Abstract
The human intestinal microbiota is one of the most densely populated ecosystems on Earth, containing up to 1013 bacteria/g and in some respects can be considered an organ itself given its role in human health. Bacteriophages (phages) are the most abundant replicating entities on the planet and thrive wherever their bacterial hosts exist. They undoubtedly influence the dominant microbial populations in many ecosystems including the human intestine. Within this setting, lysogeny appears to be the preferred life cycle, presumably due to nutrient limitations and lack of suitable hosts protected in biofilms, hence the predator/prey dynamic observed in many ecosystems is absent. On the other hand, free virulent phages in the gut are more common among sufferers of intestinal diseases and have been shown to increase with antibiotic usage. Many of these phages evolve from prophages of intestinal bacteria and emerge under conditions where their bacterial hosts encounter stress suggesting that prophages can significantly alter the microbial community composition. Based on these observations, we propose the “community shuffling” model which hypothesizes that prophage induction contributes to intestinal dysbiosis by altering the ratio of symbionts to pathobionts, enabling pathobiont niche reoccupation. The consequences of the increased phage load on the mammalian immune system are also addressed. While this is an area of intestinal biology which has received little attention, this review assembles evidence from the literature which supports the role of phages as one of the biological drivers behind the composition of the gut microbiota.
Keywords: gut, microbiota, bacteriophages, phages, prophages, induction, community shuffling
Introduction
Bacteriophages (phages or bacterial viruses) are the most abundant replicating entities on the planet with an estimated population size of ≥ 1030 phage particles, a notion supported by the finding that total viral abundance exceeds that of total prokaryotic abundance in aquatic systems by a factor of 10.1,2 It has been estimated that approximately 108−109 tonnes of carbon are released per day from the biological pool as a consequence of ~1029 viral infections per day in the ocean.3 Within aquatic systems, it has furthermore been proposed that phages are responsible for 20–80% of total bacterial mortality in freshwater and marine ecosystems, representing a significant limiting factor within many bacterial populations.2 Hence, phages play a significant role in influencing the microbial composition of marine communities.3 While such estimates are taken from aquatic environments, phages undoubtedly exert similar influences on other complex microbial ecosystems. Indeed, it has been said that ‘phages are found where bacteria thrive.’4
The human intestine is one of the most densely populated ecosystems on Earth, harbouring 1013−1014 cells/g of fecal matter in the large intestine.5 A healthy intestinal microbiota is composed of many bacterial classes, some of which act as symbionts with proven health-promoting functions, some are commensals - permanent residents which provide no obvious benefit or detriment to the host and some are pathobionts - permanent residents which have the potential to cause disease.6 Consequently, the intestinal microbiota can affect various host processes from gastrointestinal immunity to patho-physiological processes such as inflammatory bowel disease (IBD) and colorectal cancer. In conditions of dysbiosis (any condition in which there is a perturbation in the ‘normal’ intestinal microbiota ecology), the composition of the intestinal microbiota is altered, resulting in either a reduction in the number of symbionts and/or an increase in the number of pathobionts.6 Dysbiosis can be a major forerunner in the pathogenesis of chronic disorders such as IBD; Crohn’s disease, ulcerative colitis and irritable bowel syndrome (IBS).7,8 Dietary strategies to manipulate the composition and function of the intestinal microbiota through the use of probiotics and prebiotics continue to receive much support for preventing or attenuating the symptoms of these diseases.8-11 However, in many cases there are subsets of individuals who do not respond to such interventions. Phages undoubtedly exert significant selective pressure on the members of the human intestinal microbiome, a phenomenon which has been observed in some animal species.12 It is therefore tempting to suggest that “commensal phages” within certain individuals may play a role by infecting some of the beneficial flora, explaining the “non-responders” in some trials. The role of phages in the human intestine is an area which has received little attention and as a result, knowledge on intestinal phage diversity, numbers and functionality is limited.
The aim of this review is to shed light on the potential role that phages may play in human intestinal physiology by examining the literature with respect to phage effects on microbial population dynamics, bacterial chromosome evolution and the host immune system. We consider the phage-host dynamic which occurs in the healthy intestine where microbes may be nutritionally deprived in the lumen. In addition, it is likely that many gut microbes are embedded in biofilms and we theorize how this dynamic could be disrupted to the detriment of the host. We also review the latest evidence of how phages can actually interact directly with the immune system of mammals. In many cases, we can only extrapolate from studies performed in other microbial ecosystems. However, the limited number of studies which have been performed directly on phages of the human intestine, together with phage metagenomic studies make a powerful argument that phages are one of the driving forces shaping the functionality and diversity of the human intestinal microbiome.
Bacteriophage Abundance and Diversity in the Human Intestine
Phages can replicate by means of four lifecycles; lytic, lysogenic, chronic infection and pseudolysogeny.13 This flexibility accounts in part for their biological success rate in terms of natural abundance and diversity. During the lytic lifecycle, the phage infects the cell and redirects cellular metabolism toward phage replication, leading to the production of progeny phages. These progeny phages are then released during cell lysis and continue the infectious cycle in the presence of sensitive hosts. In contrast, in a lysogenic cycle the phage genome becomes incorporated into the host chromosome as a prophage (temperate phage) following infection of the cell and replicates with the host chromosome until such time as a lytic cycle is induced. Interestingly, prophages in marine bacteria have been described as ‘dangerous molecular time bombs’ which are capable of bursting their hosts in response to environmental perturbations.14 However, prophages can also confer immunity to the host strain against infecting phages (homo-immunity). Chronic infection occurs when an infected cell releases progeny phage by budding or extrusion but the cell remains intact in the process.2Lastly, pseudolysogeny has been suggested as a common name for different phage life cycles such as carrier-state (plasmid-like prophages) which is discussed in more detail by Weinbauer.2
Electron microscopy (EM), culture-based phage ecology and most recently, metagenomic approaches are continuing to provide significant insights into phage diversity within the human intestine. As early as 1974, Flewett and coworkers15 demonstrated that tailed phages were the most abundant virus-like particles (VLPs) observed in the feces of humans using transmission electron microscopy (TEM) (Fig. 1). More recently, TEM studies of human biopsies of intestinal mucosal samples concurred with this finding, demonstrating that the dominant VLPs of human feces belonged to the order Caudovirales, displaying morphotypes consistent with the tailed phages Siphoviridae, Myoviridae and Podoviridae, being present at 1.2 × 108 VLPs per biopsy in healthy subjects.16 Long-tailed phages presumably possess an adaptive advantage in the viscous intestinal environment, being able to “seek-out” bacterial host receptors in a more efficient manner, benefiting from rotational movement of the particle.12 In addition, prophages induced from human fecal bacteria upon induction with DNA damaging agents have been detected by EM. Calderaro et al.17 detected phages by EM after experimental induction with mitomycin C in weakly β-hemolytic human intestinal spirochaetes isolated from patients with intestinal disturbances.
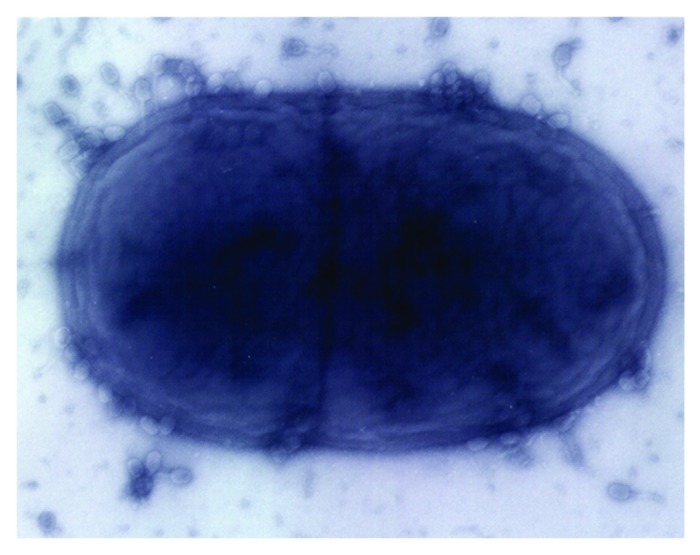
Figure 1. Electron micrograph of tailed phages infecting a dividing bacterial cell.
Culture-based approaches allow direct isolation of intact replicating phage particles and can provide insight into phage-host interactions at bacterial strain level. However, as the host-specificity of most phages is generally narrow or even strain-specific for many phages, the selection of suitable hosts is a major limiting factor when studying phage diversity within complex ecosystems such as the intestine. For example, a three-week survey to isolate Escherichia coli phages from stool samples of Bangladeshi children hospitalized with diarrhea resulted in almost selective isolation of T4-related phages when the E. coli host strain K803 was used as the indicator.18 The E. coli host strain 0127:K63 yielded a different set of phages, showing the morphology of Jersey and β-4-like phages.18 This highlights the level of phage specificity which exists in this environment. Despite this limitation, culture-based methods have resulted in the isolation of numerous phages from human fecal samples using cultivable microorganisms at concentrations ranging from 0–105 plaque forming units (PFU/ml)/g of dry feces.19-25 In some cases, phages have been isolated with a goal to providing possible indicators of fecal contamination in water and as such have generated important insight into phage ecology in the intestine. Phages which infect E. coli, Salmonella, Bacteroides and Enterococcus faecalis among others have been isolated directly from human sewage samples or polluted waters.26-30 A study to determine the occurrence and concentrations of somatic coliphages (phages which infect via the cell membrane) and Bacteroides fragilis phages in the stools of a human population residing in Eastern France for use as possible indicators of viral contamination of water showed that no correlation exists between phage concentration and the age or sex of the human subject.23 With respect to the RNA-containing F-specific (F-RNA) coliphages, approximately 10–20% of human feces have been found to harbour them.12 Of the four genetic groups which have been identified, groups II and III are much more prevalent in human fecal samples than in animal fecal waste.29,31,32 In developing a low-cost method for fecal source identification, Ebdon et al.27 reported that phages which infect the strain Bacteroides (GB-124) are specific to human feces. Most recently, a virulent phage which infects the probiotic cheese strain Lactobacillus paracasei NFBC338 of human intestinal origin was isolated from a sewage sample indicating the potential presence of this and other similar phages in the intestine,33 (Fig. 2). The genome sequence of the 142 kb phage Lb338-1 revealed 199 open reading frames, although only 22% had significant homology to genes found in other Gram positive-associated phages. No significant genomic similarity was observed between this phage and the probiotic host strain suggesting that it has not evolved from that source. Intestinal phages have also been isolated against human pathogenic strains with a view to being exploited for phage therapy purposes. Figure 3 presents a range of tailed phages with lytic activity against the pathogens Clostridium difficile, E. coli 0157:H7 and Salmonella enterica.34-36
Figure 2. Electron micrograph images of phage ϕLb338–1 stained with phosphotungstic acid 0.2% (A) and close-up views of Lb338- (B) (reprinted from Gene, Vol. 448, Alemayehu D, Ross RP, O' Sullivan O, Coffey A, Stanton C, Fitzgerald GF, McAuliffe, O., Genome of a virulent bacteriophage Lb338–1 that lyses the probiotic Lactobacillus paracasei cheese strain, 29–39, © 2009, with permission from Elsevier).
