Abstract
The use of biomarkers to infer drug response in patients is being actively pursued, yet significant challenges with this approach, including the complicated interconnection of pathways, have limited its application. Direct empirical testing of tumor sensitivity would arguably provide a more reliable predictive value, although it has garnered little attention largely due to the technical difficulties associated with this approach. We hypothesize that the application of recently developed microtechnologies, coupled to more complex 3-dimensional cell cultures, could provide a model to address some of these issues. As a proof of concept, we developed a microfluidic device where spheroids of the serous epithelial ovarian cancer cell line TOV112D are entrapped and assayed for their chemoresponse to carboplatin and paclitaxel, two therapeutic agents routinely used for the treatment of ovarian cancer. In order to index the chemoresponse, we analyzed the spatiotemporal evolution of the mortality fraction, as judged by vital dyes and confocal microscopy, within spheroids subjected to different drug concentrations and treatment durations inside the microfluidic device. To reflect microenvironment effects, we tested the effect of exogenous extracellular matrix and serum supplementation during spheroid formation on their chemotherapeutic response. Spheroids displayed augmented chemoresistance in comparison to monolayer culturing. This resistance was further increased by the simultaneous presence of both extracellular matrix and high serum concentration during spheroid formation. Following exposure to chemotherapeutics, cell death profiles were not uniform throughout the spheroid. The highest cell death fraction was found at the center of the spheroid and the lowest at the periphery. Collectively, the results demonstrate the validity of the approach, and provide the basis for further investigation of chemotherapeutic responses in ovarian cancer using microfluidics technology. In the future, such microdevices could provide the framework to assay drug sensitivity in a timeframe suitable for clinical decision making.
INTRODUCTION
Epithelial ovarian cancer (EOC) is the leading cause of death from gynecological malignancies largely due to its late diagnosis resulting from the unspecific symptomatology associated with the early stages of this cancer.1, 2, 3, 4, 5 Ovarian carcinomas are highly heterogeneous and can be subdivided according to their cell type: serous, mucinous, endometrioid, clear cell, Brenner, mixed subtypes, or undifferentiated.6, 7, 8 Patients can present with ascites,9, 10 a voluminous effusion with a cellular fraction consisting mainly of epithelial ovarian cancer cells, lymphocytes, and mesothelial cells. While more than 80% of the patients will initially respond to treatment, recurrence is common but is generally observed within variable time intervals.11, 12, 13, 14 The high rate of recurrence and mortality of this disease (>80%) underscores the need for a greater understanding of the molecular basis of the disease and the subsequent development of new clinical tools for both detection and patient management. While platinum/taxane-based treatment is currently the gold standard in first line therapy, failure of this treatment in a significant portion of patients remains a serious problem.15, 16, 17 Identifying non-responders, thereby reducing costs and toxicities associated with no clinical benefit, while offering these individual alternative first line treatments, remains one of the most important aspects in the initial clinical management of the ovarian cancer patient, and the focus of future clinical trials.18, 19, 20
Over the past decade, therapeutic strategies against cancer have experienced a framework shift from broad cytotoxic drugs to a more personalized level of medicine-based targeted therapeutics, largely due to the realization that a panacea to cancer may not exist, given the genetic-epigenetic diversity of tumors and their variable responses to chemotherapy.21, 22 Strategies to predict tumor responses to treatment, especially for targeted therapies, have relied either on biomarkers or tumor-based assays.23 However, personalized medicine has suffered from somewhat disappointing outcomes, in part due to the imperfect predictive potential of biomarkers as well as the inability of empirical methods to fully appreciate the complexity of the cancer microenvironment.24 In addition, both approaches have been hampered by intratumoral heterogeneity (ITH), where sub-populations within a tumor may respond differently from the major oncogenic cellular component.25, 26, 27 ITH has been identified in a number of human cancer types including acute lymphoblastic leukemia, prostate, melanoma, pancreatic, gastric, uterine, and ovarian carcinomas.28, 29, 30, 31, 32, 33, 34
In the past, most common in vitro methods to test tumor chemotherapeutic responses have relied on dissociating solid tumors explanted into monolayer (2D) cultures, but this approach has proven unsatisfactory as only a limited and highly selected cell population is able to explant and grow. In addition, there is ample evidence to suggest that 2D monolayers lack many of the physical characteristics found in tissues, resulting in dramatically different therapeutic responses to that observed in patients.35 An alternative approach would be to develop in vitro systems that rely on 3-dimensional (3D) cultures such as spheroids. The advantage of such an approach is that spheroids provide a more appropriate approximation of cell-cell interactions within the tumor, and their drug response may better reflect patient response than 2D monolayer cultures.36, 37 3D spheroids have been shown to mimic gene expression profiles seen in tissues and present intermediate levels of complexity in terms of cellular organization and sample handling.38 In particular, spheroid models mimic the microenvironment of avascular solid tumors and in the case of EOC, naturally formed spheroids are abundant in ascitic fluid.39, 40, 41, 42 It has also been demonstrated that it is possible to grow large numbers of 3D spheroids from tumor-explanted cells thus determining chemoresponse diversity in a large array of samples originating from single source tumor.42, 43 Moreover, due to the geometric, dimensional, and functional homologies between spheroids and solid tumor biopsy sections, assay methods performed on spheroids may be more easily extended to tissue microsections.38
We hypothesize that the application of microfluidic technologies on 3D spheroid-based sampling could provide multiplexed real time monitoring of drug sensitivity within a timeframe suitable for clinical decision making. As a proof of concept, we present a novel low-cost microfluidic system that can be loaded with multiple 3D spheroids derived from the TOV112D EOC cell line44, 45 obtained using our previously reported hanging droplet spheroid culture technique.43, 46 Using vital dyes coupled with confocal microscopy, we were able to monitor cell death in response to chemotherapeutic agents directly within the microfluidic device. Here, we detail the design and fabrication of the microfluidic system, the parameters related to its use for chemotherapeutic testing, and for characterizing the spheroids' biological response within the system during drug exposure.
MATERIALS AND METHODS
Design, fabrication, and surface modification of the microfluidic system
The device layout was designed using the computer-aided-design (CAD) program (Solidworks, Dassault Systemes, France). The mold was manufactured using micromachining poly-methyl methacrylate (PMMA). PMMA is thermally stable at the processing temperature (∼70 °C) of soft-lithography and unlike, silicon molds, it does not require to be silanized. The microfluidic device was fabricated in polydimethylsiloxane (PDMS) using soft-lithographic methods.47, 48 A mixture of PDMS base to cross-linker (Sylgard 184, Dow Corning, USA) in a 10:1 ratio w/w was poured onto the PMMA mold, degassed, heated at 70 °C for 30 min, and was subsequently peeled off from the mold and was reheated again at 70 °C for another 30 min. Inlet and outlet ports were punched in the PDMS slab by 4 mm and 2 mm diameter biopsy punches. The microchannel surface of the slab and a medical grade glass-slide were oxidized by air-plasma for 50 s and immediately pressed together (for at least 30 s) for bonding which formed the enclosed microchannel system. Last, a trimmed-end 200 μl micropipette tip was inserted into the inlet port and the outlet port was completed by inserting an 18-gauge blunt needle end in the punched hole. Dimensions of the different parts of the channels are given in Fig. 1. In particular, the outlet dimension was adjusted to prevent backflow. As this backflow is caused by the height difference between outlet and inlet liquid column, it is directly proportional to , where hi and h0 are liquid column heights at the inlet and the outlet, respectively, and D0 is the outlet diameter (Fig. 2). To prevent cell adhesion to the microchannel walls, which could disrupt the spheroid structure, microchannel surfaces were treated with 10 mg/ml poly(ethylene glycol)-block-poly(propylene glycol)-block-poly(ethylene glycol) (Pluronic F108, Sigma-Aldrich, USA) dissolved in sterilized phosphate buffered saline (PBS, pH 7.4) for 30 min at 37 °C. Following treatment, excess pluronic F108 was purged out and the fluidic system was washed with PBS, 70% ethanol and ovarian surface epithelium (OSE) cell culture medium (Wisent, QC, Canada), respectively. Finally, channels were filled with OSE medium (Wisent, QC, Canada) and the device was stored at room temperature under aseptic conditions.
