Abstract
This work presents a microfluidic chamber array that generates soluble gradients using sequentially pulsed fluid delivery (SPFD). SPFD produces stable gradients by delivering flow pulses to either side of a chamber. The pulses on each side contain different signal concentrations, and they alternate in sequence, providing the driving force to establish a gradient via diffusion. The device, herein, is significant because it demonstrates the potential to simultaneously meet four important needs that can accelerate and enhance the study of cellular responses to signal gradients. These needs are (i) a scalable chamber array, (ii) low complexity fabrication, (iii) a non-shearing microenvironment, and (iv) gradients with low (near zero) background concentrations. The ability to meet all four needs distinguishes the SPFD device from flow-based and diffusion-based designs, which can only achieve a subset of such needs. Gradients are characterized using fluorescence measurements, which reveal the ability to change the curvature of concentration profiles by simple adjustments to pulsing sequence and flow rate. Preliminary experiments with MDA-MB-231 cancer cells demonstrate cell viability and indicate migrational and morphological responses to a fetal bovine serum gradient. Improved and expanded versions of this technology could form the basis of high-throughput screening tools to study cell migration, development, and cancer.
INTRODUCTION
Concentration gradients of biomolecular signals play an important role in many biological processes, both in normal physiology and in disease. Such processes include embryonic development,1, 2 tissue repair,3 immune surveillance,4, 5 and cancer metastasis.6, 7 To improve our understanding of cellular responses to signal gradients, a variety of microfluidic gradient-generating devices have been developed. Microfluidic systems are useful because they exhibit laminar flows that can generate predictable and stable concentration gradients, which can surmount limitations of traditional gradient assays (e.g., transwell, Dunn, under-agarose) or enable entirely new lines of study.8, 9 Also, some microfluidic systems have demonstrated the potential for parallelization and greatly reduced sample usage, leading to higher throughput and lower cost experimentation, respectively.10, 11 Microfluidic gradient-generating devices take numerous forms and approaches, and they are highlighted in reviews.12, 13, 14, 15
This work is motivated by investigators' need for a method to study cellular responses to signal gradients that simultaneously offers (i) higher throughput/array scalability,10, 13 (ii) low fabrication complexity, (iii) negligible shear stresses on cells,16, 17, 18 and (iv) gradients presented with low background concentrations19 for certain studies. These four needs are reflective of the overarching desire for cell-based assays that provide higher throughput and greater physiological relevance, which are critically needed in basic science and to improve research productivity in biopharmaceutical lead discovery.20, 21
Flow-based and diffusion-based systems are the two predominant categories of microfluidic gradient-generating cell-based assays.14 Both categories demonstrate unique advantages and limitations, causing them to satisfy subsets of the four abovementioned needs. However, neither category is designed to meet all four. Flow-based systems rely on splitting and re-combining of multiple flow streams over several generations, resulting in a gradient perpendicular to the direction of flow.22, 23, 24, 25, 26 Flow-based gradients exhibit excellent stability; however, potential limitations are unintended shear stresses on cells, a spatially changing gradient, and difficulty in scaling arrays. Alternatively, diffusion-based designs produce gradients via diffusion from a source to a sink, which are separated from a main chamber by porous barriers, such as microcapillaries,27, 28 membranes,29, 30, 31, 32 hydrogels,33, 34 and packed microparticles.35 These gradients typically present a low shear microenvironment; however, potential limitations are reduced diffusion speeds, restricted background concentrations, and complex fabrication.
Recently, Atencia et al.36 pursued a diffusion-based design referred to as a “microfluidic palette,” consisting of a circular chamber with access ports at its edge. The access ports are flushed with medium of different concentrations at specific intervals (replenishment), forming a gradient across the chamber from port to port. The microfluidic palette surmounts limitations of both flow-based and other diffusion-based designs by producing gradients that simultaneously exhibit negligible shear stress, span low background concentrations, and achieve steady-state at moderately low time scales (relative to other diffusion-based systems). However, the microfluidic palette is a single chamber device, which would be difficult to scale into an array due to a vacuum channel network that must be interspersed among the active flow channels. In addition, fabrication is complex, consisting of a 4-layer stack of glass-glass-PDMS-PMMA that requires drilling and/or channel patterning in all three materials and two alignment steps. Also, the gradient can be sensitive to pressure differences across the ports, which requires careful matching of flow rates and fluidic resistances for each port's supporting flow network; thus, perturbations in pressure could result in cross-convection that disrupts the gradient. A detailed discussion of flow-based and diffusion-based capabilities is found in the supplementary material.37
In this work, we present a microfluidic gradient-generating device based on sequentially pulsed fluid delivery (SPFD). The device is significant because it demonstrates the potential to concurrently meet all four abovementioned needs, making it unique among flow-based and diffusion-based designs (Table TABLE I.). In SPFD, fluid flows alternate on either side of a chamber in discrete pulses with coordinated timing. By pulsing different signal concentrations on each side, a concentration gradient is formed across the chamber by diffusion. SPFD is notably similar to the microfluidic palette with respect to its alternating flows; however, SPFD is significantly different in that its design and architecture enable scalable arrays, low complexity fabrication, and robustness against pressure differences through use of integrated valves. Thus, SPFD achieves gradients that concurrently exhibit full stability, negligible flow-induced shear stresses, and a wide range of background concentrations (i.e., near-zero background concentrations), while also exhibiting a scalable chamber array that is fabricated via a single alignment step.
TABLE I.
Comparison of relative capability levels of three microfluidic gradient device categories against specific needs (low, med, and high). Relative fabrication complexity of SPFD device ranked as med-level due to the requirement for an alignment step.
| NEED | Flow-based | Diffusion-based (incl. microfluidic palette) | This work (SPFD) |
|---|---|---|---|
| Scalability to arrays | Med | Low | High |
| Low complexity fabrication | High | Low-Med | Med |
| Low/no shear | Med | High | High |
| Presentation of low background conc. | High | Low | High |
A prototype device is designed and fabricated with an in-line array of six chambers. Fluorescence measurements are performed to investigate the effects of pulsing sequence and pulse flow rate on steady-state concentration profiles and rise times. In doing so, we demonstrate that adjustments in the pulsing sequence and flow rates can change the curvature of concentration profiles from linear to concave. Also, we perform preliminary cell experiments, where migrational and morphological responses of human breast cancer cells (MDA-MB-231) are monitored under non-gradient and fetal bovine serum (FBS) gradient conditions.
DEVICE DESIGN
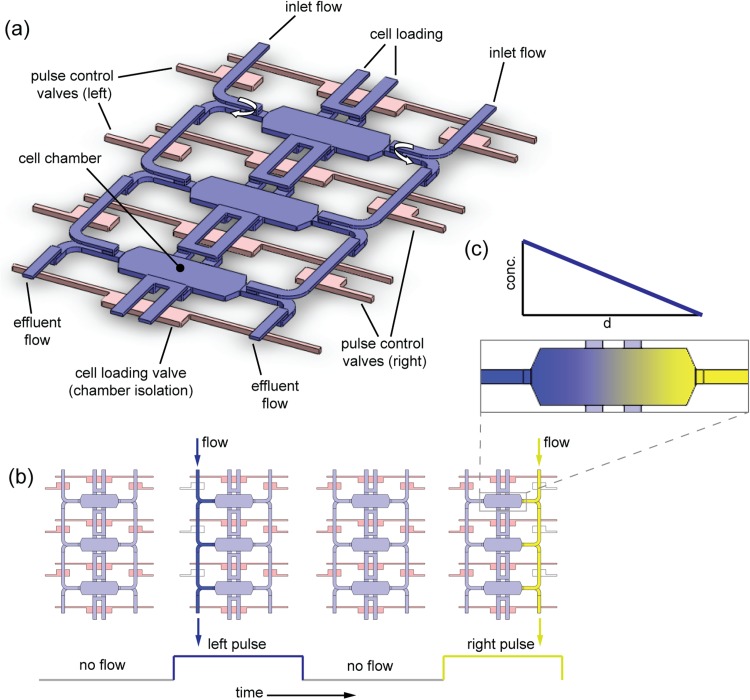
The device is designed with an in-line array of six chambers. Figure 1a is a schematic of three chambers in the array, as well as their supporting channel network and integrated valves, which implement SPFD. The design makes use of a 3-layer architecture (containing vertical flow interconnections between layers) and compression-molding fabrication techniques from a previous work.38 Although it exploits previous tri-layer architecture and fabrication methods, this work is distinct by virtue of its channel routing design, which produces “wrap-around” flows (explained below) at the sides of each chamber. Wrap-around flows are critical to forming gradients with low background concentrations without the use of porous barriers. Additionally, this work is distinct in its experimental application and mechanism: the previous work was designed to elicit epithelial-to-mesenchymal transitions (EMT) in cells exposed to uniform signal fields (no gradients) and with continuous flow, while this work is designed to elicit directional migration responses from cells exposed to a gradient using pulsed flow.