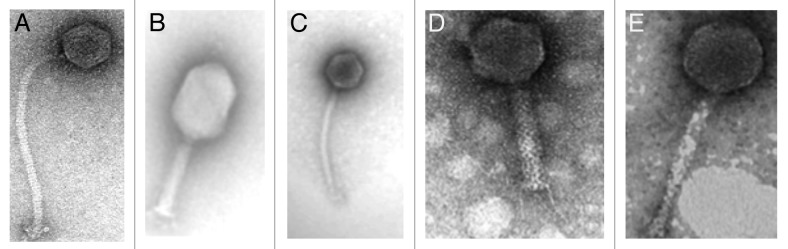
Figure 3. Electron micrographs of intestinal phages (A) Clostridium difficile phage (reprinted from Gene, Vol. 462, Horgan M, O'Sullivan O, Coffey A, Fitzgerald GF, van Sinderen D, McAuliffe O, Ross RP, Genome analysis of the Clostridium difficile phage PhiCD6356, a temperate phage of the Siphoviridae family, 34–43, © 2010, with permission from Elsevier); (B) and (C) Escherichia coli 0157: H7 phages (reprinted from O’ Flynn et al., 2004, © 2004, American Society for Microbiology, Applied and Environmental Microbiology, 70: 3417–3424, DOI: 10.1128/AEM.70.6.3417-3424.2004 and amended with permission from American Society for Microbiology); (D) and (E) Salmonella enterica phages (reprinted from O' Flynn et al., 2006, Journal of Applied Microbiology, Vol. 101, 521–259, John Wiley and Sons, © The Society for Applied Microbiology).
Only a few metagenomic studies of the phage community in the human intestine have been performed, but those which have been completed provide a great deal of information regarding the diversity and abundance of phages in the gastrointestinal tract. These studies not only detect phage sequences but also detect eukaryotic viral genomes, for this reason the collective term viral community is used. Breitbart et al.37 performed the first metagenomic analysis of an uncultured viral community from human feces using partial shotgun sequencing and found that most of the sequences were unrelated to anything previously reported. This suggests a wide diversity of phages in this environment, which is largely uncharacterized. The recognizable viruses were mostly siphophages and prophages within bacterial genomes and the community contained an estimated 1200 viral genotypes, where the most abundant virus made up 4% of the total community. An average viral genome size of 30 kb was assumed for the population modeling based on pulsed field gel electrophoresis.
The fecal viromes of four pairs of adult female monozygotic twins and their mothers were recently characterized,38 which demonstrated that while co-twins and their mothers share a significant degree of similarity in their fecal bacterial communities compared with unrelated individuals, viromes are unique to individuals regardless of their degree of genetic relatedness. The same individual was found to harbour very similar fecal viral communities over at least a one year period which varied by less than 5%. Interestingly, 81% of reads from the VLP-derived metagenomic data sets did not match to any known viruses. Most of the identifiable viruses were prophages or phages generally classified as temperate whose predicted hosts were members of the Firmicutes and Bacteroidetes. These were the most abundant bacterial taxa in the sampled fecal microbial communities. Prophages found in at least one VLP sample were observed at high abundance in 13 different sequenced human intestinal microbial genomes demonstrating that temperate phages are dominant members of the fecal VLP preparations. The presence of integrases in the assembled VLP contigs also supports the hypothesis that temperate phages are the dominant phages in the fecal microbiota. A number of proteins whose functions are involved in processes associated with anaerobic intestinal microbiota were found to be encoded by the VLP-derived contigs including a reductant for the de novo anaerobic synthesis of nucleotides, three oxidative-stress-related proteins and a group of proteins involved in peptidoglycan synthesis, among others. The low level of intrapersonal diversity, the dominance of a few temperate phages and the lack of apparent genetic changes at critical sites such as the clustered regularly interspaced short palindromic repeats (CRISPR) indicate that Red Queen dynamics (predator/prey co-adaptation) which have been described in other environmental ecosystems are absent in the very distal intestine.38 The authors38 postulate that phage production in the fecal community occurs through induction of prophages caused by energy limitation in the feces, which was supported by in vivo studies using gnotobiotic mice. In this case, human gut symbionts, which had colonized mice for two weeks, yielded active prophages from fecal and cecum samples.
Investigation of the gut viromes of six human subjects on one-week controlled feeding regimens revealed changes in the virome community to a new state in response to dietary intervention.39 The study also highlighted that the variation observed in virome samples between individuals before intervention became more alike as a result of similar diet. A subset of 39 VLP contigs (from a total of 7175 contigs) was found to change in association with the intervention revealing a trend toward enrichment in Siphoviridae and depletion of Myoviridae. Of the > 7000 complete and partial phage genomes compiled, the majority were found to be temperate contigs consisted of the usual viral functions required for lytic and lysogenic growth as well as novel functionalities including antibiotic resistance and phage-encoded CRISPR arrays. As indicated in other studies, evidence of predator/prey co-adaptation was absent in the gut microbial community as oscillation of phage and bacterial abundances was not detected. It is suggested that the observed changes in the virome communities could be a consequence of changes in the abundance of the bacterial host populations, but this remains to be confirmed. However, the authors do state that initial results indicate lysogenic induction as a contributing factor.
Breitbart et al.40 presented the first description of the DNA viral community within an infant’s intestine using metagenomic sequencing. Direct epifluorescent microscope counts did not detect any viral particles in the meconium; however, by the end of the first week after birth there were approximately 108 virus particles/g wet weight of feces, most of which appeared to be phages. The majority of sequences had no significant hits, suggesting that the fecal viral community is mostly novel. The most common matches were to phages (double-stranded DNA phage groups: Podoviridae, Siphoviridae and Myoviridae), but the majority (72%) were similar to Siphoviruses and prophages and over 25% of the phage-like sequences were similar to phages that infect lactic acid bacteria (LAB) including Lactobacillus, Lactococcus, Streptococcus and Enterococcus. Several sequences showed homology to genes involved in lysogeny, including integrases and anti-repressors, suggesting that prophages are an important and dynamic component of the intestinal microbiota. The most abundant fecal viral sequences were not identified in breast milk or formula milk, suggesting a non-dietary initial source of viruses. The authors suggested that the earliest infant fecal viruses may be the result of induction of prophages from the newly colonized microbial flora.40 In general, the infant intestinal viral community exhibited a low level of diversity, consisting of only 8 viral genotypes, with the most abundant genotype comprising 43.6% of the total viral community. Microarray experiments indicated that the overall viral community composition changed between 1 and 2 weeks of age, a phenomenon also observed for the infant intestinal microbiota.41,42
A metagenomic study of the RNA-containing viral community in human feces of healthy individuals indicated the prevalence of plant pathogenic RNA viruses, presumably acquired via dietary sources.43 Letarov and Kulikov12 suggested that ingestion of these particles with food occurs at a much higher rate than the internal production of RNA-containing phages.
Bacteriophage Diversity Differs with Physiological Status of the Human Host
The intestinal microbiota is a dynamic “organ”, prone to fluctuations in composition and functionality, with significant consequences for the host as apparent from comparative studies among different groups of individuals. Likewise, studies comparing the diversity and distribution of phages between healthy subjects and patients suggest that the phage population is equally dynamic and begs the question as to whether phages are responding to changes in microbiota composition or are fundamental elements driving bacterial dynamics in the intestine. Studies examining phage levels in different groups of individuals based on physiological status remarkably demonstrate that the abundance and composition of phages differs between healthy subjects and patients, where the latter group have been shown to harbor much higher phage numbers.
In determining the number of coliphages and coliphage strains present in fecal samples collected from both healthy individuals and patients with certain internal and leukemic diseases, Furuse et al.22 noted that qualitative and quantitative differences existed between the phages isolated from both groups. The fecal coliphage titers in healthy subjects were found to be low and the pools of free virions in the feces were represented mainly by temperate phages, presumably maintained in the human intestine through spontaneous induction in lysogenic bacteria. In contrast, some patients harboured a substantial amount of virulent phages of various serotypes. Phage titers increased in several patients when the severity of the clinical symptoms increased. Moreover, Serratia phage titers were markedly increased from the feces of a patient who had developed serious clinical symptoms as a result of infection with the bacterium Serratia.
In a more recent study, Lepage et al.16 measured the total viral community associated with the intestinal mucosa and compared viral abundance between healthy subjects and patients with Crohn disease. Using epifluorescence microscopy, remarkably high numbers of VLPs were observed in the intestinal mucosa of Crohn disease patients with an average of 1.2 × 109 VLPs/biopsy, with dominant morphotypes consistent with Siphoviridae, Myoviridae and Podoviridae. Each individual seemed to be colonized by one dominant family, a phenomenon which was confirmed also by pulsed field gel electrophoresis of viral genomic DNA. Crohn disease patients harboured significantly more VLPs than healthy subjects (2.9 × 109 vs. 1.2 × 108 VLPs/biopsy), but decreased amounts of VLPs were detected in Crohn disease ulcerated mucosa with an average of 2.1 × 109 VLPs/biopsy compared with 4.1 × 109 VLPs/biopsy from Crohn’s disease non-ulcerated mucosa. The authors hypothesized that in non-ulcerated mucosa more viruses are produced or can survive longer. Based on the overall results from the study, Lepage et al.16 postulated that phages might also play a role in the dysregulated immune response of IBD patients to the mucosal-associated bacterial population. However, to fully appreciate the role of phages in the pathogenesis of IBD, the authors16 suggested the need for more detailed studies on larger cohorts relating phage populations to disturbances in bacterial populations.