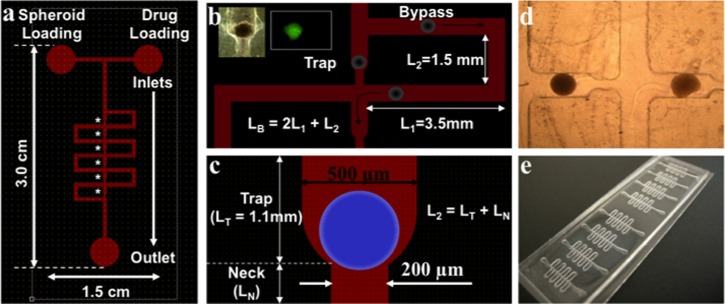
Figure 1.
Schematic representation of the microfluidic device for spheroid entrapment. (a) Layout of the device showing six serially located traps, indicated by the asterisks. (b) Mechanistic elaboration of serial spheroid entrapment. Dimensions of bypass and trapping sections are shown. Height of the microchannels = 500 μm. Black arrows indicate the bypass flow following spheroid entrapment and subsequent flow through the device. The inset in panel (b) demonstrates a trapped spheroid and its imaging with CTG. (c) Detailed illustration of a trapping section. L2 is the total length of the through channel, i.e.,sum of the lengths of trap and neck sections. Spheroid is depicted by the blue circle. (d) Phase-contrast image (4×objective) of two TOV112D spheroids trapped in a microfluidic device. (e) Image of a device containing 8 microchannel systems in a single glass slide, for parallelized analysis.
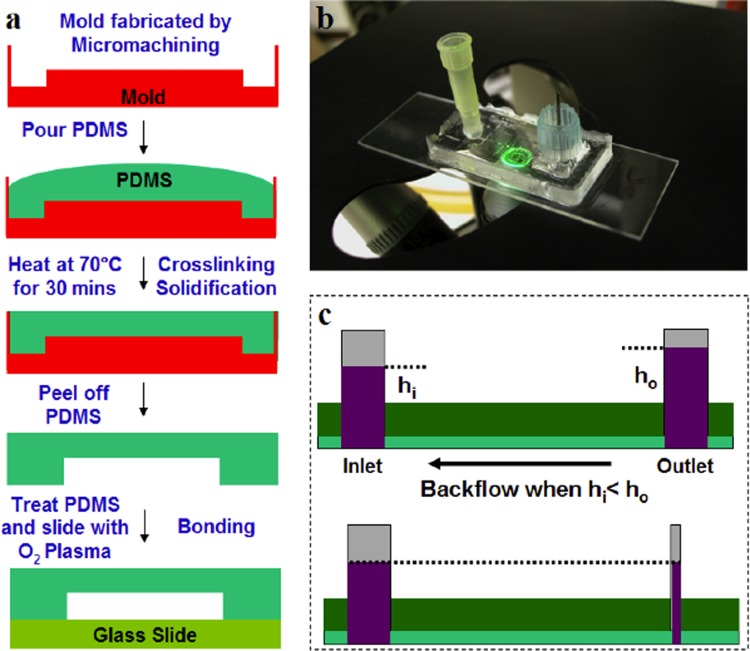
Figure 2.
Critical aspects of device fabrication. (a) Schematic representation of the device fabrication process by soft-lithography or PDMS-based rapid prototyping. (b) Representative image of the microfluidic devices. (c) Illustration detailing the necessity for a narrow outlet to reduce backflow. Backflow is due to the height difference between outlet and inlet liquid column and it is directly proportional to , where hi and h0 are liquid column heights at the inlet and the outlet, respectively, and D0 is the outlet diameter. Therefore, by reducing the diameter of outlet, it is possible to reduce the strength of backflow, although it cannot be completely eliminated in this system.
Spheroid culture of TOV112D cell line
TOV112D cells were cultured in OSE medium supplemented with 10% fetal bovine serum (FBS), 2.5 μg/ml amphotericin B (Wisent, QC, Canada), and 50 μg/ml gentamicin (Invitrogen, ON, Canada).44, 45 Cells were maintained as monolayer cultures in 100 mm tissue culture petri plates in 5% CO2 at 37 °C. At 70%–80% confluence, cells were harvested in suspension by treating them with trypsin (0.05% trypsin, 53 mM EDTA, Wisent, QC, Canada) for 2 min at 37 °C. Two ml of serum-supplemented media were added to neutralize the trypsin and the suspension was then collected in a 15 ml tube and centrifuged at 2000 rpm for 5 min. The supernatant was discarded and the cell pellet was resuspended in 2 ml of fresh serum-supplemented media. Cells were then counted using a haemocytometer and adjusted to a concentration of 250 000 cells/ml (i.e., 4000 cells/16 μl drops). To initiate the spheroid culture by the hanging droplet method,43 16 μl drops were transferred onto the inner surface of the cover of a 150 mm polystyrene vacuum gas plasma-treated petri dish (BD Labware, NJ, USA). To avoid droplet coalescence and to maintain integrity of the spheroids, 110-120 droplets were transferred to each petri dish cover. The cover was then placed over the petri dish which contained 10 ml of sterilized PBS solution in order to reduce the dehydration of the droplets. In this “hanging droplet” arrangement, spheroids grew with increasing compactness for 6 days. At the seventh day from the initiation, the spheroids were harvested by washing the petri dish cover with 5 ml PBS. Spheroid suspensions were then centrifuged for 2 min at 2000 rpm. The supernatant was carefully discarded and the spheroids were resuspended in 2 ml of fresh serum supplemented media. We also tested the effect of extra serum and extracellular matrix (ECM) supplements by forming the spheroids in OSE medium with 20% serum or 10% serum + 2.5% matrigel (BD Biosciences, Canada) or 20% serum + 2.5% matrigel (BD Biosciences, Canada).
Spheroid loading and assay
For each device, between 8 and 10 spheroids were taken out in 150 μl media by a 200 μl pipette and were dispensed into the inlet of the microfluidic device. A flow of liquid was then initiated by applying a small suction pressure on the outlet. As a result of the flow, spheroids were serially loaded into each trapping section. The underlying principle of the serial loading of spheroids is described in Sec. 3A. For investigating the effect of cytotoxic agents, after the spheroids are entrapped in specified segments, fluid within the microchannel system was replaced by OSE medium containing appropriate concentrations of drug and the system was incubated for 6–24 h at 5% CO2 at 37 °C. Finally, the response of the drug-treated samples, at appropriate time points, was compared to the response of control samples. In controls, microchannels were perfused with media without drug to ensure that spheroids were exposed to same level of fluid stress as in the experimental arm. For this study, we selected two common ovarian cancer specific drugs namely carboplatin (stock solution—2.25 mg/ml in 5% dextrose, Novopharm, QC, Canada) and paclitaxel (stock solution—0.18 mg/ml in 5% dextrose, Hospira, QC, Canada). In a separate set of experiments, IC50 values of carboplatin and paclitaxel for 2D monolayer culture of TOV112D cell line were determined to be 13.96 and 1.89 μM, respectively. While probing responses of the same drugs on TOV112D spheroids, we used the aforementioned IC50 values as references and varied the drug concentration by dilution in serum supplemented OSE media in the range 0.1-100 IC50. After the desired incubation period, the microchannel system was washed with PBS, replacing the drug-containing medium. The entrapped spheroids were labeled with 5 μM CellTracker Green (CTG, Molecular Probes, Invitrogen, USA) for 1 h and 1.5 μM propidium iodide (PI, Molecular Probes, Invitrogen, USA) for 20 min at 37 °C. While CTG labels all live cells, PI can permeate through the membranes and label the nucleic acids of dead cells only. The CTG-PI dye combination has been used previously in live-dead imaging or in viability assays.49 Spheroids were then imaged by a confocal microscope (TCS SP5 II, Leica Microsystems, Germany). Spheroid height was found to be in the range of 70–80 μm and all spheroids were imaged in 15 different z-sections (∼5 μm step). Three dimensional shape of the spheroids was found to be similar to what had been reported previously.50 For each z-section, the mortality fraction (number of dead cells/total cells) was determined using an image processing program (matlab, MathWorks, USA) or by manual live-dead cell counting.
