Abstract
Pathogenic autoinflammatory responses triggered by dysregulated microbial interactions may lead to intestinal disorders and malignancies. Previously, we demonstrated that a lipoteichoic acid (LTA)-deficient Lactobacillus acidophilus strain, NCK2025, ameliorated inflammation-induced colitis, significantly reduced the number of polyps in a colonic polyposis cancer model and restored physiological homeostasis in both cases. Nonetheless, the regulatory signals delivered by NCK2025 to reprogram the gastrointestinal microenvironment, and thus resist colonic cancer progression, remain unknown. Accumulating evidence suggest that epigenetic changes, in the presence and absence of pathogenic inflammation, can result in colorectal cancer (CRC). To test possible epigenetic modifications induced by NCK2025, the expression of epigenetically regulated, CRC-associated genes was measured with and without bacterial treatment. In vivo and in vitro, NCK2025 enhanced the expression of tumor suppressor genes that may regulate CRC development. Therefore, differential epigenetic regulation of CRC-related genes by NCK2025 represents a potential therapy against colitis-associated and sporadic CRC.
Keywords: colorectal cancer, aberrant epigenetic alterations, DNA methylation, lipoteichoic acid, inflammation
Introduction
Maintenance of mucosal barrier integrity and low responsiveness to the large numbers of commensal microbes that inhabit the gastrointestinal tract are crucial for intestinal health. The intestinal immune system is able to accurately and efficiently discriminate between invading pathogens and commensal bacteria, thereby attacking only those that are detrimental to the host while deterring an inflammatory reaction in their absence.1,2 However, a breach of commensal-specific tolerance can lead to uncontrolled inflammation in the gut and subsequent inflammatory disorders, such as inflammatory bowel diseases (IBD). Chronic inflammation, characterized by activated immune cells, generation of reactive oxygen species (ROS) and DNA damage, predisposes individuals to developing cancer.3 Consequently, IBD patients have been found to be at increased risk for the development of a subtype of colorectal cancer (CRC) termed colitis-associated cancer (CAC).4 In CAC, genetic instability resulting from proinflammatory insults promotes epithelial cell transformation and colon cancer progression through the formation of polyps, adenomas and carcinomas. On the other hand, sporadic CRC is thought to arise independent of preexisting inflammation and the majority of cases are attributed to environmental toxins and irritants, as well as the composition of the microbiome.5,6 Nonetheless, inflammatory anti-tumor responses in sporadic CRC can further induce DNA alterations and tumor-promoting mutations.7
In our recent publication,8 successful treatment of colonic polyposis was achieved via oral treatment with a genetically modified Lactobacillus acidophilus NCFM strain, NCK2025, that reduced localized inflammation and enhanced regulatory responses. The mouse model of colonic polyposis used for our studies (APClox468 × TS4-Cre) possesses conditional abnormalities in the adenomatous coli (APC) gene and demonstrates extensive polyposis throughout the distal ileum and colon.8 APC mutations account for the onset of Familial Adenomatous Polyposis, a hereditary form of CRC, as well as for the majority of sporadic colorectal tumors.9 Inactivating mutations of the APC protein disrupt the formation of the Wnt signaling pathway destruction complex and subsequent degradation of β-catenin, a transcriptional coactivator of target genes that contribute to oncogenesis.9 Thus, by decreasing cancer progression in APClox468 × TS4-Cre mice through the anti-inflammatory properties of NCK2025, our data provided further evidence for the deleterious role of inflammation in CRC, including subtypes with genetic predisposition. Here, we explore additional regulatory mechanisms orchestrated by lipoteichoic acid (LTA)-deficient L. acidophilus that may help counterbalance mucosal epithelial cell damage and genetic instability caused by pathogenic inflammation.
Aberrant Epigenetic Modifications, Inflammation and Colon Cancer
The view that the accumulation of genetic changes results in cancer has evolved to include the unfavorable role of epigenetic modifications of pre-neoplastic cells.10,11 Epigenetics, as opposed to mutations in the DNA sequence, refers to hereditable changes in gene expression levels due to structural alterations that influence a gene’s ability to be activated or silenced. Epigenetic mechanisms that have been proposed to contribute to cancer include DNA methylation, histone modifications, nucleosome positioning and non-coding RNAs.10 Together, genetic mutations and epigenetic alterations are thought to promote carcinogenesis through the inactivation of tumor suppressor genes and the activation of oncogenes. The most extensively studied dysregulated epigenetic mechanism in cancer is DNA methylation. In fact, many more methylated genes than genetic mutations have been found in primary CRC tumor samples when compared with normal colon samples.12 DNA methylation plays an essential role in development13 and involves the addition of a methyl group to the 5′ position of cytosines. In cancer, CG rich sequences in the promoter regions of tumor suppressor gene become targets for aberrant DNA methylation. Uncontrolled hypermethylation can lead to transcriptional silencing by (1) directly interfering with the binding of transcription factors or by (2) the recruitment of methyl-CpG binding domain (MBD) proteins. DNA-bound MBD proteins provide docking sites for histone deacetylases and other chromatin remodeling complexes resulting in a chromatin structure that is repressive and inaccessible.