Figure 1.
Chamber array architecture and sequentially pulsed fluid delivery. (a) Chambers (blue), flow channels (blue), and valves (pink) form a 3-dimensional network. Media (with or without stimulus) are input along sides, while cells are loaded via central channels. Flow travels along either side of the chambers and undergoes “wrap-around” flow (curved arrows), leaving the central part of the chambers undisturbed by flow. Integrated valves regulate flow in top layer channels. (b) Four phases of pulsing sequence. Sequence begins with all valves closed, then left side valves open to release flow pulse with high concentration (dark blue). All valves close again; then right side valves open to release a flow pulse with low concentration (dark yellow). The sequence repeats throughout the experiment. (c) Flow pulses replenish each side with their respective input concentrations, establishing effectively constant boundary conditions. A gradient forms via diffusion-dominated mass transport.
The concept of sequentially pulsed delivery is depicted in Fig. 1b. Two input flows with different signal concentrations are positioned on each side of the chambers. Four stages of a pulsing sequence are shown, illustrating how flows alternate on each side. Flows are stopped at the specified times by integrated monolithic valves.39 The valves actuate and de-actuate in a manner such that flow pulses never occur at the same time; when a pulse occurs on one side of the chambers, the valves on the other side are closed. Hence, the trajectory of each flow pulse is confined to its respective side, and the flow wraps from the upper to the lower layer at the side of the chamber (“wrap-around” flow indicated by the curved arrows in Fig. 1a). As a result, this four-stage sequence leaves the main part of the chambers undisturbed and makes the device particularly robust against sources of cross-convection that would disrupt gradient formation, such as asymmetries in pressure or in fluidic resistance.
The purpose of alternating flows is to periodically replenish each side with its input concentration (Fig. 1c). Replenishments are frequent enough so that the concentration on each side is effectively constant, which provides a steady driving force for diffusion and stable gradient formation across the chamber.
MATERIALS AND METHODS
Fabrication
Devices are fabricated using soft lithography,40 multi-layer PDMS thermal bonding,39 and through-hole processing. Fabrication requires two master molds (masters A and B). Each master is a silicon wafer (100 mm diameter) with a photoresist pattern forming device features in positive relief. Photoresist patterns are realized using standard photolithographic methods.41
Master A contains a two-layer pattern of SU-8 (MicroChem Corp., Newton, MA, USA), a negative-type photoresist. The first layer is 50 μm tall, and its features define the bottom device layer, containing chambers and parts of the channel network (flow and valve). The second layer, which is patterned on the first layer, is 30 μm tall with features defining the middle device layer (i.e., through-holes that enable vertical interconnection of flow paths between layers).
Master B contains patterns made of AZ P4620 (AZ Electronic Materials Corp., Branchburg, NJ, USA), a positive-type photoresist, that define flow and valve channels (25 μm tall). These channels have a semi-circular cross-section, allowing them to be fully closed by push-up valves located throughout the device.
Devices are made by aligning and thermally bonding two molded layers of PDMS (Sylgard 184, Dow Corning Corp., Midland, MI, USA) derived from masters A and B. Through-holes are realized by adapting the compression-molding techniques of Folch et al.42 and Kim et al.43 with certain modifications. The resulting two-layer PDMS device and a glass slide are then exposed to air plasma for 30 s, and both are brought into contact and irreversibly bonded. The process is illustrated in Fig. 7 (in the supplementary material)37 and further described in a previous work.38
Experimental setup
Image acquisition is performed on a DM IRB/E inverted microscope (Leica Microsystems GmbH, Wetzlar, Germany) at 10× magnification in phase contrast mode (Fig. 11, supplementary material).37 The device is mounted to a motorized x-y stage (Ludl Electronic Products Ltd., Hawthorne, NY, USA) to translate it through a 6-position circuit to image each chamber in the array. Hollow stainless steel pins (L-shaped, 21G) are press-fitted into device ports and are connected to flexible plastic tubing (PE-60) for fluid and pressure delivery. Flow is driven by a syringe pump (PHD 2000, Harvard Apparatus, Holliston, MA, USA). Pressurization of on-chip valves is regulated by off-chip miniature solenoid valves (Series 188, ASCO Valve Inc., Florham Park, NJ, USA) attached to a pressure source at 30 psig. Images are acquired using a digital CCD camera (ORCA C4742-95-12, Hamamatsu Photonics K.K., Hamamatsu City, Japan). Valve actuation, stage movement, and image acquisition are computer-controlled using a custom-designed interface written in the labview programming environment (National Instruments Corp., Austin, TX, USA).
Gradient characterization
Soluble concentration gradients in the chambers are verified by way of fluorescence measurements. Dextran (70 000 MW) labeled with Oregon Green (D-7172, Molecular Probes Inc., Eugene, OR, USA) is dissolved in aqueous buffer (phosphate buffered saline (PBS)) and pulsed along one side of the chambers. A sequence of fluorescent images (GFP filter set) is acquired to record the evolution of the dextran concentration profile and formation of the gradient. Sequences are acquired at different flow rates and flow pulsing schedules to determine their effect on profile formation and steady-state curvature.
Cell culture off-device
MDA-MB-231 cells (HTB-26, ATCC, Manassas, VA, USA), a human breast cancer cell line, are maintained in Leibovitz's L-15 medium (Gibco 11415, Invitrogen, Carlsbad, CA, USA) with 10% FBS and 1% penicillin/streptomyocin. Cells are incubated at 37 °C in humidified air, and medium is changed every 48–72 h. Cells are passaged with trypsin-EDTA solution (0.25% w/v trypsin and 0.53 mM EDTA, Gibco 25200, Invitrogen, Carlsbad, CA, USA), and the morphology and proliferation rate of cells are monitored over passages to verify consistent behavior.
Prior to each trial, cells are serum-deprived by changing the medium to L-15 with 0.1% w/v bovine serum albumin (BSA) (A9418, Sigma, St. Louis, MO, USA) (hereafter referred to as base medium) for 12 h. Then, cells are detached from their culture flask using EDTA solution (Versene, Gibco 15040, Invitrogen, Carlsbad, CA, USA). Once the cells are detached, excess base medium is added, and the suspension is centrifuged. The supernatant is aspirated, and the cells are resuspended in base medium to a density of ∼1 × 106 cells/ml and loaded into the device.
Device preparation, cell loading, and incubation in-device
Each device is pre-filled with PBS. To prevent on-chip valves from introducing gas bubbles into the device during experiments, each valve is filled with water. Fibronectin (Fn) (Gibco 33016, Invitrogen, Carlsbad, CA, USA) at concentration 8 μg/ml (in PBS) is introduced through the cell loading port. The chamber array is filled with Fn solution, and it is incubated at 37 °C for 1 h to allow matrix molecules to adsorb to chamber surfaces. Following adsorption, the device is filled with BSA solution (2% w/v in PBS) and incubated at room temperature for 45 min to block surfaces from non-specific binding. Base medium (L-15 with 0.1% w/v BSA) is then introduced to the input ports and flushed through the device.
Cell suspension is then introduced into the cell loading channel and flowed through the chamber array by gentle pressure. Cells are stopped and confined to their chambers by closing the cell loading and pulse control valves.
The device is transferred from the microscope to a humidified air incubator at 37 °C. Cells are incubated in the device for 12 h to allow full attachment and spreading on the substratum prior to image acquisition.
