Abstract
The present work demonstrates the use of a dielectrophoretic lab-on-a-chip device in effectively separating different cancer cells of epithelial origin for application in circulating tumor cell (CTC) identification. This study uses dielectrophoresis (DEP) to distinguish and separate MCF-7 human breast cancer cells from HCT-116 colorectal cancer cells. The DEP responses for each cell type were measured against AC electrical frequency changes in solutions of varying conductivities. Increasing the conductivity of the suspension directly correlated with an increasing frequency value for the first cross-over (no DEP force) point in the DEP spectra. Differences in the cross-over frequency for each cell type were leveraged to determine a frequency at which the two types of cell could be separated through DEP forces. Under a particular medium conductivity, different types of cells could have different DEP behaviors in a very narrow AC frequency band, demonstrating a high specificity of DEP. Using a microfluidic DEP sorter with optically transparent electrodes, MCF-7 and HCT-116 cells were successfully separated from each other under a 3.2 MHz frequency in a 0.1X PBS solution. Further experiments were conducted to characterize the separation efficiency (enrichment factor) by changing experimental parameters (AC frequency, voltage, and flow rate). This work has shown the high specificity of the described DEP cell sorter for distinguishing cells with similar characteristics for potential diagnostic applications through CTC enrichment.
INTRODUCTION
Malignant tumors are characterized by the migration of tumor cells (metastasis) into distant regions of the body during disease progression. Patients with metastatic cancer have significantly worse prognoses with very low survival rates. The survival rate for patients with a localized breast cancer is estimated to be 98.6%.1 In fact, even when the disease has spread to regional lymph nodes, the survival rate is relatively high at 83.8%. However, after the spread of the disease into distant organs during metastasis, the survival rate plummets to 23.4%.1 Metastasis begins as the cancer cells detach from the initial growth site. Tumor angiogenesis, growth of new vasculature, provides a route for cancerous cells to enter the blood stream and lymph fluid, facilitating transport to distant organs.2 The early seeding of cancer cells in distant organs contributes to metastatic relapse which is difficult to detect even with a range of diagnostic tools.3 After surgery, approximately 50% of patients come out of remission due to undetected metastasis.4 The main cause of secondary tumor growth is single or clustered cancer cells traveling through the blood stream. The number of circulating tumor cells (CTCs) is expected to be low in the early stages of the cancer, on the order of 1 cell/ml.5, 6, 7 Therefore, very sensitive monitoring equipment must be used to ensure early detection and aggressive treatment.
Current diagnostic techniques for tumor metastasis include imaging studies (computerized and magnetic resonance tomography) or serum marker assays (carcinoma antigen 15.3 and carcinoembryonic antigen).8 However, such detection methods result in a high rate of false negatives as they lack the specificity to detect signals at such small orders of magnitude. Another method of detecting metastasis directly is the detection and analysis of CTC which offers a promising alternative to invasive biopsies.9, 10, 11, 12, 13 CTC detection is a diagnostic model that has been extensively investigated. For example, the FDA-approved CellSearch® detects CTCs by targeting cell markers, such as EpCAM and Cytokeratins (CK) 8, 18, and 19.14 Studies have previously analyzed the efficacy of CK+ tests and found a direct correlation between the number of CK+ cells and the prognosis of a breast cancer patient.15 CellSearch has been shown to successfully isolate cells using ferrofluid nanoparticles at a sensitivity of 1 cell per 7.5 ml of blood.14 While these results are promising, the utilized biomarkers are general to most tumors. Therefore, it cannot identify the type and origin of that tumor cell or differentiate it from other CTC's that might simultaneously flow in the blood. The possibility of various types of CTCs being present requires a more specific identification of the cells. This step is important for patients faced with the spread of disseminated tumor cells after resection of the solid tumor.16 Furthermore, some markers used by CellSearch like CK19, EGF receptor, and mammaglobin have been shown to be expressed in normal mitogen-stimulated peripheral blood mononuclear cells.17
An alternative paradigm for CTC detection is through leveraging differences in physical characteristics of tumor cells, such as size, charge, density, and biomarker expression which can ultimately lead to target-cell isolation. For instance, the herringbone-chip developed by Mehemt Toner et al.18 utilizes antibody-biomarker binding to capture CTCs from the blood. While it has been demonstrated that this approach is highly effective in capturing the general CTC population, it lacks the ability to differentiate between CTCs with similar biomarker expression such as breast and colon cancers. For example, breast cancer (MCF-7) and colon cancer (HCT 116) cells have an upregulation of MMP-9, while genitourinary tract, lung, and thyroid cancers do not.19 Additionally, both forms of cancer have an increased expression of FAK and CR-1.20, 21 In addition to expressing similar biomarkers, morphologically both MCF-7 (Ref. 22) and HCT 116 (Ref. 23) have a dense network of microvilli on their surface.
A developing technology for CTC enrichment is dielectrophoresis (DEP)24 in a microfluidics device, which offers solutions to many of the shortcomings previously described.25, 26 DEP allows for the directed movement of polarizable particles. Because mammalian cells in suspension meet this criterion, it is possible to use DEP to manipulate cell movement. DEP has been used for enriching small populations of cells from cell mixtures,27 cell trapping in nuclear fusion11 and even progenitor-daughter cell characterization.12 For biological cells, the DEP-induced behavior is governed by surface morphology, extracellular biomarkers, and intracellular events.28, 29 This work shows the potential of this technique in isolating CTCs from multiple origins in the same sample and identifying each origin.
The techniques previously mentioned are effective in identifying the presence of metastatic events; however, they have a difficulty in providing information for locating the origin of the cancerous mass. Recognition of different cancer cell types can provide helpful insight for clinical treatment, since we can identify the origin and degree of metastasis in CTCs. If there are multiple types of cancer cells, it could be difficult to separate them since most cancer cells are epithelial and have common biomarkers, similar sizes, and morphologies. While many types of cancer cells have been isolated from noncancerous cells, the isolation of a specific type of cancer cell from other cancer cells using DEP has, to our knowledge, not been explicitly shown.
