regions was inferred by the method of Nei and Gojobori [21]–[23] with PAML 4.0. The L1 and L2 gene sequences of the HPV-16, and 58 viral strains studied were deposited into the NCBI GenBank database (http://www.ncbi.nim.nih.gov/GenBank/index.html): JX313693-JX313793. The reference viral sequences that were used to construct the distinct phylogenetic branches were collected from the GenBank sequence database under the following accession numbers: HPV-16: AY686580, FJ006723 (China, Xinjiang), AY686581, EU118173, AF534061, U89348, AF536179, AF472508, AF536180, AF472509, and AY686579 [16]; HPV-58: D90400 (Japan), EU918765, HQ537760 (isolate AS347), HQ537762, HQ537763, HQ537776, HQ537774, and HQ537777.
Results
Phylogenetic and amino acid mutations analysis of HPV-16 L1 sequences
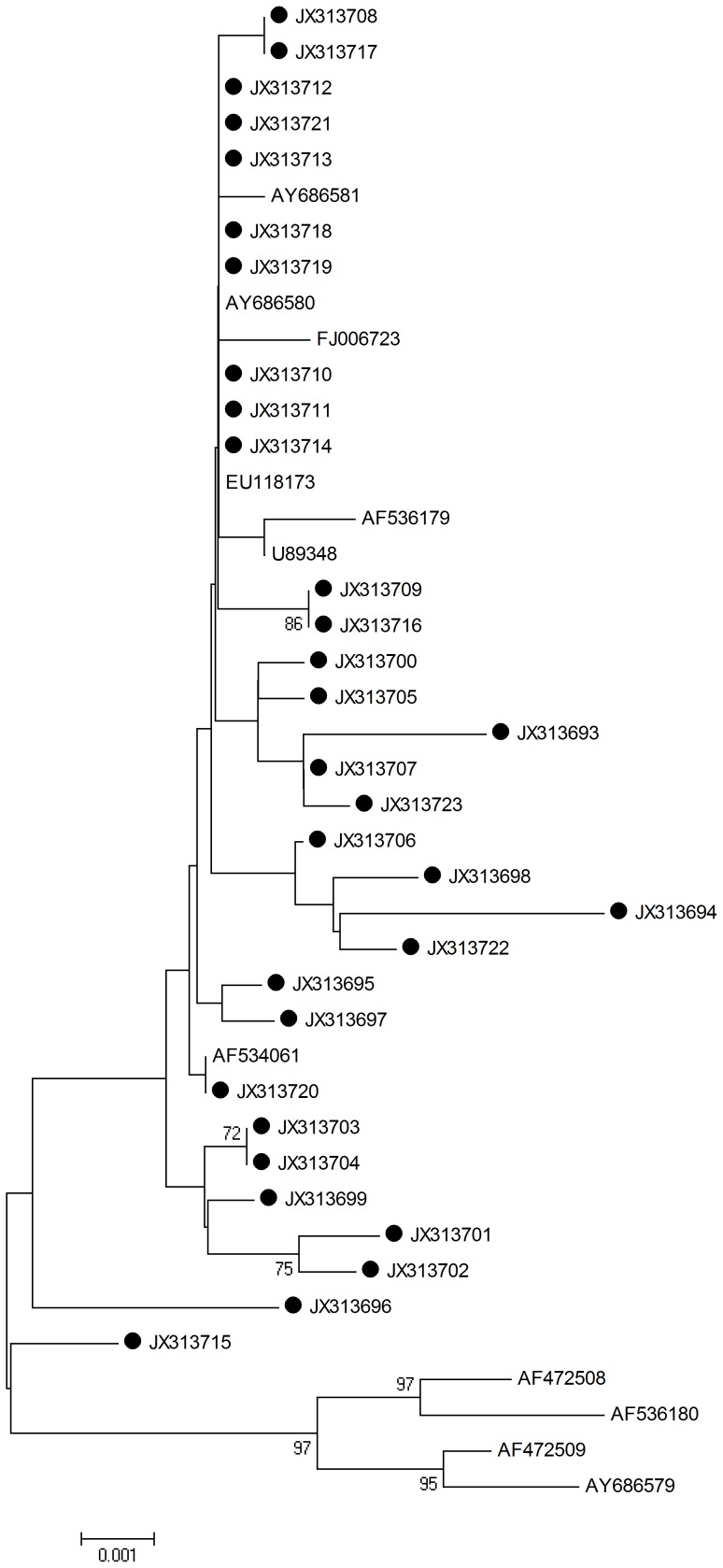
L1 HPV-16 sequences were determined and analyzed by aligning L1 1596 nucleotide sequences from all viral strains (n = 31; including the reference sequences). The neighbor joining phylogenetic tree can be seen in Fig. 1.
Figure 1. Neighbor joining phylogenetic tree generated using the nucleotide sequences of HPV-16 L1 gene.
Study sequences are labeled in black. Others are standard sequences, including: AY686580, FJ006723 (China, Xinjiang), AY686581, EU118173, AF534061, U89348, AF536179, AF472508, AF536180, AF472509, and AY686579. Phylogenetic trees were constructed by the Neighbor-Joining method and the Kimura 2-Parameter model by MEGA package.
Twenty-nine single nucleotide changes were identified among the sequences studied. Specifcally, 13/29 (44.8%) were synonymous mutations and 16/29 (55.2%) were non-synonymous mutations. Of the 6 amino acid mutations observed in the sequences encoding the alpha helix, only one was a non-synonymous mutation. 2 non-synonymous mutations were observed in the sequences encoding the beta turn (glycine to arginine) (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.). 1 of 31 sequences did not belong to any standard type branch (Fig. 1). 7 samples were found to have the same mutation from A to C at the position of 979. The detected mutations are summarized in Table 2.