Studies aiming to understand the contribution of proinflammatory mediators to changes in the epigenetic landscape are still in their early stages; nonetheless, recent research has shed some light on this association.14 In the context of IBD, the DNA methylation status of the colonic mucosa positively correlated with the extent of inflammation of the colon sections analyzed (active distal vs. quiescent proximal).15 Moreover, interleukin-6 (IL-6), a key tumor-promoting proinflammatory cytokine linked with CAC,16,17 has been shown to increase and stabilize the protein levels of DNA methyltransferase-1 (DNMT-1), a CpG island methylator, which then led to the methylation of gene promoter regions and targeted-gene silencing.18 Of interest, our work indicated that clinical improvement of APClox468 × TS4-Cre mice after NCK2025 treatment correlated with lower levels of serum IL-6.8 Another study found that in vitro treatment of an intestinal epithelial cell line with interferon-γ (IFNγ) resulted in enhanced DNA methylation corresponding with increases in the expression levels of DNMT-1.19 On the other hand, proinflammatory cytokines may promote elevated levels of methyltransferases to prevent the transcription of damaging gene products. For instance, in a mouse model of chemically induced colitis, in vivo inhibition of inflammation-induced DNA methylation of the colonic mucosa exacerbated disease.19 Consequently, in some cases, inflammation-mediated DNA hypermethylation may arise as a protective mechanism that could then become pro-tumorigenic and detrimental to the host if inflammation persists.
Genetic Modifications Induced by NCK2025
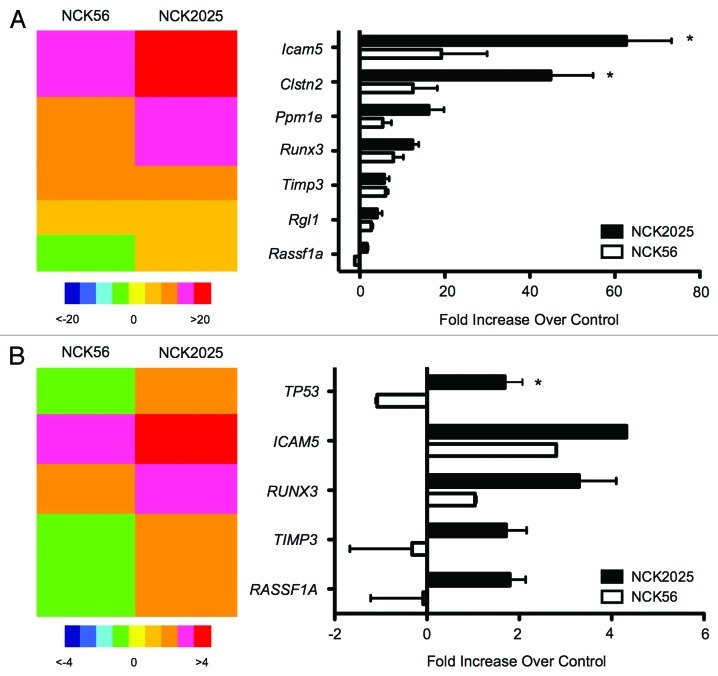
Taken together, these observations prompted us to investigate whether our L. acidophilus NCFM strain lacking the gene responsible for the biosynthesis of LTA20 could not only change the proinflammatory milieu in CRC but also prevent aberrant DNA methylation of the mucosal epithelium. Before measuring cytosine methylation levels in the disease state, we simply tested the expression levels of genes commonly methylated in CRC11,21,22 after treatment with different L. acidophilus NCFM strains. In some experiments, healthy C57B/L6 mice were orally treated with the wild-type strain (NCK56), the protective LTA-deficient strain (NCK2025), or PBS, and gene expression analyses of the distal colon took place 24 h later (Fig. 1A). Additionally, we incubated the human colon carcinoma cell line, HT-29 (kindly provided by Dr. Yi Qiu, University of Florida), with NCK56 or NCK2025 for 48 h and again measured gene expression differences compared with untreated controls in vitro (Fig. 1B). Our in vivo and in vitro observations indicate that NCK56 was able to induce the expression of some of the genes tested; however, our LTA-deficient protective strain, NCK2025, consistently promoted higher transcriptional levels of genes that have been found to be silenced through DNA methylation in colonic cancer (Fig. 1).
