Abstract
Despite being invasive within surrounding brain tissues and the central nervous system, little is known about the mechanical properties of brain tumor cells in comparison with benign cells. Here, we present the first measurements of the peak pressure drop due to the passage of benign and cancerous brain cells through confined microchannels in a “microfluidic cell squeezer” device, as well as the elongation, speed, and entry time of the cells in confined channels. We find that cancerous and benign brain cells cannot be differentiated based on speeds or elongation. We have found that the entry time into a narrow constriction is a more sensitive indicator of the differences between malignant and healthy glial cells than pressure drops. Importantly, we also find that brain tumor cells take a longer time to squeeze through a constriction and migrate more slowly than benign cells in two dimensional wound healing assays. Based on these observations, we arrive at the surprising conclusion that the prevailing notion of extraneural cancer cells being more mechanically compliant than benign cells may not apply to brain cancer cells.
INTRODUCTION
Primary brain tumors occur at an incidence of approximately 0.006% per capita, however, in children brain tumors account for approximately 23% of all cancers, ranking second to leukemia. Although these tumors rarely metastasize outside of the central nervous system, they frequently invade nearby tissues via a diffuse local invasion pattern which makes effective therapy difficult.1 In some rare cases, primary brain tumors have been observed to metastasize via the cerebro-spinal fluid pathways,2 and blood vessels.3 Previous studies of brain tumor invasiveness have examined the role of three dimensional extracellular matrix rigidity in regulating brain tumor cell migration and proliferation,4, 5 the role of genetic expression on cell migration ability,1, 6, 7 and the invasiveness of primary brain tumor cell lines have been compared using animal models.8 The mechanical properties of mammalian brain tissues have been previously studied using a variety of techniques including shear rheometry,9, 10, 11 scanning force microscopy, and optical stretching,12 however, none of these studies have examined the mechanical properties of brain tumor cells in comparison with healthy human glial cells.
The mechanical properties of extraneural cancer cells in comparison with benign cells have been previously studied using several techniques. Using atomic force microscopy (AFM), Cross et al. have demonstrated that lung and breast cancer cells taken from the pleural fluids of cancer patients are nearly four times softer than their normal counterparts.13 Jonas et al., using scanning force microscopy, have also demonstrated that cancerous human breast, lung, skin, and colon cells grown in culture are softer than their non-invasive counterparts.14 Likewise, Wu et al., using micropipette aspiration, have demonstrated that cancerous liver cells are softer than healthy cells grown in culture.15 Thus, all of these methods indicate that extraneural cancer cells are softer than benign cells. While accurate, the measurement techniques used in these studies are relatively low throughput, in that tens to hundreds of cells could have been analyzed in a single day.
Microfluidic techniques for measuring cell mechanical properties have recently emerged that are higher throughput than the aforementioned studies, in that hundreds to thousands of cells can be characterized in a single hour.16, 17, 18, 19 One such study characterised the invasive ability of breast cancer cells, as well as classified different breast cancer cell lines' metastatic potential based on the cells' capability to stretch under the application of optical forces.20 Similarly, breast cancer and non-cancerous cells have been characterized by their time to enter a narrow channel constriction while in flow, and their deformation while passing through the channel. In these studies, it was found that highly metastatic breast cancer cells are more easily deformed than less metastatic and healthy cells, and that metastatic cells enter the confined microchannel more quickly.21 It has also been demonstrated that highly metastatic breast cancer and osteosarcoma cells are softer than less cancerous cells based on their shear-induced inertial migration in flow through a microchannel.22 The stretching of cancer cells in strong extensional flows generated in microfluidic devices has also been used to correlate cell deformability to disease state.23 It is apparent that several microfluidic techniques have been used to characterize deformability of individual extraneural cancer cells, however, the ability of microfluidic devices to probe the mechanical properties of brain cancer cells remains to be addressed.
Our goals in this study are twofold: first we develop a device called the microfluidic cell squeezer (MCS) and test the ability of the device to perform rapid mechanical measurements on individual cells. This tool relies on observing the flow-induced passage of individual tumor cells through a narrow microchannel with simultaneous measurement of mechanical resistance offered by the confined flowing cell. Using the MCS device, in addition to measuring the mechanical resistance via the excess pressure drop offered by the cell, other parameters such as cell velocity, shape deformation, and entry time into the squeezer are simultaneously characterized, which could be potentially indicative of cellular mechanical properties. Second, we use this device to characterize the differences in the mechanical properties of cancerous glioblastoma and astrocytoma cells in comparison with benign human glial cells. We also conduct wound closure assays to characterize cellular migration in cancerous and benign glial cells. We find that, of the measured parameters, the entry time has the strongest correlation with the malignancy of glial cells. Moreover, we find that healthy glial cells have a shorter entry time in the MCS and migrate more quickly on wounded substrates compared to brain tumor cells. Based on these observations, we arrive at the surprising conclusion that the prevailing notion of extraneural cancer cells being more compliant than benign cells may not apply to brain cancer cells.
WORKING PRINCIPLE OF THE MICROFLUIDIC CELL SQUEEZER
In addition to observing the transport of individual cells through a narrow channel in the MCS device, we integrate a microfluidic comparator to measure temporal variations in pressure drop due to the flow of cells. This microfluidic manometer has been previously demonstrated to be a sufficiently sensitive means to measure the excess pressure drop associated with the hydrodynamic flow of confined red blood cells24, 25 and aqueous droplets26 through a microchannel. This microfluidic manometer technique has also been demonstrated to be able to differentiate between healthy and chemically rigidifed confined red blood cells, as the stiffened red blood cells have a larger excess pressure drop associated with their passage through a constriction than healthy red blood cells.24 We expect that the pressure drops of the confined stiffer objects are larger since the cells are not able to conform to the applied shear stress as much as the healthy cells, and therefore, occupy the channel more fully offering a higher hydrodynamic resistance than healthy cells.