Experiments and image acquisition
After incubation, the device is re-mounted to the microscope stage, and experimental media are introduced to the input ports. The medium entering the ports is either base medium or base medium loaded with FBS (10% v/v) (16000, Invitrogen, Carlsbad, CA, USA), depending on desired conditions. The device is covered by an optically transparent enclosure (made of PDMS) into which is fed humidified air to prevent evaporation and gas bubble nucleation in the device (Fig. 11, supplementary material).37 A heating fan maintains the system at 37 °C.
Each experiment is made up of two stages. In the first stage, the flow pulsing sequence is engaged but there is no FBS gradient. Images of each chamber are acquired in 3 min intervals for 6 h. The second stage begins immediately after, wherein the flow pulsing sequence continues and a gradient of FBS is presented to the cells. The gradient is turned on by changing the selected input flows at the flow routers. Images are also acquired in 3 min intervals for 6 or more hours, depending on experiment. Photos for each chamber are then compiled into a sequence spanning stages 1 and 2 for image processing. Control experiments are also performed, where there is no gradient during both stages (first and second).
Data for gradient experiments are derived from 3 replicate experiments (3 devices) with 6 identical chambers per device. Migrational and morphological data are collected from 205 cells.
Cell segmentation and data analysis
Cells are segmented in each image from their respective backgrounds for morphological and tracking analysis. We apply a heuristic involving image registration, image division, flattening, dilation, superimposition, flood-filling, and erosions using matlab (The MathWorks Inc., Natick, MA, USA) and Image-Pro 6.1 (Media Cybernetics Inc., Bethesda, MD, USA) algorithms.
First, the chemotactic index (CI) (defined in the supplementary material)37 of cells for the entire region of interest (ROI) for each trial is calculated. Then, the ROI is divided into 4 spatial sections, and each data point for CI, speed, area, and circularity is calculated from non-overlapping subsequences of 7 consecutive photo frames, and each data point is binned into one of the 4 sections in the chamber. Area and circularity are calculated as the average from each set of 7 photos, and speed is calculated by dividing the root-mean-square displacement by the time interval spanning the 7 photos (18 min).
All statistical analyses are performed by anova using Prism software (GraphPad Software Inc., La Jolla, CA, USA). Tukey's posthoc tests are performed. The supplementary material and Fig. 12 (in the supplementary material)37 contain further details regarding image processing and data analysis.
RESULTS AND DISCUSSION
Demonstration of gradients—Effects of pulsing sequence and flow rates
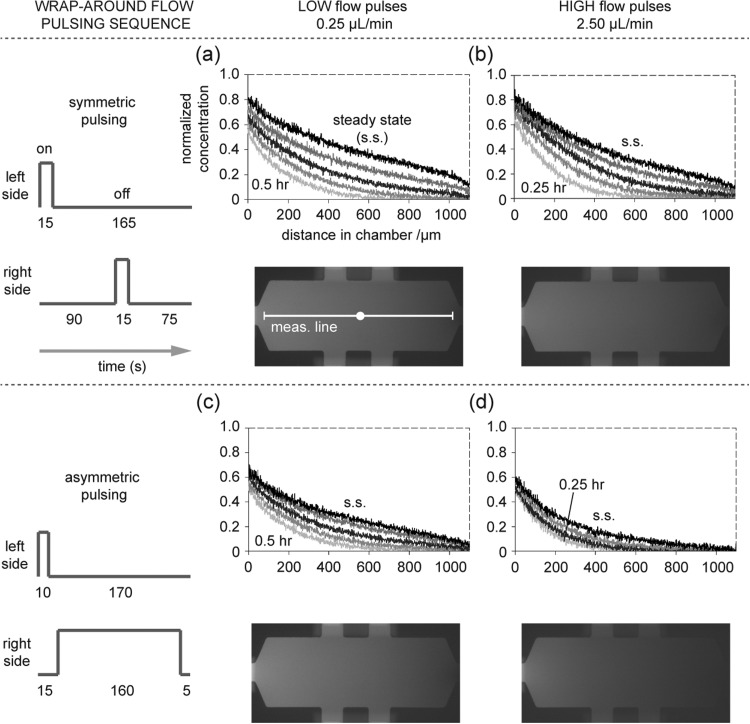
Results from gradient characterization are in Fig. 2, where concentration profiles from four different combinations of pulsing sequence and flow rate are shown. Figures 2a, 2b show results for a pulsing sequence where the left and right pulses are equal in duration (i.e., symmetric pulsing), while Figs. 2c, 2d show results where the pulses on the right side (non-dextran side) are significantly longer than those on the left side (i.e., asymmetric pulsing). Within each subfigure are plots at two flow rates (0.25 μl/min and 2.5 μl/min). In each of the four graphs, there are multiple profiles at different time points as the gradient forms, starting from a zero concentration initial condition. The topmost profile in each graph represents the steady-state. Comparison of the steady-state profiles reveals differences in their shapes, some of which are near-linear, while others (particularly the lower right curve) show noticeable curvature (exponential shape).
Figure 2.
Gradient formation measured via fluorescence imaging. Top-most profile in each graph is the stable, steady-state (s.s.) profile. Medium loaded with fluorescently labeled dextran (70 000 MW) is pulsed along one side of the chambers. Results of four combinations of flow pulsing sequence and flow rate are shown. (a) A pulsing sequence with equal pulse times (15 s per pulse) generates a near linear steady-state profile at the lesser flow rate (0.25 μl/min) and (b) a slightly curved steady-state profile at the greater flow rate (2.5 μl/min). Fluorescence photographs of chambers at steady-state are shown; measurement line (1100 μm) indicates where intensity is measured. (c) A pulsing sequence with unequal pulse times generates a slightly curved steady-state profile at the lesser flow rate and (d) a noticeably curved (concave) steady-state profile at the greater flow rate. Profiles for the lesser flow rate (two left side plots) correspond to 0.5 h, 1 h, 2 h, 4 h, and s.s. (bottom to top profiles, respectively). Profiles for greater flow rate and equal pulsing (upper right plot): 0.25 h, 0.5 h, 1 h, 2 h, and s.s. Profiles for greater flow rate and unequal pulsing (lower right plot): 0.25 h, 0.5 h, 1 h, and steady-state.
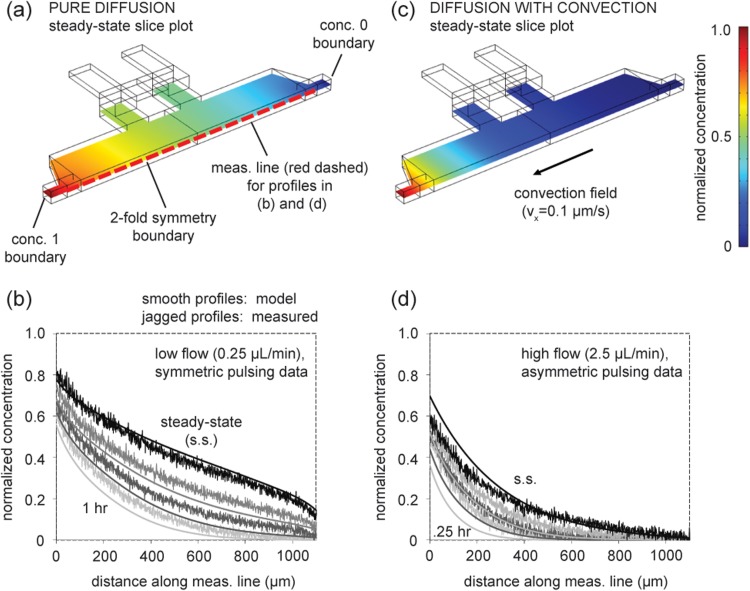
A linear profile would be expected across geometries with constant cross-section if the transport is only diffusion-based. In this work, the chamber cross-section is generally constant, with the exception of the chamber narrowing at its left and right sides (as it tapers to the channel width) and the cell loading channels. Therefore, the near linear steady-state profile for the symmetric pulsing sequence with low flow (Fig. 2a) suggests diffusion-dominated transport. For comparison, a 3D finite element model of the chamber geometry that simulates only diffusion-based transport is built (Figs. 3a, 3b) (Multiphysics 4.2a, COMSOL AB, Stockholm, Sweden). The model simulates transient gradient formation, starting with a zero concentration initial condition.