RESULTS
Microfluidic system design, fabrication, and operation
Design
The microfluidic system for spheroid entrapment was designed according to the principles of serial entrapment for beads, as reported previously.51 In this design, the microchannel system is composed of a series of trapping sections with a narrower section, or neck. Each neck is arranged in parallel with a longer bypass channel (Figs. 1a, 1b). For our purpose, channel dimensions are fixed in such a way that in the absence of a trapped spheroid, the bulk of the flow passes through the trapping section carrying the leading spheroid towards the trapping area. The design was adjusted to ensure that the system dimensions allowed proper spheroid entrapment while preventing them from squeezing through the trap's neck under the hydrodynamic pressures induced by the flow.52
We analyzed the flow mechanics through trap and bypass channels as described below. From the perspective of lump fluid mechanical analysis, and neglecting the flow perturbations due to the presence of edges and bends, we used the hydraulic/electric circuit analogy to model pressure driven flow within our system, i.e., Q = ΔP/R where Q is the volumetric flow rate, ΔP is the applied pressure difference, and R is the hydraulic resistance.52 In the present case, the hydraulic resistance ratio of two fluidic paths—connected in parallel—governs the relative flow rates through the fluidic circuits. R is related to the geometric parameters of the channel, with the length (L), the width (W), and the height (H) of the channel by the well-known expression for the hydraulic resistance of rectangular channel53
| (1) |
where η is the fluid viscosity.
In the present circuit (Figs. 1b, 1c), ratio of the flow through the trap and the neck region (QTN) to the flow though the bypass (QB) is governed inversely by the ratio of corresponding hydrodynamic resistances, yielding to
| (2) |
and in the absence of spheroids, the majority of the flow should be through the trap, implying
| (3) |
In Eqs. 2, 3, subscripts T, N, B, 1, and 2 refer to trap, neck, bypass, transverse section of bypass and longitudinal section of bypass, respectively. Importantly, Eqs. 1, 2, along with the constraint provided by Eq. 3, indicate towards a limit to which the neck region could be narrowed down without requiring significant increase in the length of the bypass channel and hence, the device dimension. To elaborate, narrowing down the neck leads to an increase in RN. In order to meet all constraints simultaneously, one then needs to increase R2 by increasing its length. Hence, one can observe that narrowing down the neck leads to stretching of the transverse section of the bypass channel. The aforementioned relation thus highlights an important constraint on the physical dimension of the device, especially if one considers upscaling in an array of microfluidic traps (Fig. 1e).
Once a spheroid becomes entrapped, it partially blocks the channel, thus, considerably raising the hydrodynamic resistance of the trap. As a result, most of the flow is diverted to the bypass channel, which carries the subsequent spheroid towards the next available trap (Figs. 1a, 1b). This design allowed serial and separate entrapment of multiple spheroids to be loaded on the chip simultaneously. It should be noted that the percentage of flow diversion will vary with the size of the spheroid being trapped as well as on the elasticity of the spheroid. However, the device has shown consistent spheroid trapping for a wide range of spheroid sizes.
Fabrication
The mold is fabricated by micromachining in PMMA considering the feature sizes of our design (∼500 μm). This process has the advantage of being fast and cheap compared to conventional photolithographic methods. Once the mold is fabricated, replication of hundreds of PDMS devices can be generated (Fig. 2a). PDMS is a non-cytotoxic material and optically transparent, making it suitable for the biological applications.48 Moreover, the PDMS surfaces can be chemically modified allowing it to be adapted to specialized needs. Here we coated the microchannel walls with Pluronic F108, which is known to prevent cell adhesion.54
Operation—spheroid loading
Spheroids were cultured by the hanging droplet method as previously described43 and loaded into the microfluidic system. For the TOV112D ovarian cancer cell line, we observed that spheroids grown up to 7 days in hanging droplets were optimally suitable for the loading into the microfluidic device. Before day 7, spheroids are not compact enough to withstand the shear stresses during the loading and were found to fragment with little flow. After day 9, spheroids remained compact but the mortality fraction (i.e., number of dead cells/total number of cells) within the spheroid increased significantly. This induces high background mortality preventing assessment of the effect of a specific drug with a proper control. These observations indicate that the most suitable experimental window is between day 7 and 9, and for all experiments, spheroids grown up to day 7 in hanging droplet condition were subsequently loaded into the microchannel system. At day 7, the mortality fraction within the spheroid was found to be approximately 20%, which corresponds to the mortality fraction observed in certain stages of avascular tumors.55 To prevent backflow, the width of the outlet column was adjusted, taking into account the height of both the inlet and outlet columns (Figs. 2b, 2c). We found that spheroids made from the TOV112D cell line, at day 7, were distributed in narrow size range (radius = 150–200 μm). Surprisingly, this size was maintained within the range of seeding density of 1000–4000 cells/droplet. Above this density range, either multiple spheroids formed, or a spheroid with bud-like protrusions appeared, while below it, spheroids were not compact. This observation indicated a characteristics length scale of spheroid formation, although the reason behind this remains unknown.
Monitoring chemotherapeutics in EOC microfluidics entrapped spheroids
After spheroid loading into the microfluidic system (Fig. 3a), they were treated with either carboplatin or paclitaxel, at different concentrations and for different durations. Carboplatin is a platinum-based drug, like cisplatin, which has been used specifically in chemotherapy of ovarian, lung, head, and neck cancers.56 It cross-links DNA and hence, interferes with the mitotic cell division. Cells with damaged DNA then undergo apoptosis through diverse intracellular signaling pathways.57 Paclitaxel also inhibits mitotic cell division although by a different mechanism.58 It binds and stabilizes microtubule polymers and prevents the formation of mitotic spindle assembly and chromosome segregation, which are mandatory for the cells to undergo mitosis. Prolonged cell-cycle arrest at the G2/M checkpoint then induces pro-apoptotic signaling.59
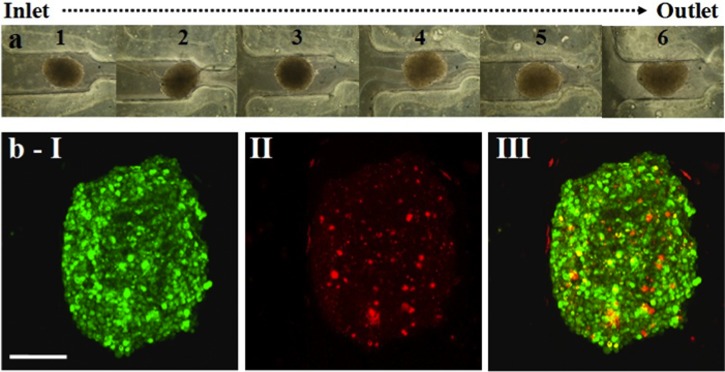
Figure 3.
Spheroid loading and live-dead cell imaging. (a) Phase contrast images of six TOV112D spheroids entrapped in a single microfluidic device. These spheroids were subjected to live-dead imaging by dual fluorescent staining technique. (b) Confocal image of a CTG-PI stained spheroid, as projected over 15 confocal z-sections across the spheroid height. (b-I): CTG (green fluorescent). (b-II) PI (red fluorescent). (b-III) CTG-PI merged channels. Note that PI monitored cell death is minimal and is evenly distributed spatially. Scale bar represents 120 μm.