Our microfluidic comparator, shown in Figure 1a, consists of two identical channels, each of which has a wide entry zone followed by a long constriction with a width of 15 μm, a length of 286 μm (A172 cells) or 238 μm (1321N1 and normal glial cells), and a height of 11 μm. Both these constrictions are connected downstream to form a comparator region. One of the channels is referred to as the test channel where the cells are introduced, and the other, the reference channel. The liquid in the reference channel is coloured with a food dye (McCormick black) to visualize the laminar interface between the two fluid flows. When equal driving pressures () are imposed at the inlets of both the channels and if there are no cells present in the constriction the interface between the two fluids is balanced at the center of the comparator region since the hydrodynamic resistance of the two channels are the same, as shown in Figure 1a. However, when a cell passes through the constriction as shown in Figure 1b, the interface between the two fluids moves upward toward the test channel, because of the partially obstructing cell that reduces the flow rate in the test channel.
Figure 1.
Working principle of the microfluidic cell squeezer. (a) Image of the MCS with no cell present in the test channel's squeezer, resulting in a balanced interface in the comparator region between the fluids in the reference and test channels, when equal driving pressures () are imposed. (b) Image of the MCS with an A172 glioblastoma cell present in the test channel's squeezer, resulting in an interface displacement in the comparator region towards the channel with the higher hydrodynamic resistance or excess pressure drop.
By measuring the displacement of the fluid interface , and comparing this displacement with calibrated interface displacements for known excess pressure drops (), the due to the passage of a cell through the constriction can be determined quantitatively. Figures 2a, 2b show the positions of the fluid interface for small (78 Pa) and large (1250 Pa) applied excess pressure drops respectively. Quantitative interface location measurements are obtained from such images by plotting the greyscale values of a vertical row of pixels 4.6 μm from the tip in the comparator region, which corresponds to the “y direction” at the location of the Cartesian axis in Figure 1a. The position of the interface is determined with sub-pixel accuracy by fitting a sigmoid erf function to the greyscale data, shown in Figure 2c as a dashed curve, numerically rendering the fitted curve at gradations one thousand times finer than the pixel gradations. The true interface displacement () is deemed to be the midpoint between the lowest and highest greyscale regions of the fitted curve. These calculations were performed using the Matlab curve-fitting toolbox.
Figure 2.
Calibration of the microfluidic cell squeezer. Images of the MCS with a 4651 Pa driving pressure and an excess pressure of: (a) 76.06 Pa, and (b) 1250 Pa. (c) Plots of greyscale values 10 μm off the tip in the comparator region corresponding to figures (a) and (b) above, where the dashed curves correspond to sigmoid erf fits. (d) Calibration plot of interface displacement for known applied excess pressure drops . The solid line corresponds to a linear fit.
Calibration curves for a known driving pressure were determined by measuring interface displacements for a wide range of applied excess pressure drops, and obtaining a linear-least-squares fit to find a relation of the form where m is the slope and b is the y-intercept. From this linear relation, the pressure drop associated with the passage of a cell through a constriction can be determined from measurements of the interface displacement. Calibration curves and data for vs are shown in Figure 2d where the error in these fits is . The experimentally obtained calibration values correspond to an average over a minimum of 3 runs, and the error bars correspond to the range of the data over these repeated runs.
EXPERIMENTAL METHODS
Cell culture
A172, 1321N1, and normal human astrocyte lines L0329 and L0367 were obtained from Peter Syapin (TTU Health Sciences Center, Department of Pharmacology and Neuroscience). A172 glioblastoma cells were cultured in Dulbecco's modified eagle medium (DMEM) supplemented with 1.1% 10 mM Nonessential Amino Acid mix, 1.1% of 200 mM l-glutamine and 11% fetal bovine serum. 1321N1 astrocytoma cells were cultured in DMEM supplemented with 1.1% 200 mM l-glutamine stock, 1.1% Penicillin/Streptomycin (Gibco 15140-148), and 5.4% fetal bovine serum. Normal human astrocyte cell lines L0329 and L0367 were cultured in flasks treated with poly- l-lysine (ScienCell 0403) as per the manufacturer's instructions, the flasks were rinsed, and the cells were incubated in Astrocyte Medium (ScienCell 1801). All cells were cultured at in a 5% atmosphere. The cells were harvested for the experiments when approximately 80% confluent by treatment with Trypsin/EDTA solution (ScienCell 0103).