Figure 3.
Comparison of concentration profiles from numerical modeling and measured data. (a) Oblique view of 3D model geometry (half chamber) for diffusion-only simulation with concentration boundaries set at 1 and 0. Slice plot shows steady-state (s.s.) solution (at z = 25 μm, half thickness of chamber). (b) Formation of gradient from zero concentration initial condition. Transient and s.s. diffusion-only model solutions compared to measured data from low flow, symmetric pulsing experimental conditions. Profiles represent 1, 2, 4 h, and s.s. (from bottom to top), and modeled and measured profiles share corresponding grayscale shading. (c) Steady-state solution when convection is introduced. (d) Transient and s.s. diffusion-with-convection model solutions compared to measured data from high flow, asymmetric pulsing conditions. Profiles represent 0.25, 0.5, 1 h, and s.s. (from bottom to top). Simulations use diffusion coefficient of 70 kDa dextran (same MW as in experiments) as estimated in literature (∼3 × 10−7 cm2/s).44 Measurement line is 1100 μm. Note: profiles in (b) and (d) do not begin with normalized concentration 1 because measurement line does not span the entire length of the chamber.
The diffusion-only simulation and measured data show rough agreement at three time points and steady-state (Fig. 3b), supporting the suggestion that the device exhibits diffusion-dominated transport under symmetric pulsing and low wrap-around flow conditions.
When the pulsing schedule is asymmetric, the flow rate is higher, or both, the experimental observations show noticeable curvature in steady-state profiles (Figs. 2b, 2c, 2d). Curvature is particularly evident in Fig. 2d under asymmetric pulsing and high flow conditions. Steady-state profiles can exhibit curvature when convection is present. This is apparent when considering the continuity equation of mass conservation under incompressible flow (Eq. 1a) and its reduction to convective and diffusive components under the steady-state, no reaction condition (Eq. 1b),
| (1a) |
| (1b) |
where C is concentration, u is velocity, D is diffusivity (assumed constant), and r is reaction rate. A non-zero ∇C component would be expected in the device, due to the driving force provided by the replenishment of the source and sink by pulsed flows. Thus, a non-zero convection (u) would force the existence of curvature in the ∇2C component. To model the effect of convection on the profiles, a small velocity field is simulated in the model of the chamber (Figs. 3c, 3d). The steady-state profile of the model shows curvature similar to that of the measured steady-state (Fig. 3d). In addition, the transient curves of the model at three time points are similar in shape to corresponding measurements.
The similarities between the modeled and measured profiles in Figure 3 suggest that (i) transport with low wrap-around flow rates and symmetric pulsing is analogous to pure diffusion and (ii) transport with high wrap-around flow rates and asymmetric pulsing is analogous to diffusion with small convective drift. Although these illustrative models and measured data are similar, they do not match exactly because the models in Figure 3 do not capture the complex mechanism of action of alternating flow pulses. To confirm this, a full-scale model of the device that simulates alternating flow pulses and replenishments at source and sink would be required. The model could estimate where, when, and to what magnitude convection takes place during an experiment; these estimates could potentially be validated via particle tracking. Building, properly bounding, and automating the model through decades of pulsing cycles, as well as tracking experiments, could form the basis of a future work to mature the SPFD concept. If asymmetric pulsing and high wrap-around flow rates, in fact, contribute to a convective drift, Figure 3 indicates the drift would be miniscule from a physiological perspective. The modeled flow velocity (0.1 μm/s) is comparable to low shear interstitial flow.45
In summary, the results demonstrate the ability to present stable concentration profiles with curvatures that can vary from linear to exponential by changing the pulsing sequence and/or flow rate. Such unique flexibility may prove useful to experimentalists who wish to measure the response of cells to concentration profiles with different curvatures without needing to redesign the device. Curved (concave) profiles are particularly physiologically interesting, as they produce a microenvironment that mimics diffusion from a point source (e.g., signal release) into an infinite reservoir (e.g., interstitial tissue space). Although curved concentration profiles have been engineered using flow-based devices,23, 46 shear stresses are created due to the convection required for gradient generation. At the same time, diffusion-based devices are limited only to linear profiles at steady-state. Using SPFD, a curved concentration profile can be stably maintained while simultaneously exhibiting low shear, which may serve as a suitable representation for certain physiological contexts and enable new types of studies. Our experimental results and simplified model also suggest that it is possible to further shape the concentration gradient by applying asymmetric pulsing of the source and sink buffer.
Further discussion on the selection of flow rates and pulsing sequences can be found in the supplementary material.37
Array scalability
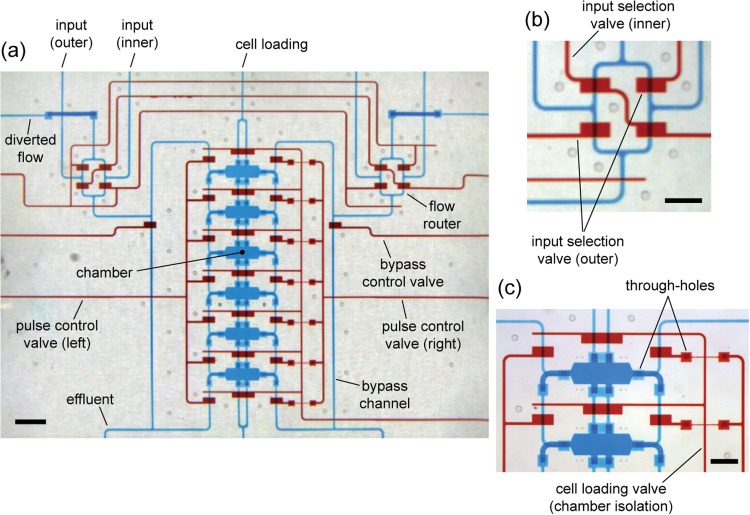
Images of a fully fabricated device are shown in Fig. 4. The device consists of the chamber array with supporting fluid bussing channels, cell loading channels, and integrated valves. Gradients are turned on and off by flow selection routers that use valves to select which input flows are directed into the chamber array (Fig. 4b).
Figure 4.
Fully fabricated device loaded with colored dyes for visualization of chamber array and 3D channel network. Blue dye is loaded in flow channels; red dye is loaded in valve control channels. (a) Overview of the device. Flow of inputs is selected by routers—one pair of inputs (inner or outer pair) is directed into the chamber network, while the other is diverted. Pulse control and bypass valves work synchronously to pass flow along chamber sides or to effluent (preventing over-pressurization). Pulse control comprises 7 interconnected valves on each side of the chamber array. (b) Magnified view of the right flow router, indicating which valves direct inputs into the chamber network. (c) Magnified view of chambers. Chamber isolation valves are permanently closed after cell loading (note: chamber isolation control channels weave underneath pulse control channels. Scale bar is 1000 μm in (a) and 500 μm in (b) and (c).
Examination of Fig. 4a and Fig. 7 (in the supplementary material)37 shows that the chamber array could be efficiently scaled-out to larger in-line or 2D arrays. More chambers could be added to the existing column of six chambers or adjacent columns could be added, which would share the same bypass channels, control valves, and effluent channels. This would minimize the number of world-to-chip input/output ports and enable chambers to be more closely positioned, both of which would save device real-estate. Efficient scalability is facilitated by the 3D flow network, which resembles roadway bridges and underpasses (bridge-and-underpass architecture).38 The architecture makes it possible to construct intricate bussing networks that can weave vertically around various obstacles (i.e., valves, chambers, other channels) to feed the proper flows to rows and columns of chambers.
All chambers in the array are designed to present the same gradient (Fig. 9, supplementary material),37 which would enable replicate conditions and the potential for large-scale parallelization. Chambers with the same gradients are intended to address the aforementioned limitation of spatially changing gradients in flow-based designs, and they result from the SPFD approach that confines the flows to either side of the chambers.