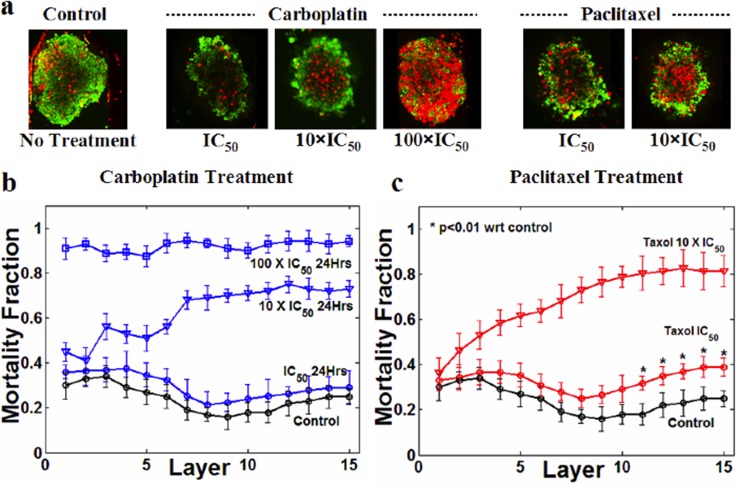
After drug treatment, cells in the spheroids were labeled with CTG and PI (Fig. 3b) for dual fluorescent staining-based live-dead imaging.49 Upon 3D confocal imaging, we noted that spheroids were sickle-shaped rather than spherical (Fig. 4b). Such shape of spheroids has been reported previously.50 Therefore, it was important to distinguish the mortality fraction as a function of the confocal microscopy spheroid height (Z-axis), as this creates differences in cell density. In particular, we determined that image layers 1-6 defined cell-poor layers while the lower image layers 7-15 were cell-rich (Fig. 4). As expected, the mortality fraction increased when spheroids were exposed to a chemotherapeutic agent, with more cell death noted at 24 versus 6 h of exposure (Fig. 4). No significant changes in the mortality fraction were observed after 24 h in this system.
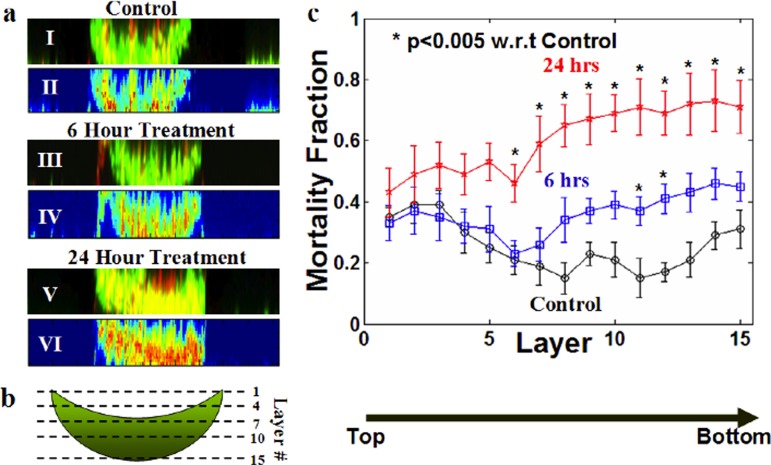
Figure 4.
Distribution and temporal evolution of the mortality fraction. (a) Representative side view images of CTG-PI labeled TOV112D spheroids after 0, 6, and 24 h of incubation with 10× the TOV112D reference IC50 of carboplatin. I, III, and V illustrate merged confocal images while II, IV, and VI are corresponding pseudo-color images highlighting the distribution of mortality, in which red represents high mortality. Note that red intensity is more prominent with increased treatment time (compare 6 to 24 h). (b) Graphical representation of the imaging scheme. From top to bottom, 15 different cross-sectional images (termed image layers) were acquired for each spheroid. (c) Distribution of mortality fraction (number of dead cells/total number of cells in a single section) across 15 image layers comparing controls and spheroids treated with 10× the TOV112D monolayer IC50 of carboplatin for 6 or 24 h. Data are averaged over three independent experiments (total number of samples, n = 16) and shown as mean ± standard deviation.
We have previously reported on the TOV112D IC50 for both paclitaxel and carboplatin in 2D monolayer cultures,60 which served as the reference IC50. Based on these references, trapped spheroids were treated for 24 h with different multiples of the IC50 values (0.1, 1, 10, and 100×) for either paclitaxel or carboplatin and monitored for cell death. Data for 0.1 × IC50 of both drugs, not illustrated in the figure for the sake of clarity, were found to be essentially identical to the data from the control samples, indicating there was no effect of the drugs at this concentration. For carboplatin, we observed that the reference IC50 values did not yield any significant increase in mortality fraction in the spheroid model as compared to controls (Fig. 5) although some cell death was noted at the appropriate IC50 with paclitaxel. By linear interpolation, we found that IC50 values of carboplatin and paclitaxel for TOV112D spheroids were 8.9 ± 1.2 and 7.1 ± 0.6 times the monolayer IC50 values of these two drugs, respectively (Figs. 5b, 5c).
Figure 5.
Effect of carboplatin and paclitaxel on TOV112D spheroids. The reference IC50 is based on drug responses in TOV112D monolayer cultures. (a) Merged CTG-PI confocal section (image layer #13) of spheroids subjected to carboplatin and paclitaxel at different concentrations. No image is provided for the 100 × IC50 paclitaxel treated spheroids, as under these conditions they lost their structural integrity and were fragmented during the vital dye staining. (b) Variation in the distribution of mortality fractions across all z-sections with varying carboplatin concentrations (1, 10, and 100 × IC50). (c)Variation in mortality distribution with changes in paclitaxel concentration (1 and 10 × IC50). Treatment time is 24 h for all samples. Each data point is averaged over three independent experiments (with total number of samples, n = 16) and shown as mean ± standard deviation.
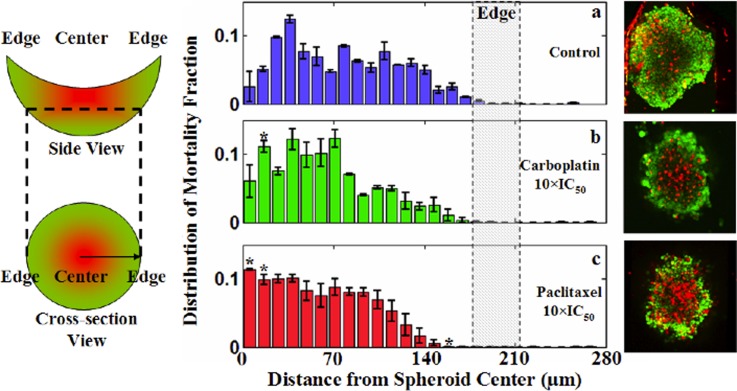
Finally, analyzing the mortality distribution across the spheroid cross-section (XY plane), it was found that effects of the two drugs were most prominent in the center of the spheroids (Fig. 6). In Fig. 6, the probability distribution of the mortality fraction was plotted for samples exposed to carboplatin and paclitaxel versus untreated samples. The phenomenon of greater cell death at the center of the spheroid was found to be most prominent for the paclitaxel treatment (Fig. 6c).
Figure 6.
Radial distribution of the mortality fraction. (Left) Graphical delineation of the image analysis scheme to determine the radial distribution of the mortality fraction. (Middle, panels (a)–(c)) Averaged mortality distribution function of image layers 10-13. (Right) Representative sample images corresponding to the quantitative results. For each sample, distribution function has been normalized (dead cells within a radius range/the number total dead cells) so that integration of it over the whole radial domain remains one. The symbol * in (b) and (c) represents a point for which an increase or decrease in the mortality fraction is statistically significant (p < 0.01) with respect to controls. Each data point is averaged over three independent experiments (with total number of samples, n = 16) and shown as mean ± standard deviation. Note that treatment with carboplatin and paclitaxel showed a more prominent rise in mortality fraction closer to the spheroid center than near the edges.