Wound healing assay
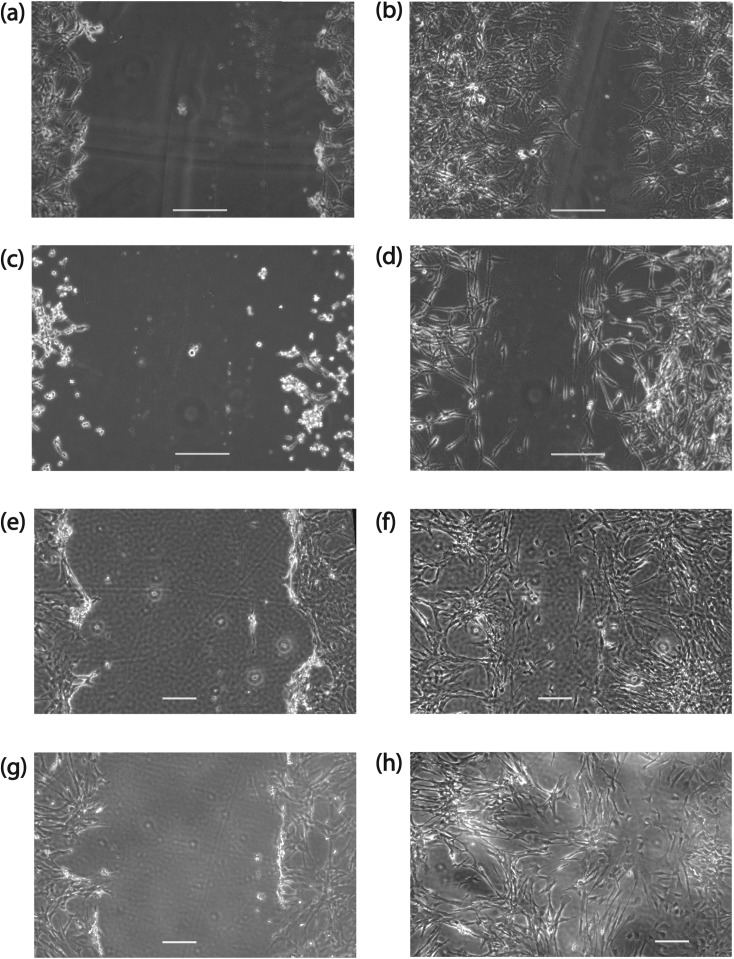
Wound healings invasiveness assays were conducted following the protocols of Liang et al.27 Cells were removed from their culture flasks on their second passage by application of Trypsin/EDTA solution and subcultured in petri dishes with a 60 mm diameter and 15 mm depth. When the cells were confluent, a scratch was made on the monolayer of the cells using a P200 pipet tip. The scratch was monitored over the course of approximately 24 h for all cell lines using an Olympus CK-41 inverted microscope in phase contrast mode with a 10× magnification objective and a PCO 1200 CMOS camera (A172 and 1321N1 cell lines) or a 4× magnification objective and a SVSi Streamview CCD camera (L0329 and L0367 cell lines). 3 replicates were conducted for each cell line. The wound closing velocity was determined by manually finding the average distance between the edges of each scratch at known time intervals using imagej software,28 plotting these distances against time, and fitting to determine the slope. Representative images of an initial wound for each cell line, and a subsequent image taken approximately 22–24 h later are shown in Figures 3a, 3b, 3c, 3d, 3e, 3f, 3g, 3h.
Figure 3.
Wound scratch assay, where A172 cells are shown (a) at the time of scratch creation and (b) 1320 min after scratch creation. 1321N1 cells are shown (c) at the time the scratch was made and (d) 1300 min later. L0329 cells: (e) initial scratch and (f) 1376 min later. L0367 cells: (g) initial scratch and (h) 1407 min later. The length of the scale bars is 200 μm.
Fabrication of the microfluidic cell squeezer
Microfluidic devices were fabricated using standard soft lithography techniques.29 A master mould was fabricated by spin-coating a SU8-5 negative photoresist (Microchem) on a 76.2 mm diameter silicon wafer, the thickness of which corresponds to the height of the channels (11 μm). Degassed polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning) was poured into the mould and baked for 2 h at . The PDMS was cut and peeled from the mould, and inlet and outlet holes were punched (Harris Uni-core, 0.75 mm hole). The devices were then bonded to glass cover slides using air plasma (Plasma Cleaner PDC-32G, Harrick Plasma) and baked at for 5 min to obtain a permanent bond. Prior to experiments, the chips were incubated with a mixture of 0.01 M phosphate-buffered saline solution and 4 wt. % bovine serum albumin (BSA) for 1 h at to reduce the non-specific adhesion of cells to the channel walls.
Microfluidic cell squeezer experiments
Cell growth medium containing cells and trypan blue dye (viscosity , density , Cellgro), and medium containing McCormick black food dye (viscosity , density ) were loaded into two liquid reservoirs affixed to vertically mounted optical rails (Edmund Optics). The trypan blue dye was used to account for dead cells that might be entering the test channel and the black food dye was used to visualize the displacement of the interface. The liquid reservoirs were connected to the microfluidic device with 20 gauge stainless steel blunt tips, 0.5 mm inner diameter tygon tubing and 20 gauge hollow blunt pins, which were inserted into the inlet holes of the device. The outlet was left open to atmospheric pressure. The pressure drop associated with the tubing was calculated to be 0.1% of the pressure drop in the chip. The experiments were conducted with a driving pressure of 4651 Pa which closely mimics intracranial pressure.30 The shear stress corresponding to this driving pressure in our MCS device ranges from 0.3 to 3 Pa. The Reynolds numbers in all of the experiments are less than 0.3.
Bright-field imaging was conducted using an Olympus IX-70 inverted microscope with a 40× magnification objective and a Phantom v310 12-bit CMOS camera with pixels using a reduced 1008 × 304 pixel sensor size. With this optical setup, the effective pixel size is 0.46 μm. Typically exposure times of 15 μs and frame rates of 9000 frames per second were used to observe the motion of cells passing through the squeezer and the associated interface displacement of the microfluidic manometer.
36 A172 cells, 18 1321N1 cells, and 11 normal human astrocytes were analyzed in the squeezer experiments. The throughput of benign cells in this device was lower than the tumor cells, as the benign cells tended to stick to the cells' entry channel upstream of the constriction (despite passivation of channel surface with BSA) and reduce the throughput. Our observation is consistent with a previous study that demonstrated that benign MCF10A breast epithelial cells can be differentiated from MCF7 breast cancer cells by their higher adhesion to label-free micro-patterned surfaces in microfluidic chips.31
Measured quantities from the MCS experiments
We quantify a number of parameters pertaining to the transport of both brain tumor and healthy glial cells in the MCS device.
Peak excess pressure drop ()
The excess pressure drop of the cells increases as the cell enters the constriction, reaches a plateau, and then decreases as the cell exits the constriction—see Figure 4. We classify the excess pressure drop of the cells by the peak value obtained over the course of their passage through the constriction. The uncertainty associated with the measurement of is taken as 10% of the peak value, as given by the calibration curve.
Figure 4.
Left panel: excess pressure drop as a function of time for a single A172 cell passing through the MCS. Right panel: the A172 cell corresponding to the excess pressure drop in the left panel, shown in the squeezer at several times indicated in the left panel.
