Abstract
The honey bee, Apis mellifera, harbors a characteristic gut microbiota composed of only a few species which seem to be specific to social bees. The maintenance of this stable and distinct microbial community depends on the social lifestyle of these insects. As in other animals, the bacteria in the gut of honey bees probably govern important functions critical to host health. We recently sequenced a metagenome of the gut microbiota of A. mellifera, assigned gene contents to bins corresponding to the major species present in the honey bee gut, and compared functional gene categories between these species, and between the complete metagenome and those of other animals. Gene contents could be linked to different symbiotic functions with the host. Further, we found a high degree of genetic diversity within each of these species. In the case of the gammaproteobacterial species Gilliamella apicola, we experimentally showed a link between genetic variation of isolates and functional differences suggesting that niche partitioning within this species has emerged during evolution with its bee hosts. The consistent presence of only a few species, combined with strain variation within each of these species, makes the gut microbiota of social bees an ideal model for studying functional, structural, and evolutionary aspects of host-associated microbial communities: many characteristics resemble the gut microbiota of humans and other mammals, but the complexity is considerably reduced. In this addendum, we summarize and discuss our major findings and provide a detailed perspective on future research.
Keywords: Gilliamella, Snodgrassella, gut microbiome, insects, pectin, symbionts
Introduction
The gut microbiota of animals typically contributes to development, immunity, and nutrition1-4 and thus, plays an important role in the host’s health status. However, many bacterial gut communities display complex composition5-9 and temporal/spatial dynamics10-12 that present a challenge for disentangling functional roles, unraveling bacterial interactions, and understanding evolutionary trajectories of individual lineages.3,7,9 In particular, animals with a social lifestyle harbor characteristic and highly specialized gut microbiota.13,14 Social interactions between host individuals, such as maternal care, food sharing or grooming, facilitate transmission of gut-associated bacteria resulting in persistent associations, coevolution with hosts and the emergence of specialized symbiont lineages, and the competitive exclusion of unspecific colonizers. In many invertebrates, progeny develop independently of parents and must acquire gut bacteria from the environment each generation. However, transmission in social insects, such as termites, ants, and certain bees and wasps, resembles that in humans and other mammals, in which mother-offspring contact and other social interactions are a source of symbiont acquisition. This makes the gut microbiota of social animals suitable models to study functional and evolutionary aspects of host-associated microbial communities.
In the social insect A. mellifera (honey bee), independent studies of bacterial community profiles based on 16S rRNA sequences showed that female worker bees harbor a distinctive gut microbiota.15-19 With only nine species (i.e., closely related strains with ≥ 97% sequence identity in their 16S rRNA gene), the composition of this bacterial assemblage is relatively simple compared with other gut-associated communities. The main bacterial members of the community are two gammaproteobacterial species, one recently assigned the formal name Gilliamella apicola and the other one referred to as Gamma-2, one Betaproteobacterium recently named Snodgrassella alvi, two distinct Firmicutes, one Bifidobacterium, two Alphaproteobacteria, and one species within Bacteroidetes. The prospects for future experimental work on these species are enhanced by recent success in growing most of them in axenic culture, which also enables the establishment of official nomenclature.20 A subset of these species has also been found in the guts of various Bombus species (bumble bees), a genus of social bees closely related to A. mellifera.21 Based on non-culture-based surveys, non-social insects have gut bacteria mostly falling within known bacterial genera common in numerous environments and having erratic distribution among host individuals within a species, suggesting that the microbiota is acquired via repeated colonization from environmental sources.22-26 Interestingly, most of the bacterial species present in Apis and Bombus have neither been found in solitary bees nor elsewhere in the environment and occur as deep-branching phylogenetic lineages.17 Within these lineages, taxa isolated from honey bees and bumble bees seem to constitute distinct sister clades.17,18,27 These findings indicate long-standing association between these bacteria and their hosts, potentially reflecting long-term coevolution, and suggesting the existence of specific symbiotic interactions relevant for the characteristic lifestyle of these insects.
In our recently published study,28 we sequenced a metagenomic DNA sample from the pooled guts of 150 A. mellifera worker bees originating from the same colony. Our analysis provided insights into the evolution and the functional gene content of this characteristic gut microbiota and allowed us to predict the symbiotic capabilities of these bacteria in the host. In the following, we will give a short summary of the results and provide a perspective for future research.