Effect of extracellular matrix and elevated serum supplementation during spheroid formation on chemotherapeutic response
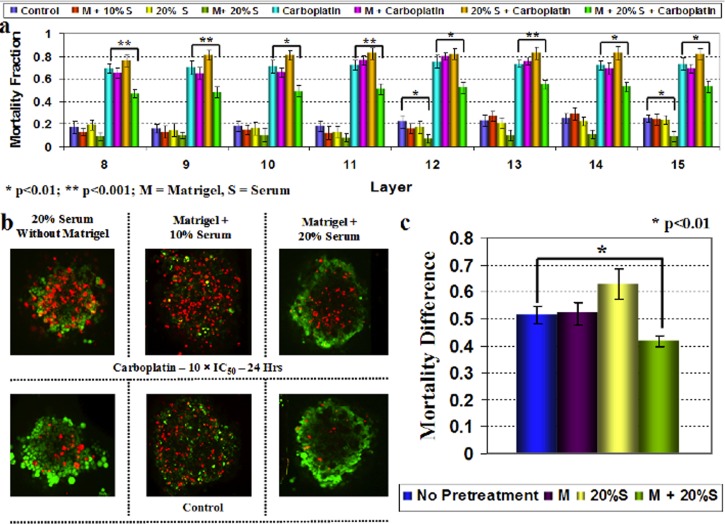
The tumor microenvironment is thought to alter chemotherapeutic responses. To see whether we could mimic some of these events in our in vitro model, we tested the effects of extracellular matrix (through the use of matrigel) and altered growth factors (through elevated serum) on the chemoresponse of TOV112D spheroids (Fig. 7). Excess serum and matrigel supplementation have been shown to increase the compactness of cancer cell spheroids, possibly by providing essential extracellular matrix proteins.61, 62, 63 Here, we observed that the addition of matrigel and high serum during spheroid formation significantly decreased the mortality fraction in response to 10 × IC50 of carboplatin in all relevant cross-sections (Fig. 7a). Applied individually, serum and matrigel supplementations had no effect on the response characteristics of TOV112D spheroids (Fig. 7a). This observation was found to be true for both exact (Figs. 7a, 7b) and differential (i.e., drug induced—control) mortality fractions (Fig. 7c) upon carboplatin treatment.
Figure 7.
Effect of extracellular matrix (matrigel) and increased serum supplementation during spheroid formation on the chemotherapeutic response. (a) Distribution of the mortality fraction in image layers 8-15 of confocal z-sections and its dependence on matrigel and increased serum supplementation. (b) Representative merged CTG-PI confocal sections (image layer 13) of spheroids showing the variation of mortality with different culture conditions. In this panel, control corresponds to cells not exposed to carboplatin. (c) Response to carboplatin treatment was variable depending on the culture conditions, as accessed by the average morality fraction over image layers 8-15 of samples. Note that the simultaneous addition of matrigel and increased serum (20%) supplementation showed a statistically significant increase in chemoresistance, while neither had a significant effect alone. In addition, in the absence of drug, addition of matrigel and 20% serum appears tosignificantly diminish basal cell death within the spheroids (statistically significant in image layers 12 and 15). Each datapoint is averaged over three independent experiments (with total number of samples, n = 16) and shown as mean ±standard deviation.
DISCUSSION
The ability to predict the response of a patient's tumor to chemotherapy before initiating treatment should allow for better clinical management of these individuals. Ideally, this would entail direct testing of patient samples in a time-frame suitable for clinical decision making. Here, we have presented a prototype of a microfluidic system where one can monitor the chemotherapeutic response of ovarian cancer spheroids. In our methodology, we focused on 3D spheroids as they are known to reproduce functional, histomorphological, volume growth kinetics, and mass transport properties present in solid tumor tissue. Additionally, spheroids can directly be isolated from ovarian ascites, or be formed from tumoral disaggregates41, 42 where no culture amplification step is required, thus, minimizing in vitro selection bias. Utilizing a microfluidics platform provides several technical advantages that make it an attractive system in chemoresponse testing. The platform's ability to precisely control sample drug exposure both spacially and temporally, the high resolution imaging, and the experimental reproducibility within the same device are some of the main advantages. In addition, the ability for parallel testing of several chemotherapeutics, using small quantities of expensive drugs, suggests that this approach might be practical for multiple analyses with small samples. Microfluidic systems have been used previously to generate spheroids on-chip.64, 65, 66, 67 However, unlike the present device, they start from single cells, which then form aggregates within the device. The current design, however, offers the possibility to entrap and test other biologically relevant samples such as tissue sections from biopsies.
By relying on a confocal-based live-dead cell imaging protocol, we can quantify cell death while improving our understanding of the temporal-spatial aspects of cell death within a 3D architecture. We were initially puzzled by the observation that image layers 1-5 of the spheroids were devoid of cellular material at the center of the image. After image processing, it was determined that using the hanging droplet method to generate spheroids generated 3D sickle-shaped structures which could be attributed to forces within the droplets, namely, surface tension and gravity. Similarly shaped spheroids grown as hanging droplets have been independently described.50 The analysis also allowed us to observe the spatial distribution of cell death. Although we have previously shown that cell proliferation occurs throughout the spheroid diameter,43 we noted that cell death was most prominent in the center of the spheroid. Some studies have reported a centripetal decrease in metabolic activity within spheroids, and that such metabolically quiescent cells show increased plasma membrane permeability.36, 37, 68, 69 Therefore, while precise metabolic profiles of TOV112D spheroids remain to be investigated, it is possible that the preferential chemosensitivity of the spheroid core may be linked to reduced metabolic activity. Another possibility is whether hypoxia contributes to the phenomena, although initial results looking at hypoxia-inducing genes within the spheroid have not identified the existence of such a gradient.43
Our results confirm previous observations that 3D cultures are associated with an increase in drug resistance as compared to their monolayer counterparts. This increased resistance has been attributed correlatively or causally to multiple genetic and physical factors such as core necrosis by hypoxia, drug permeability, inhibition of apoptosis, altered surface receptor expression, altered overall gene expression profile, and changes in metabolic activity.35 Moreover, it has been shown previously that in relation to these factors, spheroid models provide a better approximation of in vivo solid tumors than 2D monolayer cultures, which partially explains the reduced in vivo efficacy of in vitro tested therapeutics.36, 37, 69 In the present case, given that the typical radius of TOV112D spheroids ranges between 150–200 μm, which is approximately 2-fold smaller than the characteristic thickness (>400 μm) necessary for eliciting significant hypoxia or heterogeneous drug distribution,35 we do not believe that these two factors are directly linked to increases in drug resistance. A more plausible explanation may be that gene expression is significantly altered in the spheroid context. In support of this notion, we have previously identified differential gene expression when comparing monolayer and spheroid cultures from several different ovarian cancer cell lines.43, 70 Among genes showing significant variations were genes associated with cytoskeletal protein binding. Further experiments to clarify gene expression in the context of drug exposure would need to be conducted. In addition, a recent genome-wide gene expression analysis in lung and squamous cancer cells revealed that growing cells in 3D cultures mainly affected genes that alter the composition of extracellular matrix and were associated with tissue development, intercellular and cell-ECM adhesion, and the immune response.71 Findings of this study correlate with a previous study that showed that 3D culture altered the expression patterns of integrin heterodimers, specifically upregulating mesenchymal markers such as α5β1.72 Using either an overexpression or a silencing approach, it would be possible to directly test the impact of candidate genes for their contribution to the altered response we see in the two model systems, and further determine their effect on metabolic activity, the ECM, and cell-surface receptor mediated sequestration of drug molecules.73
One major advantage of the developed system is the ability to dissect the contribution of the microenvironment to drug response. It has been shown previously that additional supplementations such as high serum, ECM proteins, and matrigel all affect the compactness of 3D cellular aggregates.63, 74, 75 Such artificial supplements can allow cell lines lacking inherent spheroid forming abilities to acquire them. In addition, and as demonstrated by our results, manipulating the microenvironment can affect the inherent background mortality. Any supplementation should bear some relevance to in vivo conditions and should be systemically incorporated in order to eliminate artifactual outcomes. Previously, it has been shown that many ovarian cancers present with naturally growing spheroids in ascitic fluid and that such in vivo spheroid formation depends on the concentration of laminin, collagen IV and fibronectin within the fluid.61, 62 Large and compact spheroids form only when the aforementioned proteins are abundant in ascitic fluid. Extrapolating these observations, and noting that matrigel possesses high concentrations of laminin and collagen IV and that serum contains fibronectin, suggests that these supplements are relevant.63
The spheroid-based system, while powerful, does have its limitations. In particular, spheroids are simplified models that lack the extensive vasculature often associated with high stage solid tumors. Vascularization can alter the transport and the penetration of the therapeutic agents.76 In addition, active signaling from tumor-infiltrating endothelial cells can alter the motility of cancer cells and facilitate tissue invasion and metastasis.77 Besides contributing to diffusion, the nature of the interstitial flow (IF) within tumor tissue can also influence drug penetration. Pressure effects due to IF in a tumor tissue has been found to delay drug penetration, and relieving this pressure has been shown to augment therapeutic efficacy.78 Currently there is no spheroid model that takes IF into consideration. Finally, tumor microenvironment governs the process of oncogenesis in a crucial way. In addition to cancer cells, a typical solid tumor may contain several other cell types such as cancer-associated fibroblasts, tumor-promoting inflammatory cells, endothelial cells, and pericytes.79 Importantly, all of these have regulatory or promoting effects on tumor progression. Though it is possible to make spheroids out of multiple cell types, and thus, to mimic the tumor microenvironment to some extent, such models generally lack the complexity of tumor tissues. Many of these limitations may be overcome by replacing in vitro generated spheroids with spheroids present in the ascites of ovarian cancer patients, or from appropriately sized tumor tissues.
