Abstract
Tumor microenvironment is a highly complex system consisting of non-cancerous cells, soluble factors, signaling molecules, extracellular matrix, and mechanical cues, which provides tumor cells with integrated biochemical and biophysical cues. It has been recognized as a significant regulator in cancer initiation, progression, metastasis, and drug resistance, which is becoming a crucial component of cancer biology. Modeling microenvironmental conditions of such complexity in vitro are particularly difficult and technically challenging. Significant advances in microfluidic technologies have offered an unprecedented opportunity to closely mimic the physiological microenvironment that is normally encountered by cancer cells in vivo. This review highlights the recent advances of microfluidic platform in recapitulating many aspects of tumor microenvironment from biochemical and biophysical regulations. The major events relevant in tumorigenesis, angiogenesis, and spread of cancer cells dependent on specific combinations of cell types and soluble factors present in microenvironmental niche are summarized. The questions and challenges that lie ahead if this field is expected to transform the future cancer research are addressed as well.
INTRODUCTION
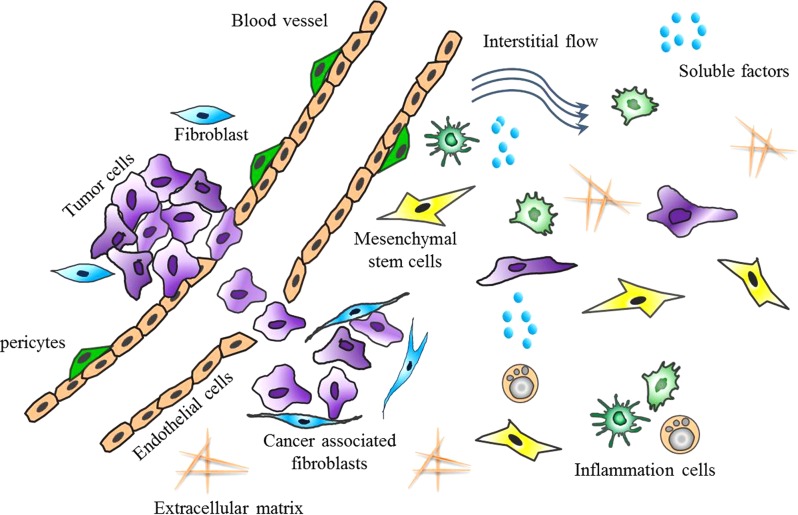
Tumor microenvironment is a crucial component of cancer biology, which is increasingly recognized as a significant factor involved in cancer initiation, progression, metastasis, and drug resistance.1, 2 It is mainly composed of a variety of soluble factors, multiple types of non-malignant cell components, extracellular matrix (ECM), and mechanical cues, in which the cell-cell, cell-matrix, autocrine, paracrine, and hormonal interactions signaling are characterized by high levels of spatio-temporal complexity (Fig. 1). Over the past decades, researchers have gradually realized that the tumor growth is not just determined by malignant cancer cells themselves but also by tumor microenvironment due to the genetic and cell biological development.3, 4, 5 While the normal cellular microenvironment can inhibit malignant cell growth, the modifications that occur in the tumor microenvironment synergistically support cell proliferation. Therefore, intensive understanding of the contribution of tumor microenvironment to tumor progress and development is beneficial in guiding the successful design of anti-cancer therapy and new clinical cancer diagnostics.
Figure 1.
Schematic of the tumor microenvironment consisting of non-cancerous cells, soluble factors, signaling molecules, ECM, mechanical cues, and so on.
In vivo, tumor microenvironment is a highly complex system, which provides tumor cells with integrated biochemical and biophysical cues.1 Several biological properties, including tissue hypoxia, soluble factors secretion, proteolytic enzyme production, and immune response co-constitute the special hallmarks of tumor microenvironment, demonstrating the critical roles in tumor growth, metastasis, and angiogenesis.6, 7, 8 It is becoming increasingly apparent that the malignant tumor progression is maintained by dynamic interplay between the tumor cells and many distinct cell types existing in the adjacent microenvironment, including endothelial cells, fibroblasts, inflammation cells, and so on.9 For example, tumor cells could recruit stroma cells through production and secretion of stimulatory growth factors and cytokines, and so on.10 The locally activated tumor microenvironment in turn could remodel ECM arrangement and modify the proliferative and invasion behavior of tumor cells. In addition to the biochemical factors, some biophysical factors have been increasingly observed to enable the regulation of various cell behaviors and physiological process, such as cell adhesion, migration, proliferation, and tumor invasion.11 As such, the very nature of tumor microenvironment is a heterogenic system with complexity, which is based on the dynamic molecular signaling events, providing malignant cells with the ability adaptive to microenvironmental changes. Such kind of complexity requires the approach to investigate many variables in microenvironment niche dynamically and precisely.