Results and Discussion
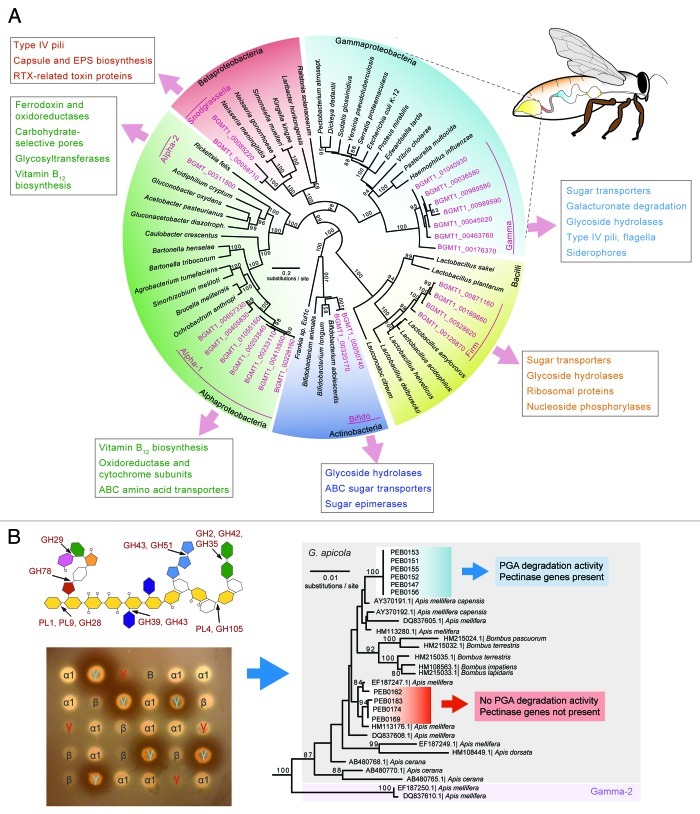
We assembled ~81 million Illumina shotgun reads from a 500 bp insert library into 76.6 Mb of total sequence. Taxonomic profiles generated from this data based on conserved protein sequences confirmed the simple composition of the honey bee gut microbiota on the species level: most assembled sequences fell into a few deep-branching phylogenetic lineages corresponding to the nine previously identified species. Interestingly, we found a high degree of genetic diversity within each of these lineages, as indicated by the presence of closely related, but divergent homologs of single-copy genes (Fig. 1A). Analysis of variable sites in a conserved set of ribosomal protein-encoding genes confirmed the presence of many divergent sequences in each lineage. These findings indicate that genetically divergent variants of these species coexist in a single colony; and a study focusing on rRNA sequences showed coexistence of divergent strains within individual bees.18
Figure 1. (A) Sequences from the honey bee gut metagenome (pink color) fall into a few phylogenetic lineages distinct from other bacteria. Although there can be more than one species per lineage (Gamma and Firm), the presence of several distinct sequences per lineage indicates genetic diversity within these bee-specific bacteria. Functions belonging to COG categories enriched in one lineage relative to the others are shown in boxes. Colors in the digestive tract of the bee illustration match colors of the different bacterial phyla in the tree and indicate regions where the corresponding bacteria are most abundant.27,48 (Localization of the two Alphaproteobacteria and the Bifidobacterium are not known.) (B) Genetic differences are linked to functional differences, as shown for the example of pectin degradation by G. apicola. The scheme of the pectin molecule shows glycoside hydrolase (GHs) and polysaccharide lyase (PLs) families identified in the honey bee metagenome with their respective cleavage sites. Different colors represent different sugar molecules. Degradation of polygalacturonate (PGA), the major component of pectin, by a subset of G. apicola isolates is demonstrated by clearance zones forming around bacterial patches on an agar plate. γ, Gilliamella; β, Snodgrassella; α1, Alpha-1; and B, Bifido. G. apicola isolates differing in capacity to degrade pectin (depicted by blue and red coloring) belong to distinct phylogenetic clades as shown by a 16S rRNA gene tree. Pectinase genes were absent in isolates negative for PGA degradation.