In ovarian cancer, ITH was estimated to occur in 54% of patients with advanced disease as determined by flow cytometry analyses on cells isolated from solid tumors.80 Disparities in DNA content and K-Ras mutations have been noted between primary ovarian tumors and corresponding lymph-node and peritoneal metastases, further supporting the importance of ITH.27, 81, 82 Significant differences have also been observed in immunohistochemical staining of mismatch repair proteins, including high mMSH2 staining in metastatic versus primary tumors. Additional evidence comes from Khalique et al. who confirmed ITH in 16 cases of grade 3 serous EOCs using microsatellite and single nucleotide polymorphism (SNP) analyses.26 This study has not only identified ITH in chromosomes 13 and 17 in regions that harbor important regulatory genes including p53, BRCA1 and BRCA2, but also suggested a common origin of all clonal variations within a single tumor. Similarly, Cooke et al. have used comparative genomic hybridization (CGH) strategies including 24-color fluorescence in situ hybridization and SNP analysis to delineate ITH among cell lines derived from three high-grade serous ovarian cancer cases.25 Interestingly, the CGH study also revealed that cisplatin sensitive and resistant cells share a common ancestor. How ITH affects overall drug response remains an intense field of study. Going forward, it will be possible to incorporate different cell populations in our assay system and to track them using specific fluorochromes. This would allow the determination of chemotherapeutic responses when drug sensitive and resistant cells are co-cultures while providing a system to assess the contribution of ITH to the ultimate host response to a specific agent.
From the technological perspective, there were several challenges that needed to be addressed. First of all, multicellular spheroids show more deformability and vulnerability to fluidic shear and compressive stresses than single cellular entities. For example, with little compressive pressure (>100 Pa, i.e., no more than 1 cm increase in pressure-head), a spheroid measuring 400 μm in diameter was observed to squeeze through a 200 μm wide constriction, thus, avoiding entrapment and incurring the undesired loss of samples that became permanently deformed and disintegrated in the process. Consequently, it is necessary to reduce the neck region as much as possible to avoid this compression phenomenon. However, as mentioned above (Sec. 3A1), narrowing the neck region under the current design constraints would mean elongation of the bypass section in order to keep the appropriate hydraulic resistance ratio for trapping. This constraint could potentially limit the scope of miniaturization and any future objective of large scale integration of such assay systems in a monolithic platform. For samples with high heterogeneity, a conservative approach with a very narrow neck region would be preferred. In that case, widening of the device dimension could be prevented by replacing linear bypass channels with serpentine alternatives typically used in micromixing experiments.83 Such alternatives would increase the bypass hydraulic resistance while still allowing spheroids to move through. Nevertheless, the current design has the advantage that the trapping properties only depend on the relative dimensions of the channels and not on their absolute sizes, which is convenient as different cell lines produce spheroids with different properties.43 In most cases, the design could therefore be scaled up or down to adapt to the typical diameter, shape and elasticity of the samples studied. Our results do support that the overall approach is feasible, and with further modification could be adapted to real-time monitoring of drug sensitivity in a clinically relevant setting.
CONCLUSION
Our purpose, in this work, was to delineate a microfluidic system capable of serially confining 3D spheroids in specific sections with well-defined geometry, in which controlled therapeutic treatment can be performed and monitored. While we choose spheroids over actually tissue samples for the proof-of-concept, we anticipate that upon successful implementation of the design its scope could be extended to actual tissue samples from tumor biopsy. Indeed, such microfluidic platforms would require minute amount of tissue samples which would enable testing a large number of specimens without necessitating the dissection of a large chunk of tissue mass.23 While, in the current investigation, we emphasize on a specific epithelial ovarian cancer model, the current protocol is sufficiently generic to be used in investigating the chemoresponse of spheroids or tissue sections of other solid tumors.
ACKNOWLEDGMENTS
We thank Luke Masson for corrections to the manuscript. A.-M.M.-M. is a Researcher of the Centre de recherche du Centre hospitalier de l'Université de Montréal (CRCHUM)/Institut du cancer de Montréal, which receives support from the Fonds recherche Québec-Santé (FRQS). The ovarian tumor banking and cell lines was supported by the Banque de tissus et de données of the Réseau de recherche sur le cancer of the FRQS affiliated with the Canadian Tumor Repository Network (CTRNet). A.-M.M.-M. and O.G. received support for this work through a Collaborative Health Research Grant from the National Sciences and Engineering Research Council of Canada (NSERC) and the Canadian Institutes of Health Research (CIHR).
References
- Bowtell D. D., Nat. Rev. Cancer 10(11), 803–808 (2010). 10.1038/nrc2946 [DOI] [PubMed] [Google Scholar]
- Farley J., Ozbun L. L., and Birrer M. J., Cell Res. 18(5), 538–548 (2008). 10.1038/cr.2008.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns E. M. and Bowtell D. D., Cancer Res. 72(11), 2701–2704 (2012). 10.1158/0008-5472.CAN-11-3911 [DOI] [PubMed] [Google Scholar]
- Haruta S., Furukawa N., Yoshizawa Y., Tsunemi T., Nagai A., Kawaguchi R., Tanase Y., Yoshida S., and Kobayashi H., Oncol. Rep. 26(6), 1347–1356 (2011). 10.3892/or.2011.1456 [DOI] [PubMed] [Google Scholar]
- Ouellet V., Ling T. H., Normandin K., Madore J., Lussier C., Barres V., Bachvarov D., Rancourt C., Tonin P. N., Provencher D. M., and Mes-Masson A. M., BMC Cancer 8, 346 (2008). 10.1186/1471-2407-8-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soslow R. A., Int. J. Gynecol. Pathol. 27(2), 161–174 (2008). 10.1097/PGP.0b013e31815ea812 [DOI] [PubMed] [Google Scholar]
- McCluggage W. G., Pathology 43(5), 420–432 (2011). 10.1097/PAT.0b013e328348a6e7 [DOI] [PubMed] [Google Scholar]
- Normandin K., Peant B., Le Page C., de Ladurantaye M., Ouellet V., Tonin P. N., Provencher D. M., and Mes-Masson A. M., Clin. Exp. Metastasis 27(1), 55–69 (2010). 10.1007/s10585-009-9303-6 [DOI] [PubMed] [Google Scholar]