Presently, several cell-based, organotypic/explant, and animal models have been adopted to investigate the complicated interactions between diverse types of cells and signal molecules in the tumor microenvironment.12, 13, 14, 15 A number of cytokine networks and signal pathways mediated the tumor invasion and metastasis have been identified by using various approaches including immunochemistry, polymerase chain reaction (PCR) assay, and western blotting.16, 17, 18, 19 Various gene mutation and genetic variation in many types of cancer during progression were gradually determined by gene profile analysis.20, 21, 22 However, as a heterogenic and dynamic ecosystem, the tumor microenvironment exhibits a high level of spatial-temporal complexity, in which the cancer cells response to therapy are influenced by the chemical and physical cues and cell-cell communications. Therefore, the construction of microenvironmental conditions with such complexity is very difficult and technically challenging.
The microfluidic technology is emerging as an attractive platform for a variety of applications in cell biology and bioengineering fields.23, 24, 25, 26, 27 This microscale technology allows for the flexible design of the microchannel network with inherent comparable dimensions to the size of cells and blood vessels, and the realization of controllable laminar fluidic flow with ultralow volumes at microscale. In addition, it enables the experimental conditions parallelization, the measurement of cellular dynamics in real-time, and the delivery of reagents, nutrients, and other stimuli to cells precisely.28, 29, 30, 31 Particularly, it facilitates the recreation of the micro-scale cell confinement with critical cell-cell interfaces, spatiotemporal chemical gradients, and dynamic mechanical microenvironment of cells with a higher similarity to physiological conditions, which is not possible by conventional approach. These new capabilities that emerged from the convergence of microfluidics with microengineering and cell biology have led to the progresses to reconstitute the physiologically relevant tumor microenvironment on a chip.32, 33, 34, 35, 36
In this article, we provide an overview of progress made in this field over the past few years with a focus on the applications of microfluidic platform in recapitulating many aspects of tumor microenvironment. We review the capabilities of this platform to offer well-defined and controllable chemical and physical cues with a similarity to tumor microenvironment. We also discuss the potential use of this platform for cancer biology, and drug testing applications, as well as challenges for the field that must be addressed to translate this technology into useful tool in near future.
BIOCHEMICAL REGULATION OF TUMOR MICROENVIRONMENT
Both biochemical and biophysical properties of the tumor microenvironment can regulate various cellular behaviors, such as growth, differentiation, morphogenesis, and migrations of cancer cells. The major biochemical factors in local host tissue mainly include non-malignant cell components, soluble factors, ECM components, and so on. Based on the capabilities of microfluidic technology with highly spatial and temporal control, it is feasible to reconstitute the essential elements of physiologically relevant tumor microenvironment with biochemical properties. As fewer works are reported regarding to the ECM components study using microfluidic approach, here we mainly focus on the work to explore the roles of soluble factors gradients and non-malignant cell components involved in the microenvironmental niche.