Table 2. Nucleotide sequence mutations of HPV-16 L1.
| Domain: HPV-16 L1 sequence | ||||||||||||||||||||||||||||
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 1 | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 3 | 4 | 7 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 1 | 2 | 3 | 4 | 4 | 5 | 5 | 5 | ||
| 5 | 0 | 0 | 0 | 0 | 6 | 6 | 9 | 6 | 9 | 2 | 1 | 4 | 7 | 7 | 7 | 8 | 8 | 8 | 9 | 3 | 6 | 2 | 7 | 9 | 0 | 2 | 6 | |
| 2 | 2 | 3 | 4 | 7 | 5 | 8 | 9 | 6 | 9 | 3 | 4 | 5 | 7 | 8 | 9 | 1 | 2 | 3 | 0 | 2 | 6 | 9 | 8 | 5 | 7 | 5 | 6 | |
| NC001526 | G | G | G | C | C | A | C | G | G | A | G | A | A | T | T | A | C | T | C | C | G | C | T | A | G | A | G | A |
| JX313693 | . | C | T | A | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | G | . | . | . | . | . |
| JX313694 | A | . | T | A | . | . | . | T | A | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | C | C |
| JX313695 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . |
| JX313696 | . | . | . | . | . | . | . | . | . | . | A | . | . | C | C | C | . | . | . | . | . | T | . | . | . | . | . | . |
| JX313697 | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . |
| JX313698 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | C | . | . | . | . | A | . | . | . | . | . | C | C |
| JX313699 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | C | . | . | . | . | . | . | . | G | . | . | . | . |
| JX313700 | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . |
| JX313701 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | C | T | C | . | . | . | T | G | . | . | . | . | . |
| JX313702 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | C | T | C | T | . | . | . | . | . | . | . | . | . |
| JX313703 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | C | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313704 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | C | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313705 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | T | . | . | . | . | . | . |
| JX313706 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | C |
| JX313707 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | G | . | . | . | . | . |
| JX313708 | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313709 | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . |
| JX313710 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313711 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313712 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313713 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313714 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313715 | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . |
| JX313716 | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . |
| JX313717 | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313718 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313719 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313720 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313721 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313722 | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | C | C |
| JX313723 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | G | . | C | . | . | . |
| AA mutation | . | E-D | A-Y(35) | A-Y(35) | T-N | R-L | V-A | T-P(327) | T-P(327) | S-P | S-L | E-K | G-R | K-Q | G-R | |||||||||||||
| second structure | Alpha helix | Alpha helix | Alpha helix | Alpha helix | Alpha helix | Alpha helix | Beta turn | Beta turn | ||||||||||||||||||||
Compared to prototype HPV sequences, insertion and deletion events were not identified and there was no evidence of premature stop codons or nucleotide deletions in the L1 HPV-16 sequences analyzed.
Phylogenetic and amino acid mutations analysis of HPV-16 L2 sequences
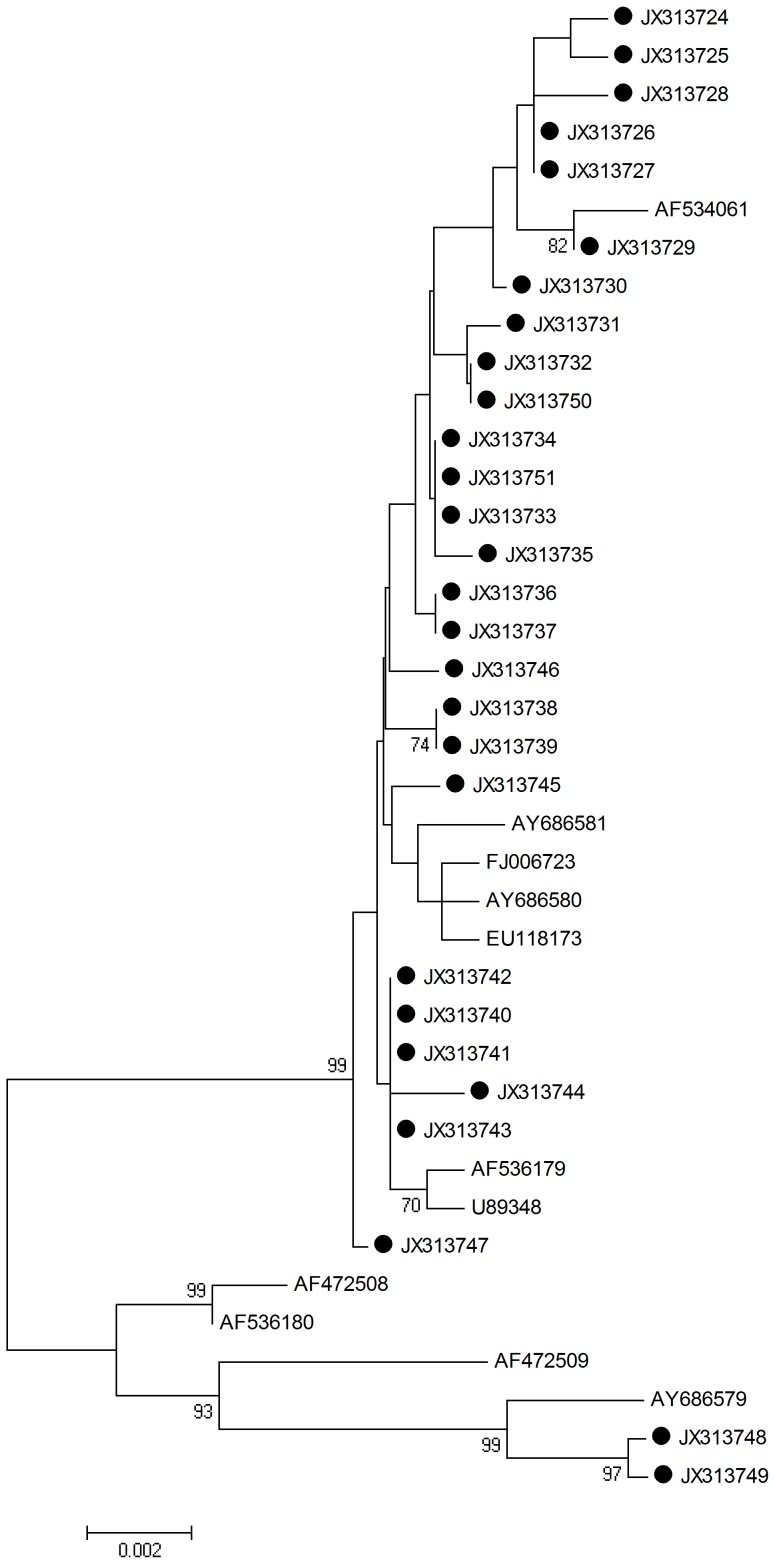
L2 HPV-16 sequences were determined and analyzed by aligning L2 1422 nucleotide sequences from all viral strains (n = 28; including the reference sequences). The neighbor joining phylogenetic tree can be seen in Fig. 2.