- Press J. Z., Reyes M., Pitteri S. J., Pennil C., Garcia R., Goff B. A., Hanash S. M., and Swisher E. M., Int. J. Gynecol. Cancer 22(4), 546–552 (2012). 10.1097/IGC.0b013e318241d9b9 [DOI] [PubMed] [Google Scholar]
- Lane D., Matte I., Rancourt C., and Piche A., J. Ovarian Res. 3, 1 (2010). 10.1186/1757-2215-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CGARN, Nature 474(7353), 609–615 (2011). 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankoo P. K., Shen R., Schultz N., Levine D. A., and Sander C., PLoS One 6(11), e24709 (2011). 10.1371/journal.pone.0024709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R. C.Bast, Jr., Hennessy B., and Mills G. B., Nat. Rev. Cancer 9(6), 415–428 (2009). 10.1038/nrc2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog T. J. and Pothuri B., Nat. Clin. Pract. Oncol. 3(11), 604–611 (2006). 10.1038/ncponc0637 [DOI] [PubMed] [Google Scholar]
- Miller D. S., Blessing J. A., Krasner C. N., Mannel R. S., Hanjani P., Pearl M. L., Waggoner S. E., and Boardman C. H., J. Clin. Oncol. 27(16), 2686–2691 (2009). 10.1200/JCO.2008.19.2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong D. K., Bundy B., Wenzel L., Huang H. Q., Baergen R., Lele S., Copeland L. J., Walker J. L., and Burger R. A., N. Engl. J. Med. 354(1), 34–43 (2006). 10.1056/NEJMoa052985 [DOI] [PubMed] [Google Scholar]
- Konner J. A., Grabon D. M., Gerst S. R., Iasonos A., Thaler H., Pezzulli S. D., Sabbatini P. J., Bell-McGuinn K. M., Tew W. P., Hensley M. L., Spriggs D. R., and Aghajanian C. A., J. Clin. Oncol. 29(35), 4662–4668 (2011). 10.1200/JCO.2011.36.1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebarth A. J., C. N.Landen, Jr., and Alvarez R. D., Clin. Obstet. Gynecol. 55(1), 156–172 (2012). 10.1097/GRF.0b013e31824b1699 [DOI] [PubMed] [Google Scholar]
- Gardner G. J. and Jewell E. L., Cancer Control 18(1), 44–51 (2011). [DOI] [PubMed] [Google Scholar]
- Musrap N. and Diamandis E. P., Mol. Cancer Res. 10(10), 1254–1264 (2012). 10.1158/1541-7786.MCR-12-0353 [DOI] [PubMed] [Google Scholar]
- Bild A. H., Yao G., Chang J. T., Wang Q., Potti A., Chasse D., Joshi M. B., Harpole D., Lancaster J. M., Berchuck A., J. A.Olson, Jr., Marks J. R., Dressman H. K., West M., and Nevins J. R., Nature 439(7074), 353–357 (2006). 10.1038/nature04296 [DOI] [PubMed] [Google Scholar]
- Mullighan C. G., Phillips L. A., Su X., Ma J., Miller C. B., Shurtleff S. A., and Downing J. R., Science 322(5906), 1377–1380 (2008). 10.1126/science.1164266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Veer L. J. and Bernards R., Nature 452(7187), 564–570 (2008). 10.1038/nature06915 [DOI] [PubMed] [Google Scholar]
- Schilsky R. L., Nat. Rev. Drug Discov. 9(5), 363–366 (2010). 10.1038/nrd3181 [DOI] [PubMed] [Google Scholar]
- Cooke S. L., Ng C. K., Melnyk N., Garcia M. J., Hardcastle T., Temple J., Langdon S., Huntsman D., and Brenton J. D., Oncogene 29(35), 4905–4913 (2010). 10.1038/onc.2010.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalique L., Ayhan A., Weale M. E., Jacobs I. J., Ramus S. J., and Gayther S. A., J. Pathol. 211(3), 286–295 (2007). 10.1002/path.2112 [DOI] [PubMed] [Google Scholar]
- Geisler J. P., Rose S. L., Geisler H. E. et al. , CME J. Gynecol. Oncol. 7, 25–28 (2002). [Google Scholar]
- Barry W. T., Kernagis D. N., Dressman H. K., Griffis R. J., Hunter J. D., Olson J. A., Marks J. R., Ginsburg G. S., Marcom P. K., Nevins J. R., Geradts J., and Datto M. B., J. Clin. Oncol. 28(13), 2198–2206 (2010). 10.1200/JCO.2009.26.7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubele M., Mattis A., Zitzelsberger H., Walch A., Kremer M., Hutzler P., Hofler H., and Werner M., Cancer Genet. Cytogenet. 110(2), 94–102 (1999). 10.1016/S0165-4608(98)00205-2 [DOI] [PubMed] [Google Scholar]
- Yasui H., Konishi E., Urasaki K., Yasukawa S., and Yanagisawa A., Gastric Cancer 9(3), 223–228 (2006). 10.1007/s10120-006-0386-3 [DOI] [PubMed] [Google Scholar]
- Bachtiary B., Boutros P. C., Pintilie M., Shi W., Bastianutto C., Li J. H., Schwock J., Zhang W., Penn L. Z., Jurisica I., Fyles A., and Liu F. F., Clin. Cancer Res. 12(19), 5632–5640 (2006). 10.1158/1078-0432.CCR-06-0357 [DOI] [PubMed] [Google Scholar]
- Cottu P. H., Asselah J., Lae M., Pierga J. Y., Dieras V., Mignot L., Sigal-Zafrani B., and Vincent-Salomon A., Ann. Oncol. 19(3), 595–597 (2008). 10.1093/annonc/mdn021 [DOI] [PubMed] [Google Scholar]
- Liu W., Laitinen S., Khan S., Vihinen M., Kowalski J., Yu G., Chen L., Ewing C. M., Eisenberger M. A., Carducci M. A., Nelson W. G., Yegnasubramanian S., Luo J., Wang Y., Xu J., Isaacs W. B., Visakorpi T., and Bova G. S., Nat. Med. 15(5), 559–565 (2009). 10.1038/nm.1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke S. L., Temple J., Macarthur S., Zahra M. A., Tan L. T., Crawford R. A., Ng C. K., Jimenez-Linan M., Sala E., and Brenton J. D., Br. J. Cancer 104(2), 361–368 (2011). 10.1038/sj.bjc.6605971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich J., Seidel C., Ebner R., and Kunz-Schughart L. A., Nat. Protoc. 4(3), 309–324 (2009). 10.1038/nprot.2008.226 [DOI] [PubMed] [Google Scholar]
- Kunz-Schughart L. A., Freyer J. P., Hofstaedter F., and Ebner R., J. Biomol. Screening 9(4), 273–285 (2004). 10.1177/1087057104265040 [DOI] [PubMed] [Google Scholar]
- Gottfried E., Kunz-Schughart L. A., Andreesen R., and Kreutz M., Cell Cycle 5(7), 691–695 (2006). 10.4161/cc.5.7.2624 [DOI] [PubMed] [Google Scholar]
- Kunz-Schughart L. A., Cell Biol. Int. 23(3), 157–161 (1999). 10.1006/cbir.1999.0384 [DOI] [PubMed] [Google Scholar]
- Seton-Rogers S., Nat. Rev. Cancer 11(8), 538–539 (2011). 10.1038/nrc3112 [DOI] [PubMed] [Google Scholar]
- Burleson K. M., Boente M. P., Pambuccian S. E., and Skubitz A. P., J. Transl. Med. 4, 6 (2006). 10.1186/1479-5876-4-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier L., Puiffe M. L., Le Page C., Filali-Mouhim A., Chevrette M., Tonin P. N., Provencher D. M., and Mes-Masson A. M., Transl. Oncol. 3(4), 230–238 (2010); available at http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2915414/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puiffe M. L., Le Page C., Filali-Mouhim A., Zietarska M., Ouellet V., Tonin P. N., Chevrette M., Provencher D. M., and Mes-Masson A. M., Neoplasia 9(10), 820–829 (2007). 10.1593/neo.07472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietarska M., Maugard C. M., Filali-Mouhim A., Alam-Fahmy M., Tonin P. N., Provencher D. M., and Mes-Masson A. M., Mol. Carcinog. 46(10), 872–885 (2007). 10.1002/mc.20315 [DOI] [PubMed] [Google Scholar]