Elongation index (EI)
The elongation index of the cells is determined by taking the ratio of the major axis of the cells inside the constriction to the major axis prior to entering the constriction. The major axis was determined from elliptical fits manually performed to images of the cells during their passage through the constriction using imagej software.28 The reported value is the maximum, typically occurring within a few cell diameters from entry into the constriction. The uncertainty associated with this measurement corresponds to a 1-pixel measurement error in manually determining the elliptical fits to the cells prior to entering and while in the constriction.
Cell speed ()
The centroid of the cells was also determined from the elliptical fits, and the centroids were used to calculate the cells' speeds as they passed through the squeezer. The value reported is the mean cell speed, and the measurement uncertainty corresponds to the range of all of the speeds measured.
Entry time ()
The time elapsed between the point of first contact of the cells with the constriction and the point of full containment within the constriction. The uncertainty in this measurement corresponds to half of the time interval between successive images.
RESULTS
Typical hydrodynamic response of a cell in the MCS device
The pressure drop associated with a cell's passage through a constriction was measured for each cell line; a representative example of the pressure drop signal is shown in the left panel of Figure 4 for the entire time an A172 cell is passing through the constriction. The position of the cell is associated with the evolution of in the right panel of Figure 4. We typically observe that begins increasing as the cell enters the constriction, reaches a maximal value when the cell is approximately 2 cell diameters in the channel, and forms a plateau as it continues to traverse the channel. As the cell exits the constriction rapidly decreases, and becomes negative when the cell enters the comparator region and displaces the interface between the test and reference fluids.
We find that the elongation index and cell speeds follow trends similar to that of during the passage of the cells through the constriction, where typically the elongation index attains a plateau value and does not appreciably decrease until the cell exits the constriction. The cells' speeds tend to display some fluctuations, hence we use the range of speeds as the uncertainty in these measurements. We note that because we measure EI and manually (for every fifth image in the several hundred captured images), we do not obtain the time evolution of EI or at the same time resolution as .
Excess pressure drop due to the passage of cells through the MCS device
We characterize the pressure drop associated with the passage of each cell through the constriction by the peak value of and refer to it as . A histogram of for normal human astrocytes (L0329 and L0367 cell lines), A172 glioblastoma cells, and 1321N1 astrocytoma cells are shown in Figure 5a. In order to determine the statistical significance between the pressure drops of these cell lines, we performed two-sample Kolmogorov-Smirnov tests on all pairs of pressure drop data sets. For all cases, the null hypothesis is that the distributions for pairs of data sets are the same. For the benign cells and the A172 cancer cells, the p-value we obtained was 0.094, for benign cells and 1321N1 cancer cells it was 0.1424, and for the A172 and 1321N1 cancer cells it was 0.8608. In the case of the benign and A172 cancer cells, the p-value is low enough to reject the null hypothesis with 10% significance, however, the p-value for the benign and 1321N1 cell comparison was not low enough to reject the null hypothesis. We also note that the p-value we obtained in comparing the two different cancer cell lines is significantly higher than comparisons between normal and cancerous cells. A summary of our statistical tests is shown in Table TABLE I..
Figure 5.
(a) Histogram of the peak pressure drop , and (b) dependence of on the cells' confinement as parametrized by the ratio of the cell's radius to the hydraulic radius of the squeezer.
TABLE I.
Statistical significance of differences between cell lines for measured distributions of excess pressure drop , EI, cell speed in constriction, and entry times into the constriction. The statistical significance was determined using two-sample Kolmogorov-Smirnov tests. In all cases, the null hypothesis is that the distributions for pairs of data sets are the same.
| Measured quantity | Cell line 1 | Cell line 2 | Null hypothesis rejected (10% significance) | Statistical significance (p-value) |
|---|---|---|---|---|
| Benign | A172 | Yes | 0.0904 | |
| Benign | 1321N1 | No | 0.1424 | |
| 1321N1 | A172 | No | 0.8608 | |
| EI | Benign | A172 | No | 0.9999 |
| EI | Benign | 1321N1 | No | 0.9770 |
| EI | 1321N1 | A172 | No | 0.5384 |
| Speed | Benign | A172 | No | 0.2490 |
| Speed | Benign | 1321N1 | No | 0.9770 |
| Speed | 1321N1 | A172 | Yes | 0.0425 |
| Entry time | Benign | A172 | Yes | 0.0065 |
| Entry time | Benign | 1321N1 | Yes | 0.0669 |
| Entry time | 1321N1 | A172 | No | 0.7061 |
In order to assess the influence of confinement on the pressure drop, we plot against the confinement parameter in Fig. 5b, where is the cell's unconfined radius and is the hydraulic radius of the confined channel. Here, W and H are the constriction's width and height, respectively. We expect that large and highly confined cells will have larger pressure drops associated with their passage through a constriction than small weakly confined cells with equal stiffness, since their increased surface area in proximity with the channel walls will result in a larger hydrodynamic resistance than for small cells. We find that as expected for all three cell types, larger cells tend to yield higher values. In particular, we observe that when increases from 1 to 1.5, the value increases by a factor of ten, indicating small increase in confinement can lead to large increases in the mechanical resistance for glial cells. We also observe considerable differences amongst cells of similar sizes. For example, varies by about an order of magnitude (nearly from 200 to 2000 Pa) at a confinement of approximately 1.25 for the cancer cell lines. We expect that this large variation amongst cells with similar sizes is due to the different mechanical properties of the cells, and not solely due to cell size differences. The statistical analysis in Table TABLE I., shows that the value for benign and A172 cells is statistically different, but not between benign and 1321N1 cells. Therefore, it is possible that with larger sample sizes, values can distinguish between benign and cancerous cells.