Gradient of soluble factors involved in cancer cell migration
Many biological processes in cancer such as cell migration and invasion can occur in the tumor progression, with the presence of various growth factors, chemoattractants, and other biological agents. In vivo, these soluble factors can be driven towards the vicinity of tumor by the interstitial flow through ECM in a gradient mode.37 The microfluidic approach can offer a robust way to generate the dynamic soluble factor gradient reproducibly in the diverse physiologically relevant conditions.38
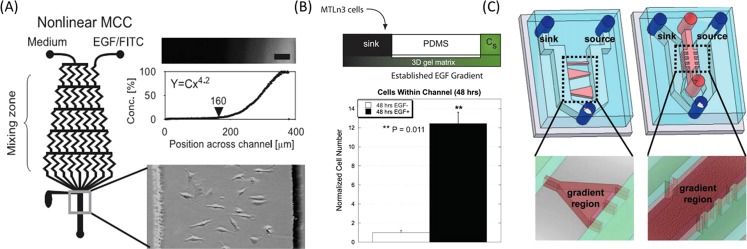
Currently, there are two ways to produce the soluble factors gradients on a microfluidic platform such as flow-based and diffusion-based methods. The former is largely dependent on the presence of fluid flow over the gradient regions, utilizing the convection in the laminar flow streams to form molecule gradient. While, the latter is mainly dependent on the free-diffusion of soluble molecules through the microchannels with high fluidic resistance or within three-dimensional (3D) matrix.39, 40 By intensive investigations of the cell migrations under soluble factors gradient, the mechanism by which the tumor cell senses and interprets gradients around the cell body is gradually well understood. Epidermal growth factor (EGF), as a common soluble factor, is known to activate a cascade of multiple signaling pathways that can facilitate tumor growth, angiogenesis, and metastasis. Also, enhanced overexpression of EGF-receptor is proved to be closely associated with tumor progression and poor survival in various malignancies, such as lung,41 head and neck,42, 43 breast,44 and colorectal cancer.45 Up to now, several works have been reported to generate flow-based EGF gradient with various concentration ranges and profile shapes by using the premixing microfluidic network and study the chemotaxis behaviors of cancer cells.46, 47, 48 Metastatic breast cancer cells were observed to exhibit marked directional movement toward higher EGF concentration in nonlinear gradient profile, indicating the specific migration behavior of cancer cells dependent on the shape of gradient profile and the range of EGF concentrations46 (Fig. 2a). Meanwhile, the effect of antibody against EGF receptor on the chemotaxis of breast cancer cells (MDA-MB-231) was evaluated quantitatively,47 and anti-EGF receptor-treated cells were randomly polarized, indicating the prominent role of EGF receptor in regulating the breast cancer cells migration and chemotaxis behaviors.
Figure 2.
Gradient of soluble factors regulated the invasion of tumor cell. (a) Flow-based gradient generator with a premixing microchannel network.46 Reprinted with permission from S. J. Wang et al., Exp. Cell Res. 300, 180 (2004). Copyright 2004 Elsevier. (b) Diffusion-based gradient generated between sink reservoir and source microchannel.49 Reprinted with permission from V. V. Abhyankar et al., Lab Chip 8, 1507 (2008). Copyright 2008 The Royal Society of Chemistry. (c) Similar gradient generation method on 2D plate and in 3D matrix by varying the media between the two channels.50 Reprinted with permission from B. Mosadegh et al., Langmuir 23, 10910 (2007). Copyright 2007 American Chemical Society.
In addition, other researchers reported the formation of stable gradient by diffusion of soluble molecule through the porous 3D ECM, aiming to provide more realistic and reliable biological process49, 50 (Figs. 2b, 2c). Beebe et al. developed an accessible microfluidic structure to generate long-term EGF gradient in the straight channel within a 3D microenvironment, the numbers of cancer cells existing in stimulated channels were greatly higher than that from unstimulated channel49 (Fig. 2b). Alternatively, by designing the microfluidic device with two parallel perfusion channels connected by elliptic cell culture chambers, a stable EGF gradient can be generated by diffusion of the molecules.51 Based on protrusion index, the migration of tumor cell can be measured quantitatively, thus, facilitating the characterization of cell behaviors in response to chemical gradients in the tumor microenvironment.