To characterize isolation and separation of different types of cells, fluorescent dyes are often used in post-experimental analysis.28 For instance, DEP isolation techniques (e.g., DEP-FFF and HLDEP-FFF) are evaluated through staining the elute of post-separation or RT-PCR,30, 31 and can, therefore, be more expensive and tedious. In contrast, another study used dyes to examine the effects of DEP on cells during or after the experiment but lacked evidence that the DEP behavior was unaltered as a consequence of the dye.32
The objective of this work is to explore highly specific separation of two different types of cancer cells (breast cancer cell MCF-7 and colorectal cancer cell HCT-116) and the characterization of separation efficiency (isolation and enrichment) for future application of CTC detection in whole blood. Cells are labeled for monitoring the separation of the two type cancer cells under the case where the labeling will have almost no effect for separation. The present DEP technique reports a highly specific method for separation of two types of cancer cells and represents a promising method for the noninvasive and label free detection of pre-metastatic and metastatic cancers through the isolation of different types of CTCs from the blood.
Principle of the method
DEP is the phenomenon of controlled particle movement as the result of an applied nonuniform electric field, which induces a dipole moment on the particle due to the electrical polarization at the particle's membrane with the surrounding solution.33, 34, 35, 36 Subsequently, the particle is translated in the field to achieve electrostatic equilibrium. This movement results from a DEP force.
In the low frequency region (roughly lower than 10 MHz), cell DEP behavior is largely determined by extracellular factors, including membrane-bound proteins, cell size, solution conductivity, and electric permittivity. The DEP behavior of the particles is dominated by the difference between the particle conductivity and that of the surrounding medium. In this region, the DEP force is described as37
| (1) |
where Re, ε, σ, a and Erms are real part, permittivity, conductivity, particle radius, and electric field, respectively, and the subscript m and p denote the medium and particle values. The vacuum permittivity is ε0. In the high frequency region (higher than 10 MHz), DEP is under the effects of the particle and medium dielectric properties. Thus, the DEP behavior of the particles is dominated by the difference between the particle permittivity and that of the surrounding medium. In this dielectric region, the DEP force is shown as
| (2) |
DEP force on a particle is dependent upon particle radius a, the dielectric constant of the medium εm, and the electric field Erms. With a constant electric field, applied on a medium with a defined dielectric constant, particles with slightly different radii or dielectric properties, will exhibit different DEP behaviors. Selective isolation of a specific particle can be achieved by applying an additional non-electrical force into the system.
In the low frequency region, the conductivity terms compose what is referred to as the Claussius-Mossotti factor, K(ω)38
| (3) |
where σm is the medium conductivity and σp is the particle conductivity. Positive DEP (pDEP) generally occurs when K(ω) > 1, which means the polarizability of the particle is larger than the suspending medium, inducing movement towards the regions of high electric field. In contrast, negative DEP (nDEP) occurs when the polarizability of the particle is smaller than the suspending medium, inducing movement towards the regions of low electric field (away from the electrode) or K(ω) < 1.39
Our strategy for separating different types of cancer cells is to find an AC frequency, where the target cells experience nDEP, while the other type cells undergo almost no DEP force, i.e., under the first cross-over frequency (where the DEP force becomes zero). However, for a given media conductivity, the two types of cells may have the cross-over frequency close to each other, and the nDEP force difference of the target cells and other cells present in a mixture can be very small, and hence, it is difficult to separate them. However, the difference between the cross-over frequencies of the two types of cells can be increased by changing the media conductivity.
The relationship between the first cross-over frequency, particle radius, membrane capacitance, and solution conductivity can be described by Eq. 4. The value of this cross-over frequency is directly proportional to the solution conductivity. As the conductivity increases, the DEP spectra should shift to the right (i.e., increasing frequency).40
| (4) |
where Cm is capacitance of the cell membrane. Since MCF-7 and HCT-116 should have different values of Cm, their difference in fx01 can be increased by changing the media conductivity. A nDEP-based cell separation device was designed on the principle that a DEP force and an applied hydrodynamic force can be balanced to direct particle movement through microchannels, as shown in Fig. 1.
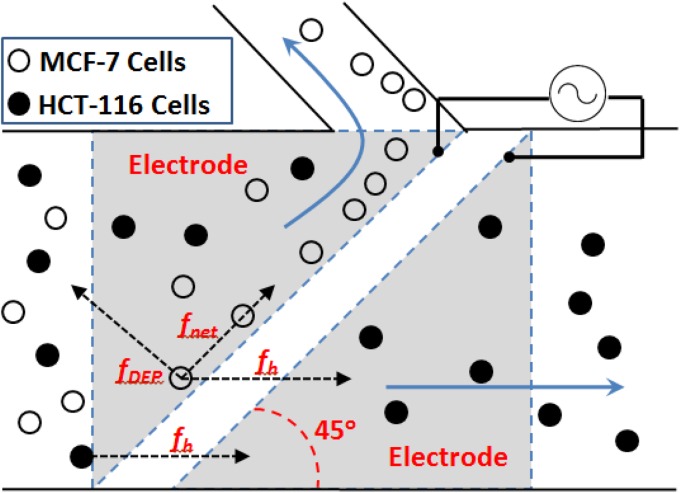
Figure 1.
Schematic of the DEP cell sorter. One particle is represented as a free-body diagram with the hydrodynamic, DEP force, and resultant force vectors. Arrows indicate flow through the main channel (80% infusion rate) and through the side channel (20% infusion rate), respectively.