Figure 1. LTA-deficient L. acidophilus induces the expression of genes commonly silenced through DNA methylation in CRC. (A) Healthy C57B/L6 mice were treated orally with wild-type L. acidophilus NCFM (NCK56), protective LTA-deficient L. acidophilus NCFM (NCK2025), or PBS, and colonic tissue samples were collected 24 h after treatment. Expression levels of epigenetically regulated, CRC-associated genes were measured by Real-time PCR and normalized to PBS-treated levels. (B) HT-29 colon carcinoma cells were treated for 48 h and analyzed as above. * denotes statistical significance p < 0.05.
Control of the immune response in the gut by nonpathogenic and pathogenic microorganisms begins with detection of the bacteria through the interaction of pattern recognition receptors (PRR) expressed on resident innate immune cells and microbe-associated molecular patterns (MAMPS) found on the cell surface of the bacteria. Similarly, intestinal epithelial cells not only protect the host by providing a physical barrier against invading microbes, but can also be intimately involved in the maintenance of gut homeostasis by sensing microorganisms through their own PRR and regulating subsequent innate responses.23 It is not surprising then that microbiota-mucosal interactions could also alter the epigenetic status of intestinal epithelial cells. The two L. acidophilus NCFM strains tested in our studies differ in their expression of LTA on their cells surface. Therefore, our data suggest that the gene expression changes observed are due to other L. acidophilus NCFM cell surface components. LTA has been postulated to induce proinflammatory immune responses in the gut.8,20,24 Conversely, the absence of the surface layer protein A (SlpA) upregulates the production of proinflammatory cytokines,25 indicating an anti-inflammatory effect of this protein. However, contributions of SlpA to epigenetic modifications and auto-inflammation in the host have not been explored. In this respect, the current focus of our laboratory centers on the role of Slps of L. acidophilus, including SlpA, in intestinal immune modulation.
Concluding Remarks
Although the importance of gut microbiota in mucosal immune maturation is appreciated,26 the early results presented here indicate a potential link between commensal and pathogenic bacteria in the colon and the epigenetic status of the intestinal epithelium, which may consequently result in protection from or susceptibility to inflammatory diseases and CRC (Fig. 2). Indeed, this area of research is presently gaining significant interest.27,28 As our previous work demonstrates that NCK2025 protects against precancerous colonic polyps in an animal model of the disease,8 future work will focus on the molecular mechanisms responsible for protection beyond changes in the immune response. Namely, we will explore the DNA methylation landscape of diseased mice with and without oral treatment with our bacterial strain of interest. Our current hypothesis is that NCK2025 contributes to protection from CRC by normalizing pathogenic inflammation in the gut, which in turn prevents further tissue damage, genetic instability and propensity to carcinogenesis (Fig. 2). Alternatively, NCK2025 may directly influence the mucosal epithelium to avoid aberrant epigenetic modifications that may inactivate tumor suppressor genes. We have generated a model of invasive colon cancer that, in addition to the APC abnormality, also harbors a conditional deletion of the phosphatase and tensin homolog deleted on chromosome 10 (PTEN) gene29 (APClox468 × PTENlox × TS4-Cre). This animal model provides an additional tool to dissect the molecular events taking place during all stages of colon cancer progression and/or prevention with NCK2025 therapy. Further understanding of the immunomodulatory effects of NCK2025 and the molecular consequences of its treatment in the host will help move this probiotic agent one step closer to clinical application.
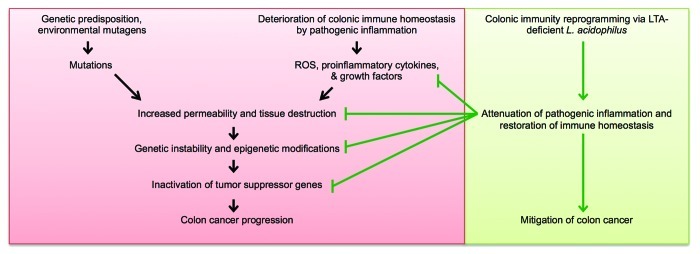
Figure 2. Schematic summary of the potential role of LTA-deficient L. acidophilus in CRC mitigation. In sporadic CRC and CAC, tissue destruction and increased permeability lead to genetic instability and aberrant epigenetic alterations, thereby activating a deleterious cascade that culminates in colon cancer progression. Our previous studies demonstrate that NCK2025 dampens pathogenic inflammation in the gut and restores immunological homeostasis. As a result, proinflammatory mediators no longer promote oncogenesis. Data presented here suggest that in addition to its immunomodulatory role, NCK2025 may also be involved in the prevention of epigenetic modifications that are conducive to CRC development. Consequently, LTA-deficient L. acidophilus may not only be helpful in the treatment of CAC, but also in the prevention of sporadic CRC.