The honey bee lifestyle might have contributed to the unique composition of its microbiota. Because honey bee colonies are founded by swarms of many individuals, no severe transmission bottleneck restricts strain diversity. Opportunities for inter-colony exchange are also frequent, when beekeepers introduce new queen bees into existing hives (a regular apicultural practice) or via forager bees robbing the food resources from neighboring hives. By promoting colonization of individual bees with multiple strains, social interactions, facilitate inter-strain recombination. Although the metagenomic assembly is not suited for assessing extent of recombination, inter-strain recombination was detected in analyses of cloned 16S rRNA sequences from S. alvi and G. apicola.18
Besides providing novel insights into the taxonomic diversity of this characteristic microbial consortium and its evolution beyond the 16S rRNA level, our metagenomic study enabled the identification of gene contents involved in symbiotic functions with the host. Compared with the gut microbiota of other animals the honey bee gut metagenome is generally enriched in carbohydrate-related functions, most likely reflecting specific adaptation to the carbohydrate-rich diet of their host. Nectar and honey mainly contain the two monosaccharides glucose and fructose and the disaccharide sucrose. These sugars provide an abundant energy source for the host,29,30 as well as for its gut microbiota, and sugar uptake systems belonging to different phosphotransferase system (PTS) families were enriched in our metagenomic data set. Many of these transporters belonged to the mannose family of PTSs. Typically, mannose is present only in trace amounts in nectar.31 However, when present in higher concentrations, it is highly poisonous for honey bees.32 Whether honey bee gut bacteria are able to specifically metabolize mannose, and thus detoxify certain nectar components, remains to be experimentally tested. Alternatively, PTSs of the mannose family can show broad substrate specificity,33 so these systems could function in the metabolism of various non-toxic sugars derived from nectar, pollen, or host glycans.
Another carbohydrate-related function enriched in our data set was the “arabinose efflux permease” family. These transporters are involved in the export/import of compounds such as antimicrobials, amino acids, and sugars.34 The large diversity of this family in our sequencing data suggests that these proteins confer a plethora of different functions in the bee gut microbiota. A fraction of these transporters showed similarity to known drug efflux pumps34 including closely related homologs of tetracycline resistance genes. A diverse set of such export functions might confer protection for the bacteria against a range of pesticides applied in apiculture and agriculture or against naturally occurring antimicrobials taken up by bees via their plant-derived diet or produced by the microbiota itself.35,36
We also analyzed functional gene content differences between the distinct species or lineages present in the honey bee gut microbiome (Fig. 1A). To this end, we assigned assembled metagenomic sequences to bins representing the different species or lineages and compared among them. One rationale behind this analysis came from a previous study which showed that different species localize to different gut regions and thus are likely to occupy distinct functional niches.27 The analysis showed that bins corresponding to the two Gammaproteobacteria (G. apicola and Gamma-2) and to the Betaproteobacterium (S. alvi) encoded a relatively high number of functions related to biofilm formation and host interaction, such as Type IV pili, outer membrane proteins, and secretion (Fig. 1A). This was in agreement with previously published observations from fluorescence microscopy showing that exactly these species associate closely with the host epithelium where they form biofilm-like structures.27
Carbohydrate-related functions including the previously mentioned sugar uptake systems were predominantly present in the Gammaproteobacteria, the Firmicutes, and the Bifidobacterium (Fig. 1A). To test whether these species have the genetic potential to metabolize complex polysaccharides (besides the mono- and dissaccharides typically present in nectar), we identified and classified all glycoside hydrolases (GHs) and polysaccharide lyases (PLs) present in the honey bee gut metagenome. Major cellulases and hemicellulases, which are typical enzymes involved in degradation of plant cell walls, were absent from our data set. Instead, we found genes encoding pectin-degrading enzymes (Fig. 1B). Pectin is a heteropolysaccharide building up a jelly-like matrix in which the cellulose/hemicellulose network of plant cell walls is embedded.37 Pectin is also a major constituent of the primary cell wall of pollen and is highly abundant in pollen tubes during pollen maturation.38 This is particularly interesting, as pollen is the sole source of protein in the bee diet, but breakdown of pollen in the honey bee gut is not fully understood. Pollen consists of a rigid exine structure,39 which is thought to be resistant to enzymatic degradation. It has been hypothesized that in the gut of honey bees the exine structure is either burst open by osmotic shock or that pollen maturation results in the extrusion of the primary plant cell wall.40 In either case, the pectin-containing primary cell wall would be accessible for enzymatic degradation by the gut microbiota. This might result in the disintegration of the pollen cell wall providing host access to the nutrient-rich pollen germ. Strikingly, in a histochemistry-based study focusing on pollen digestion in the honey bee gut, pectin degradation was found to occur in the midgut region.41 Since genomes of honey bees (and other animals) are not known to encode enzymes for pectin degradation, it was hypothesized that bacterial enzymes mediate this activity.