As previously mentioned, scalability is an important unrealized need in the field of microfluidic gradient-generating devices that would offer greater experimental throughput. Some works have addressed this by creating impressive chamber arrays in flow-based systems where the same flow streams cross directly over multiple chambers.25 Although useful in certain studies, communication between rows/columns could confound results, depending on the purpose of the study. In contrast, this work demonstrates the potential for efficient scalability of gradient-producing arrays made up of chambers that could be fully independent by use of the 3D channel network, thereby removing the possibility of inter-row/column communication.
Low complexity fabrication
The ultimate hope for microfluidic technologies is that they become practical and commercially available. An important aim in this is to minimize fabrication complexity, which can speed the adoption of microfluidics (especially in non-microfluidics laboratories) and eventually lead to low-cost, high-volume production. Recently, it has been demonstrated that multilayer PDMS systems, requiring multiple alignment steps in fabrication, can be successfully commercialized47 or can be reversibly sealed to pre-existing cell cultures to improve ease-of-use.48 This work reduces fabrication to a single alignment step for an all-PDMS device (Fig. 8, supplementary material).37 By minimizing fabrication complexity, the likelihood of realizing future versions of the device with scaled-out chamber arrays is increased.
Also, we avoid the complexity of hybrid structures (e.g., separate membranes or hydrogel components embedded in PDMS), which is the characteristic of many diffusion-based designs.29, 32, 33, 49, 50, 51 Hybrid structures are potentially susceptible to manufacturing variation due to (i) the necessity for careful and precise handling of fragile membrane components, (ii) repeated alignment steps (i.e., base-to-membrane, membrane-to-layer), and/or (iii) batch-to-batch variation in hydrogel properties or errors in the gel-forming process. Furthermore, we avoid drilling36 and micromachining/etching25, 36 on glass, which has been utilized to fabricate microfluidic palette and flow-based gradient devices. Instead, this work employs a modified compression-molding technique and an all-PDMS thermal bonding process (see Sec. 3).
One must acknowledge that the fabrication method in this work is not as simple as rapid prototyping methods of most single-layer flow-based gradient devices;24 nevertheless, the intent of this work is to minimize fabrication complexity to a sustainable level, while at the same time significantly augmenting capabilities (i.e., scalability, no shear, low background concentrations).
Low shear stress microenvironment—Simulation results
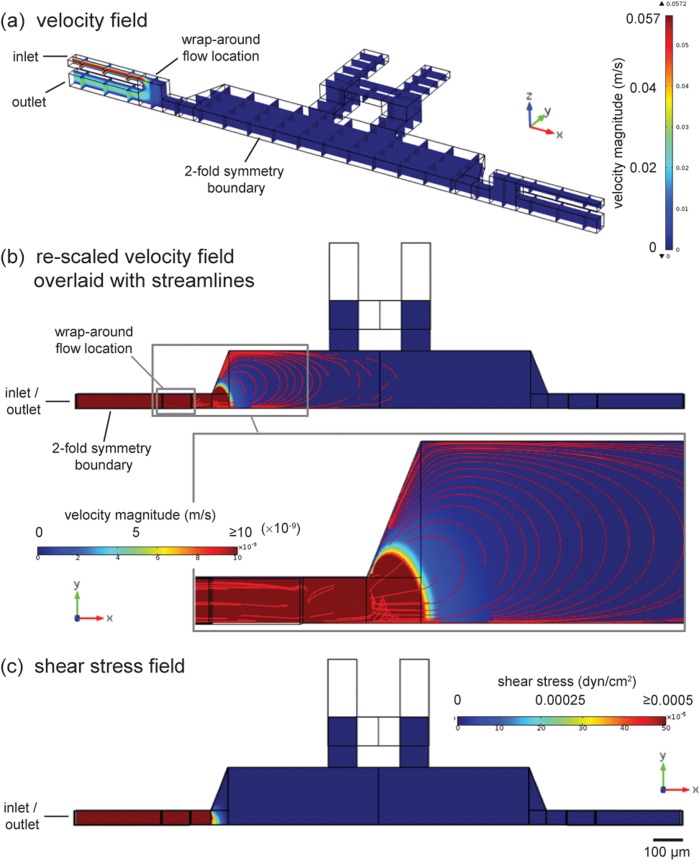
The device is designed to produce negligible flow-induced shear stresses. To assess the velocity field and shear levels, a 3D finite element model of the chamber is constructed using the comsol software package (Multiphysics 4.2a, COMSOL AB, Stockholm, Sweden). The model simulates wrap-around flow along the side of a chamber that occurs during a flow pulse (Fig. 5). The model geometry consists of half of a chamber with side channels and connecting vertical channels. All boundary conditions are specified as no-slip, except for inlet and outlet boundaries, as well as a two-fold symmetry boundary. Wrap-around flow is driven by imposing a 2.5 μl/min flow rate at the inlet boundary and arbitrary zero pressure at the outlet; the flow rate is chosen to match the upper bound of experimental flow rates. The other side of the chamber is fluidically isolated (no-slip condition) to simulate integrated on-chip valves that are closed to prevent flow across the chamber. The Navier-Stokes equations are solved to obtain the velocity field at steady-state, which is used to calculate shear stresses on the chamber surface.
Figure 5.
Simulation of chamber flow field during a flow pulse. (a) Oblique view of 3D model geometry with solution (velocity field) shown in slices. Heat plot visualizes how greatest velocity exists in wrap-around flow region at the side of the chamber. (b) Overhead view with velocity field solution in plane at z = 25 μm (half thickness of chamber) and 3D streamlines. Inset shows close-up of low-magnitude convective field (velocity legend re-scaled) penetrating into chamber. Transition from well-organized (contoured) to random streamlines indicates the extent of penetration into the chamber by the convective field. (c) Shear stress at the surface of the chamber is less than 0.0005 dyn/cm2. Simulation solved at highest experimental flow rate by imposing a 2.5 μl/min flow rate at the inlet boundary.
The resulting velocity plot (Fig. 5a) shows that convection is mostly within the wrap-around flow path. In Figure 5b, the velocity heat plot is re-scaled, indicating that the wrap-around flow introduces a convective field that penetrates into the chamber. Streamlines are plotted (Fig. 5b and inset) to visualize the trajectory of convection and estimate the extent of penetration by such convection. The distribution of the streamlines indicates that the convective field penetrates roughly halfway into the chamber, after which the streamlines become randomly oriented. The convective field could influence the curvature of concentration profiles, as discussed earlier in the Sec. 4A). Despite the existence of a convective field, the magnitude of it is low from the physiological perspective (order of 10 nm/s or less), and the corresponding shear stress field on the chamber floor (Fig. 5c) shows shear stresses less than 0.0005 dyn/cm2 across the chamber.
The motivation for engineering a microenvironment with minimal shear stresses is to provide a reasonable model for physiological contexts where fluid flow is very small. Such contexts may occur in interstitial spaces throughout connective tissue and in the brain. This is one of many shear contexts throughout the body. For example, in vivo shear stresses of 10–100 dyn/cm2 have been reported on vascular endothelial cells in blood vessels (a high shear context), while less than 2 dyn/cm2 has been reported for hepatocytes (mid-shear context).52 In interstitial spaces, shear stresses are expected to be smaller, although the fibrous 3D extracellular matrix makes precise estimates of shear stress difficult.53 Interstitial fluid velocities of less than 1 μm/min have been measured, which indicates extremely low flow (and correspondingly low shear).45 As well, human neural stem cells have been successfully cultured in microenvironments with shear stresses estimated at 0.0005 dyn/cm2.22 The results of the model in Fig. 4 match such low shear stresses, and therefore the device in this work could potentially be applied to perform gradient experiments that mimic interstitial and other low flow contexts.
Gradients with low background concentration
The gradients demonstrated in this device exhibit a background concentration that approaches zero at the non-source side (Fig. 2). Although this observation appears matter-of-fact, it is important to note that near-zero background concentrations are inherently difficult to achieve in diffusion-based systems, since a significant fraction of the concentration change from source to sink occurs within the porous barriers. Thus, the background concentrations that can be presented to cells in the main chamber are restricted to roughly 0.35 to 0.70 on a normalized basis in diffusion-based designs.28, 33 While flow-based systems are capable of presenting gradients with low background concentrations, they do so at the risk of inducing cellular responses to flow-induced shear stress.