Acknowledgments
This work was supported in part by NIH Grant 1R01AI098833–01, DoD Grant CA111002 and NIH/NCRR Clinical and Translational Science Award to the University of Florida (UL1 RR029890).
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/22822
References
- 1.Rossi O, van Baarlen P, Wells JM. Host-Recognition of Pathogens and Commensals in the Mammalian Intestine. Curr Top Microbiol Immunol. 2011 doi: 10.1007/82_2011_191. [DOI] [PubMed] [Google Scholar]
- 2.Kelsall BL, Leon F. Involvement of intestinal dendritic cells in oral tolerance, immunity to pathogens, and inflammatory bowel disease. Immunol Rev. 2005;206:132–48. doi: 10.1111/j.0105-2896.2005.00292.x. [DOI] [PubMed] [Google Scholar]
- 3.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubin DC, Shaker A, Levin MS. Chronic intestinal inflammation: inflammatory bowel disease and colitis-associated colon cancer. Front Immunol. 2012;3:107. doi: 10.3389/fimmu.2012.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson AJ, Collins PD. Colon cancer: a civilization disorder. Dig Dis. 2011;29:222–8. doi: 10.1159/000323926. [DOI] [PubMed] [Google Scholar]
- 6.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–82. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 7.Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–14, e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 8.Khazaie K, Zadeh M, Khan MW, Bere P, Gounari F, Dennis K, et al. Abating colon cancer polyposis by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci U S A. 2012;109:10462–7. doi: 10.1073/pnas.1207230109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 10.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuebel KE, Chen W, Cope L, Glöckner SC, Suzuki H, Yi JM, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiefer JC. Epigenetics in development. Dev Dyn. 2007;236:1144–56. doi: 10.1002/dvdy.21094. [DOI] [PubMed] [Google Scholar]
- 14.Hartnett L, Egan LJ. Inflammation, DNA methylation and colitis-associated cancer. Carcinogenesis. 2012;33:723–31. doi: 10.1093/carcin/bgs006. [DOI] [PubMed] [Google Scholar]
- 15.Saito S, Kato J, Hiraoka S, Horii J, Suzuki H, Higashi R, et al. DNA methylation of colon mucosa in ulcerative colitis patients: correlation with inflammatory status. Inflamm Bowel Dis. 2011;17:1955–65. doi: 10.1002/ibd.21573. [DOI] [PubMed] [Google Scholar]
- 16.Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–13. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grivennikov SI, Karin M. Inflammatory cytokines in cancer: tumour necrosis factor and interleukin 6 take the stage. Ann Rheum Dis. 2011;70(Suppl 1):i104–8. doi: 10.1136/ard.2010.140145. [DOI] [PubMed] [Google Scholar]
- 18.Foran E, Garrity-Park MM, Mureau C, Newell J, Smyrk TC, Limburg PJ, et al. Upregulation of DNA methyltransferase-mediated gene silencing, anchorage-independent growth, and migration of colon cancer cells by interleukin-6. Mol Cancer Res. 2010;8:471–81. doi: 10.1158/1541-7786.MCR-09-0496. [DOI] [PubMed] [Google Scholar]
- 19.Kominsky DJ, Keely S, MacManus CF, Glover LE, Scully M, Collins CB, et al. An endogenously anti-inflammatory role for methylation in mucosal inflammation identified through metabolite profiling. J Immunol. 2011;186:6505–14. doi: 10.4049/jimmunol.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohamadzadeh M, Pfeiler EA, Brown JB, Zadeh M, Gramarossa M, Managlia E, et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4623–30. doi: 10.1073/pnas.1005066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varley KE, Mitra RD. Bisulfite Patch PCR enables multiplexed sequencing of promoter methylation across cancer samples. Genome Res. 2010;20:1279–87. doi: 10.1101/gr.101212.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi JM, Dhir M, Van Neste L, Downing SR, Jeschke J, Glöckner SC, et al. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res. 2011;17:1535–45. doi: 10.1158/1078-0432.CCR-10-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4607–14. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zadeh M, Khan MW, Goh YJ, Selle K, Owen JL, Klaenhammer T, et al. Induction of intestinal pro-inflammatory immune responses by lipoteichoic acid. J Inflamm (Lond) 2012;9:7. doi: 10.1186/1476-9255-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci U S A. 2008;105:19474–9. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Licciardi PV, Wong S-S, Tang ML, Karagiannis TC. Epigenome targeting by probiotic metabolites. Gut Pathogens 2010; 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shenderov BA. Gut indigenous microbiota and epigenetics. Microb Ecol Health Dis. 2012;23:17195. doi: 10.3402/mehd.v23i0.17195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–50. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]