Phage-Host Dynamics in the Intestine: Ecological Assumptions from the Gut and other Environments
In many environmental ecosystems the population dynamics of phage-bacteria interactions is predatory, following Lokta-Volterra or “kill-the-winner” dynamics.44 This essentially means that phages expand on the fastest growing host population and the phage epidemic ceases when the diminished host population no longer supports phage replication.4 Interestingly, Wiggins and Alexander45 reported that a population density of approximately 104 cells/ml is required for exponential phage growth. Thus ‘kill-the-winner’ dynamics ensures that microbial diversity is maintained and that no particular species can dominate an ecosystem.
The recently proposed “constant-diversity” (CD) dynamics model suggests that ‘kill-the-winner’ dynamics is responsible for maintaining diversity even among closely related lineages.46 For example, the high level of coliform diversity observed in horse feces has been linked to the selective pressure of coliphage infection.47 Two distinct phages were shown to drive the diversification of a single strain, namely the marine microorganism Flavobacterium, resulting in a multi-strain and multi-resistant population.48 This can be explained by the fact that Lokta-Volterra interactions display evidence of Red Queen co-evolution.38 This evolutionary race between phages and their hosts is particularly apparent with changes in phage tail fibers over time (which change to adapt to different bacterial recognition sites), changes in bacterial phage receptors (which change to prevent phage attachment), or CRISPR elements.49 These regions in the host are referred to as metagenomic islands (MGIs) since they are predicted to be unique to individual cell types being under-represented in all of the genomes of one species.46
A comparison by Rodriguez-Valera et al.46 of MGIs (greater than 10 kb) in all sequenced marine bacterial species to the marine metagenome demonstrated that specific functional categories were over-represented. These included genes encoding products that are exposed extracellularly such as the variable O chain of the lipopolysaccharide (LPS), exopolysaccharide biosynthesis clusters and genes involved in sugar modifications of extracellular structures. All the genes identified encoded potential phage recognition sites, suggesting a role in phage avoidance.46 Thus, the authors46 concluded that phage-interacting genes are often strain specific and when shared between strains they tend to be more highly divergent.
“Kill-the-winner” or CD dynamics is not expected to occur in physically constrained microbial communities such as biofilms.46 We have already seen that ‘kill-the-winner’ dynamics was not reported for phage-host interactions in the very distal intestine38 suggesting that such interactions in the intestine are different. Obviously, the physiological status and the spatial location of the cells are important elements determining which type of interaction will ensue. For example, it has been reported that E. coli in the lumen of the colon are nutritionally deprived and non-replicating50 and are thus a poor target for phages.4 The metabolically active intestinal E. coli cells are found in microcolonies associated with the mucus layer of the intestinal mucosa51,52 and as suggested by Chibani-Chennoufi et al.4 are physically shielded from phages. The bacteria in the large intestine exist in a multiplicity of different microhabitats and metabolic niches in the mucus layer lining the intestine, the mucosa and on the surfaces of digestive residues in the intestinal lumen.53 These biofilms are usually multispecies consortia and many species in the biofilm exhibit greater resistance to antibiotics and other deleterious environmental factors that are inhibitory to free living cells.54-56 Cell lysis in biofilms may occur as a result of cell aging or through the action of bacteriocins and phages.57
In terms of access, many biofilms contain water-filled channels which would enable phages to penetrate the biofilm interior.57 However, biofilms can be encased in a hydrated extracellular polymeric substances matrix which is composed of polysaccharides, proteins, nucleic acids and lipids.58,59 Certain phages do produce polysaccharases or polysaccharide lyases which degrade bacterial capsular polysaccharides to allow access to bacterial cell surfaces which have been shown to assist in biofilm disruption.60,61 These enzymes are very specific, seldom acting on more than a few closely related polysaccharide structures.62,63
Many bacteria in the biofilm will present sub-optimal environments for phage infection and growth (as suggested by Sutherland et al.57). The tight cell-cell binding between genetically distinct bacteria known as co-aggregation, which is observed in bacterial biofilms, may also conceal bacterial phage receptors.57 Some studies have suggested that phages and bacteria can co-exist stably within a biofilm,64-66 although presumably would not display the same phage-host dynamics observed for planktonic bacteria. For example, using fluorescence correlation spectroscopy to study diffusion and reaction of phages inside biofilms, Briandet et al.67 demonstrated that phages could diffuse within the biofilm, could be immobilized, amplified and released by a lytic cycle in the biofilm and could interact with their specific binding sites on the bacteria, even in the absence of lytic activity.
The “spatial refuge hypothesis” of Schrag and Mittler68 may provide some explanation for the long-term persistence of bacteria and phages within biofilms. In a study with coliphages, the authors observed that the persistence of phages was facilitated by the attachment of the bacteria to glass, resulting in phage densities of 105−108 PFU/ml, as opposed to extinction in liquid cultures. The final percentage of sensitive bacteria was significantly higher in walls than in liquid populations, suggesting that the glass surfaces acted as a spatial refuge for sensitive bacteria. Weinbaur2 suggests that spatial refuges in the environment could be a result of attachment to particles. Likewise, spatial refuge in the intestine could be linked to protection of bacterial cells in biofilms, or in inaccessible areas such as crypts or the appendix for example, preventing phage from driving all sensitive bacteria to extinction. It would thus appear that the phage-host dynamic in the intestine is more markedly affected by prophages rather than lytic viruses and may play a significant role in shaping the intestinal microbial community. This also presumably explains the lack of any study to date reporting a virus-to-bacterium ratio in the human intestine approaching the 10:1 ratio observed in eutrophic estuarine waters.4
Prophages are the Major Drivers of Microbial Evolution and Diversity in Intestinal Bacteria
It has been estimated that 60–70% of sequenced bacterial genomes contain prophages14,69,70 with many bacteria being polylysogenic, containing multiple prophages.71 This is hardly surprising given that two-thirds of all sequenced γ-proteobacterial and low GC Gram-positive bacteria have been found to harbour prophage sequences.72 It has even been suggested that the earliest infant fecal viruses may be the result of induction of prophages from newly colonized microbial flora.40 In terms of bacterial genome location, prophages tend to integrate into transfer RNA (tRNA) genes and as a result a number of prophages also carry tRNA genes. Rearrangements of the bacterial genome across prophage sequences can also occur for lysogens carrying multiple sequence-related prophages.72 The prophages of pathogenic bacterial genomes have been identified as major contributors to strain-specific DNA,73,74 a phenomenon which is also seen in intestinal commensal bacteria. Interestingly, microarray analysis of the intestinal commensal Lactobacillus johnsonii NCC533 revealed that prophages represented about 50% of the strain-specific DNA.75 Likewise, two closely related but independent isolates of the intestinal commensal Lb. johnsonii LJ771 were found to differ mainly in prophage content.76 Presumably, in the intestine, lysogeny is a much more advantageous state from the perspective of the phage, being shielded from low host abundance due to nutrient limitations or the inability to seek out suitable hosts which may be protected in biofilms. This has also been observed in the marine environment whereby slow growth was shown to favor lysogeny in the Listonella pelagia phage-host system.77
Prophages have also been shown to benefit bacterial host fitness78 and six mechanisms have been postulated toward accomplishing this14,78: (1) prophages serve as anchor points for genome rearrangements, often functioning as transposons, (2) prophages can interrupt genes through insertion, producing metabolic losses through silencing non-essential gene functions, (3) prophages usually confer homo-immunity to related phage infections, (4) prophages can cause lysis of closely related nearby strains (“kill-the-relatives”), (5) prophages can introduce new fitness factors by conversion or transduction and (6) phage repressors can potentially silence host genes.
Fitness factors are additional genes which are not essential for the phage life cycle and are often termed lysogenic conversion factors or morons (for more DNA, or genes that are not required to complete the phage lytic cycle.)78 Unlike virulence factors, fitness factors are broader in scope, are not limited to pathogenic bacteria and have been divided into three classes: survival factors (e.g., nutrient uptake systems); defensive factors (e.g., masking of antigen); offensive factors (e.g., toxins).78 An example of prophage regulation of host genes was recently observed when it was shown that the phage lambda cI repressor (which inhibits the expression of the genes required for the lytic cycle) binds to the operator of the pckA gene, which encodes phosphoenolpyruvate carboxykinase, a critical gene in gluconeogenesis.79 The authors argued that the increased fitness of the lysogen was derived from the slower growth of the microorganism in energy-poor environments. Prophage maintenance was also shown to result in a reduction in substrate utilization capability in a pseudolysogen of Listonella pelagia compared with its uninfected counterpart.14 Phage Vibrio harveyi myovirus-like (VHML) infection of five V. harveyi strains also resulted in altered phenotypes as determined by substrate utilization and one strain was so dramatically altered that it assumed the profile of Vibrio campbelli.80 In addition, E. coli lysogens of laboratory phages, P1, P2, lambda and Mu exhibit a higher metabolic activity and faster growth rate than non-lysogenic counterparts.81,82 ‘Kill-the-relatives’ dynamics was aptly observed when spontaneous prophage induction in a few lysogenic cells within Salmonella co-cultures was shown to enhance the competitive fitness of the lysogen as a whole, ensuring the maintenance and spread of viral DNA.83 The biological cost to the bacterial population appears minimal as the fraction of phage-producing cells can be estimated to be less than 10−5 of the population.83
Are Prophages “Molecular Time Bombs” of the Human Intestinal Microbiota?