Cell elongation
We also measured the cells' EI, defined as the ratio of the squeezed to pre-squeezed major axes, and show the histogram of each cell line in Figure 6a and confinement dependence in Figure 6b. We have found that the null hypothesis that pairs of these distributions are the same cannot be rejected with a 10% significance level, as shown in Table TABLE I., which implies that at the same driving pressure the cancerous as well as benign cells undergo similar shape deformations. We have also probed how EI varies with confinement for all three cell types, as shown in Figure 6b. We observe that EI tends to increase with cell confinement within the scatter of the data for all three cell types, indicating that smaller cells have elongated less than the larger ones in the constriction. Consistent with Fig. 6a, we observe in Fig. 6b that there is no discernible difference in the EI between the cancer cell lines and normal glial cells.
Figure 6.
(a) Histogram of the EI, and (b) dependence of EI on the cells' confinement.
Cell speed in the squeezer
We have additionally examined the speed of the cells in the narrow constriction, and show histograms of the cells' speed for the three cell lines studied in Figure 7a. Here we find that a two-sample Kolmogorov-Smirnov test can differentiate between the speeds of A172 and 1321N1 cancer cells with 10% significance level, however, comparisons between the cancer cell lines and benign cells cannot reject the null hypothesis that the distributions are the same as shown in Table TABLE I.. We also examine the dependence of cell speed on confinement in Fig. 7b. Interestingly, the data show that unlike the strong dependence of on confinement, no obvious dependence of cell speed on confinement was observed within the scatter of the data.
Figure 7.
(a) Histograms of the cell speed associated with the passage of a cell through a narrow microfluidic constriction, and (b) the dependence of cell speed on the cell's confinement.
The speed, or velocity, of a soft deformable confined object in a microchannel is dictated by the ability of surrounding liquid to push the deformable object as well as the degree of frictional resistance with the channel walls. In our case, we observe that the surrounding liquid can move much more slowly than the cell, about 20%–80% slower, with no obvious differences between cancer cells and benign cells or the degree of cell confinement. At least, in the range of confinement values investigated (), the lack of dependence of cell speed on cell size indicates that frictional contact with the channel walls is similar for all cells and cell lines studied. In our study, the microfluidic devices were pre-treated with a BSA solution to reduce non-specific adhesion. It is possible that this pre-treament reduced the friction coefficient of the cells with the surfaces of the chips and thus all cell lines have similar speeds relative to their degree of confinement. A similar observation was made by Hou et al. who found that the velocities of MCF-7 breast cancer cells and MCF-10 A breast epithelial cells speeds' did not differ in the flow-induced passage through a narrow microchannel.21
Cell entry time into the squeezer
Hou et al. were able to correlate the entry times—or the time taken from the moment the cell touches the constriction to fully entering into it—with cancerous and benign breast cancer cells.21 They found that larger cells had longer entry times, and that the non-cancerous MCF-10 A cells on average had a longer entry time in comparison with cancerous MCF-7 cells. Conclusion of Hou et al. about increased entry times being reflective of a cell line's increased stiffness are supported by AFM measurements performed on these cell lines by Li et al.32 who have determined that the apparent elastic moduli of MCF-10 A cells are approximately twice as large as the apparent elastic moduli of MCF-7 cells.
Another study of cell deformability in the passage through a narrow constriction has determined that cell passage times through the constriction similarly depend on cell size, where the cell passage time includes the entry time that we consider here.33 This study has also demonstrated that when the cells' cytoskeleton is made compliant by application of a chemotherapy drug which interferes with actin polymerization and reduces cell stiffness, cell transit times decrease in such a manner that the decrease of travel time is more apparent for larger cell sizes. Thus, findings from prior literature indicate that cell entry times can indeed reflect mechanical properties despite variability in cell size.
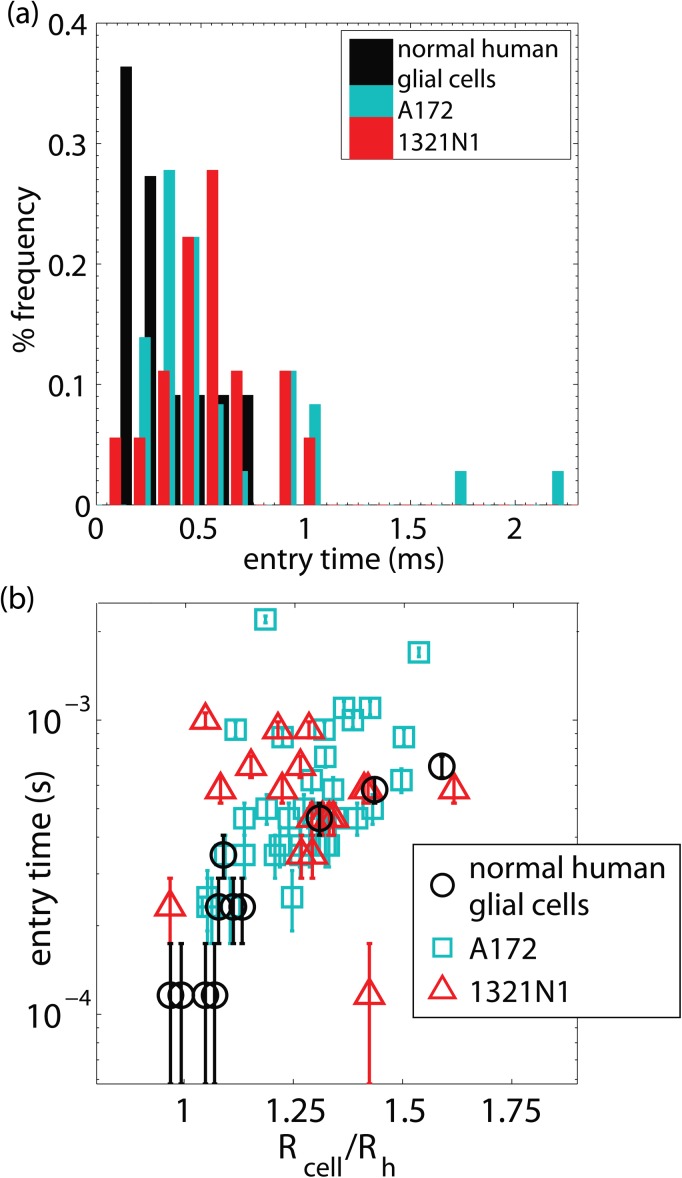
We examined the cell entry times into the narrow squeezer for our cancerous and non-cancerous glial cell lines, shown as histograms in Figure 8a and plotted against confinement in Figure 8b. We also find that larger cells have longer entry times, and that the normal human astrocyte cell lines have a shorter entry time than the cancerous cell lines (two-sample Kolmogorov-Smirnov test with a 10% significance level, see Table TABLE I.), however, the null hypothesis that A172 and 1321N1 cell lines are drawn from the same distribution cannot be rejected (two-sample Kolmogorov-Smirnov test with a 10% significance level). The finding that normal human astrocytes take less time to squeeze through the constriction than cancerous brain tumor cells is in striking contrast to what has been observed with breast cancer cells. This indicates that the normal human astrocyte cell lines are softer than the cancerous glioblastoma A172 and astrocytoma 1321N1 cell lines.