In our study, we experimentally tested different honey bee gut isolates for their capability to degrade polygalacturonic acid, the main component of pectin. As predicted from the metagenomic gene content, we exclusively found isolates of the gammaproteobacterial species G. apicola to harbor pectin-degradation activity. However, not all G. apicola isolates could degrade pectin, suggesting functional differences among isolates. PCR screening for pectinase genes in combination with phylogenetic analysis, indeed, revealed that distinct phylogenetic clusters of G. apicola differ in their capability to degrade pectin (Fig. 1B). Thus, genetic variation at the strain level in the gut microbiota might be linked to differences in ecologically relevant functions.
Perspective
With its simple and characteristic composition, the honey bee gut microbiota provides an excellent model to study functional and structural aspects of gut-associated bacterial communities. Furthermore, their specific association with social bees and their deep-branching phylogenetic positions make the major bacterial taxa of the honey bee gut community interesting candidates for evolutionary analysis. While several previous studies have focused on using 16S rRNA to assess the bacterial composition of the microbiota and its localization in the honey bee gut, our metagenomic analysis provides first insights into the functional capabilities of these bacteria, and thus provides a basis for future experimental work. Based on current knowledge, most members of the microbiota can only be found in social bees. This indicates a highly specific adaptation to this characteristic environment, which has likely resulted in the evolution of symbiotic interactions with the host. Indeed, in bumble bees, it was recently shown that the socially transmitted gut microbiota protects against a common tryposomatid pathogen, Crithidia bombus.42 Whether the honey bee gut microbiota also confers such protection against corresponding pathogens remains to be tested. However, common symbiotic functions are likely, in view of the similar composition of their gut microbiota and the close relationship of common pathogens; possibly, symbionts and their functions in hosts were established in an ancestor of these two bee genera. From the metagenomic data alone, we can only hypothesize how such defensive functions might be mediated. However, gene functions linked to host interaction, biofilm formation, or secretion are good candidates for involvement in protective mechanisms. By modulating the immune system of the host, some of these functions probably have an indirect role in defense. Adult worker bees largely lack gut bacteria at eclosion from the pupal stage and acquire their symbionts within the colony during the first few days as adults.27 This initial colonization might prime the immune system and could be a prerequisite for providing protection against parasites. Nutrition is another important aspect of health and is likely supported by these bacteria. Extracellular enzymes may help to breakdown dietary components, and bacterial metabolism and biosynthetic capabilities may augment nutrient supplies. Food consumption of young worker bees is regulated via social interactions (trophallaxis), and induces important behavioral changes related to labor division in the hive.43,44 Thus, bacterial functions related to food processing could possibly influence social behaviors of their hosts.
For elucidating such functional aspects, experimental approaches will be extremely helpful. Functions in hosts could be identified by comparing bees that lack their specific gut microbiota to bees re-colonized with bacteria grown in the laboratory. By removing bees from the hive before emergence, microbiota-free individuals can be raised and subsequently colonized with isolates of interest. Culturing of bacteria will also enable the establishment of genetic tools, so that specific hypotheses, for example pectin breakdown by G. apicola isolates, could be experimentally tested in vivo. Such experimental approaches could be used to measure the fitness effects of individual bacterial genotypes or defined bacterial communities on the host.
Another intriguing issue concerns the causes and consequences of the genetic diversity identified in species of the honey bee gut microbiota. The pattern of high levels of strain variation but fewer deep lineages seems to be a signature feature of gut microbial communities and parallels observations for the mouse and the human gut microbiota.45 Two questions of central importance are (1) whether the genetic diversification is a result of the divergent adaptation to different niches in the gut and (2) to what degree it is linked to the social lifestyle of the host. Comparing taxonomic composition and genetic diversity of the gut microbiota among bee species with different social lifestyles could enable unprecedented insights into the evolution of gut-associated bacterial communities. Novel approaches such as single-cell sequencing combined with comparative and evolutionary genomics will further help to elucidate the underlying genetic mechanism of the diversification within distinct species.