This apparatus consists of a main channel for cell mixtures and a side channel for collecting target cells. Two electrodes on the bottom surface of the microchannel are aligned 45° to the main channel with an AC electric signal applied to both electrodes at a phase difference of 180°. As the target particle approaches the gap between the electrodes, this particle will experience a DEP force. In this system, appropriate voltage and frequency are chosen to induce nDEP force against the hydrodynamic force at an angle, resulting in a net force parallel to the electrodes. The hydrodynamic force is defined as41
| (5) |
The net force of DEP (fDEP) and hydrodynamic force (fh) repels the target particles to the side channel, which is described by
| (6) |
The chip design allows 80% of fluid to flow through the main channel and 20% fluid to flow to the side channel. Other particles in the mixture (non-target cells) should experience either no DEP force or a minimal nDEP or pDEP force which is easily overcome by the hydrodynamic force, allowing convective transport through the main channel and eventual elution. The core principle of this isolation technique is to determine the optimal frequency at which the target cell's fDEP is relatively high (fDEP > fh1), while the fDEP of the non-target cell is minimal (fDEP ≈ 0).
MATERIALS AND EXPERIMENTAL DESIGN
Sample preparation
The human breast cancer cell line MCF-7 (ATCCTM) was the target cell type (to be isolated from main stream of sample), while the human colorectal cancer cell line HCT-116 (ATCCTM) served as the non-target cell. Both cell lines were cultured in a 10% fetal bovine serum (FBS), Mediatech Celllgro RPMI 1640 media (Fischer Scientific) with L-glutamine, and penicillin-streptomycin. The cells were incubated for 3 days between each passage at 37 °C and 5% CO2. Trypsin was used to detach cells from their flasks. Prior to DEP spectral measurements, cells were suspended in solutions of various PBS concentration levels (0.025X, 0.05X, and 0.1X) to determine the correlation between the solution conductivity and characteristic DEP behavior (i.e., spectra). Experiments were run within two hours of receiving the fresh cells.
Hoechst 33342 (InvitrogenTM), a positively charged, fluorescent, nuclear dye was used to quantify cell separation. The dye was used to label the HCT-116 cells which were suspended in a 0.1X PBS. This simplified the visual identification of HCT and MCF cells during and after separation experiments. The dye was selected only for visualization in contrast with other studies which employ alternate dyes for the post-experiment analysis.28
Microfabrication of microfluidic cell sorter
The separation chip was made using lamination microfabrication. A transparent acrylic plastic substrate of 24 mm × 20 mm × 1.25 mm was used as the top (cap) layer. One inlet and two outlet wells (one for the main channel, and the other for the side channel) with a diameter of 1.6 mm were drilled on the top layer. Channels were carved on a middle layer with a height of 40 μm. The widths of the main and side channels were 550 μm and 150 μm, respectively. Indium tin oxide (ITO) thin film electrodes were placed on a glass substrate with a gap of 20 μm. ITO was chosen because of its unique property to be optically transparent (to facilitate visualization through microscopy) yet electrically conductive. Finally, pressure-sensitive adhesive was used to bond all the layers together and prevent any leakage of solution. These materials were selected to maximize cost-efficiency and quality while minimizing cell-material interactions which could confound experimental results.
Experimental setup
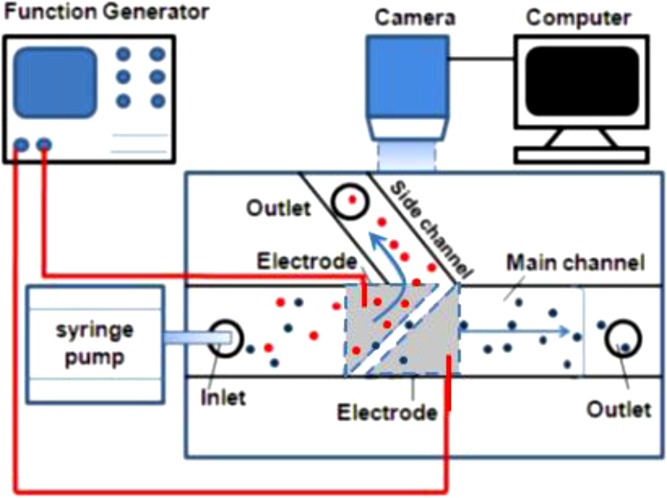
A schematic of the setup used for the DEP separation is shown in Fig. 2. The DEP cell sorter was placed on the test bed of an inverted epi-fluorescent microscope (Olympus-IX70). A syringe pump (Harvard PHD 2000) was used to deliver the sample to the chip. A function generator (Tektronix, Model AFG3102) was used to supply AC electrical signal to the two ITO electrodes. A 10X objective lens (NA = 0.25) was used for magnification of the image. The cell motion was captured by a high-resolution CCD camera (SensiCam-QE, Cooke Corp.).
Figure 2.
Schematic of experimental setup for conducting the separation of cell mixtures.