There is a need for devices that present stable gradients with both low background concentration and low shear stress, since it is shown that cells respond to gradients across a wide range of background concentrations and to emerging gradients (shallow gradients with low background concentrations).19, 54 These responses are demonstrated by cells that typically exist in contexts with low flow-induced shear, such as immune system cells, fibroblasts, and cancer cells.55 The device in this work has the potential to meet this need because it demonstrates gradients with low background and low shear, while at the same time exhibiting scalability and relatively low fabrication complexity.
Preliminary cell experiments
Preliminary cell experiments are performed to optimize device preparation and cell loading protocols, achieve characteristic attachment and spreading of the cells, and verify cell viability during device operation (i.e., valve actuation, flow routing, and sequential pulsing) (see Sec. 3 and also the supplementary material).37 Additionally, gradient experiments are performed to test the device's potential to elicit cellular response. MDA-MB-231 cells, a human mammary adenocarcinoma, are chosen for experiments because they are shown to respond to various signals, including specific growth factors, cytokines, and sera.56, 57 Also, cancer cells of this type are suspected of migrating through low flow interstitial spaces of the stroma during metastasis,58 which corresponds with the low shear intent of the device.
Chemotactic index is an important measure of response in gradient experiments, since it captures both the direction and magnitude of cells' migrational trajectory relative to the gradient source. Positive CI indicates net movement toward the source, while negative CI indicates movement away (see CI definition in supplementary material).37 The resulting CI from experiments is shown in Fig. 6b, which compares CI for stages 1 and 2. The CI of stage 1 (no gradient) shows no evidence of directional bias, while the CI of stage 2 (FBS gradient) shows a noticeable bias toward the FBS source. Migrational bias toward the FBS source is expected, since chemotactic cancer cells, particularly MDA-MB-231, are known to migrate toward sources of serum and its components (i.e., epidermal growth factor).46, 59
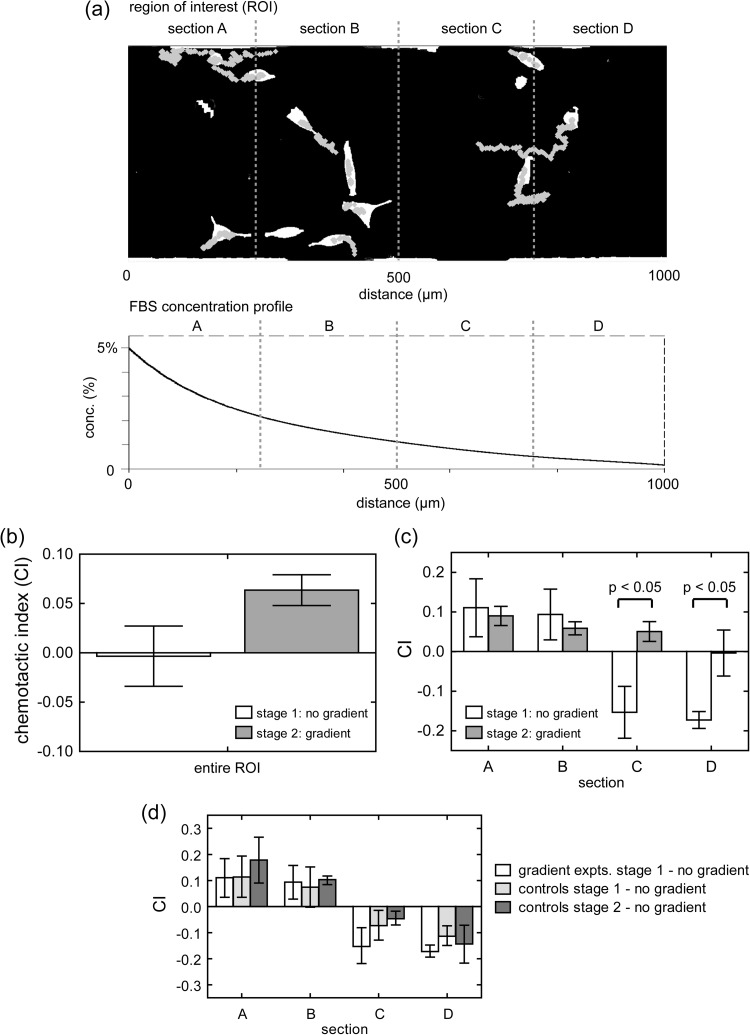
Figure 6.
CI resulting from cell experiments. Positive CI indicates net movement toward the FBS source, while negative CI indicates movement away. (a) Representative segmented image (ROI) shows cells colored white and migration tracks in gray. The FBS concentration profile used in experiments is plotted as an exponential fit (for clarity). Cell centroids are tracked over two stages of each experiment. Stage 1 is 6 h with no gradient, and stage 2 is 6 h with the gradient. (b) CI for stage 1 (non-gradient) and stage 2 (gradient) condition for entire ROI. (c) CI for stages 1 and 2, when the ROI is divided into four equal sections into which the data are binned. Statistically significant differences occur in sections C and D. (d)No gradient control experiments show similar CI between the stages 1 and 2. Total 205 cells are tracked. Error bars areSEM.
To further examine the migrational bias and the components of data that contribute to it, chambers are divided into 4 sections (shown in Fig. 6a), and CI data are binned based upon the section of the chamber in which they are observed. Fig. 6c shows significant changes in CI in sections C and D after the gradient is applied. In particular, it appears that cells in section C change direction when the gradient is applied and migrate, on average, up the gradient toward the FBS source. In addition, cells in section D appear to change from a net rightward migration (negative CI) to a CI of roughly zero. The changes in sections C and D appear to be the primary drivers that contribute to the overall change in CI from stage 1 to stage 2 in Fig. 6b.
A notable observation is that the cells appear to be dispersing in both directions before the gradient is applied. CI values in Fig. 6c indicate that cells in sections A and B tend to migrate to the left (positive CI), while cells in sections C and D tend to migrate to the right (negative CI) (see Fig. 6c stage 1 results). Such behavior could be caused by the way cells are loaded into chambers. Cell loading channels are positioned near the middle of the chambers (Fig. 1a). Therefore, the cell population is seeded more densely in the middle of the chamber when experiments begin (i.e., clustered in sections B and C). In the absence of a directional stimulus (i.e., no gradient), motile cells will engage in a persistent random walk characterized by direction changes that are spontaneous or prompted by cell-cell collisions.60, 61 The random walk, collisions, and ability of highly motile cells to resist contact inhibition would cause cells to disperse outwardly from the densely populated starting area similar to molecular diffusion.62 This dispersion resembles a radial monolayer cell migration assay,63, 64 where cells are densely seeded in a circular region, and their radial migration outward from the initial region is tracked. Modifications to the cell loading channels so they distribute cells more uniformly could mitigate such dispersive migration.
Another potential cause of dispersive migration is if additional gradients are formed, which could be made up of nutrients or secreted molecules due to cellular metabolism, respiration or signaling. Such gradients could be chemoattractive or chemorepellant, and therefore they could confound the analysis of migration in response to the applied gradient (from the device). Further discussion on confounding gradients and ways to mitigate their effect is in supplemental material (section on potential limitations of approach and topics of further investigation).37
Nevertheless, when the gradient is applied during stage 2, responses from sections C and D suggest that the initial directional bias (away from the serum source) is reversed and nullified, respectively. As a result, the overall CI shifts toward the FBS source.
CI results from sections A and B during stage 2 (Fig. 6c) could be interpreted in light of certain phenomena. When there is no gradient (stage 1), the positive CI could be due to the abovementioned dispersion from the middle of the chamber. When the gradient is applied, cells continue to show positive CI, which could indicate a continuation in their random-walk-like dispersive migration. Another possibility is that cells sense the gradient, but the level of background concentration causes additional responses that change the cells' morphology (area and circularity), as shown in Fig. 13 (supplementary material).37 The reduction in area and rise in circularity suggest a contractile morphology indicative of cells that have partially detached from the substratum, which could suggest that cell-substratum adhesion is reduced or intracellular tension is increased.62 Therefore, a reduction of adhesion could interfere with efficient chemotaxis, resulting in the slightly lower CI for sections A and B during FBS gradient stimulation (stage 2). One way to test for this is to perform the same experiment, but coat the substrate with a higher concentration of fibronectin (Fn). The presumption is that the increased Fn concentration would increase adhesion, enabling cells in sections A and B to migrate more efficiently when the gradient is applied.