Figure 2. Neighbor joining phylogenetic tree generated using nucleotide sequences of the HPV-16 L2 gene.
Study sequences are labeled in black. Others are standard sequences, including: AY686580, FJ006723 (China, Xinjiang), AY686581, EU118173, AF534061, U89348, AF536179, AF472508, AF536180, AF472509, and AY686579. Phylogenetic trees were constructed by the Neighbor-Joining method and the Kimura 2-Parameter model by MEGA package.
Seventeen single nucleotide changes were identified among the sequences studied. Specially, 9/17 (52.9%) were synonymous mutations and 8/17 (47.1%) were non-synonymous mutations. No amino acid changes were discovered at residues 65–71 and 112–120, which play a important role in inducing neutralizing antibodies [24]. No amino acid mutations occurred in the sequences encoding the alpha helix. Only one non-synonymous mutation was observed in the sequences encoding the beta turn (aspartic acid to glutamic acid). Both JX313748 and JX313749 L2 sequences fell into the same branch of AY686579 (Fig. 2). The detected mutations are summarized in Table 3.
Table 3. Nucleotide sequence mutations of HPV-16 L2.
| Domain: HPV-16 L2 sequence | |||||||||||||||||
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||
| 1 | 3 | 7 | 7 | 7 | 7 | 8 | 9 | 9 | 9 | 0 | 0 | 0 | 1 | 2 | 2 | 3 | |
| 9 | 6 | 4 | 7 | 8 | 8 | 7 | 1 | 5 | 9 | 1 | 5 | 7 | 1 | 3 | 3 | 9 | |
| 2 | 3 | 7 | 4 | 1 | 3 | 6 | 6 | 4 | 5 | 8 | 9 | 4 | 3 | 0 | 9 | 6 | |
| NC001526 | G | A | T | A | G | T | A | A | A | C | G | A | T | T | A | T | T |
| JX313724 | . | . | . | . | A | . | . | . | . | . | C | . | . | . | . | . | . |
| JX313725 | . | . | C | . | A | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313726 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313727 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313728 | . | . | . | T | . | . | T | . | . | . | . | . | . | . | . | . | . |
| JX313729 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313730 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313731 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . |
| JX313732 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . |
| JX313733 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313734 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313735 | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . |
| JX313736 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313737 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313738 | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . |
| JX313739 | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . |
| JX313740 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313741 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313742 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313743 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313744 | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | C |
| JX313745 | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . |
| JX313746 | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . |
| JX313747 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313748 | T | C | . | . | . | . | . | . | . | . | . | C | A | . | . | A | . |
| JX313749 | C | C | . | . | . | . | . | . | . | . | . | C | A | . | . | A | . |
| JX313750 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . |
| JX313751 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| AA mutation | E-D | D-N | D-E | K-N | T-N | E-Q | D-E | F-L | |||||||||
| second structure | Random coil | Random coil | Extended strand. | Random coil | Extended strand. | Extended strand. | Random coil | Random coil | Random coil | Random coil | Extended strand. | Random coil | Random coil | Beta turn | Extended strand. | Random coil | Extended strand. |
Insertion and deletion events were not present and there was no evidence of premature stop codons or nucleotide deletions within the L2 HPV-16 analyzed sequences.
Phylogenetic and amino acid mutations analysis of HPV-58 L1 sequences
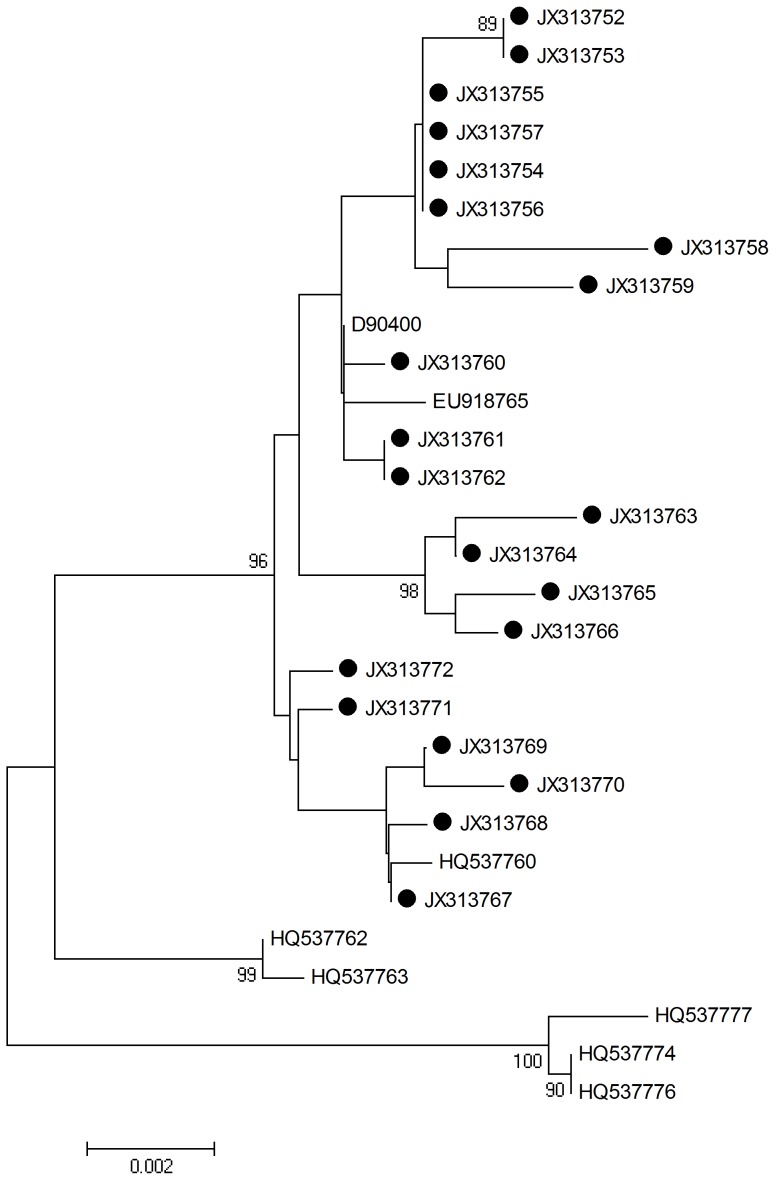
L1 HPV-58 sequences were determined and analyzed by aligning L1 1575 nucleotide sequences from all viral strains (n = 21; including the reference sequences). The neighbor joining phylogenetic tree can be seen in Fig. 3.