EXPERIMENTAL RESULTS AND DISCUSSION
DEP spectra of MCF-7 and HCT-116
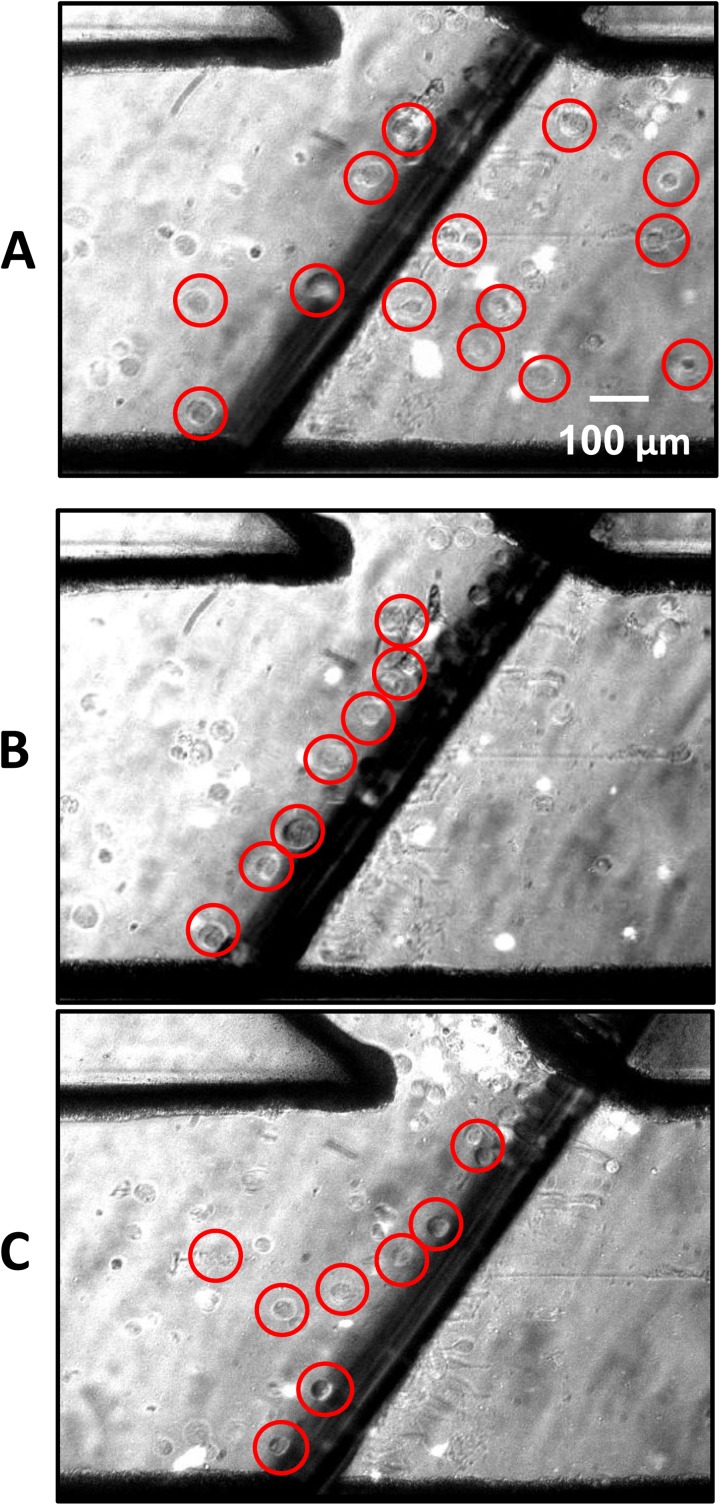
Each type of cell was studied separately to determine the relationship between AC electric field frequency, solution conductivity, and the cell's unique DEP properties. In particular, the low frequency region was of special interest. This was performed to determine the frequencies and the medium conductivity at which the two types of cell experienced sufficiently different DEP forces. The DEP characteristics of the MCF-7 and HCT-116 cells were studied using a simple wedge microchip previously described by Yang et al.27 Fig. 3 shows an example of DEP spectrum measurement using a wedge microchip. Cell motion was monitored immediately after the AC power was supplied. The cells begin at rest and, after applying the electric current, the motion of the cell is correlated with the DEP force. The circled cells shown in Fig. 3 indicate the motion of the cells due to DEP force. The cells experienced an nDEP force and moved away from the center, where the electric field was highest.
Figure 3.

MCF-7 displacement due to negative DEP force, highlighted cells shown at differing times after AC current was applied to the two electrodes shown as the two dark areas. (a) Cells position right prior to application of current. (b) Cells moved away from electrode tips 2 s after the electrode activation. (c) Cells continuing negative DEP path at 4 s.
The DEP force was quantified in terms of cellular acceleration, which was in turn determined by the DEP velocity (displacement of the cells per number of frames). The cell suspension was loaded into the wedge chip and the cells were allowed to come to rest. Because a DEP spectrum is influenced by the medium conductivity, the DEP spectra were measured for both cell types in three different PBS concentrations: 0.025X, 0.05X, and 0.1X PBS. These concentrations correspond to conductivities of 600 μS/cm, 1000 μS/cm, and 3000 μS/cm, respectively. As previously discussed, in the low frequency regime, DEP behavior is highly dependent upon medium conductivity. The purpose of quantifying the spectra for each cell type in media with three different conductivities was to determine the optimal medium conductivity for relevant differences in DEP behaviors.
Fig. 4 shows the DEP spectra. Fig. 4a shows the DEP spectra for HCT-116 in the three media. Data of special interest are the first cross-over frequency in the low frequency region. These experimental results support the predicted trend from Eq. 4 as there is a direct relationship between increasing conductivity and increasing the first cross-over frequency. In Fig. 4b, the same trend is evident for the MCF-7 cells as the conductivity is increased. (Note, due to their complexity of the DEP spectra, placing all spectra of MCF-7 and HCT116 cells into one figure could make it difficult to read.) In both the 0.025X and 0.05X PBS media, both cell types have extremely close values for the first cross-over frequency: 0.2 MHz and 0.5 MHz, respectively. These similar frequency values would make separation of different types of cells difficult. In contrast, the 0.1X PBS medium presents a scenario where the first cross-over frequency for HCT-116 is approximately 4 MHz, while MCF-7's is at 10 MHz. It was posited that increased conductivity amplified the difference in the cross-over frequency values.
Figure 4.