Figure 6d compares stage 1 of gradient experiments with both stages (1 and 2) of control experiments. Migrational bias (CI) is similar (no statistical difference) among the three stages within each of the four sections (A-D) of the chamber.
Speed, area, and circularity for non-gradient and gradient stages are reported in Fig. 13 and compared to controls in Fig. 14 (in the supplementary material).37 In general, speed and circularity increase when the FBS gradient is applied, while area decreases. In the gradient condition, both area and circularity show a slight trend, where the area increases from left to right (source to sink), while circularity decreases in the same direction.
Potential limitations and topics of further investigation are discussed in supplemental material, including nutrient transport simulations (Fig. 15, supplementary material)37 to assess the likelihood of confounding gradients of small molecules in the device.
CONCLUSIONS
This work introduces a microfluidic chamber array that utilizes sequentially pulsed fluid delivery to generate soluble concentration gradients. The device is intended to address the need for gradient-generating devices that exhibit (i) array scalability, (ii) relative simplicity in fabrication, (iii) a no-shear microenvironment, and (iv) gradients with low background concentrations. The combination of all four capabilities is unique among microfluidic devices for the study of cellular response to soluble gradients. The combination of low shear and low background is a physiologically relevant context (e.g., interstitial space), and experimentalists continually seek higher-throughput (scalable) platforms that are relatively simple to fabricate or can be manufactured en-masse and purchased commercially.65 Although flow-based and diffusion-based designs are able to address a subset of such capabilities, none are able to meet all four simultaneously.
The device in this work demonstrates the capability to meet the four needs through the realization of a 6-chamber array and fluorescent characterization of gradients using different pulsing sequences. This is enabled by a 3D channel architecture with wrap-around flows, as well as a simplified, all-PDMS fabrication process with a single alignment step. Preliminary cell experiments demonstrate cell viability and migrational response to a FBS gradient by a location-dependent subset of cells.
Further experiments are required to fully validate the device as a cell-based gradient assay. Such experiments would include different pulsing sequences, different cell types and signals, and modified designs that mitigate the potential for confounding gradients. Improved and expanded versions of this array technology could be directly applied to studies in cancer, development, tissue regeneration/repair, and immunology, as well as serve as a tool to screen potential therapeutic compounds to treat related diseases in a parallel fashion.
ACKNOWLEDGMENTS
The authors thank Dr. Kwanghun Chung and Dr. Lino Gonzalez for helpful discussions and Dr. Jeffrey N. Stirman for LabVIEW automation programming. E.S.P. was supported by a National Defense Science and Engineering Graduate (NDSEG) fellowship from the U.S. Department of Defense, and H.L. was supported by a DuPont Young Professor grant and a Sloan Research Fellowship. The authors also thank the US National Institutes of Health and the U.S. National Science Foundation for financial support.
References
- Aman A. and Piotrowski T., Dev. Biol. 341(1), 20–33 (2010). 10.1016/j.ydbio.2009.11.014 [DOI] [PubMed] [Google Scholar]
- Hatten M. E., Annu. Rev. Neurosci. 22, 511–539 (1999). 10.1146/annurev.neuro.22.1.511 [DOI] [PubMed] [Google Scholar]
- Martin P., Science 276(5309), 75–81 (1997). 10.1126/science.276.5309.75 [DOI] [PubMed] [Google Scholar]
- Luster A. D., Alon R., and von Andrian U. H., Nat. Immunol. 6(12), 1182–1190 (2005). 10.1038/ni1275 [DOI] [PubMed] [Google Scholar]
- Zhelev D. V. and Alteraifi A., Ann. Biomed. Eng. 30(3), 356–370 (2002). 10.1114/1.1477446 [DOI] [PubMed] [Google Scholar]
- Friedl P. and Wolf K., Nat. Rev. Cancer 3(5), 362–374 (2003). 10.1038/nrc1075 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H., Wyckoff J., and Condeelis J., Curr. Opin. Cell Biol. 17(5), 559–564 (2005). 10.1016/j.ceb.2005.08.002 [DOI] [PubMed] [Google Scholar]
- El-Ali J., Sorger P. K., and Jensen K. F., Nature 442(7101), 403–411 (2006). 10.1038/nature05063 [DOI] [PubMed] [Google Scholar]
- Takayama S., Ostuni E., LeDuc P., Naruse K., Ingber D. E., and Whitesides G. M., Nature 411(6841), 1016 (2001). 10.1038/35082637 [DOI] [PubMed] [Google Scholar]
- Dittrich P. S. and Manz A., Nat. Rev. Drug Discov. 5(3), 210–218 (2006). 10.1038/nrd1985 [DOI] [PubMed] [Google Scholar]
- Gomez-Sjoberg R., Leyrat A. A., Pirone D. M., Chen C. S., and Quake S. R., Anal. Chem. 79(22), 8557–8563 (2007). 10.1021/ac071311w [DOI] [PubMed] [Google Scholar]
- Toetsch S., Olwell P., Prina-Mello A., and Volkov Y., Integr. Biol. 1(2), 170–181 (2009). 10.1039/b814567a [DOI] [PubMed] [Google Scholar]
- Keenan T. M. and Folch A., Lab Chip 8(1), 34–57 (2008). 10.1039/b711887b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim H. J., and Jeon N. L., Integr. Biol. 2(11-12), 584–603 (2010). 10.1039/c0ib00055h [DOI] [PubMed] [Google Scholar]
- Irimia D., Annual Review of Biomedical Engineering (Annual Reviews, Palo Alto, 2010), Vol. 12, pp. 259–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korin N., Bransky A., Dinnar U., and Levenberg S., Lab Chip 7(5), 611–617 (2007). 10.1039/b702392h [DOI] [PubMed] [Google Scholar]
- Walker G. M., Sai J. Q., Richmond A., Stremler M., Chung C. Y., and Wikswo J. P., Lab Chip 5(6), 611–618 (2005). 10.1039/b417245k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. M., Zeringue H. C., and Beebe D. J., Lab Chip 4(2), 91–97 (2004). 10.1039/b311214d [DOI] [PubMed] [Google Scholar]
- Van Haastert P. J. M. and Devreotes P. N., Nat. Rev. Mol. Cell Biol. 5(8), 626–634 (2004). 10.1038/nrm1435 [DOI] [PubMed] [Google Scholar]
- Mayr L. M. and Bojanic D., Curr. Opin. Pharmacol. 9(5), 580–588 (2009). 10.1016/j.coph.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Whitesides G. M., Nature 442(7101), 368–373 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- Chung B. G., Flanagan L. A., Rhee S. W., Schwartz P. H., Lee A. P., Monuki E. S., and Jeon N. L., Lab Chip 5(4), 401–406 (2005). 10.1039/b417651k [DOI] [PubMed] [Google Scholar]
- Herzmark P., Campbell K., Wang F., Wong K., El-Samad H., Groisman A., and Bourne H. R., Proc. Natl. Acad. Sci. U.S.A. 104(33), 13349–13354 (2007). 10.1073/pnas.0705889104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon N. L., Baskaran H., Dertinger S. K. W., Whitesides G. M., Van de Water L., and Toner M., Nat. Biotechnol. 20(8), 826–830 (2002). 10.1038/nbt712 [DOI] [PubMed] [Google Scholar]
- Wang C. J., Li X., Lin B., Shim S., Ming G. L., and Levchenko A., Lab Chip 8(2), 227–237 (2008). 10.1039/b713945d [DOI] [PubMed] [Google Scholar]
- Yang C.-G., Wu Y.-F., Xu Z.-R., and Wang J.-H., Lab Chip 11(19), 3305–3312 (2011). 10.1039/c1lc20123a [DOI] [PubMed] [Google Scholar]
- Keenan T. M., Hsu C. H., and Folch A., Appl. Phys. Lett. 89(11), 114103 (2006). [Google Scholar]
- Shamloo A., Ma N., Poo M. M., Sohn L. L., and Heilshorn S. C., Lab Chip 8(8), 1292–1299 (2008). 10.1039/b719788h [DOI] [PubMed] [Google Scholar]
- Cate D. M., Sip C. G., and Folch A., Biomicrofluidics 4(4), 044105–044110 (2010). 10.1063/1.3490784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lokuta M. A., Huttenlocher A., and Beebe D. J., Lab Chip 9(12), 1797–1800 (2009). 10.1039/b901613a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel M., Galas J.-C., Dahan M., and Studer V., Lab Chip 12(7), 1340–1346 (2012). 10.1039/c2lc20994b [DOI] [PubMed] [Google Scholar]
- Mosadegh B., Agarwal M., Tavana H., Bersano-Begey T., Torisawa Y., Morell M., Wyatt M. J., O'Shea K. S., Barald K. F., and Takayama S., Lab Chip 10(21), 2959–2964 (2010). 10.1039/c0lc00086h [DOI] [PubMed] [Google Scholar]
- Cheng S. Y., Heilman S., Wasserman M., Archer S., Shuler M. L., and Wu M. M., Lab Chip 7(6), 763–769 (2007). 10.1039/b618463d [DOI] [PubMed] [Google Scholar]
- Kothapalli C. R., van Veen E., de Valence S., Chung S., Zervantonakis I. K., Gertler F. B., and Kamm R. D., Lab Chip 11(3), 497–507 (2011). 10.1039/c0lc00240b [DOI] [PubMed] [Google Scholar]
- Choi E., Chang H.-k., Young Lim C., Kim T., and Park J., Lab Chip 12(20), 3968–3975 (2012). 10.1039/c2lc40450h [DOI] [PubMed] [Google Scholar]
- Atencia J., Morrow J., and Locascio L. E., Lab Chip 9(18), 2707–2714 (2009). 10.1039/b902113b [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4774313 for a review of flow-based and high resistance gradient devices, device layout, illustration of fabrication process, chamber-to-chamber comparisons, time-to-steady-state (rise time) discussion, further results from preliminary cell experiments, methods for image processing and data analysis, nutrient transport models, and movie of cell experiment.