Figure 3. Neighbor joining phylogenetic tree generated using nucleotide sequences of the HPV-58 L1 gene.
Study sequences are labeled in black. Others are standard sequences, including: D90400 (Japan), EU918765, HQ537760 (isolate AS347), HQ537762, HQ537763, HQ537776, HQ537774, and HQ537777. Phylogenetic trees were constructed by the Neighbor-Joining method and the Kimura 2-Parameter model by MEGA package.
Twenty-four single nucleotide changes were identified among the sequences studied. Specifically, 14/24 (58.3%) were synonymous mutations and 10/24 (41.7%) were non synonymous mutations. 8 amino acid mutations (four non-synonymous) occurred in the sequences encoding the alpha helix, and 4 mutations were observed in the sequences encoding the beta turn with 2 being non-synonymous. 8 samples were found to have the same mutation from C to A at position 1124. The detected mutations are summarized in Table 4.
Table 4. Nucleotide sequence mutations of HPV-58 L1.
| Domain: HPV-58 L1 sequence | ||||||||||||||||||||||||
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||
| 2 | 2 | 2 | 3 | 7 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 9 | 9 | 9 | 0 | 0 | 1 | 2 | 2 | 3 | 4 | 1 | |
| 4 | 6 | 7 | 0 | 7 | 2 | 2 | 3 | 5 | 5 | 6 | 7 | 7 | 3 | 5 | 9 | 2 | 7 | 2 | 3 | 9 | 1 | 3 | 4 | |
| 5 | 4 | 0 | 9 | 5 | 6 | 7 | 0 | 1 | 2 | 2 | 0 | 7 | 6 | 7 | 6 | 9 | 7 | 4 | 3 | 6 | 7 | 7 | 2 | |
| HQ537759 | A | A | C | T | G | C | T | G | G | A | A | T | C | C | A | A | A | G | C | G | A | A | A | A |
| JX313752 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | A | A | . | . | C | . |
| JX313753 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | A | A | . | . | C | . |
| JX313754 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | A | . | . | . | . | . |
| JX313755 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | A | . | . | . | . | . |
| JX313756 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | A | . | . | . | . | . |
| JX313757 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | A | . | . | . | . | . |
| JX313758 | . | . | . | . | A | . | . | A | A | . | . | . | . | . | . | G | . | . | A | . | . | . | . | . |
| JX313759 | . | . | . | . | . | T | C | . | A | . | T | . | . | . | . | G | . | . | A | . | . | . | . | . |
| JX313760 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . |
| JX313761 | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . |
| JX313762 | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . |
| JX313763 | C | . | A | . | . | . | . | . | . | G | . | C | . | . | . | . | . | A | . | . | . | . | . | C |
| JX313764 | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | A | . | . | . | . | . | C |
| JX313765 | . | . | . | G | . | . | . | . | . | G | . | C | . | . | G | . | . | A | . | . | . | . | . | . |
| JX313766 | . | G | . | . | . | . | . | . | . | G | . | . | . | . | G | . | . | A | . | . | . | . | . | . |
| JX313767 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| JX313768 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . |
| JX313769 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . |
| JX313770 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | G | . | . |
| JX313771 | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . |
| JX313772 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . |
| AA mutation | N-T | D-N | L-S(276) | L-S(276) | P-K | R-K | N-Y | T-N | K-T | K-T | ||||||||||||||
| second structure | Beta turn | Beta turn | Alpha helix | Alpha helix | Alpha helix | Alpha helix | Beta turn | Alpha helix | Beta turn | Alpha helix | Alpha helix | Alpha helix | ||||||||||||
Compared to prototype HPV sequences, neither frame shifts, premature stop codons, insertions nor deletions were observed in the L1 HPV-58 analyzed sequences.
Phylogenetic and amino acid mutations analysis of HPV-58 L2 sequences
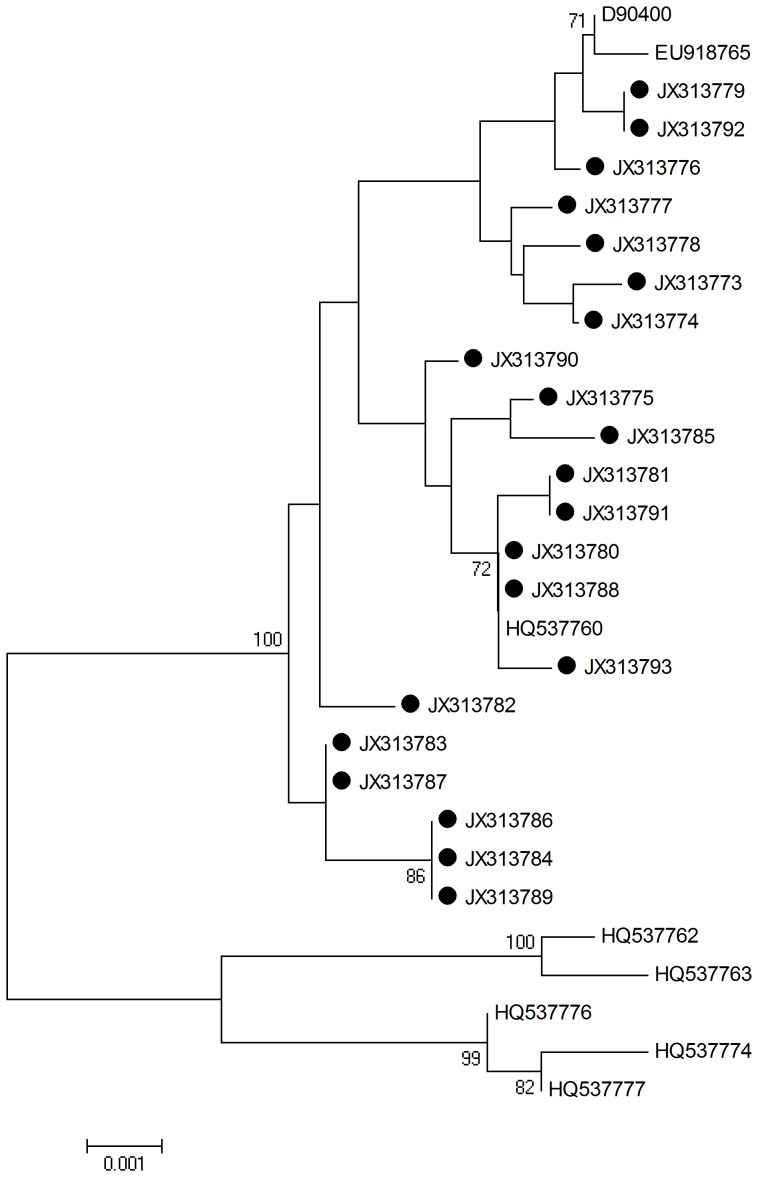
L2 HPV-58 sequences were determined and analyzed by aligning L2 1419 nucleotide sequences from all viral strains (n = 21; including the reference sequences). The neighbor joining phylogenetic tree can be seen in Fig. 4.