Figure 8.
(a) Histograms of the cells' entry times into the narrow constriction, and (b) the dependence of entry time on the cell's confinement.
Cell migration assay
To further correlate cell deformability with invasiveness, we determined the migration ability of these cell lines using wound healing assays.27 In order for a cancer cell to change its position within surrounding tissues, it can use migration mechanisms that are similar to those that occur in healthy tissues during wound healing.34 For instance, wound healing assay studies have demonstrated that increased migration coincides with increased cancer invasiveness in animal models,6 and the increased metastatic potential of breast cancer cell lines.35
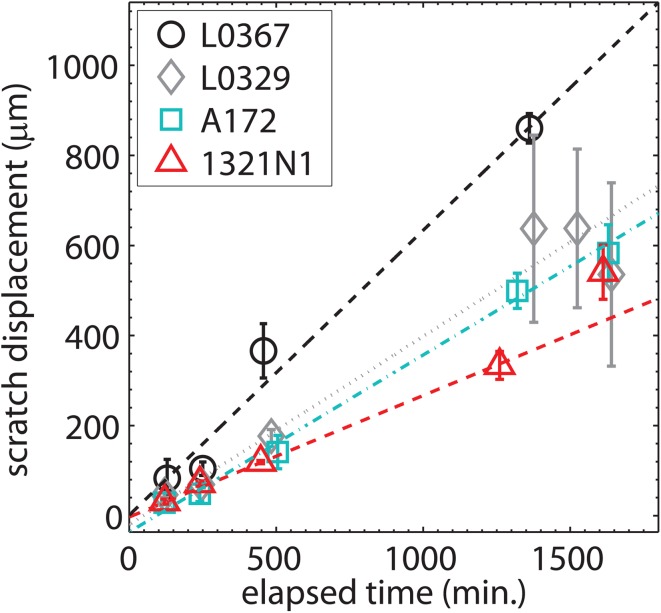
To determine the migration velocity, in Figure 9, we plot the scratch wound displacement (rather than the wound width) against the time elapsed since the scratch was made. We find that the wound in normal human astrocyte cell lines L0329 and L0367 close more rapidly than for A172 glioblastoma or 1321N1 astrocytoma cell lines. Linear fits to the displacement vs. time data yield the following migration rates: L0367 cells migrate at a rate of 0.6 ± 0.1 μm/min, L0329 cells migrate at a rate of 0.42 ± 0.06 μm/min, A172 cells migrate at a rate of 0.39 ± 0.04 μm/min, and 1321N1 cells migrate at a rate of 0.26 ± 0.03 μm/min. Uncertainties in these migration rates are determined from 95% confidence bounds on the slope of the linear fit.
Figure 9.
Wound scratch displacement against time for normal healthy asctrocytes (L0367 and L0329 cell lines) and brain cancer cells (A172 glioblastoma and 1321N1 astrocytoma cell lines). The data points correspond to the mean over 3 runs, and the error bars correspond to the range of the data.
DISCUSSION
Here, we have presented the first measurements of the peak pressure drop due to the passage of glioblastoma, astrocytoma, and normal human glial cells through confined micro channels, as well as the elongation, speed, and entry time of the cells in the confined channels. Our results show that: (i) the similarity of cancerous and benign brain cells cannot be rejected based on speeds and elongation, (ii) entry time into a narrow constriction appears to be a more sensitive indicator of the differences between malignant and healthy glial cells than pressure drop measurements, and (iii) brain tumor cells take a longer time to squeeze through the constriction and migrate slower suggesting they are stiffer compared to their normal counterparts—an observation that is strikingly different compared to the mechanical properties of malignant extraneural cells. Below we discuss possible explanation for these results and their implications.
We expect that the peak excess pressure drop for cells, similar to other deformable particles such as drops, would depend on the ability of the fluid to flow around the cell through lubricating films and gutter flows in the corners of the channels with rectangular cross-section, the advecting flow field ahead of and behind the soft object, as well as the flow field within the soft object, which is a complex situation not fully described by any quantitative model.26 If all of the cell types examined occupy the constriction channel in a similar manner, we would expect the lubricating and gutter flows to be similar producing similar values; recall that the EI was found to be indistinguishable for all cell lines studied. However, we found a significant dependence of on cell size, indicating stronger confinement makes it difficult for the fluid to pass around the cells.