While bacteria and their integrated prophages can co-exist in a “passive” state under the appropriate conditions, prophages can be induced in response to a variety of environmental factors or when host survival is threatened. This may have significant consequences for microbial community composition. Moreover, released prophages may become completely virulent; for example, serial passage of a temperate Streptococcus thermophilus phage resulted in its replacement by a virulent derivative deletion mutant after only a few days.4
In the marine setting, many environmental pollutants have been shown to cause prophage induction of indigenous marine lysogenic bacteria.84,85 Sunscreen products were even shown to increase virus production through prophage induction in marine bacterioplankton.86 Bacterial lysogens of mammalian hosts also undergo induction in response to different agents or conditions. Some antibiotics have been shown to induce prophages, even in intestinal bacteria. For example, the antibiotic ASP250 (chlortetracycline, sulfamethazine and penicillin) was shown to cause prophage induction in the swine gut microbiota which resulted in significant population shifts in phage and bacterial communities.87 Carbadox and metronidazole were found to induce a prophage-like gene transfer agent designated VSH-1 from the etiologic agent of swine dysentery Brachyspira hyodysenteriae.88
Antibacterial growth promoters including the quinoxaline-1,4-dioxide type antibiotics olaquindox and carbadox enhanced the release of shiga-toxin (stx) converting phage particles from shiga-toxin producing E. coli (STEC), suggesting a mechanism for the spread of STEC and development of new STEC pathotypes.89 The antibiotic norflaxicin and H2O2 were also both shown to induce stx-bearing prophages from STEC.90 Los et al.91 have postulated that oxidative stress, which may occur during intestinal colonisation and neutrophil-produced H2O2, may increase production of Shiga toxin as a result of stx-converting prophages in STEC bacteria.
The phage-inducing factor for Streptococcus pyogenes was identified as a small molecule (< 1 kDa) in secretions from mammalian cells92 and suggests that phages have evolved to sense their environment to more precisely time the induction of prophages in a setting which presumably has greater chance of containing sensitive recipient bacterial hosts.93 Trace amounts of benzo[a]pyrene diol (BPDE) from cigarette smoke, which can be found in the vaginal secretion of women who smoke, significantly increased prophage induction in vaginal lactobacilli.94 Certain species of bacteria have been shown to cause prophage induction in neighboring organisms as a form of interspecies competition. H2O2 production by Streptococcus pneumoniae in the nasopharynx has been shown to kill lysogenic (but not non-lysogenic) Staphylococcus aureus by inducing the SOS response.95 The authors postulate that the production of an SOS-inducing signal to activate prophages in neighboring organisms may be a widespread strategy for bacterial interference. The lactococcal narrow spectrum bacteriocin lactococcin 972, which inhibits the incorporation of cell wall precursors in the septum area thereby inhibiting cell division, has also been shown to induce prophages in the lysogenic strain Lactococcus lactis IPLA 513 in a concentration-dependent manner.96
The essential signaling molecules of quorum sensing in many Gram-negative bacteria, the acyl-homoserine lactones (AHL), have been shown to trigger phage production in soil and groundwater bacteria via a mechanism that does not involve the SOS response.97 AHL production from Pseudomonas aeruginosa triggered lambda prophage induction from E. coli. Thus, the prophage elegantly uses a signal indicative of high host cell density to maximise the probability of finding sensitive hosts for subsequent infection. While the authors state that cell-density dependant prophage induction appears to be in conflict with the generalized model of homo-immunity in pure bacterial cultures, a broad host range may be the rule rather than the exception in highly diverse microbial ecosystems such as soil or complex biofilms. This phenomenon was observed among a total of 67 prophages induced from vaginal lactobacilli isolated from women of the United States and Turkey.98 All 67 phages were infective against lactobacilli from both collections and the host range of most phages was broad, including multiple Lactobacillus species. It has been suggested that phages isolated from nutrient-poor marine environments showed a trend toward polyvalency.4
Many phages have also developed strategies to alter their host range using sophisticated molecular mechanisms. For example, Bordetella phage BPP-1 can switch its host specificity by generating mutations in the ligand-binding surface of its major tropism-determinant (mtd), the gene responsible for host recognition, resulting from the activity of a phage-encoded diversity-generating retro-element.99 Phages of Streptococcus thermophilus can alter their host range by exchanging the variable domains of the anti-receptor gene (responsible for recognition of host receptor) which are flanked by conserved collagen-like repeats that serve as target sites for homologous recombination.100 The invertible G segment in phage Mu DNA controls the host range of the phage and depending on the orientation of the G segment, two types of phage particles are produced which recognize different cell surface receptors.101
Many prophages carry key virulence factors, as already observed for stx-bearing prophages. As a result, prophages have been identified as prime suspects in the emergence of new pathogens and epidemic clones, which has led to a conceptual shift from host-pathogen interactions to host-pathogen-phage interactions as reviewed by Brussow et al.78 For example, more than 500 bacterial species have been reported to produce Shiga toxin. The majority of stx-bearing prophages have been shown to recognize an essential and highly conserved bacterial surface protein termed YaeT which has promoted the rapid spread of shigatoxigenic potential throughout multiple E. coli serogroups and related bacterial species.102
Overall, prophage induction in the intestine may be another contributing trigger in dysbiosis, leading to changes in the composition of the intestinal microbiota by changing the ratio of symbionts to pathobionts, a hypothesis which we have termed “community-shuffling” (Fig. 4). For example, prophage induction from a symbiont would very likely lead to lytic infection of closely related symbionts. This would presumably lead to an imbalance in the microbial population, providing an opportunity for pathobiont occupation of the symbiont niche, thus increasing the ratio of pathobionts to symbionts. In support of this theory, the antibiotic associated prophage induction observed by Allen et al.87 in the swine gut microbiota also coincided with a decrease in the lactic acid bacterium Streptococcus and an increase in Escherichia populations. The observed decrease in Streptococcus spp and the abundance of Streptococcus phages also supported “kill-the-winner” population dynamics in this environment. Furthermore, the authors suggest that the immunosuppressive effects observed for antibiotics in mice103,104 may be connected to the abundance of LABs.
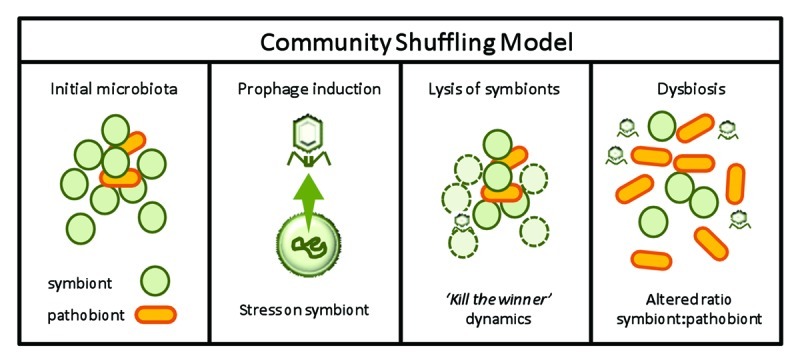
Figure 4. Model for “community shuffling” hypothesis demonstrating potential consequences of prophage induction among the intestinal microbiota.
Prophage excision has also been linked to cell death in biofilms, a phenomenon observed for P. aeruginosa, with a resultant impact on biofilm architecture.105,106 It has also been postulated that dispersing biofilm cells revert to the planktonic mode of growth.56 This would enable “kill-the-winner” dynamics to ensue for a period of time and may even lead to the extinction of some microbial lineages as observed for certain algal blooms in the marine environment.107
Through prophage induction as a result of an environmental trigger, subsequent “kill-the-winner” dynamics and niche re-occupation, the “community shuffling” model may explain the increased number of VLPs and the population shifts which have been observed in patients suffering from ailments of the gastrointestinal tract as opposed to healthy individuals. Assuming that planktonic mode of growth ensues for a period of time, it is also important to consider bacterial strategies to overcome phage predation which can have consequences for cell functionality and phage diversity.
Bacteriophage Resistance and Host Avoidance Mechanisms
In response to phage predation, bacteria have evolved strategies to evade phage attack but, following typical Red Queen-dynamics, phages continuously adapt to overcome such mechanisms. These phage resistance mechanisms have been very well characterized for the industrially important LAB which serve as starter cultures in the food industry, but are undoubtedly widespread in nature.108,109 For example, phage adsorption can be impeded through the masking of phage receptors, while membrane-associated proteins have been associated with prevention of phage DNA injection. Within the cell, restriction/modification (R/M) systems can degrade phage-injected DNA or specialized systems may interfere with various stages of phage replication, transcription and translation such as CRISPR or abortive infection mechanisms.110
Metagenomic studies of salterns and sludge ecosystems have indicated that many of the apparent genetic changes over time occur at loci that prevent phage attachment, such as outer membrane proteins and polysaccharides,38 indicating the significance of this form of resistance on bacterial evolution. However, the extent of phage resistance in the intestine is unclear given that the Red Queen dynamics of planktonic bacteria do not appear to apply in this setting. In the development of a genetic bar-code to define an organism’s niche, O’Sullivan et al.111 demonstrated that R/M genes specific to dairy lactobacilli were not found in intestinal-associated lactobacilli. The authors postulated that dairy bacteria may be more susceptible to phage attack than those found in the intestinal environment. In contrast, R/M systems were deemed an enriched component in the set of laterally transferred genes among the intestinal-associated Bacteroidetes.112 Moreover, a total of 48 (out of 74) human intestinal bacterial species and the prominent intestinal archeon, Methanobrevibacter smithii, were found to harbor CRISPR elements.38 The lateral transfer of a CRISPR locus of the Lhe1 family from low GC-Firmicutes (e.g., Streptococcus or Lactobacillus), to a high GC-Actinobacteria (Bifidobacterium) has been hypothesized to have occurred as a result of Bifidobacterium phage predation in the human intestinal environment,113 but active CRISPR dynamics were not apparent for fecal phages of healthy adults, again highlighting the prevalence of prophages rather than lytic phages in the fecal microbiota.38
Natural phage resistance mechanisms have been implicated in escalating phage population diversity through recombination events between phage genomes and native prophage sequences or host chromosomal sequences. This strategy has enabled phages to overcome resistance mechanisms, a phenomenon that has been observed for dairy starter cultures in a factory setting.114-116 For example, lactococcal phage 50 became completely resistant to the type II R/M system encoded on the lactococcal plasmid pTR2030 after it acquired a copy of the plasmid-encoded methylase gene through an in vivo genetic exchange event.114 Four phage mutants resistant to the abortive infection mechanisms AbiK, AbiT or both emerged after extensive genome re-shuffling with resident prophage sequences.116 Thus, potential phage resistance mechanisms in the intestine may serve to amplify the phage population. However, Morgan et al.117 recently demonstrated that co-evolving enemies may escape from Red-Queen dynamics using mutator bacteria (mutation rate approximately 50 times higher than the wild-type) and phages as the model. Indeed, it appeared that the phages could not keep up as they hit their “evolutionary speed limit”.