DEP spectra. (a) HCT116 DEP spectra for solutions of three different conductivities across the frequency range of 0.1 MHz to 100 MHz, and HCT-116 spectra with dyed (Hoechst 33342) samples at 0.1X PBS. In both cases (without and with dye), they have their first cross-over frequency at virtually the same value. (b) MCF-7 DEP spectra for solutions of three different conductivities across the frequency range of 0.1 MHz to 100 MHz. The dashed line intersects at 3.2 MHz, where the cell separation was most efficient.
Figure 4a also compares the DEP spectra for undyed and dyed HCT 116 cells with Hoechst 33342, in 0.1X PBS media. There are slight differences in DEP force in the region prior to the cross-over frequency. Despite these low frequency variances, the dyed and undyed cells have their first cross-over point at approximately the same frequency, suggesting a similar behavior at that point during the experiment. The difference in behaviors at higher frequency regions was not relevant to this study. Having confirmed that the spectra of the HCT-116 dyed and undyed cells were similar near the first cross-over frequency, the 0.1X PBS solution was used for separating HCT 116 and MCF-7 cells.
In the frequency region of 3–4 MHz, the DEP force of HCT-116 was nearly zero, but MCF-7 experienced nDEP force, providing an almost ideal condition for separation of these two cell types. The region from 6–10 MHz was excluded because the pDEP force experienced by the HCT-116 would lead to cell aggregation along the electrode, blocking flow through the main channel.
Separation of MCF-7 cells and HCT116 cells in the DEP cell sorter
The dyed HCT 116 cells were mixed with MCF-7 in a 0.1X PBS solution and the cell mixture was pumped into the cell sorter. The fluid flowed from the inlet to the outlet at the main channel (80% of the inlet flow rate) and the side channel (20% of the inlet flow rate) with a total inlet flow rate of 0.15 μl/min. The flowing cell mixture was visualized as the frequency was adjusted from 1-5 MHz to determine which frequency repelled the most MCF-7 cells into the side channel while allowing HCT-116 cells to continue flow through the main channel. The most efficient frequency in this case was 3.2 MHz. An example of separation is in Fig. 5 which shows the images of the separation of the MCF-7 cells from the HCT-116. The cells which appear bright are labeled HCT-116 cells while the darker cells are MCF-7.
Figure 5.
Separation of MCF-7 (encircled) and HCT-116 (fluorescent). The circles are used to mark MCF-7. (a) Shows flow of HCT-116 and MCF-6 through the main channel prior to activation of the AC electric field. (b) Reflects flow through the channel after activation at 3.2 MHz where undyed cells (MCF-7) are aligned parallel to the electrode (dark region) and are being deflected into the side channel while the HCT-116 path is unaltered. (c) Shows a low specificity scenario of separation between the MCF-7 and HCT-116 with both being deflected into the side channel under a frequency of 2 MHz.
Without AC activation of the electrodes, the MCF-7 cells moved together with HCT-116 along the main channel and passed through the electrode pair from left to right, with no separation of the MCF-7 from the HCT-116 as shown in Fig. 5a. However, after the power source was turned on, nDEP force repelled the MCF-7 cells towards the side channel and aligned them parallel to the electrode pair, as shown in Fig. 5b. There are virtually no gray MCF-7 cells downstream of the electrode pair. Meanwhile, the HCT-116 cells experienced no DEP force and remained flowing through the main channel. The MCF-7 cells and HCT-116 cells were separated and MCF-7 cells were enriched in the side channel. In addition to separating the MCF-7 cells, it was also essential that only a small number of HCT-116 cells were deflected into the side channel due to hydrodynamic force.
This separation system is very sensitive. Small changes in AC frequency will significantly change the results. This can be seen in Fig. 5c where the AC frequency is 2.0 MHz, and both the MCF-7 and HCT-116 cells can be seen repelling towards the side channel. Both cell types experienced nDEP force, preventing their separation. Fig. 5 indicates that DEP is highly specific in the separation of cells and such a separation and isolation under particular conditions (media conductivity and AC frequency) could serve as a new “biomarker” for a target cell.
Due to their numerous morphological and biological similarities, MCF-7 cell and HCT-116 cell have similar DEP spectra as shown in Figs. 45c. However, since each cell type has its unique dielectric property, one cell type can have a distinct response to the change of environment (such as media conductivity). Leveraging such a subtle difference, it is possible to increase the difference in DEP response of two similar cell types by changing their media conductivity to achieve different cross-over frequencies for the two cell types.
Quantity analysis of cell separation
Videos of the experiments were analyzed to determine the sorter performance using a metric termed: cellular enrichment factor. This is defined as the ratio of target cells deflected towards the side channel to the total number of target cells flowing through the inlet.42
| (7) |
where A is the number of target cells deflected into the side channel and B is the number of the target cells flowing through the main channel. The enrichment factor was calculated by analyzing the experimental videos in 1 min segments. One minute allowed for over 200 cells to pass through the channel. Enrichment factors calculated from each segment were averaged.