We have demonstrated that the cells' speeds cannot be used to reject the similarity between benign and cancerous cell lines within the considerable scatter of the data, therefore, we have concluded that frictional contact with the channel walls is similar for all cells in the presence of a BSA pre-treatment of the chips. Our finding that benign cells have similar elongations to cancerous cells under similar confinement was unexpected since it has been previously demonstrated that breast cancer MCF-7 cells are more deformable than non-cancerous MCF-10 breast tissue cells using an optical stretching technique.20 We have demonstrated that the ability of the cells to elongate when confronted with a constriction is similar among all cell lines studied, and that the dominant trend is a size effect, i.e., larger cells elongate more to enter a narrow constriction than smaller cells. This could occur if all of the living cells elongate just enough to enter the flow within the microchannel, and no more.
Finally, we have showed that the entry time of benign glial cells is shorter than that of tumor cells, and that this is the most sensitive measurement made in this device which is capable of differentiating benign from cancerous cells. This measure can differentiate softer from stiffer cell types, as the entry time reflects the speed of the cells' response to shear stress upon entry to the constriction. A faster entry time results from a faster rate of strain of the cell's cytoskeleton and organelles in response to the shear stress, which is indicative of a softer material. Hou et al. found the opposite trend for extraneural cancer cells; they found that MCF7 breast cancer cells had a shorter entry time into a narrow constriction than benign MCF10A breast epithelial cells.21 Despite the trends being different for various cell lines, the underlying commonality is that whole cell mechanical properties are more sensitive to step changes in fluid strain rate.
With regards to our finding that normal glial cells are softer when considering entry times into narrow microchannels, and their enhanced migration speeds in comparison with cancerous cells, we postulate that it may not be possible to apply the knowledge of the mechanical properties and invasive patterns of extra-neural cancer cells relative to healthy cells directly to primary brain tumor cells. The classical picture of how metastasis occurs includes various stages of cell detachment from a primary tumor, migration in surrounding tissues, intravasation and extravasation into and out of the vasculature, formation of micrometastases, and tumor growth at a secondary site.36, 37 All of the steps in this pattern of metastasis may not be exploited by brain tumor cells for various reasons, since brain tumor metastases outside of the central nervous system are relatively rare, and several theories have been developed to explain this rarity. One possible explanation is that physiological compensation for an expanding tumor is more difficult in the brain than other places in the body, and that may limit the time a tumor has to develop metastases.38 Another theory is that cerebral tumors do not frequently metastasize because the intracerebral environment is not hostile enough to the tumor cells to promote the growth of highly cancerous clones. The central nervous system has less connective-tissue stroma than the rest of the body, which implies that cancerous clones which are able to invade outside of the brain are not selected as having a significant growth advantage relative to less highly cancerous cells.39
CONCLUSIONS
We have presented an original application of the microfluidic manometer technique to investigations of the mechanical properties of cancerous and benign brain cells. Since microfluidic devices are increasingly being developed to characterize the mechanical properties of tumor cells, it is important to understand the relationships between cell size, hydrodynamic resistance, cell velocity, and shape deformation under confinement. Our study illustrates that such relationships can be explored in detail. Future studies should be directed towards probing the detailed micromechanics of tumor cells, including the role of the nucleus, by increasingly confining the tumor cells and exposing them to varying driving pressures. Since entry time has been identified as a key indicator of malignancy, microfluidic devices can be engineered with multiple squeezing channels to enable high throughput mechanical characterization of tumor cells.
ACKNOWLEDGMENTS
We thank Raul Martinez-Zaguillan and Souad Sennoune for stimulating discussions and Peter Syapin for cell lines and cell culture support and advice. The authors acknowledge funding by a Discovery Grant from the American Brain Tumor Association and the Cancer Prevention and Research Institute of Texas (Grant No: RP100680).
References
- Pilkington G. J., Brain Pathol. 4, 157 (1994). 10.1111/j.1750-3639.1994.tb00826.x [DOI] [PubMed] [Google Scholar]
- Darwish B., Koleda C., Lau H., Balakrishnan V., and Wickremesekera A., J. Clin. Neurosci. 11, 640 (2004). 10.1016/j.jocn.2003.12.008 [DOI] [PubMed] [Google Scholar]
- Schweitzer T., Vince G. H., Herbold C., Roosen K., and Tonn J. C., J. Neuro-Oncol. 53, 107 (2001). 10.1023/A:1012245115209 [DOI] [PubMed] [Google Scholar]