Another important consideration is that the acquisition of phage resistance could have important consequences for bacterial hosts. It has been hypothesized that as bacterial phage receptors may also correspond to bacterial virulence determinants, phage resistant derivatives (with modified phage receptors) may no longer be capable of causing disease.118 In certain instances the acquisition of phage resistance may result in reduced fitness in the bacterial cell, a phenomenon often encountered when generating phage resistant derivatives of starter cultures. Hence, phage resistant pathogens may have a lower capacity to colonize the host.118 This phenomenon was observed for phage resistant derivatives of Campylobacter jejuni which were unable to efficiently colonize the broiler chicken intestine.119 Recent studies have demonstrated that phage resistant mutants may have functional properties that differ from those of the parental strain. Phage resistant derivatives of Lactobacillus delbreuckii strains demonstrated either a preserved or enhanced ability to activate the small intestinal immune response of BALB/C mice who received pure preparations of phage sensitive or phage resistant cultures over 7 consecutive days.120
Direct Phage Effects on the Mammalian Host Immune System
It has been proposed that phages could contribute to the bio-control of bacteria by reducing bacterial numbers to levels that can be tolerated by the immune system.121 It has been shown that there is a threshold density of bacteria which causes mortality in untreated animals that is reduced by treatment with phages.118 Direct influences of phages on the immune system have also been recently documented. Virus particles of T-even phages were found to reduce reactive oxygen species production, to suppress T-cell and NF-κβ activation facilitating transplant tolerance, and to exert anti-tumor activity.122-125 In the case of phage T4, it has been proposed that binding of the phage particles to eukaryotic cells is mediated by the capsid protein gp24 containing the KGD amino acid motif, similar to the RGD motif recognized by β3 integrins.126,127 The binding is believed to be at least partially responsible for the immunosuppressive and anti-tumor activities of the phage preparation in vivo and in vitro.128 Gorski et al.129 and Kniotek et al.130 have also documented the immunosuppressive activity of phages in the intestines when administered in vivo, inhibiting both humoral and cell-mediated immunity. Thus, intestinal phages may not only eliminate harmful bacteria and reduce commensal bacteria but may also modulate local immune reactions directly themselves.121
Riley131 hypothesized that as phages are immunogenic, these particles could play a role in the pathogenesis of autoimmune and other inflammatory diseases by acting as antigens on epithelial surfaces, by binding to antibodies as immune complexes, through molecular mimicry or as superantigens. The immunogenic effect could take place when phage particles approach high numbers after lytic infection of the normal flora. In support of this hypothesis, it was found that patients with abdominal aortic aneurysms were more likely to have been infected with strains of Chlamydia pneumonia that harbour phages than strains without phages.132 We have already noted that cigarette smoke can increase prophage induction in vaginal lactobacilli and it is interesting to note that smoking is more widespread in sufferers of Crohn disease.133,134 Our theory of “community shuffling” alongside Riley’s theory of the immunological consequences of free lytic phages support the overall hypothesis that phages may have a much greater impact on the overall physiology of the intestinal tract than previously appreciated.
Perspective
The relationship between human health and the gut microbiota has become more apparent with the demonstration of an altered microbial composition in several intestinal and extra-intestinal disorders. We propose that phages play an often invisible role in shaping the composition of the human intestinal microbiota and understanding phage-host dynamics in the intestine will promote our appreciation of how the overall ecosystem in the intestine functions and responds to diet, medication and lifestyle factors.
Prophages appear to be the dominant phage-form in the intestine and in the healthy intestine the number of free lytic phages is relatively low. In addition, markers of Red Queen dynamics in this ecosystem are lacking, suggesting that phages and their bacterial hosts are not involved in the predator-prey warfare observed for planktonic bacteria. However, in the dysbiotic state, lytic phage populations increase. It appears that the accumulation of lytic phages in the intestine may be the result of prophage induction from the resident microbiota.
We propose that prophage induction may be another contributor to intestinal dysbiosis through the concept of “community-shuffling”, resulting in an alteration in the ratio of symbionts to pathobionts through “kill-the-winner” dynamics and niche reoccupation with an undesirable microbiota. Various factors have been shown to trigger prophage induction in many bacteria and recently certain antibiotics were linked to prophage induction in the intestinal microbiota of swine.87 The consequences of changing the phage-host dynamic in the intestine from a prophage-dominated environment to one which contains lytic phages would presumably have consequences for bacterial evolution and cell functionality and may even impinge on the host immune system as phage particles increase in concentration.
More studies are required to address phage numbers and diversity in the human intestine from different population subsets. The metagenomic studies which have been conducted to date on this topic have produced an impressive degree of information and further studies should revise our understanding of phage levels and diversity in the intestinal setting. However, one of the issues associated with metagenomic studies of this nature is the abundance of viral metagenomic sequences lacking any database matches, hampering the interpretation of sequencing data, the assembly of complete gut viral genomes and ultimately an understanding of viral-host biology. Yet such a finding is hardly surprising given that only 0.0002% of the global viral gene pool has been sequenced according to estimations.135 Beyond this, the rapid evolutionary capacity of viral genomes through recombination events, nucleotide substitutions, deletions and insertions, ensures that the gene pool is constantly enriched with different data. Despite such diversity at the nucleotide and even amino acid sequence levels, common three dimensional structures have been observed for multiple phage proteins encoded in syntenic regions from functionally interchangeable cassettes.136 This observation is in line with the classical modular theory of phage evolution which states that “the product and unit of phage evolution is not a given virus but a family of interchangeable genetic elements (modules), each of which is multigenic and can be considered as a functional unit”.137 Hence, DNA segments lacking any sequence similarity may fulfill homologous functions. This also explains the mosaic nature of phage genomes where horizontal gene transfer is a major contributing factor in their evolution. Minot et al.136 have recently integrated this concept into a metagenomic assembly to probe genome structure in 5.6 Gb of viral DNA from six individuals. Gene content was characterized by comparing the database of viral RefSeq proteins to the assembled contigs. This resulted in functional cassettes (composed of homologously functional genes) linking together viral contigs with a high degree of connectivity from diverse genomes. In addition, many of the ORFs could be clustered into inferred cassettes demonstrating conservation of gene order and orientation despite the considerable sequence divergence observed among genomes. The authors suggest that the extremely variable modules resulting from metagenomic assembly may represent functionally similar gene sets which have diversified to escape phage resistance systems. This recent study thus highlights the significance of connecting metagenomic analysis with the traditional notion of bacteriophage evolution to aid interpretation of sequencing data.
Metagenomic studies which sample the microbial and viral communities from different population subsets should now go a long way to defining the dynamic relationship between the viral community and its microbial hosts and ultimately the physiological status of the mammalian host. Experimental approaches which enable the study of phage activity in vivo will also permit a deeper understanding of phage diversity and behavior within the mammalian intestine. Recruiting the other “omic” technologies along with metagenomics in strategically designed experiments is undoubtedly an appealing approach. For example, by sequencing environmental mRNAs, metatranscriptomics enables the identification of the expressed repertoire of genes under a particular set of conditions. The application of this approach revealed carbohydrate metabolism, energy production and synthesis of cellular components as the main functional roles of the healthy human gut microbiota.138 Applying such an approach to study the expression of phage genes in the intestinal setting should indicate the functionality of prophage-encoded genes in microbial physiology; the environmental factors which trigger the expression of genes associated with the lytic cycle and determine the dynamic which exists between phage and microbial host under different physiological states and at different sites along the mammalian gut. While the limited sequence similarities observed across phage genomes at the nucleotide level may pose an issue in data interpretation, metaproteomics, which studies protein expression, could provide a more suitable platform for assessing phage activity, enabling the direct study of the expressed phage proteins.
Such studies may also result in novel concepts for probiotic therapy, which is an important consideration since one of the overall goals of mammalian gut microbiology is to develop suitable therapies to prevent irregularities within this ecosystem and the myriad of disorders that may result. While further work in this particular field is a prerequisite, the limited studies to date provide enough evidence to speculate the potential consequences of the viral community for probiotic therapy:
If prophage induction can alter the intestinal microbial composition, should all probiotics be cured of prophages?
Should probiotics be equipped with an arsenal of phage resistance mechanisms to evade phage attack and prevent diversification of the phage population? It is important to consider that phage resistant derivatives may have different functional attributes to the parent strain which must be considered before they can be used for therapeutic purposes.
Should probiotics be administered in rotation, as performed in the dairy setting for starter cultures? This type of approach has been recently patented for the administration of probiotics to animals.139
Should probiotic therapy be complemented with phage therapy? Using such an approach phage preparations against pathobionts could be administered to eliminate niche-reoccupation with an undesirable microbiota.