Frequency effect
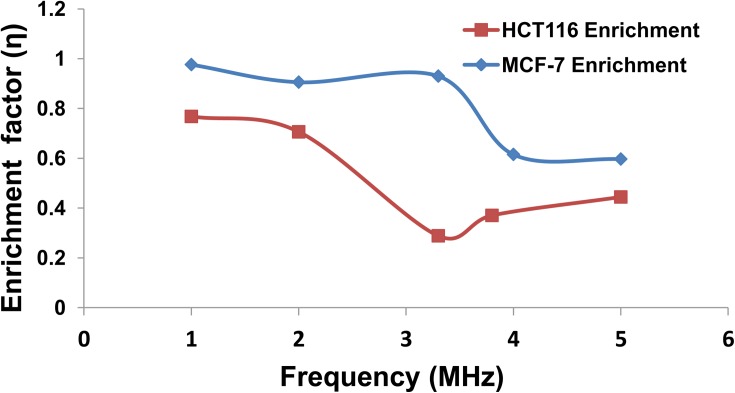
The enrichment factor was used to (1) determine the optimal frequency of separation at which there is low enrichment of HCT-116 while retaining high enrichment of MCF-7 as shown in Fig. 6; and (2) demonstrate the high specificity of DEP for cell separation. The enrichment was measured from a frequency of 1 MHz to approximately 5 MHz. It was determined that the region of highest enrichment (where η = 1) for MCF-7 was between 1.0 and 3.2 MHz, while the region of highest enrichment for HCT-116 was between 1.0 and 2.0 MHz. In the region of 2.0-3.2 MHz, η of HCT-116 decreased rapidly to 0.25 due to the fast decrease of nDEP force. However, beyond 3.2 MHz, the enrichment factor of HCT-116 increased again rapidly. For instance, at 5 MHz, η of HCT-116 reached to approximately 0.45. It is only near 3.2 MHz that the difference in the enrichment factor between the MCF-7 and HCT-116 reached its maximum. The difference in enrichment at 3.2 MHz for each cell type is shown in Figure 6. This supports our selection of 3.2 MHz as the most efficient separation frequency.
Figure 6.
Relation between the enrichment factor and frequency for MCF-7 and HCT-116. At 3.2 MHz, the enrichment factor for MCF-7 and HCT116 is 93% and 28%, respectively. The relationship confirms the high specificity and optimum frequency of 3.2 MHz for the cell separation.
Voltage effect
The enrichment factor was measured as a function of voltage under different flow rates. The voltage ranged from 1 Vpp to 16 Vpp as shown in Fig. 7 under three flow rates (0.1 μl/min, 0.2 μl/min, and 0.3 μl/min) at a frequency of 3.2 MHz for analysis. From inspection of Eq. 1, it is clear that DEP force increases with increased gradient of the squared electric field, therefore η should be positively correlated with the magnitude of the electric field E. E is equal to the ratio of voltage to distance between electrodes. In this system, the distance between the electrodes remains constant, therefore increasing the applied voltage will directly increase the magnitude of E, and η. Fig. 7 shows that higher voltages have higher enrichments; however, it quickly reaches a limit. At a flow rate of 0.1 μl/min, enrichment factor increases but begins to plateau around 8 Vpp to a value of 0.93. Similarly, the 0.2 μl/min flow rate plateaus around the same enrichment factor, but reaches saturation at about 12 Vpp. Fig. 7 indicates that increasing voltage is necessary and more efficient for separation cells, when the flow rate is high.
Figure 7.
Relationship between the enrichment factor and voltage for MCF-7 under different flow rates. Voltage and flow rate were modified to optimize the process and to show how the efficiency of the system was impacted by changing these parameters.
Flow rate effect
The flow rate effect is also displayed in Fig. 7 at each given voltage. As the flow rate was increased, the hydrodynamic force increased, and the DEP force became relatively weak and the enrichment factor decreased. At low flow rates, the hydrodynamic force is small, and the low voltages can generate sufficient DEP force to isolate the MCF-7 from the solution. For instance, at a voltage of 12 Vpp, the enrichment factor was 0.93 for the flow rate of 0.1 μl/min. However, increasing the flow rate to 0.3 μl/min yielded η of 0.8 at the same voltage of 12 Vpp. At relatively low and high voltages, the flow rate effect was relatively weak, but at other values, the flow rate strongly influenced enrichment values. At low voltages, the DEP force was too low to repel the cells. At high voltages, the DEP force overwhelmed the hydrodynamic force and dominated the cell motion. In the middle range of the voltage, however, the DEP force and hydrodynamic force competed with each other and increasing the flow rate would increase the hydrodynamic force and decrease the enrichment factor, Therefore, in this region, the enrichment factor was very sensitive to the flow rate as displayed in Fig. 7.
The presence of a maximum enrichment factor less than one is related to the nature of the designed sorter. The width of the main channel limits the enrichment factor of the target cell as cells furthest from the side channel are deflected from the wall which has a minimal velocity (∼0 under no-slip condition) to the centerline of the channel where velocity is the highest. After this deflection has occurred, there is insufficient time for continued deflection in to the side channel. Future work will focus on optimizing channel width to cell size to mitigate this issue.
Potential application in cancer research and diagnostics
In cancer research and diagnostics, label-free isolation and separation of cells is highly useful. It is especially useful if the separation method facilitates the retrieval of target live cells for downstream analysis. It is only due to recent progress in microfluidic technology that has opened a new opportunity for bioscience, that DEP has become a popular technique for biological applications, although it was first described by Pohl as early as 1951.30, 43 DEP is frequently used as a label-free method in particle separation and isolation since DEP events are specifically dictated by inherent particle and media properties.38, 44, 45, 46
For biological cells, the DEP-induced behavior is governed by surface morphology, extracellular proteins, and intracellular events. This DEP property has been leveraged to separate and enrich cancer cells from other types of cells. For instance, breast CTCs have been successfully isolated from human blood by exploiting this cell type's unique DEP behavior as compared to hematic components (e.g., erythrocytes and T-lympohocytes).47 However, separation of two different types of cancer cells using DEP has not been studied until this work. For CTC application, high sample throughput and target cell purity of the enriched sample is required. This can be realized using a cascaded and staggered DEP sorter (demonstrated by this group) for increasing target cell purity and throughput.42
Additionally, enriched target cells remain unaltered and can be easily cultured for further testing and analysis. Providing an improved method for early detection of pre-metastatic tumor is an important clinical goal to achieve because progression of metastatic cancer rapidly decreases survival rates. Also, providing a means to determine the origin of CTCs can assist clinicians in targeted treatment and eradication of malignant growths.