Figure 4. Neighbor joining phylogenetic tree generated using nucleotide sequences of the HPV-58 L2 gene.
Study sequences are labeled in black. Others are standard sequences, including: D90400 (Japan), EU918765, HQ537760 (isolate AS347), HQ537762, HQ537763, HQ537776, HQ537774, and HQ537777. Phylogenetic trees were constructed by the Neighbor-Joining method and the Kimura 2-Parameter model by MEGA package.
Seven single nucleotide changes were identified among the sequences studied. Specifically, 3/7 (42.9%) were synonymous mutations and 4/7 (57.1%) were non-synonymous mutations. No amino acid changes were observed at residues 65–71 and 112–120, which play an important role in inducing neutralizing antibodies. No amino acid mutations occurred in the sequences encoding the alpha helix and beta turn, but 6 mutations were found in the random coil, and one in the extended strand. The detected mutations are summarized in Table 5.
Table 5. Nucleotide sequence mutations of HPV-58 L2.
| Domain: HPV-58 L2 sequence | |||||||
| 3 | 6 | 6 | 7 | 7 | 7 | 9 | |
| 7 | 1 | 9 | 0 | 2 | 5 | 6 | |
| 8 | 6 | 9 | 0 | 7 | 7 | 3 | |
| HQ537759 | A | G | A | C | T | G | A |
| JX313773 | . | . | C | A | . | . | G |
| JX313774 | . | . | C | . | . | . | G |
| JX313775 | . | . | C | A | . | . | . |
| JX313776 | . | . | . | . | . | . | G |
| JX313777 | . | . | . | . | . | . | G |
| JX313778 | . | . | . | . | A | . | G |
| JX313779 | . | . | . | . | . | . | G |
| JX313780 | . | . | . | . | . | . | . |
| JX313781 | . | . | . | . | A | . | . |
| JX313782 | . | . | . | . | . | . | . |
| JX313783 | . | . | . | . | . | . | . |
| JX313784 | C | A | . | . | . | . | . |
| JX313785 | . | . | C | A | . | . | . |
| JX313786 | C | A | . | . | . | . | . |
| JX313787 | . | . | . | . | . | . | . |
| JX313788 | . | . | . | . | . | . | . |
| JX313789 | C | A | . | . | . | . | . |
| JX313790 | . | . | . | . | . | . | . |
| JX313791 | . | . | . | . | A | . | . |
| JX313792 | . | . | . | . | . | . | G |
| JX313793 | . | . | . | . | . | A | . |
| AA mutation | L-N | Q-H | Q-K | D-N | |||
| second structure | Random coil | Random coil | Random coil | Random coil | Random coil | Random coil | Extended strand. |
Insertion and deletion events were not identified and there was no evidence of premature stop codons or nucleotide deletions in the L2 HPV-58 sequences analyzed.
Selective pressure analysis of all sequences
We tested for variable dN/dS rate ratios among various lineages using the PAML4.0 [25]. There was no evidence of negative selection in the sequence alignment of HPV-16 and HPV-58 L1 and L2 genes (P-value <0.1). The selective pressure analysis results are summarized in Table 6,7,8,9.
Table 6. Site-specific tests for positive selection on HPV-16 L1.
| Models | lnL | Estimates of parameters | 2Δl | Positively selected sites |
| M7 | −2491.42 | p = 0.005, q = 0.046 | NA | |
| M8 | −2479.10 | p0 = 0.985, p = 0.005, q = 0.047, p1 = 0.015, ω = 13.30 | 24.64 P<0.05 | 35Y**, 207N**, 327T**, 328S*,493K, 503K |
Table 7. Site-specific tests for positive selection on HPV-16 L2.
| Models | lnL | Estimates of parameters | 2Δl | Positively selected sites |
| M7 | −2159.05 | p = 0.005, q = 0.020 | NA | |
| M8 | −2150.14 | p0 = 0.998, p = 0.009, q = 0.029, p1 = 0.002, ω = 188.75 | 17.82 P<0.05 | 330F**, 378F* |
Table 8. Site-specific tests for positive selection on HPV-58 L1.
| Models | lnL | Estimates of parameters | 2Δl | Positively selected sites |
| M7 | −2392.27 | p = 0.005, q = 0.020 | NA | |
| M8 | −2389.02 | p0 = 0.991, p = 16.456, q = 99.000, p1 = 0.009, ω = 14.706 | 6.50 P<0.05 | 5L, 150L*, 276L, |
Table 9. Site-specific tests for positive selection on HPV-58 L2.
| Models | lnL | Estimates of parameters | 2Δl | Positively selected sites |
| M7 | −2089.36 | p = 0.005, q = 0.047 | NA | |
| M8 | −2076.71 | p0 = 0.982, p = 2.308, q = 99.000, p1 = 0.018, ω = 24.49 | 25.30 P<0.05 | 231T*, 233H*, 234K*, 243L*, 446M** |
Table 6 , 7 , 8 , 9: lnL, the log-likelihood difference between the two models; 2Δl, twice the log-likelihood difference between the two models; The positively selected sites were identified with posterior probability ≥0.9 using Bayes empirical Bayes (BEB) approach. One asterisk indicates posterior probability ≥0.95, and two asterisks indicate posterior probability ≥0.99. NA means not allowed. NS means the sites under positive selection but not reaching the significance level of 0.9.