In conclusion, future compositional studies of the intestinal microbiota should include the viral community as well. We now know that the viral community is a dynamic component of the intestinal microbiota. While it is highly diverse, composed primarily of unidentified genetic sequences, it is clear that it is unique to its mammalian host, is capable of responding to environmental stimuli and may provide valuable biomarkers for structure and functionality of the intestinal microbiota and above all, mammalian host health. Its study will ultimately lead to an improved understanding of the intestinal microbiota but most importantly should also result in innovative strategies to reduce harmful imbalances and help biologists delineate the best possible therapies for sufferers of intestinal diseases.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/22371
References
- 1.Bergh O, Børsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–8. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 2.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–81. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Suttle CA. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 4.Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brüssow H. Phage-host interaction: an ecological perspective. J Bacteriol. 2004;186:3677–86. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchesi J, Shanahan F. The normal intestinal microbiota. Curr Opin Infect Dis. 2007;20:508–13. doi: 10.1097/QCO.0b013e3282a56a99. [DOI] [PubMed] [Google Scholar]
- 6.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins SM, Denou E, Verdu EF, Bercik P. The putative role of the intestinal microbiota in the irritable bowel syndrome. Dig Liver Dis. 2009;41:850–3. doi: 10.1016/j.dld.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 8.Reiff C, Kelly D. Inflammatory bowel disease, gut bacteria and probiotic therapy. Int J Med Microbiol. 2010;300:25–33. doi: 10.1016/j.ijmm.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Bosscher D, Breynaert A, Pieters L, Hermans N. Food-based strategies to modulate the composition of the intestinal microbiota and their associated health effects. J Physiol Pharmacol. 2009;60(Suppl 6):5–11. [PubMed] [Google Scholar]
- 10.O'Toole PW, Cooney JC. Probiotic bacteria influence the composition and function of the intestinal microbiota. Interdiscip Perspect Infect Dis. 2008;175285:1–9. doi: 10.1155/2008/175285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quigley EM. Prebiotics and probiotics; modifying and mining the microbiota. Pharmacol Res. 2010;61:213–8. doi: 10.1016/j.phrs.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Letarov A, Kulikov E. The bacteriophages in human- and animal body-associated microbial communities. J Appl Microbiol. 2009;107:1–13. doi: 10.1111/j.1365-2672.2009.04143.x. [DOI] [PubMed] [Google Scholar]
- 13.Ackermann HW, DuBow MS. General properties of bacteriophages Viruses of prokaryotes, CRC Press, Boca Raton, 1987:202. [Google Scholar]
- 14.Paul JH. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008;2:579–89. doi: 10.1038/ismej.2008.35. [DOI] [PubMed] [Google Scholar]
- 15.Flewett TH, Bryden AS, Davies H. Diagnostic electron microscopy of feces. I. The viral flora of the feces as seen by electron microscopy. J Clin Pathol. 1974;27:603–8. doi: 10.1136/jcp.27.8.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lepage P, Colombet J, Marteau P, Sime-Ngando T, Doré J, Leclerc M. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut. 2008;57:424–5. doi: 10.1136/gut.2007.134668. [DOI] [PubMed] [Google Scholar]
- 17.Calderaro A, Dettori G, Collini L, Ragni P, Grillo R, Cattani P, et al. Bacteriophages induced from weakly beta-haemolytic human intestinal spirochaetes by mitomycin C. J Basic Microbiol. 1998;38:323–35. doi: 10.1002/(SICI)1521-4028(199811)38:5/6<323::AID-JOBM323>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 18.Chibani-Chennoufi S, Sidoti J, Bruttin A, Dillmann ML, Kutter E, Qadri F, et al. Isolation of Escherichia coli bacteriophages from the stool of pediatric diarrhea patients in Bangladesh. J Bacteriol. 2004;186:8287–94. doi: 10.1128/JB.186.24.8287-8294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calci KR, Burkhardt W, 3rd, Watkins WD, Rippey SR. Occurrence of male-specific bacteriophage in feral and domestic animal wastes, human feces and human-associated wastewaters. Appl Environ Microbiol. 1998;64:5027–9. doi: 10.1128/aem.64.12.5027-5029.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornax R, Moriñigo MA, Gonzalez-Jaen F, Alonso MC, Borrego JJ. Bacteriophages presence in human feces of healthy subjects and patients with gastrointestinal disturbances. Zentralbl Bakteriol. 1994;281:214–24. doi: 10.1016/S0934-8840(11)80572-4. [DOI] [PubMed] [Google Scholar]
- 21.Dhillon TS, Dhillon EK, Chau HC, Li WK, Tsang AH. Studies on bacteriophage distribution: virulent and temperate bacteriophage content of mammalian feces. Appl Environ Microbiol. 1976;32:68–74. doi: 10.1128/aem.32.1.68-74.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuse K, Osawa S, Kawashiro J, Tanaka R, Ozawa A, Sawamura S, et al. Bacteriophage distribution in human feces: continuous survey of healthy subjects and patients with internal and leukaemic diseases. J Gen Virol. 1983;64:2039–43. doi: 10.1099/0022-1317-64-9-2039. [DOI] [PubMed] [Google Scholar]
- 23.Gantzer C, Henny J, Schwartzbrod L. Bacteroides fragilis and Escherichia coli bacteriophages in human feces. Int J Hyg Environ Health. 2002;205:325–8. doi: 10.1078/1438-4639-00152. [DOI] [PubMed] [Google Scholar]
- 24.Havelaar AH, Pot-Hogeboom WM, Furuse K, Pot R, Hormann MP. F-specific RNA bacteriophages and sensitive host strains in feces and wastewater of human and animal origin. J Appl Bacteriol. 1990;69:30–7. doi: 10.1111/j.1365-2672.1990.tb02908.x. [DOI] [PubMed] [Google Scholar]
- 25.Kai M, Watanabe S, Furuse K, Ozawa A. Bacteroides bacteriophages isolated from human feces. Microbiol Immunol. 1985;29:895–9. doi: 10.1111/j.1348-0421.1985.tb00891.x. [DOI] [PubMed] [Google Scholar]
- 26.Bonilla N, Santiago T, Marcos P, Urdaneta M, Domingo JS, Toranzos GA. Enterophages, a group of phages infecting Enterococcus fecalis and their potential as alternate indicators of human fecal contamination. Water Sci Technol. 2010;61:293–300. doi: 10.2166/wst.2010.815. [DOI] [PubMed] [Google Scholar]
- 27.Ebdon J, Muniesa M, Taylor H. The application of a recently isolated strain of Bacteroides (GB-124) to identify human sources of fecal pollution in a temperate river catchment. Water Res. 2007;41:3683–90. doi: 10.1016/j.watres.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 28.Gino E, Starosvetsky J, Armon R. Bacteriophage ecology in a small community sewer system related to their indicative role in sewage pollution of drinking water. Environ Microbiol. 2007;9:2407–16. doi: 10.1111/j.1462-2920.2007.01355.x. [DOI] [PubMed] [Google Scholar]
- 29.Schaper M, Jofre J, Uys M, Grabow WO. Distribution of genotypes of F-specific RNA bacteriophages in human and non-human sources of fecal pollution in South Africa and Spain. J Appl Microbiol. 2002;92:657–67. doi: 10.1046/j.1365-2672.2002.01600.x. [DOI] [PubMed] [Google Scholar]
- 30.Tartera C, Lucena F, Jofre J. Human origin of Bacteroides fragilis bacteriophages present in the environment. Appl Environ Microbiol. 1989;55:2696–701. doi: 10.1128/aem.55.10.2696-2701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brion GM, Meschke JS, Sobsey MD. F-specific RNA coliphages: occurrence, types and survival in natural waters. Water Res. 2002;36:2419–25. doi: 10.1016/S0043-1354(01)00547-4. [DOI] [PubMed] [Google Scholar]
- 32.Cole D, Long SC, Sobsey MD. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl Environ Microbiol. 2003;69:6507–14. doi: 10.1128/AEM.69.11.6507-6514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alemayehu D, Ross RP, O’Sullivan O, Coffey A, Stanton C, Fitzgerald GF, et al. Genome of a virulent bacteriophage Lb338-1 that lyses the probiotic Lactobacillus paracasei cheese strain. Gene. 2009;448:29–39. doi: 10.1016/j.gene.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 34.O’Flynn G, Coffey A, Fitzgerald GF, Ross RP. The newly isolated lytic bacteriophages st104a and st104b are highly virulent against Salmonella enterica. J Appl Microbiol. 2006;101:251–9. doi: 10.1111/j.1365-2672.2005.02792.x. [DOI] [PubMed] [Google Scholar]
- 35.O’Flynn G, Ross RP, Fitzgerald GF, Coffey A. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl Environ Microbiol. 2004;70:3417–24. doi: 10.1128/AEM.70.6.3417-3424.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horgan M, O’Sullivan O, Coffey A, Fitzgerald GF, van Sinderen D, McAuliffe O, et al. Genome analysis of the Clostridium difficile phage PhiCD6356, a temperate phage of the Siphoviridae family. Gene. 2010;462:34–43. doi: 10.1016/j.gene.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 37.Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–3. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, et al. Viruses in the fecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–8. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–25. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159:367–73. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 41.Tapiainen T, Ylitalo S, Eerola E, Uhari M. Dynamics of gut colonization and source of intestinal flora in healthy newborn infants. APMIS. 2006;114:812–7. doi: 10.1111/j.1600-0463.2006.apm_488.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang M, Ahrné S, Antonsson M, Molin G. T-RFLP combined with principal component analysis and 16S rRNA gene sequencing: an effective strategy for comparison of fecal microbiota in infants of different ages. J Microbiol Methods. 2004;59:53–69. doi: 10.1016/j.mimet.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SW, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thingstad TF. Elements of a theory for the mechanism controlling adundance, diversity and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;46:1320–8. doi: 10.4319/lo.2000.45.6.1320. [DOI] [Google Scholar]