- Ulrich T. A., de Juan Pardo E. M., and Kumar S., Cancer Res. 69, 4167 (2009). 10.1158/0008-5472.CAN-08-4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak A. and Kumar S., Integr. Biol. 3, 267 (2011). 10.1039/c0ib00095g [DOI] [PubMed] [Google Scholar]
- Hu J. and Verkman A. S., FASEB J. 20, 1228 (2006). 10.1096/fj.06-5930fje [DOI] [PubMed] [Google Scholar]
- Soda Y., Marumoto T., Friedmann-Morvinski D., Soda M., Liu F., Michiue H., Pastorino S., Yang M., Hoffman R. M., Kesari S., and Verma I. M., Proc. Natl. Acad. Sci. U.S.A. 108, 4274 (2011). 10.1073/pnas.1016030108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicoine M. R. and Silbergeld D. L., J. Neurosurg. 82, 615 (1995). 10.3171/jns.1995.82.4.0615 [DOI] [PubMed] [Google Scholar]
- Bilston L. E., Liu Z., and Phan-Thien N., Biorheology 34, 377 (1997). 10.1016/S0006-355X(98)00022-5 [DOI] [PubMed] [Google Scholar]
- Bilston L. E., Liu Z., and Phan-Thien N., Biorheology 38, 335 (2001). [PubMed] [Google Scholar]
- Darvish K. K. and Crandall J. R., Med. Eng. Phys. 23, 633 (2001). 10.1016/S1350-4533(01)00101-1 [DOI] [PubMed] [Google Scholar]
- Lu Y.-B., Franze K., Seifert G., Steinhäuser C., Kirchhoff F., Wolburg H., Guck J., Janmey P., Wei E.-Q., Käs J., and Reichenbach A., Proc. Natl. Acad. Sci. U.S.A. 103, 17759 (2006). 10.1073/pnas.0606150103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross S. E., Jin Y. S., Rao J., and Gimzewski J. K., Nat. Nanotechnol. 2, 780 (2007). 10.1038/nnano.2007.388 [DOI] [PubMed] [Google Scholar]
- Jonas O., Mierke C. T., and Käs J. A., Soft Matter 7, 11488 (2011). 10.1039/c1sm05532a [DOI] [Google Scholar]
- Wu Z.-Z., Zhang G., Long M., Wang H.-B., Song G.-B., and Cai S.-X., Biorheology 37, 279 (2000). [PubMed] [Google Scholar]
- Di Carlo D., J. Lab. Autom. 17, 32 (2012). 10.1177/2211068211431630 [DOI] [PubMed] [Google Scholar]
- Qi D., Hoelzle D. J., and Rowat A. C., Eur. Phys. J. Spec. Top. 204, 85 (2012). 10.1140/epjst/e2012-01554-x [DOI] [Google Scholar]
- Zheng Y. and Sun Y., Micro Nano Lett. 6, 327 (2011). 10.1049/mnl.2011.0010 [DOI] [Google Scholar]
- Vanapalli S. A., Duits M. H. G., and Mugele F., Biomicrofluidics 3, 012006 (2009). 10.1063/1.3067820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guck J., Schinkinger S., Lincoln B., Wottawah F., Ebert S., Romeyke M., Lenz D., Erickson H. M., Ananthakrishnan R., Mitchell D., Käs J., Ulvick S., and Bilby C., Biophys. J. 88, 3689 (2005). 10.1529/biophysj.104.045476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H. W., Li Q. S., Lee Y. H., Kumar A. P., Ong C. N., and Lim C. T., Biomed. Microdevices 11, 557 (2009). 10.1007/s10544-008-9262-8 [DOI] [PubMed] [Google Scholar]
- Hur S. C., Henderson-MacLennan N. K., McCabe E. R. B., and Di Carlo D., Lab Chip 11, 912 (2011). 10.1039/c0lc00595a [DOI] [PubMed] [Google Scholar]
- Gossett D. R., Tse H. T. K., Lee S. A., Ying Y., Lindgren A. G., Yang O. O., Rao J., Clark A. T., and Di Carlo D., Proc. Natl. Acad. Sci. U.S.A. 109, 7630 (2012). 10.1073/pnas.1200107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abkarian M., Faivre M., and Stone H. A., Proc. Natl. Acad. Sci. U.S.A. 103, 538 (2006). 10.1073/pnas.0507171102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abkarian M., Faivre M., Horton R., Smistrup K., Best-Popescu C. A., and Stone H. A., Biomed. Mater. 3, 034011 (2008). 10.1088/1748-6041/3/3/034011 [DOI] [PubMed] [Google Scholar]
- Vanapalli S. A., Banpurkar A. G., ven den Ende D., Duits M. H., and Mugele F., Lab Chip 9, 982 (2009). 10.1039/b815002h [DOI] [PubMed] [Google Scholar]
- Liang C. C., Park A. Y., and Guan J.-L., Nat. Protoc. 2, 329 (2007). 10.1038/nprot.2007.30 [DOI] [PubMed] [Google Scholar]
- Rasband W. S., ImageJ, National Institutes of Health, Bethesda, Maryland, USA, 1997, http://rsb.info.nih.gov/ij/. [Google Scholar]
- Xia Y. and Whitesides G. M., Annu. Rev. Mater. Sci. 28, 153 (1998). 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- Franze K., Reichenbach A., and Käs J., Mechanosensitivity of the Nervous System, edited by Kamkin A. and Kiseleva I. (Springer Science+Business Media B.V., 2009), Vol. 2, pp. 173–213. [Google Scholar]
- Kwon K. W., Choi S. S., Lee S. H., Kim B., Lee S. N., Park M. C., Kim P., Hwang S. Y., and Suh K. Y., Lab Chip 7, 1461 (2007). 10.1039/b710054j [DOI] [PubMed] [Google Scholar]
- Li Q. S., Lee G. Y. H., Ong C. N., and Lim C. T., Biochem. Biophys. Res. Commun. 374, 609 (2008). 10.1016/j.bbrc.2008.07.078 [DOI] [PubMed] [Google Scholar]
- Adamo A., Sharei A., Adamo L., Lee B., Mao S., and Jensen K. F., Anal. Chem. 84, 6438 (2012). 10.1021/ac300264v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P. and Wolf K., Nat. Rev. Cancer 3, 362 (2003). 10.1038/nrc1075 [DOI] [PubMed] [Google Scholar]
- Sennoune S. R., Bakunts K., Martínez G. M., Chua-Tuan J. L., Kebir Y., Attaya M. N., and Martínez-Zaguilán R., Am. J. Physiol. Cell Physiol. 286, C1443 (2004). 10.1152/ajpcell.00407.2003 [DOI] [PubMed] [Google Scholar]
- Wirtz D., Konstantopolous K., and Pearson P. C., Nat. Rev. Cancer 11, 512 (2011). 10.1038/nrc3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Harris A., Piggott K., Shieff C., and Bradford R., Lancet Oncol. 3, 498 (2002). 10.1016/S1470-2045(02)00819-7 [DOI] [PubMed] [Google Scholar]
- E. C.Alvord, Jr., Arch. Neurol. 33, 73 (1976). 10.1001/archneur.1976.00500020001001 [DOI] [PubMed] [Google Scholar]
- Pansera F. and Pansera E., Med. Hypotheses 39, 88 (1992). 10.1016/0306-9877(92)90146-4 [DOI] [PubMed] [Google Scholar]