This work presents another example of the specificity and power of DEP coupled with microfluidics to isolate biological cells based upon intrinsic properties. The wide-range of existing and potential applications for this technology provides the opportunity for future cancer-cancer separations. This work may potentially be expanded to create an entire database of DEP spectra for different cancer types, providing the scientific community with a new standard for characterizing CTCs through the use of DEP as a new biomarker.
CONCLUSION
This work demonstrates the ability of a contactless, label free, highly specific, DEP-based, microfluidic separation system to isolate cancerous epithelial cells of one type (MCF-7) from another similar cell type (HCT-116). Additionally, the performance of the DEP sorter for separating MCF-7 from HCT-116 was characterized by measuring the relationship between the enrichment factor and operational parameters, such as AC frequency, voltage, and flow rate attaining enrichment efficiencies as high as 93% (0.1 μl/min, 9 Vpp, 3.2 MHz). The results of this work show the DEP cell sorter as a potentially label free, real-time, cost-effective option for cancer-cancer cell separation with high specificity in cancer research and diagnosis of CTC.
ACKNOWLEDGMENTS
This project was funded by the Magellan Scholars Program at the University of South Carolina. It was also partially supported by NIH Grant Nos. RR 017698 and P20 RR-016461, respectively, from the National Center for Research Resources.
References
- Howlader N., Noone A. M., Krapcho M., Neyman N., Aminou R., Waldron W., Altekruse S. F., Kosary C. L., Ruhl J., Tatalovich Z., Cho H., Mariotto A., Eisner M. P., Lewis D. R., Chen H. S., Feuer E. J., Cronin K. A., and Edwards B. K., editors, SEER Cancer Statistics Review, 1975–2008, National Cancer Institute, Bethesda, MD, 2011. [Google Scholar]
- Jiang W., Martin T., and Mansel R., Crit. Rev. Oncol. Hematol. 43, 13–31 (2002). 10.1016/S1040-8428(01)00181-0 [DOI] [PubMed] [Google Scholar]
- Muller V. and Pantel K., Am. J. Cancer 2, 77–86 (2003). 10.2165/00024669-200302020-00001 [DOI] [Google Scholar]
- Diel I., Kaufmann M., Goerner R., Costa S., Kaul S., and Bastert G., J. Clin. Oncol. 10, 1534–1539 (1992). [DOI] [PubMed] [Google Scholar]
- Lucci A., Hall C. S., Lodhi A. K., Bhattacharyya A., Anderson A. E., Xiao L., Bedrosian I., Kuerer H. M., and Krishnamurthy S., Lancet Oncol. 13(7), 688–695 (2012). 10.1016/S1470-2045(12)70209-7 [DOI] [PubMed] [Google Scholar]
- Braun S. and Pantel K., Oncologist 6, 125–132 (2001). 10.1634/theoncologist.6-2-125 [DOI] [PubMed] [Google Scholar]
- Zieglschmid V., Hollmann C., and Bocher O., Crit. Rev. Clin. Lab. Sci. 42, 155–196 (2005). 10.1080/10408360590913696 [DOI] [PubMed] [Google Scholar]
- Gradilone A., Gazzaniga P., Silvestri I., Gandini O., Trasatti L., Lauro S., Frati L., and Agliano A., Oncol. Rep. 10, 217–222 (2003). [PubMed] [Google Scholar]
- Smerage J. B. and Hayes D. F., Br. J. Cancer 94, 8–12 (2006). 10.1038/sj.bjc.6602871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clow A. L., Gaynor P. T., and Oback B. J., Biomed. Microdevices 12, 777–786 (2010). 10.1007/s10544-010-9432-3 [DOI] [PubMed] [Google Scholar]
- Flanagan L., Lu J., Wang L., Marchenko S., Jeon N., Lee A., and Monuki E., Stem Cells 26, 656–665 (2008). 10.1634/stemcells.2007-0810 [DOI] [PubMed] [Google Scholar]
- Rolle A., Günzel R., Pachmann U., Willen B., Höffken K., and Pachmann K., World J. Surg. Oncol. 3, 18 (2005). 10.1186/1477-7819-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormally E., Caboux E., Vineis P., and Hainaut P., Mutat. Res./Rev. Mutat. Res. 635, 105–117 (2007). 10.1016/j.mrrev.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Farace F., Massard C., Vimond N., Drusch F., Jacques N., Billiot F., Laplanche A., Chauchereau A., Lacroix L., Planchard D., Le Moulec S., Andre F., Fizazi K., Soria J. C., and Vielh P., Br. J. Cancer 105, 847–853 (2011). 10.1038/bjc.2011.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaforio J. J., Serrano M. J., Sanchez-Rovira P., Sirvent A., Delgado-Rodriguez M., Campos M., de la Torre N., Algarra I., Duenas R., and Lozano A., Int. J. Cancer 107(6), 984–990 (2003). 10.1002/ijc.11479 [DOI] [PubMed] [Google Scholar]
- Zhong X. Y., Dtsch. Med. Wochenschr. 125(27), 840–844 (2000). 10.1055/s-2000-7012 [DOI] [PubMed] [Google Scholar]
- Armakolas A., Panteleakou Z., Nezos A., Tsouma A., Skondra M., Lembessis P., Pissimissis N., and Koutsilieris M., Future Oncol. 6(12), 1849–1856 (2010). 10.2217/fon.10.152 [DOI] [PubMed] [Google Scholar]