Besides being a suitable model organism for studying complex gut-associated bacterial communities, the honey bee and its interaction with the environment including microbes, has drawn recent attention due to world-wide colony losses during the last few years.46,47 Whether pesticides, pathogens, or other environmental perturbations underlie these losses is not yet clear. However, based on findings to date, that honey bees possess a stable, distinctive microbiota with gene contents suggesting specific symbiotic interactions, a better understanding of the impact of the resident microbiota on honey bee health is even more urgently needed.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/22517
References
- 1.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–70. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–36. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 4.Royet J. Epithelial homeostasis and the underlying molecular mechanisms in the gut of the insect model Drosophila melanogaster. Cell Mol Life Sci. 2011;68:3651–60. doi: 10.1007/s00018-011-0828-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jami E, Mizrahi I. Composition and similarity of bovine rumen microbiota across individual animals. PLoS One. 2012;7:e33306. doi: 10.1371/journal.pone.0033306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pope PB, Denman SE, Jones M, Tringe SG, Barry K, Malfatti SA, et al. Adaptation to herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc Natl Acad Sci U S A. 2010;107:14793–8. doi: 10.1073/pnas.1005297107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullam KE, Essinger SD, Lozupone CA, O’Connor MP, Rosen GL, Knight R, et al. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol. 2012;21:3363–78. doi: 10.1111/j.1365-294X.2012.05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–5. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 10.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–84. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 11.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pédron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, et al. A crypt-specific core microbiota resides in the mouse colon. MBio. 2012;3:e00116-12. doi: 10.1128/mBio.00116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–88. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lombardo MP. Access to mutualistic endosymbiotic microbes: an underappreciated benefit of group living. Behav Ecol Sociobiol. 2008;62:479–97. doi: 10.1007/s00265-007-0428-9. [DOI] [Google Scholar]
- 15.Babendreier D, Joller D, Romeis J, Bigler F, Widmer F. Bacterial community structures in honeybee intestines and their response to two insecticidal proteins. FEMS Microbiol Ecol. 2007;59:600–10. doi: 10.1111/j.1574-6941.2006.00249.x. [DOI] [PubMed] [Google Scholar]
- 16.Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–7. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 17.Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol. 2011;20:619–28. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 18.Moran NA, Hansen AK, Powell JE, Sabree ZL. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One. 2012;7:e36393. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabree ZL, Hansen AK, Moran NA. Independent studies using deep sequencing resolve the same set of core bacterial species dominating gut communities of honey bees. PLoS One. 2012;7:e41250. doi: 10.1371/journal.pone.0041250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong WK, Moran NA. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: Snodgrassella alvi gen. nov., sp. nov., a member of the Neisseriaceae family of the Betaproteobacteria; and i gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the Enterobacteriales order of the Gammaproteobacteria. Int J Syst Evol Microbiol. 2012:In press. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- 21.Koch H, Schmid-Hempel P. Bacterial communities in central European bumblebees: low diversity and high specificity. Microb Ecol. 2011;62:121–33. doi: 10.1007/s00248-011-9854-3. [DOI] [PubMed] [Google Scholar]
- 22.Colman DR, Toolson EC, Takacs-Vesbach CD. Do diet and taxonomy influence insect gut bacterial communities? Mol Ecol. 2012 doi: 10.1111/j.1365-294X.2012.05752.x. [DOI] [PubMed] [Google Scholar]
- 23.Osei-Poku J, Mbogo CM, Palmer WJ, Jiggins FM. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol Ecol. 2012 doi: 10.1111/j.1365-294X.2012.05759.x. [DOI] [PubMed] [Google Scholar]