- Park E. S., Brown A. C., DiFeo M. A., Barker T. H., and Lu H., Lab Chip 10(5), 571–580 (2010). 10.1039/b919294h [DOI] [PubMed] [Google Scholar]
- Unger M. A., Chou H. P., Thorsen T., Scherer A., and Quake S. R., Science 288(5463), 113–116 (2000). 10.1126/science.288.5463.113 [DOI] [PubMed] [Google Scholar]
- McDonald J. C., Duffy D. C., Anderson J. R., Chiu D. T., Wu H. K., Schueller O. J. A., and Whitesides G. M., Electrophoresis 21(1), 27–40 (2000). [DOI] [PubMed] [Google Scholar]
- Madou M. J., Fundamentals of Microfabrication: The Science of Miniaturization, 2nd ed. (CRC, Boca Raton, 2002). [Google Scholar]
- Folch A., Jo B. H., Hurtado O., Beebe D. J., and Toner M., J. Biomed. Mater. Res. 52(2), 346–353 (2000). [DOI] [PubMed] [Google Scholar]
- Kim L., Vahey M. D., Lee H. Y., and Voldman J., Lab Chip 6(3), 394–406 (2006). 10.1039/b511718f [DOI] [PubMed] [Google Scholar]
- Periasamy N. and Verkman A. S., Biophys. J. 75(1), 557–567 (1998). 10.1016/S0006-3495(98)77545-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chary S. R. and Jain R. K., Proc. Natl. Acad. Sci. U.S.A. 86(14), 5385–5389 (1989). 10.1073/pnas.86.14.5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. J., Saadi W., Lin F., Nguyen C. M. C., and Jeon N. L., Exp. Cell Res. 300(1), 180–189 (2004). 10.1016/j.yexcr.2004.06.030 [DOI] [PubMed] [Google Scholar]
- See http://www.fluidigm.com/. Fluidigm is a biotech tools company that creates microfluidic-based chips and instrumentation for biological research.
- Sip C. G., Bhattacharjee N., and Folch A., Biomicrofluidics 5(2), 022210 (2011). 10.1063/1.3576931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abhyankar V. V., Lokuta M. A., Huttenlocher A., and Beebe D. J., Lab Chip 6(3), 389–393 (2006). 10.1039/b514133h [DOI] [PubMed] [Google Scholar]
- Diao J. P., Young L., Kim S., Fogarty E. A., Heilman S. M., Zhou P., Shuler M. L., Wu M. M., and DeLisa M. P., Lab Chip 6(3), 381–388 (2006). 10.1039/b511958h [DOI] [PubMed] [Google Scholar]
- Torisawa Y.-s., Mosadegh B., Bersano-Begey T., Steele J. M., Luker K. E., Luker G. D., and Takayama S., Integr. Biol. 2(11-12), 680–686 (2010). 10.1039/c0ib00041h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L., Toh Y. C., Voldman J., and Yu H., Lab Chip 7(6), 681–694 (2007). 10.1039/b704602b [DOI] [PubMed] [Google Scholar]
- Pedersen J. A., Boschetti F., and Swartz M. A., J. Biomech. 40(7), 1484–1492 (2007). 10.1016/j.jbiomech.2006.06.023 [DOI] [PubMed] [Google Scholar]
- Fuller D., Chen W., Adler M., Groisman A., Levine H., Rappel W.-J., and Loomis W. F., Proc. Natl. Acad. Sci. U.S.A. 107(21), 9656–9659 (2010). 10.1073/pnas.0911178107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoge M., Adler M., Groisman A., Levine H., Loomis W. F., and Rappel W.-J., Integr. Biol. 2(11–12), 659–668 (2010). 10.1039/c0ib00033g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A., Homey B., Soto H., Ge N. F., Catron D., Buchanan M. E., McClanahan T., Murphy E., Yuan W., Wagner S. N., Barrera J. L., Mohar A., Verastegui E., and Zlotnik A., Nature 410(6824), 50–56 (2001). 10.1038/35065016 [DOI] [PubMed] [Google Scholar]
- Bifulco K., De Chiara A., Fazioli F., Longanesi-Cattani I., Cantelmo A. R., Tirino V., Apice G., Rocco G., Lombardi M. L., and Carriero M. V., Tumori 94(4), 505–510 (2008). [DOI] [PubMed] [Google Scholar]
- Weinberg R. A., The Biology of Cancer (Garland Science, Taylor & Francis Group, LLC, New York, NY, 2007). [Google Scholar]
- Wyckoff J. B., Segall J. E., and Condeelis J. S., Cancer Res. 60(19), 5401–5404 (2000); available at http://cancerres.aacrjournals.org/content/60/19/5401.full. [PubMed] [Google Scholar]
- Cheng G., Youssef B. B., Markenscoff P., and Zygourakis K., Biophys. J. 90(3), 713–724 (2006). 10.1529/biophysj.105.063701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware M. F., Wells A., and Lauffenburger D. A., J. Cell Sci. 111, 2423–2432 (1998); available at http://jcs.biologists.org/content/111/16/2423.abstract. [DOI] [PubMed] [Google Scholar]
- Lauffenburger D. A. and Linderman J. J., Receptors: Models for Binding, Trafficking, and Signaling (Oxford University Press, Inc., New York, NY, 1993). [Google Scholar]
- Berens M. E. and Beaudry C., Cancer Cell Culture, Methods and Protocols, edited by Langdon S. P. (Humana, 2004), Vol. 88, pp. 219–224. [Google Scholar]
- Deryugina E. I. and Bourdon M. A., J. Cell Sci. 109(3), 643–652 (1996); available at http://jcs.biologists.org/content/109/3/643.full.pdf. [DOI] [PubMed] [Google Scholar]
- Berthier E., Surfus J., Verbsky J., Huttenlocher A., and Beebe D., Integr. Biol. 2(11–12), 630–638 (2010). 10.1039/c0ib00030b [DOI] [PubMed] [Google Scholar]