- 45.Wiggins BA, Alexander M. Minimum bacterial density for bacteriophage replication: implications for significance of bacteriophages in natural ecosystems. Appl Environ Microbiol. 1985;49:19–23. doi: 10.1128/aem.49.1.19-23.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasić L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–36. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 47.Golomidova A, Kulikov E, Isaeva A, Manykin A, Letarov A. The diversity of coliphages and coliforms in horse feces reveals a complex pattern of ecological interactions. Appl Environ Microbiol. 2007;73:5975–81. doi: 10.1128/AEM.01145-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Middelboe M, Holmfeldt K, Riemann L, Nybroe O, Haaber J. Bacteriophages drive strain diversification in a marine Flavobacterium: implications for phage resistance and physiological properties. Environ Microbiol. 2009;11:1971–82. doi: 10.1111/j.1462-2920.2009.01920.x. [DOI] [PubMed] [Google Scholar]
- 49.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poulsen LK, Licht TR, Rang C, Krogfelt KA, Molin S. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J Bacteriol. 1995;177:5840–5. doi: 10.1128/jb.177.20.5840-5845.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krogfelt KA, Poulsen LK, Molin S. Identification of coccoid Escherichia coli BJ4 cells in the large intestine of streptomycin-treated mice. Infect Immun. 1993;61:5029–34. doi: 10.1128/iai.61.12.5029-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poulsen LK, Lan F, Kristensen CS, Hobolth P, Molin S, Krogfelt KA. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect Immun. 1994;62:5191–4. doi: 10.1128/iai.62.11.5191-5194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macfarlane S, Dillon JF. Microbial biofilms in the human gastrointestinal tract. J Appl Microbiol. 2007;102:1187–96. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- 54.Anwar H, Dasgupta MK, Costerton JW. Testing the susceptibility of bacteria in biofilms to antibacterial agents. Antimicrob Agents Chemother. 1990;34:2043–6. doi: 10.1128/AAC.34.11.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Loosdrecht MC, Lyklema J, Norde W, Zehnder AJ. Influence of interfaces on microbial activity. Microbiol Rev. 1990;54:75–87. doi: 10.1128/mr.54.1.75-87.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webb JS, Givskov M, Kjelleberg S. Bacterial biofilms: prokaryotic adventures in multicellularity. Curr Opin Microbiol. 2003;6:578–85. doi: 10.1016/j.mib.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 57.Sutherland IW, Hughes KA, Skillman LC, Tait K. The interaction of phage and biofilms. FEMS Microbiol Lett. 2004;232:1–6. doi: 10.1016/S0378-1097(04)00041-2. [DOI] [PubMed] [Google Scholar]
- 58.Davey ME, O’toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–67. doi: 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xavier JB, Picioreanu C, Rani SA, van Loosdrecht MC, Stewart PS. Biofilm-control strategies based on enzymic disruption of the extracellular polymeric substance matrix--a modelling study. Microbiology. 2005;151:3817–32. doi: 10.1099/mic.0.28165-0. [DOI] [PubMed] [Google Scholar]
- 60.Hughes KA, Sutherland IW, Jones MV. Biofilm susceptibility to bacteriophage attack: the role of phage-borne polysaccharide depolymerase. Microbiology. 1998;144:3039–47. doi: 10.1099/00221287-144-11-3039. [DOI] [PubMed] [Google Scholar]
- 61.Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007;104:11197–202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sutherland IW. Polysaccharases for microbial polysaccharides. Carbohydr Polym. 1999;38:319–28. doi: 10.1016/S0144-8617(98)00114-3. [DOI] [Google Scholar]
- 63.Sutherland IW. Polysaccharide lyases. FEMS Microbiol Rev. 1995;16:323–47. doi: 10.1111/j.1574-6976.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 64.Lacroix-Gueu P, Briandet R, Lévêque-Fort S, Bellon-Fontaine MN, Fontaine-Aupart MP. In situ measurements of viral particles diffusion inside mucoid biofilms. C R Biol. 2005;328:1065–72. doi: 10.1016/j.crvi.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 65.Storey MV, Ashbolt NJ. Persistence of two model enteric viruses (B40-8 and MS-2 bacteriophages) in water distribution pipe biofilms. Water Sci Technol. 2001;43:133–8. [PubMed] [Google Scholar]
- 66.Tait K, Skillman LC. The efficacy of bacteriophage as a method of biofilm erradication. Biofouling. 2002;18:305–11. doi: 10.1080/0892701021000034418. [DOI] [Google Scholar]
- 67.Briandet R, Lacroix-Gueu P, Renault M, Lecart S, Meylheuc T, Bidnenko E, et al. Fluorescence correlation spectroscopy to study diffusion and reaction of bacteriophages inside biofilms. Appl Environ Microbiol. 2008;74:2135–43. doi: 10.1128/AEM.02304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schrag SJ, Mittler JE. Host-parasite coexistence: the role of spatial refuges in stabilizing bacteria-phage interactions. Am Nat. 1996;148:348–77. doi: 10.1086/285929. [DOI] [Google Scholar]
- 69.Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 70.Casjens S, Hendrix RW. Bacteriophages & the bacterial genome. The Bacterial Chromosome (Higgins NP ed) ASM Press, Washington, D.C. 2005:39-53. [Google Scholar]
- 71.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. Phage as agents of lateral gene transfer. Curr Opin Microbiol. 2003;6:417–24. doi: 10.1016/S1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 72.Canchaya C, Fournous G, Brüssow H. The impact of prophages on bacterial chromosomes. Mol Microbiol. 2004;53:9–18. doi: 10.1111/j.1365-2958.2004.04113.x. [DOI] [PubMed] [Google Scholar]
- 73.Banks DJ, Beres SB, Musser JM. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 2002;10:515–21. doi: 10.1016/S0966-842X(02)02461-7. [DOI] [PubMed] [Google Scholar]
- 74.Ohnishi M, Kurokawa K, Hayashi T. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 2001;9:481–5. doi: 10.1016/S0966-842X(01)02173-4. [DOI] [PubMed] [Google Scholar]
- 75.Ventura M, Canchaya C, Pridmore D, Berger B, Brüssow H. Integration and distribution of Lactobacillus johnsonii prophages. J Bacteriol. 2003;185:4603–8. doi: 10.1128/JB.185.15.4603-4608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Denou E, Pridmore RD, Ventura M, Pittet AC, Zwahlen MC, Berger B, et al. The role of prophage for genome diversification within a clonal lineage of Lactobacillus johnsonii: characterization of the defective prophage LJ771. J Bacteriol. 2008;190:5806–13. doi: 10.1128/JB.01802-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williamson SJ, Paul JH. Environmental factors that influence the transition from lysogenic to lytic existence in the phiHSIC/Listonella pelagia marine phage-host system. Microb Ecol. 2006;52:217–25. doi: 10.1007/s00248-006-9113-1. [DOI] [PubMed] [Google Scholar]
- 78.Brüssow H, Canchaya C, Hardt W-D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y, Golding I, Sawai S, Guo L, Cox EC. Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol. 2005;3:e229. doi: 10.1371/journal.pbio.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vidgen M, Carson J, Higgins M, Owens L. Changes to the phenotypic profile of Vibrio harveyi when infected with the Vibrio harveyi myovirus-like (VHML) bacteriophage. J Appl Microbiol. 2006;100:481–7. doi: 10.1111/j.1365-2672.2005.02829.x. [DOI] [PubMed] [Google Scholar]
- 81.Edlin G, Lin L, Kudrna R. Lambda lysogens of E. coli reproduce more rapidly than non-lysogens. Nature. 1975;255:735–7. doi: 10.1038/255735a0. [DOI] [PubMed] [Google Scholar]
- 82.Lin L, Bitner R, Edlin G. Increased reproductive fitness of Escherichia coli lambda lysogens. J Virol. 1977;21:554–9. doi: 10.1128/jvi.21.2.554-559.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bossi L, Fuentes JA, Mora G, Figueroa-Bossi N. Prophage contribution to bacterial population dynamics. J Bacteriol. 2003;185:6467–71. doi: 10.1128/JB.185.21.6467-6471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cochran PK, Kellogg CA, Paul JH. Prophage induction of indigenous marine lysogenic bacteria by environmental pollutants. Mar Ecol Prog Ser. 1998;164:125–33. doi: 10.3354/meps164125. [DOI] [Google Scholar]
- 85.Danovaro R, Armeni M, Corinaldesi C, Mei ML. Viruses and marine pollution. Mar Pollut Bull. 2003;46:301–4. doi: 10.1016/S0025-326X(02)00461-7. [DOI] [PubMed] [Google Scholar]
- 86.Danovaro R, Corinaldesi C. Sunscreen products increase virus production through prophage induction in marine bacterioplankton. Microb Ecol. 2003;45:109–18. doi: 10.1007/s00248-002-1033-0. [DOI] [PubMed] [Google Scholar]
- 87.Allen HK, Looft T, Bayles DO, Humphrey S, Levine UY, Alt D, et al. Antibiotics in feed induce prophages in swine fecal microbiomes. MBio. 2011;2:00260–11. doi: 10.1128/mBio.00260-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stanton TB, Humphrey SB, Sharma VK, Zuerner RL. Collateral effects of antibiotics: carbadox and metronidazole induce VSH-1 and facilitate gene transfer among Brachyspira hyodysenteriae strains. Appl Environ Microbiol. 2008;74:2950–6. doi: 10.1128/AEM.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Köhler B, Karch H, Schmidt H. Antibacterials that are used as growth promoters in animal husbandry can affect the release of Shiga-toxin-2-converting bacteriophages and Shiga toxin 2 from Escherichia coli strains. Microbiology. 2000;146:1085–90. doi: 10.1099/00221287-146-5-1085. [DOI] [PubMed] [Google Scholar]
- 90.Loś JM, Loś M, Wegrzyn G, Wegrzyn A. Differential efficiency of induction of various lambdoid prophages responsible for production of Shiga toxins in response to different induction agents. Microb Pathog. 2009;47:289–98. doi: 10.1016/j.micpath.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 91.Loś JM, Loś M, Wegrzyn A, Wegrzyn G. Hydrogen peroxide-mediated induction of the Shiga toxin-converting lambdoid prophage ST2-8624 in Escherichia coli O157:H7. FEMS Immunol Med Microbiol. 2010;58:322–9. doi: 10.1111/j.1574-695X.2009.00644.x. [DOI] [PubMed] [Google Scholar]