- 24.Broderick NA, Raffa KF, Goodman RM, Handelsman J. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl Environ Microbiol. 2004;70:293–300. doi: 10.1128/AEM.70.1.293-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dillon RJ, Webster G, Weightman AJ, Keith Charnley A. Diversity of gut microbiota increases with aging and starvation in the desert locust. Antonie Van Leeuwenhoek. 2010;97:69–77. doi: 10.1007/s10482-009-9389-5. [DOI] [PubMed] [Google Scholar]
- 26.Schloss PD, Delalibera I, Handelsman J, Raffa KF. Bacteria associated with the guts of two wood-boring beetles: Anoplophora glabripennis and Saperda vestita (Cerambycidae) Environ Entomol. 2006;35:625–9. doi: 10.1603/0046-225X-35.3.625. [DOI] [Google Scholar]
- 27.Martinson VG, Moy J, Moran NA. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol. 2012;78:2830–40. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A. 2012;109:11002–7. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barker RJ, Lehner Y. Influence of diet on sugars found by thin-layer chromatography in thoraces of honey bees, Apis mellifera L. J Exp Zool. 1974;188:157–64. doi: 10.1002/jez.1401880204. [DOI] [PubMed] [Google Scholar]
- 30.Barker RJ, Lehner Y. Acceptance and sustenance value of naturally occurring sugars fed to newly emerged adult workers of honey bees (Apis mellifera L.) J Exp Zool. 1974;187:277–86. doi: 10.1002/jez.1401870211. [DOI] [PubMed] [Google Scholar]
- 31.Nicolson SW, Thornburg RW. Nectar chemistry. In: Nicolson SW, Neppi M, E P, eds. Nectaries and nectar: Springer, 2007:215-65. [Google Scholar]
- 32.Sols A, Cadenas E, Alvarado F. Enzymatic basis of mannose toxicity in honey bees. Science. 1960;131:297–8. doi: 10.1126/science.131.3396.297. [DOI] [PubMed] [Google Scholar]
- 33.Zúñiga M, Comas I, Linaje R, Monedero V, Yebra MJ, Esteban CD, et al. Horizontal gene transfer in the molecular evolution of mannose PTS transporters. Mol Biol Evol. 2005;22:1673–85. doi: 10.1093/molbev/msi163. [DOI] [PubMed] [Google Scholar]
- 34.Law CJ, Maloney PC, Wang D-N. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.González-Teuber M, Heil M. Nectar chemistry is tailored for both attraction of mutualists and protection from exploiters. Plant Signal Behav. 2009;4:809–13. doi: 10.4161/psb.4.9.9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullin CA, Frazier M, Frazier JL, Ashcraft S, Simonds R, Vanengelsdorp D, et al. High levels of miticides and agrochemicals in North American apiaries: implications for honey bee health. PLoS One. 2010;5:e9754. doi: 10.1371/journal.pone.0009754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cosgrove DJ. Growth of the plant cell wall. Nat Rev Mol Cell Biol. 2005;6:850–61. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- 38.Taylor LP, Hepler PK. Pollen germination and tube growth. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:461–91. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- 39.Ariizumi T, Toriyama K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol. 2011;62:437–60. doi: 10.1146/annurev-arplant-042809-112312. [DOI] [PubMed] [Google Scholar]
- 40.Roulston TH, Cane JH. Pollen nutritional content and digestibility for animals. Plant Syst Evol. 2000;222:187–209. doi: 10.1007/BF00984102. [DOI] [Google Scholar]
- 41.Klungness LM, Peng YS. A histochemical study of pollen digestion in the alimentary canal of honeybees (Apis mellifera L.) J Insect Physiol. 1984 doi: 10.1016/0022-1910(84)90077-5. [DOI] [Google Scholar]
- 42.Koch H, Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci U S A. 2011;108:19288–92. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ament SA, Corona M, Pollock HS, Robinson GE. Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc Natl Acad Sci U S A. 2008;105:4226–31. doi: 10.1073/pnas.0800630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ament SA, Wang Y, Robinson GE. Nutritional regulation of division of labor in honey bees: toward a systems biology perspective. Wiley Interdiscip Rev Syst Biol Med. 2010;2:566–76. doi: 10.1002/wsbm.73. [DOI] [PubMed] [Google Scholar]
- 45.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 46.Evans JD, Schwarz RS. Bees brought to their knees: microbes affecting honey bee health. Trends Microbiol. 2011;19:614–20. doi: 10.1016/j.tim.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Genersch E, Evans JD, Fries I. Honey bee disease overview. J Invertebr Pathol. 2010;103(Suppl 1):S2–4. doi: 10.1016/j.jip.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 48.Vásquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, Szekely L, et al. Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS One. 2012;7:e33188. doi: 10.1371/journal.pone.0033188. [DOI] [PMC free article] [PubMed] [Google Scholar]