Discussion
Human papillomavirus (HPV) vaccines against L1 are now licensed in more than 100 countries. National and regional immunization programs aimed at young adolescent girls have been widely implemented, and include catch-up programs in some countries up to the age of 18 years or older. However these vaccines target only two of the 15 high-risk HPV types, responsible for 70–80% of cervical cancer cases. The prevention of 96% of cervical cancer would require immunity against at least 7 high risk HPV types (HPV-16, 18, 31, 33, 45, 52 and 58) [5]. L2 and other subtypes' L1 are now being used in vaccine research on a broadening scale.
Among 3000 volunteer outpatients investigated in our early study between 2009 and 2011, 646 cases were HPV positive, for a rate of 21.5%. Among the 646 positive samples, 476 cases were of the high risk type (73.7% of the total positive samples), while 170 cases were of the low risk type (26.3% of the total positive samples). The most common HPV high risk subtypes in Southwest China among female reproductive tract infections were HPV-16, 58,18, 31, 33, and 35. The most common low risk subtypes were HPV-6, 11, and 81. Among 476 high risk type positive samples, HPV-16 and 58 were the main high risk subtypes. HPV-16 comprised 217 cases (45.6% of the total high risk subtypes' samples); HPV-58 145 cases (30.5% of the total high risk subtypes' samples). HPV-16 and HPV-58 comprised 362 cases (76.1% of the total high risk subtypes' samples), while all the other high risk subtypes including HPV33,35,18,31,59,66 comprised only 114 cases (24% of the total high risk subtypes' samples) [26]. This may be attributed to the special geographic location where the samples were taken and interactions of different populations in southwest border district of China.
HPV-16 is the most prevalent high-risk types of HPV worldwide, and also the type that is most frequently associated with cancer [27]–[29]. However, the prevalence of HPV-58 and its relative contribution in the development of cervical neoplasia vary greatly in different area worldwide. HPV-58 has previously been reported to be particularly prevalent in some areas of northeastern Asian (China, Korean, Japan), some regions of central and south America, with a significant trend to increased prevalence in line with the increasing severity of lesions [30]–[38]. These data differed from those of the international study reported by Bosch et al., which did not include Chinese women [27]. HPV-58 is rarely detected in the Americas, Europe and Africa, Worldwide, HPV-58 has been found in only 2% of cervical cancers [27]. In contrast, Chan et al. [39] reported that one-third of the women with cervical cancers in Hong Kong were positive for HPV-58, and similarly high rates have also been reported for Chinese populations living in Shanghai (East of China) (30.4% in cervical cancers subgroup) [40], Jiangxi (middle of China) (18.4% in cervical cancers subgroup) [40], and Taiwan (21.0% in cervical cancers subgroup) [41]. Studies from different group suggest that HPV-58 may play a more prominent role in the development of CC in Asia than HPV types 31, 33, and 45, which are more common on other continents [31], [33], [40], [42], [43]. HPV-58 variants carrying E7 C632T (T20I) and G760A (G63S) substitutions may be associated with an increased risk for cervical cancer [39], [43].
Based on previous research of other research group and us, we chose HPV-16 (L1: n = 31, L2: n = 28) and HPV-58 (L1: n = 21, L2: n = 21) samples to explore the intratype variations, construct their phylogenetic trees and estimate selection pressures acting on the L1 and L2 genes in our current study.
It is reported that specific intratype HPV genome variations may be related to virus infectivity, pathogenicity, progression to cervical cancer, viral particle assembly and host immune response [44], [45]. However, there is still no data demonstrating if immunity to one HPV variant can protect against infection from another variant. Thus, identification of HPV genetic diversity in specific clinical settings may prove important for the rational design of diagnostic, therapeutic, and vaccine strategies [46], [47]. The main purpose of our study was to explore the nucleotide variability and phylogeny of high-risk HPV-16 and HPV-58 samples from Southwest China. To the best of our knowledge, this is the first study examining the L1 and L2 proteins of HPV-16 and -58 variants in Southwest China that have considered the complete sequences of the L1 and L2 genes. Information regarding L1 and L2 gene variations of HPV-16 and HPV-58 in the present study could have important implications for diagnosis and formulation of recommendations for the use of second-generation polyvalent HPV vaccines in China.
The neighbor joining phylogenetic tree results showed that the L1 and L2 of HPV-16 and HPV-58 are distributed in two or three standard branches, not in one specific branch. Most HPV-58 L1 and L2 sequences fell into D90400 (Japan), EU918765, HQ537760 (isolate AS347), which belong to the Asian and the European branches. Some mutations occurred in the sequences encoding the alpha helix and beta turn, which influence protein secondary structure. The function of these mutations still needs further investigations. In this study, the most common mutation of HPV-16 L1 was A979C (T327P). This position located in a major common B cell epitope peptide in both mice and humans, which might affect the immunogenicity of HPV-16 L1 [48].
From the result of selective pressure analysis we conclude that most mutations of HPV-16 and HPV-58 L1 and L2 were of positive selection, which indicated that these amino acid changes were beneficial to accommodate the human papillomavirus to its environment.
The L1 and L2 of HPV-16 and HPV-58 had a low rate of nucleotide changes, something that could be attributed to the fact that HPV uses the host cell's DNA replication machinery, which is characterized by proofreading capacity and post replication repair mechanisms [17]. Moreover, many core functions of viral proteins are very important in the viral life cycle, and this may result in selection that restricts the actual number of possible evolutionary events. Some samples did not belong to any standard type branch because of distinctive mutations or the narrow scope on choosing standard types, requiring further efforts of analysis in the future.
Nucleotide substitutions in viral genomes may affect virus assembly, carcinogenic potential, and host immunologic responses [49]. HPV-16 and HPV-58 gene diversity may help us understand the oncogenic potential of these viral strains and how polymorphisms can affect the host response following infection or vaccination.