- 92.Broudy TB, Pancholi V, Fischetti VA. Induction of lysogenic bacteriophage and phage-associated toxin from group a streptococci during coculture with human pharyngeal cells. Infect Immun. 2001;69:1440–3. doi: 10.1128/IAI.69.3.1440-1443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fischetti VA. In vivo acquisition of prophage in Streptococcus pyogenes. Trends Microbiol. 2007;15:297–300. doi: 10.1016/j.tim.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 94.Pavlova SI, Tao L. Induction of vaginal Lactobacillus phages by the cigarette smoke chemical benzo[a]pyrene diol epoxide. Mutat Res. 2000;466:57–62. doi: 10.1016/S1383-5718(00)00003-6. [DOI] [PubMed] [Google Scholar]
- 95.Selva L, Viana D, Regev-Yochay G, Trzcinski K, Corpa JM, Lasa I, et al. Killing niche competitors by remote-control bacteriophage induction. Proc Natl Acad Sci U S A. 2009;106:1234–8. doi: 10.1073/pnas.0809600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Madera C, García P, Rodríguez A, Suárez JE, Martínez B. Prophage induction in Lactococcus lactis by the bacteriocin Lactococcin 972. Int J Food Microbiol. 2009;129:99–102. doi: 10.1016/j.ijfoodmicro.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 97.Ghosh D, Roy K, Williamson KE, Srinivasiah S, Wommack KE, Radosevich M. Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction. Appl Environ Microbiol. 2009;75:7142–52. doi: 10.1128/AEM.00950-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kiliç AO, Pavlova SI, Alpay S, Kiliç SS, Tao L. Comparative study of vaginal Lactobacillus phages isolated from women in the United States and Turkey: prevalence, morphology, host range and DNA homology. Clin Diagn Lab Immunol. 2001;8:31–9. doi: 10.1128/CDLI.8.1.31-39.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu M, Deora R, Doulatov SR, Gingery M, Eiserling FA, Preston A, et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002;295:2091–4. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- 100.Duplessis M, Moineau S. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol Microbiol. 2001;41:325–36. doi: 10.1046/j.1365-2958.2001.02521.x. [DOI] [PubMed] [Google Scholar]
- 101.Sandulache R, Prehm P, Kamp D. Cell wall receptor for bacteriophage Mu G(+) J Bacteriol. 1984;160:299–303. doi: 10.1128/jb.160.1.299-303.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smith DL, James CE, Sergeant MJ, Yaxian Y, Saunders JR, McCarthy AJ, et al. Short-tailed stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes among enterobacteria. J Bacteriol. 2007;189:7223–33. doi: 10.1128/JB.00824-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–58. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–9. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bayles KW. Are the molecular strategies that control apoptosis conserved in bacteria? Trends Microbiol. 2003;11:306–11. doi: 10.1016/S0966-842X(03)00144-6. [DOI] [PubMed] [Google Scholar]
- 106.Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, et al. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2003;185:4585–92. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bratbak G, Egge JK, Heldal M. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar Ecol Prog Ser. 1993;93:39–48. doi: 10.3354/meps093039. [DOI] [Google Scholar]
- 108.Coffey A, Ross RP. Bacteriophage-resistance systems in dairy starter strains: molecular analysis to application. Antonie Van Leeuwenhoek. 2002;82:303–21. doi: 10.1023/A:1020639717181. [DOI] [PubMed] [Google Scholar]
- 109.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 110.Mills S, Ross RP, Neve H, Coffey A. Bacteriophage and anti-phage mechanisms in lactic acid bacteria. Lactic Acid Bacteria. von Wright A: CRC Press, 2011: 165-186. [Google Scholar]
- 111.O’Sullivan O, O’Callaghan J, Sangrador-Vegas A, McAuliffe O, Slattery L, Kaleta P, et al. Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. BMC Microbiol. 2009;9:50. doi: 10.1186/1471-2180-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Horvath P, Coûté-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol. 2009;131:62–70. doi: 10.1016/j.ijfoodmicro.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 114.Hill C, Miller LA, Klaenhammer TR. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J Bacteriol. 1991;173:4363–70. doi: 10.1128/jb.173.14.4363-4370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Labrie SJ, Moineau S. Abortive infection mechanisms and prophage sequences significantly influence the genetic makeup of emerging lytic lactococcal phages. J Bacteriol. 2007;189:1482–7. doi: 10.1128/JB.01111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Suárez V, Zago M, Giraffa G, Reinheimer J, Quiberoni A. Evidence for the presence of restriction/modification systems in Lactobacillus delbrueckii. J Dairy Res. 2009;76:433–40. doi: 10.1017/S0022029909990112. [DOI] [PubMed] [Google Scholar]
- 117.Morgan AD, Bonsall MB, Buckling A. Impact of bacterial mutation rate on coevolutionary dynamics between bacteria and phages. Evolution. 2010;64:2980–7. doi: 10.1111/j.1558-5646.2010.01037.x. [DOI] [PubMed] [Google Scholar]
- 118.Levin BR, Bull JJ. Population and evolutionary dynamics of phage therapy. Nat Rev Microbiol. 2004;2:166–73. doi: 10.1038/nrmicro822. [DOI] [PubMed] [Google Scholar]
- 119.Scott AE, Timms AR, Connerton PL, Loc Carrillo C, Adzfa Radzum K, Connerton IF. Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 2007;3:e119. doi: 10.1371/journal.ppat.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vinderola G, Marcó MB, Guglielmotti DM, Perdigón G, Giraffa G, Reinheimer J, et al. Phage-resistant mutants of Lactobacillus delbrueckii may have functional properties that differ from those of parent strains. Int J Food Microbiol. 2007;116:96–102. doi: 10.1016/j.ijfoodmicro.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 121.Górski A, Weber-Dabrowska B. The potential role of endogenous bacteriophages in controlling invading pathogens. Cell Mol Life Sci. 2005;62:511–9. doi: 10.1007/s00018-004-4403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Górski A, Wazna E, Dabrowska BW, Dabrowska K, Switała-Jeleń K, Miedzybrodzki R. Bacteriophage translocation. FEMS Immunol Med Microbiol. 2006;46:313–9. doi: 10.1111/j.1574-695X.2006.00044.x. [DOI] [PubMed] [Google Scholar]
- 123.Miedzybrodzki R, Switala-Jelen K, Fortuna W, Weber-Dabrowska B, Przerwa A, Lusiak-Szelachowska M, et al. Bacteriophage preparation inhibition of reactive oxygen species generation by endotoxin-stimulated polymorphonuclear leukocytes. Virus Res. 2008;131:233–42. doi: 10.1016/j.virusres.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 124.Pajtasz-Piasecka E, Rossowska J, Duś D, Weber-Dabrowska B, Zabłocka A, Górski A. Bacteriophages support anti-tumor response initiated by DC-based vaccine against murine transplantable colon carcinoma. Immunol Lett. 2008;116:24–32. doi: 10.1016/j.imlet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 125.Przerwa A, Zimecki M, Switała-Jeleń K, Dabrowska K, Krawczyk E, Łuczak M, et al. Effects of bacteriophages on free radical production and phagocytic functions. Med Microbiol Immunol. 2006;195:143–50. doi: 10.1007/s00430-006-0011-4. [DOI] [PubMed] [Google Scholar]
- 126.Dabrowska K, Switała-Jeleń K, Opolski A, Górski A. Possible association between phages, Hoc protein and the immune system. Arch Virol. 2006;151:209–15. doi: 10.1007/s00705-005-0641-7. [DOI] [PubMed] [Google Scholar]
- 127.Górski A, Kniotek M, Perkowska-Ptasińska A, Mróz A, Przerwa A, Gorczyca W, et al. Bacteriophages and transplantation tolerance. Transplant Proc. 2006;38:331–3. doi: 10.1016/j.transproceed.2005.12.073. [DOI] [PubMed] [Google Scholar]
- 128.Dabrowska K, Zembala M, Boratynski J, Switala-Jelen K, Wietrzyk J, Opolski A, et al. Hoc protein regulates the biological effects of T4 phage in mammals. Arch Microbiol. 2007;187:489–98. doi: 10.1007/s00203-007-0216-y. [DOI] [PubMed] [Google Scholar]
- 129.Gorski A, Nowaczyk M, Weber-Dabrowska B, Kniotek M, Boratynski J, Ahmed A, et al. New insights into the possible role of bacteriophages in transplantation. Transplant Proc. 2003;35:2372–3. doi: 10.1016/S0041-1345(03)00811-X. [DOI] [PubMed] [Google Scholar]
- 130.Kniotek M, Weber-Dabrowska B, Dabrowska K, Switala-Jelen K, Boratynski J, Wiszniewski M, et al. Phages as immunomodulators of antibody production. Immunology; Genomic Islands, Immune System Activation and Allergy Medimond International Proceedings Monduzzi Editors Bologna 2004:33-36. [Google Scholar]
- 131.Riley PA. Bacteriophages in autoimmune disease and other inflammatory conditions. Med Hypotheses. 2004;62:493–8. doi: 10.1016/j.mehy.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 132.Karunakaran KP, Blanchard JF, Raudonikiene A, Shen C, Murdin AD, Brunham RC. Molecular detection and seroepidemiology of the Chlamydia pneumoniae bacteriophage (PhiCpn1) J Clin Microbiol. 2002;40:4010–4. doi: 10.1128/JCM.40.11.4010-4014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hendrickson BA, Gokhale R, Cho JH. Clinical aspects and pathophysiology of inflammatory bowel disease. Clin Microbiol Rev. 2002;15:79–94. doi: 10.1128/CMR.15.1.79-94.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–29. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 135.Rohwer F. Global phage diversity. Cell. 2003;113:141. doi: 10.1016/S0092-8674(03)00276-9. [DOI] [PubMed] [Google Scholar]
- 136.Minot S, Wu GD, Lewis JD, Bushman FD. Conservation of gene cassettes among diverse viruses of the human gut. PLoS One. 2012;7:e42342. doi: 10.1371/journal.pone.0042342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brüssow H, Desiere F. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol Microbiol. 2001;39:213–22. doi: 10.1046/j.1365-2958.2001.02228.x. [DOI] [PubMed] [Google Scholar]
- 138.Gosalbes MJ, Durbán A, Pignatelli M, Abellan JJ, Jiménez-Hernández N, Pérez-Cobas AE, et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 2011;6:e17447. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sozzi T. Method for preventing the inactivation, due to specific bacteriophages, of probiotic strain mixtures used in cattle-breeding. 2006; Patent WO2006/134579A2.