- Stott S. L., Hsua C.-H., Tsukrova D. I., Yud M., Miyamotod D. T., Waltmand B. A., Rothenbergd S. M., Shaha A. M., Smas M. E., Korir G. K., Frederick J. Floyd P., Gilman A. J., Lord J. B., Winokur D., Springer S., Irimia D., Nagrath S., Sequist L. V., Lee R. J., Isselbacher K. J., Maheswaran S., Haber D. A., and Toner M., Proc. Natl. Acad. Sci. U.S.A. 107(43), 18392–18397 (2010). 10.1073/pnas.1012539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker S., Lysik R., Zarrabi M., and Moll U., Cancer Res. 53, 140–146 (1993). [PubMed] [Google Scholar]
- Bianco C., Strizzi L., Mancino M., Rehman A., Hamada S., Watanabe K., De Luca A., Jones B., Balogh G., Russo J., Mailo D., Palaia R., D'Aiuto G., Botti G., Perrone F., Salomon D., and Normanno N., Clin. Cancer Res. 12, 5158–5164 (2006). 10.1158/1078-0432.CCR-06-0274 [DOI] [PubMed] [Google Scholar]
- Cance W., Harris J., Iacocca M., Roche E., Yang X., Chang J., Simkins S., and Xu L., Clin. Cancer Res. 6, 2417–2423 (2000). [PubMed] [Google Scholar]
- Hill S. and Blask D., Cancer Res. 48, 6121–6126 (1988). [PubMed] [Google Scholar]
- de Albuquerque Garcia Redondo P., Nakamura C. V., de Souza W., and Morgado-Diaz J. A., J. Histochem. Cytochem. 52(5), 629–640 (2004). 10.1177/002215540405200507 [DOI] [PubMed] [Google Scholar]
- Moon H.-S., Kwon K., Kim S.-I., Han H., Sohn J., Lee S., and Jung H.-I., Lab Chip 11, 1118–1125 (2011). 10.1039/c0lc00345j [DOI] [PubMed] [Google Scholar]
- Daw R. and Finkelstein J., Nature 442, 367 (2006). 10.1038/442367a [DOI] [Google Scholar]
- Whitesides G. M., Nature 442(7101), 368–373 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- Yang F., Yang X., Jiang H., Bulkhaults P., Wood P., Hrushesky W., and Wang G., Biomicrofluidics 4(1), 013204 (2010). 10.1063/1.3279786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker F. F., Wang X.-B., Huang Y., Pethig R., Vykoukal J., and Gascoyne P., Proc. Natl. Acad. Sci. U.S.A. 92, 860–864 (1995). 10.1073/pnas.92.3.860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-B., Huang Y., Gascoyne P. R. C., Becker F. F., Holzel R., and Pethig R., Biochim. Biophys. Acta 1193, 330–344 (1994). 10.1016/0005-2736(94)90170-8 [DOI] [PubMed] [Google Scholar]
- An J., Lee J., Lee S. H., Park J., and Kim B., Anal. Bioanal. Chem. 394, 801–809 (2009). 10.1007/s00216-009-2743-7 [DOI] [PubMed] [Google Scholar]
- Stathopoulou A., Mavroudis D., Perraki M., Apostolaki S., Vlachonikolis I., Lianidou E., and Georgoulias V., Anticancer Res. 23, 1883–1890 (2003). [PubMed] [Google Scholar]
- Kim Y. H., An J., Lee S. H., and Kim B., J. Mech. Sci. Technol. 23, 3132–3139 (2009). 10.1007/s12206-009-0910-6 [DOI] [Google Scholar]
- Chang H.-C. and Yossifon G., Biomicrofluidics 3, 012001 (2009). 10.1063/1.3056045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher M. J., Swan E., Fiering J., Holmboe M. E., Sewell W. F., Kujawa S. G., McKenna M. J., and Borenstein J. T., J. Microelectromech. Syst. 18, 501–510 (2009). 10.1109/JMEMS.2009.2015484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz S., Holzer R., and Renaud P., Lab Chip 1(1), 29–34 (2001). 10.1039/b103896f [DOI] [PubMed] [Google Scholar]
- Munson M. S. and Yager P., Anal. Chim. Acta 507(1), 63–71 (2004). 10.1016/j.aca.2003.11.064 [DOI] [Google Scholar]
- Ermolina I. and Morgan H., J. Colloid Interface Sci. 285(1), 419–428 (2005). 10.1016/j.jcis.2004.11.003 [DOI] [PubMed] [Google Scholar]
- Hu X., Arnold W., and Zimmermann U., Biochim. Biophys. Acta 1021(2), 191–200 (1990). 10.1016/0005-2736(90)90033-K [DOI] [PubMed] [Google Scholar]
- Wang Z., Eisenberger M., Carducci M., Partin A., Scher H., and Ts'o P., Cancer 88(12), 2787–2795 (2000). [DOI] [PubMed] [Google Scholar]
- Pethig R., Biomicrofluidics 4, 022811 (2010). 10.1063/1.3456626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Pan F., Jones L., Lim M., Griffin E., Sheline Y., Mintun M., Holtzman D., and Mach R., Brain Res. 1319, 21–32 (2010). 10.1016/j.brainres.2009.12.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Yang X., Jiang H., and Wang G., Electrophoresis 32, 2377–2384 (2011). 10.1002/elps.201100039 [DOI] [PubMed] [Google Scholar]
- Pohl H., J. Appl. Phys. 22(7), 869–871 (1951). 10.1063/1.1700065 [DOI] [Google Scholar]
- Grant E. H., Sheppard R. J., and South S. P., Dielectric Behaviour of Biological Molecules in Solution (Clarendon, 1978) Vol. 7, No. 1, p. 16. [Google Scholar]
- Pethig R., Dielectric and Electronic Properties of Biological Materials (John Wiley & Sons Ltd, Chichester, UK, 1979). [Google Scholar]
- Pethig R. and Kell D., Phys. Med. Biol. 32(8), 933–970 (1987). 10.1088/0031-9155/32/8/001 [DOI] [PubMed] [Google Scholar]
- Gascoyne P. R. C., Wang X.-B., Huang Y., and Becker F. F., IEEE Trans. Ind. Appl. 33(3), 670–678 (1997). 10.1109/28.585856 [DOI] [PMC free article] [PubMed] [Google Scholar]