Acknowledgments
We would like to thank Dr. John R. Basile of University of Maryland, Dental School, Department of Oncology and Diagnostic Sciences, and Dr. Rays Jiang of Broad Institute of MIT and Harvard, for proof-reading and reviewing our manuscript.
Funding Statement
This research was supported by the National Natural Science Foundation of China (grant no. 81171946), the Natural Science Foundation of Yunnan Province (grant no. 2009ZC187M and grant no. 2011CA016), the Special Research Fund for the Doctoral Program of Higher Education of China (grant no. 20111106120055) and the Scientific Research Foundation for Returned Overseas Chinese Scholars, Ministry of Education of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. de Villiers EM, Fauquet C, Broker TR, Bernard HU, H zH (2004) Classification of papillomaviruses. Virology 324: 17–27. [DOI] [PubMed] [Google Scholar]
- 2. H zH (1996) Papillomavirus infections – a major cause of human cancers. Biochim Biophys Acta 1288: F55–F78. [DOI] [PubMed] [Google Scholar]
- 3. Tota JE, Chevarie-Davis M, Richardson LA, Devries M, EL F (2011) Epidemiology and burden of HPV infection and related diseases: implications for prevention strategies. Prev Med 53: S12–21. [DOI] [PubMed] [Google Scholar]
- 4. Garbuglia AR, Carletti F, Minosse C, Piselli P, Zaniratti MS, et al. (2007) Genetic variability in E6 and E7 genes of human papillomavirus -16, -18, -31 and -33 from HIV-1-positive women in Italy. New Microbiol 30: 377–382. [PubMed] [Google Scholar]
- 5. Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, et al. (2003) Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 348: 518–527. [DOI] [PubMed] [Google Scholar]
- 6. Buck CB, Cheng N, Thompson CD, Lowy DR, Steven AC, et al. (2008) Arrangement of L2 within the Papillomavirus Capsid. J Virol 82: 5190–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Trus BL, Roden RB, Greenstone HL, Vrhel M, Schiller JT, et al. (1997) Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 A resolution. Nat Struct Biol 4: 413–420. [DOI] [PubMed] [Google Scholar]
- 8. Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, et al. (2007) Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem 282: 31803–31811. [DOI] [PubMed] [Google Scholar]
- 9.Moscicki AB, JS S (2008) Issues in human papillomavirus vaccination in adolescents. J Adolesc Health 43: S 1–S4. [DOI] [PubMed]
- 10. Bossis I, Roden RB, Gambhira R, Yang R, Tagaya M, et al. (2005) Interaction of tSNARE Syntaxin 18 with the Papillomavirus Minor Capsid Protein Mediates Infection. J Virol 79: 6723–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kondo K, Ishii Y, Ochi H, Matsumoto T, Yoshikawa H, et al. (2007) Neutralization of HPV16, 18, 31, and 58 pseudovirions with antisera induced by immunizing rabbits with synthetic peptides representing segments of the HPV16 minor capsid protein L2 surface region. Virology 358: 266–272. [DOI] [PubMed] [Google Scholar]
- 12. Pastrana DV, Gambhira R, Buck CB, Pang YY, Thompson CD, et al. (2005) Cross-neutralization of cutaneous and mucosal Papillomavirus types with anti-sera to the amino terminus of L2. Virology 337: 365–372. [DOI] [PubMed] [Google Scholar]
- 13. Malagón T, Drolet M, Boily MC, Franco EL, Jit M, et al. (2012) Cross-protective efficacy of two human papillomavirus vaccines: a systematic review and meta-analysis. Lancet Infect Dis 12: 781–789. [DOI] [PubMed] [Google Scholar]
- 14. Pereira R, Hitzeroth II, EP R (2009) Insights into the role and function of L2, the minor capsid protein of papillomaviruses. Arch Virol 154: 187–197. [DOI] [PubMed] [Google Scholar]
- 15. Qu W, Jiang G, Cruz Y, Chang CJ, Ho GY, et al. (1997) PCR Detection of Human Papillomavirus: Comparison between MY09/MY11 and GP51/GP61 Primer Systems. J Clin Microbilo 35: 1304–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, PJ S (1995) The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papilloma-virus detection by PCR. J Gen Virol 76: 1057–1062. [DOI] [PubMed] [Google Scholar]
- 17. Frati E, Bianchi S, Colzani D, Zappa A, Orlando G, et al. (2011) Genetic variability in the major capsid L1 protein of human papillomavirus type 16 (HPV-16) and 18 (HPV-18). Infect Genet Evol 11: 2119–2124. [DOI] [PubMed] [Google Scholar]
- 18. J F (1982) Numerical methods for inferring evolutionary trees. Q Rev Biol 57: 379–404. [Google Scholar]
- 19. J F (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 20. J F (1989) PHYLIP – phylogeny inference package (version 3 2). Cladistics 5: 164–166. [Google Scholar]
- 21. Yamada T, Wheeler CM, Halpern AL, Stewart AC, Hildesheim A, et al. (1995) Human Papillomavirus Type 16 Variant Lineages in United States Populations Characterized by Nucleotide Sequence Analysis of the E6, L2, and L1 Coding Segments. J Virol 69: 7743–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nei M, T G (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426. [DOI] [PubMed] [Google Scholar]
- 23. Hamza AA, Robene-Soustrade I, Jouen E LP, Chiroleu F, Fisher-Le Saux M, et al. (2012) MultiLocus Sequence Analysis- and Amplified Fragment Length Polymorphism-based characterization of xanthomonads associated with bacterial spot of tomato and pepper and their relatedness to Xanthomonas species. Syst Appl Microbiol 35: 183–190. [DOI] [PubMed] [Google Scholar]