- Yoffou P. H., Edjekouane L., Meunier L., Tremblay A., Provencher D. M., Mes-Masson A. M., and Carmona E., PLoS One 6(6), e20705 (2011). 10.1371/journal.pone.0020705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet V., Zietarska M., Portelance L., Lafontaine J., Madore J., Puiffe M. L., Arcand S. L., Shen Z., Hebert J., Tonin P. N., Provencher D. M., and Mes-Masson A. M., BMC Cancer 8, 152 (2008). 10.1186/1471-2407-8-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Page C., Puiffe M. L., Meunier L., Zietarska M., de Ladurantaye M., Tonin P. N., Provencher D., and Mes-Masson A. M., J. Ovarian Res. 2, 4 (2009). 10.1186/1757-2215-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T., Maiti T. K., and Chakraborty S., Integr. Biol. (Cambrigde) 3(6), 684–695 (2011). 10.1039/c1ib00001b [DOI] [PubMed] [Google Scholar]
- Xia Y., and Whitesides G. M., Soft Lithography, Annu. Rev. Mater. Sci. 28, 153–184 (1998). 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- Breuls R. G., Mol A., Petterson R., Oomens C. W., Baaijens F. P., and Bouten C. V., Tissue Eng. 9(2), 269–281 (2003). 10.1089/107632703764664738 [DOI] [PubMed] [Google Scholar]
- Agastin S., Giang U. B., Geng Y., Delouise L. A., and King M. R., Biomicrofluidics 5(2), 24110 (2011). 10.1063/1.3596530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto Y., Tan W. H., Tsuda Y., and Takeuchi S., Lab Chip 9(15), 2217–2223 (2009). 10.1039/b900035f [DOI] [PubMed] [Google Scholar]
- Hattori K., Sugiura S., and Kanamori T., Lab Chip 9(12), 1763–1772 (2009). 10.1039/b816995k [DOI] [PubMed] [Google Scholar]
- Bruus H., Theoretical Microfluidics (Oxford University Press, Oxford, 2008). [Google Scholar]
- Corey J. M., Gertz C. C., Sutton T. J., Chen Q., Mycek K. B., Wang B. S., Martin A. A., Johnson S. L., and Feldman E. L., J. Biomed. Mater. Res. A 93(2), 673–686 (2010). 10.1002/jbm.a.32485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Pjesivac-Grbovic J., Cantrell C., and Freyer J. P., Biophys. J. 89(6), 3884–3894 (2005). 10.1529/biophysj.105.060640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheate N. J., Walker S., Craig G. E., and Oun R., Dalton Trans. 39(35), 8113–8127 (2010). 10.1039/c0dt00292e [DOI] [PubMed] [Google Scholar]
- Roos W. P. and Kaina B., Trends Mol. Med. 12(9), 440–450 (2006). 10.1016/j.molmed.2006.07.007 [DOI] [PubMed] [Google Scholar]
- Wang T. H., Wang H. S., and Soong Y. K., Cancer 88(11), 2619–2628 (2000). [DOI] [PubMed] [Google Scholar]
- Agarwala S. S., Cano E., Heron D. E., Johnson J., Myers E., Sandulache V., Bahri S., Ferris R., Wang Y., and Argiris A., Ann. Oncol. 18(7), 1224–1229 (2007). 10.1093/annonc/mdm088 [DOI] [PubMed] [Google Scholar]
- Samouelian V., Maugard C. M., Jolicoeur M., Bertrand R., Arcand S. L., Tonin P. N., Provencher D. M., and Mes-Masson A. M., Cancer Chemother. Pharmacol. 54(6), 497–504 (2004). 10.1007/s00280-004-0843-9 [DOI] [PubMed] [Google Scholar]
- Lengyel E., Am. J. Pathol. 177(3), 1053–1064 (2010). 10.2353/ajpath.2010.100105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleson K. M., Casey R. C., Skubitz K. M., Pambuccian S. E., T. R.Oegema, Jr., and Skubitz A. P., Gynecol. Oncol. 93(1), 170–181 (2004). 10.1016/j.ygyno.2003.12.034 [DOI] [PubMed] [Google Scholar]
- Ivascu A. and Kubbies M., J. Biomol. Screen 11(8), 922–932 (2006). 10.1177/1087057106292763 [DOI] [PubMed] [Google Scholar]
- Wu L. Y., Di Carlo D., and Lee L. P., Biomed. Microdevices 10(2), 197–202 (2008). 10.1007/s10544-007-9125-8 [DOI] [PubMed] [Google Scholar]
- Okuyama T., Yamazoe H., Mochizuki N., Khademhosseini A., Suzuki H., and Fukuda J., J. Biosci. Bioeng. 110(5), 572–576 (2010). 10.1016/j.jbiosc.2010.05.013 [DOI] [PubMed] [Google Scholar]
- Hsiao A. Y., Torisawa Y. S., Tung Y. C., Sud S., Taichman R. S., Pienta K. J., and Takayama S., Biomaterials 30(16), 3020–3027 (2009). 10.1016/j.biomaterials.2009.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Kim C., Yang J. Y., Lee H., Ahn B., Xu L., Kang J. Y., and Oh K. W., Biomicrofluidics 6(1), 14114–141147 (2012). 10.1063/1.3687409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walenta S., Doetsch J., Mueller-Klieser W., and Kunz-Schughart L. A., J. Histochem. Cytochem. 48(4), 509–522 (2000). 10.1177/002215540004800409 [DOI] [PubMed] [Google Scholar]
- Hirschhaeuser F., Menne H., Dittfeld C., West J., Mueller-Klieser W., and Kunz-Schughart L. A., J. Biotechnol. 148(1), 3–15 (2010). 10.1016/j.jbiotec.2010.01.012 [DOI] [PubMed] [Google Scholar]
- Cody N. A., Zietarska M., Filali-Mouhim A., Provencher D. M., Mes-Masson A. M., and Tonin P. N., BMC Med. Genomics 1, 34 (2008). 10.1186/1755-8794-1-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschenker O., Streichert T., Hehlgans S., and Cordes N., PloS one 7(4), e34279 (2012). 10.1371/journal.pone.0034279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. C., Zhang H., Pallavicini M., Gray J. W., Baehner F., Park C. J., and Bissell M. J., Cancer Res. 66(3), 1526–1535 (2006). 10.1158/0008-5472.CAN-05-3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tredan O., Galmarini C. M., Patel K., and Tannock I. F., J. Natl. Cancer Inst. 99(19), 1441–1454 (2007). 10.1093/jnci/djm135 [DOI] [PubMed] [Google Scholar]
- Lang S. H., Stark M., Collins A., Paul A. B., Stower M. J., and Maitland N. J., Cell Growth Differ. 12(12), 631–640 (2001). [PubMed] [Google Scholar]
- Vinci M., Gowan S., Boxall F., Patterson L., Zimmermann M., Court W., Lomas C., Mendiola M., Hardisson D., and Eccles S. A., BMC Biol. 10, 29 (2012). 10.1038/nrclinonc.2011.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoslahti E., Bhatia S. N., and Sailor M. J., J. Cell Biol. 188(6), 759–768 (2010). 10.1083/jcb.200910104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson V., Hilmarsdottir B., Sigmundsdottir H., Fridriksdottir A. J., Ringner M., Villadsen R., Borg A., Agnarsson B. A., Petersen O. W., Magnusson M. K., and Gudjonsson T., PloS one 6(9), e23833 (2011). 10.1371/journal.pone.0023833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R. K., Science 307(5706), 58–62 (2005). 10.1126/science.1104819 [DOI] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg R. A., Cell 144(5), 646–674 (2011). 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Kimball R. E., Schlaerth J. B., Kute T. E., Schlaerth A. C., Santoso J., Ballon S. C., and Spirtos N. M., Am. J. Obstet. Gynecol. 176(6), 1319–1327 (1997); discussion 1326–1327. 10.1016/S0002-9378(97)70352-1 [DOI] [PubMed] [Google Scholar]
- Sevin B. U. and Perras J. P., Am. J. Obstet. Gynecol. 176(4), 759–768 (1997); discussion 766–768. 10.1016/S0002-9378(97)70599-4 [DOI] [PubMed] [Google Scholar]
- Takeshima Y., Amatya V. J., Daimaru Y., Nakayori F., Nakano T., and Inai K., Hum. Pathol. 32(11), 1203–1208 (2001). 10.1053/hupa.2001.28956 [DOI] [PubMed] [Google Scholar]
- Capretto L., Cheng W., Hill M., and Zhang X., Top. Curr. Chem. 304, 27–68 (2011). 10.1007/128_2011_150 [DOI] [PubMed] [Google Scholar]