- 24. Karanam B, Jagu S, Huh WK, RB R (2009) Developing vaccines against minor capsid antigen L2 to prevent papillomavirus infection. Immunol Cell Biol 87: 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Z Y (2007) PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 26. Yue YF, Yang LJ, Chen JY, Pan Y, Zhao YJ, et al. (2012) Genetic Subtypes and Distribution of HPV from 3000 volunteer outpatients in Yunnan Province. Chinese Journal of Obstetrics & Gynecology and Pediatrics (Electronic Edition) 8: 467–470. [Google Scholar]
- 27. Bosch FX, Manos MM, Muñoz N, Sherman M, Jansen AM, et al. (1995) Prevalence of Human Papillomavirus in Cervical Cancer: a Worldwide Perspective. J Natl Cancer Inst 87: 796–802. [DOI] [PubMed] [Google Scholar]
- 28. Clifford GM, Smith JS, Plummer M, Munoz N, S F (2003) Human papillomavirus types in invasive cervical cancer worldwide: a meta analysis. Br J Cancer 88: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, et al. (2010) Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol 11: 1048–1056. [DOI] [PubMed] [Google Scholar]
- 30. Sun ZR, Ji YH, Zhou WQ, Zhang SL, Jiang WG, et al. (2010) Characteristics of HPV prevalence among women in Liaoning province, China. Int J Gynaecol Obstet 109: 105–109. [DOI] [PubMed] [Google Scholar]
- 31. Camara GN, Cerqueira DM, Oliveira AP, Silva EO, Carvalho LG, et al. (2003) Prevalence of human papillomavirus types in women with pre-neoplastic and neoplastic cervical lesions in the Federal District of Brazil. Mem Inst Oswaldo Cruz 98: 879–883. [DOI] [PubMed] [Google Scholar]
- 32. Bae JH, Lee SJ, Kim CJ, Hur SY, Park YG, et al. (2008) Human papillomavirus (HPV) type distribution in Korean women: A meta-analysis. J Microbiol Biotechnol 18: 788–794. [PubMed] [Google Scholar]
- 33. Sasagawa T, Basha W, Yamazaki H, M I (2001) High-risk and multiple human papillomavirus infections associated with cervical abnormalities in Japanese women. Cancer Epidemiol Biomarkers Prev 10: 45–52. [PubMed] [Google Scholar]
- 34. Chan PK, Li WH, Chan MY, Ma WL, Cheung JL, et al. (1999) High prevalence of human papillomavirus type 58 in Chinese women with cervical cancer and pre-cancerous lesions. J Med Virol 59: 232–238. [PubMed] [Google Scholar]
- 35. Wu Y, Chen Y, Li L, Yu G, Zhang Y, et al. (2006) Associations of high-risk HPV types and viral load with cervical cancer in China. J Clin Virol 35: 264–269. [DOI] [PubMed] [Google Scholar]
- 36. Liu J, Rose B, Huang X, Liao G, Carter J, et al. (2004) Comparative analysis of characteristics of women with cervical cancer in highversus low-incidence regions. Gynecol Oncol 94: 803–810. [DOI] [PubMed] [Google Scholar]
- 37. Lai CH, Chao A, Chang CJ, Chao FY, Huang HJ, et al. (2008) Host and viral factors in relation to clearance of human papillomavirus infection: a cohort study in Taiwan. Int J Cancer 123: 1685–1692. [DOI] [PubMed] [Google Scholar]
- 38. Xin CY, Matsumoto K, Yoshikawa H, Yasugi T, Onda T, et al. (2001) Analysis of E6 variants of human papillomavirus type 33, 52 and 58 in Japanese women with cervical intraepithelial neoplasia/cervical cancer in relation to their oncogenic potential. Cancer Lett 170: 19–24. [DOI] [PubMed] [Google Scholar]
- 39. Chan PK, Lam CW, Cheung TH, Li WW, Lo KW, et al. (2002) Association of Human Papillomavirus Type 58 Variant With the Risk of Cervical Cancer. J Natl Cancer Inst 94: 1249–1253. [DOI] [PubMed] [Google Scholar]
- 40. Lin QQ, Yu SZ, Qu W, Cruz Y, RD B (1998) Human papillomavirus types 52 and 58. Int J Cancer 75: 484–485. [DOI] [PubMed] [Google Scholar]
- 41. Lai HC, Sun CA, Yu MH, Chen HJ, Liu HS, et al. (1999) Favorable clinical outcome of cervical cancers infected with human papilloma virus type 58 and related types Int J Cancer. 84: 553–557. [DOI] [PubMed] [Google Scholar]
- 42. Huang S, Afonina I, Miller BA, AM B (1997) Human papillomavirus types 52 and 58 are prevalent in cervical cancers from Chinese women. Int J Cancer 70: 408–411. [DOI] [PubMed] [Google Scholar]
- 43. Ding T, Wang X, Ye F, Cheng X, Ma D, et al. (2010) Distribution of human papillomavirus 58 and 52 E6/E7 variants in cervical neoplasia in Chinese women. Gynecol Oncol 119: 436–443. [DOI] [PubMed] [Google Scholar]
- 44. Hildesheim A, Schiffman M, Bromley C, Wacholder S, Herrero R, et al. (2001) Human Papillomavirus Type 16 Variants and Risk of Cervical Cancer. J Natl Cancer Inst 93: 315–318. [DOI] [PubMed] [Google Scholar]
- 45. Pista A, Oliveira A, Barateiro A, Costa H, Verdasca N, et al. (2007) Molecular variants of human papillomavirus type 16 and 18 and risk for cervical Neoplasia in Portugal. J Med Virol 79: 1889–1897. [DOI] [PubMed] [Google Scholar]
- 46. Chaturvedi AK, Dumestre J, Gaffga AM, Mire KM, Clark RA, et al. (2005) Prevalence of human papillomavirus genotypes in women from three clinical settings. J Med Virol 75: 105–113. [DOI] [PubMed] [Google Scholar]
- 47. Stewart AC, Eriksson AM, Manos MM, Muñoz N, Bosch FX, et al. (1996) Intratype Variation in 12 Human Papillomavirus Types:a Worldwide Perspective. J Virol 70: 3127–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fukui A, Matsueda S, Kawano K, Tsuda N, Komatsu N, et al. (2012) Identification of B cell epitopes reactive to human papillomavirus type-16L1- derived peptides. Virol J 9: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cerqueira DM, de S Moraes D, Camara GN, Amaral FA, Oyama CN, et al. (2006) High HPV genetic diversity in women infected with HIV-1 in Brazil. Arch Virol 152: 75–83. [DOI] [PubMed] [Google Scholar]