Non-malignant cell components in tumor microenvironment
Fibroblasts
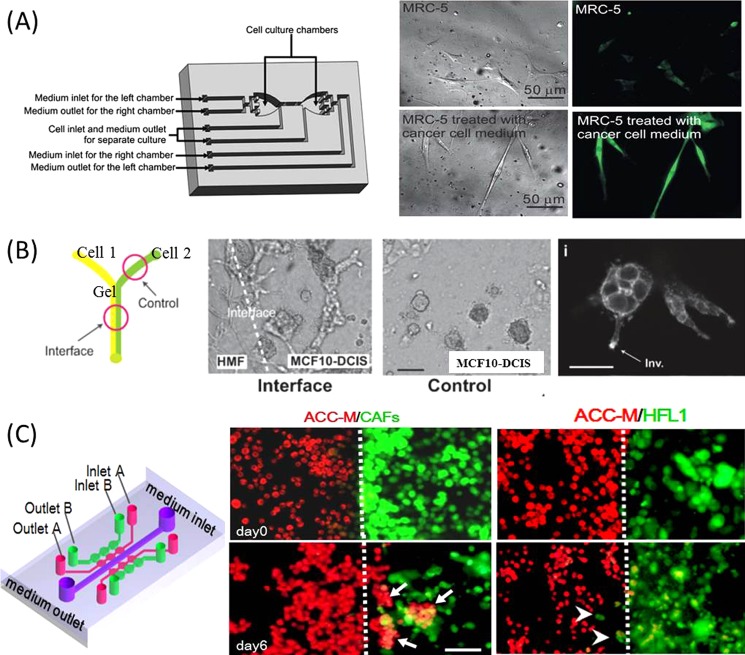
Fibroblasts are predominant cell types in the connective tissue, which have been found to be associated with tumor cells at all stages of tumor progression. They are recognized as the source of multiple growth factors that can influence the tumor cell behaviors and be responsible for the synthesis, deposition and remodeling much of ECM in tumor stroma.52 Meanwhile, fibroblasts are continuously induced by the diffusive factors secreted by tumor cells, and thus can be activated with the specific expression of α-smooth muscle actin (α-SMA).52, 53 Several microfluidic devices have been developed to investigate the interaction between fibroblasts and tumor cells within microenvironment by using separated microstructure54 or pneumatic microvalves control55 based on microscale engineering technology. Qin et al. designed a simple microfluidic device, allowing for the multiple types of cells co-culturing in the separated microchambers, with the soluble factors diffusion through the parallel cell migration regions.54 This study reproduced the biological process of fibroblasts recruitment in the presence of the tumor cells and the activation of fibroblasts with a high expression of specific marker (α-SMA). Furthermore, the molecular mechanism underlying this activated process of fibroblasts was elucidated by Lee et al.55 It was found that this paracrine loop mediated the activated process could be inhibited with the addition of transforming growth factor beta 1 (TGF-β1) receptor/ALK5 inhibitor into the medium of fibroblasts, which might support the target of cancer therapy to prevent activated fibroblast (Fig. 3a). In addition to the inducing effect of tumor cells on the fibroblasts, it was also interesting to explore the feedback effect of fibroblasts in tumor microenvironment. A laminar flow patterning on the microfluidic structure has been utilized for the loading of breast cancer cells adjacent to stromal fibroblasts, thus recapitulating the general spatial relationship between the two types of cells observed in vivo56 (Fig. 3b). It is assumed that the fibroblasts can accelerate the process of breast cancer cells transition to an invasive phenotype by secreting soluble factors and cell-cell contact. In addition, by fabricating the microstructure with precisely spatial-temporal control in the flow direction and the multi-site staying of the fluids, hepatocellular carcinoma cells have been observed to produce the spatial aggregations when co-cultured with fibroblasts on an integrated microfluidic system.57 These results suggested that the different behavior and morphology of tumor cell might be activated through the stromal signals involved in the microenvironment.
Figure 3.
Fibroblasts involved in the tumor microenvironment. (a) Fibroblasts can be activated by tumor cells in the fluidic channels of the cell culture chip with pneumatic micro-valves. The activated fibroblasts appeared as the stretched body and high expression of α-SMA under the treatment of the cancer cell medium.55 Reprinted with permission from T.-H. Hsu et al., Lab Chip 11, 1808 (2011). Copyright 2011 The Royal Society of Chemistry. (b) The invasive transition of breast cancer cells can be seen after co-cultured with fibroblasts on the Y-shaped microfluidic chip.56 Reprinted with permission from K. E. Sung et al., Integr. Biol. 3, 439 (2011). Copyright 2011 The Royal Society of Chemistry. (c) Carcinoma-associated fibroblasts promoted tumor invasion in the spheroid mode on a microfluidic 3D co-culture device.62 Reprinted with permission from T. Liu et al., Lab Chip 10, 1671 (2010). Copyright 2010 The Royal Society of Chemistry.
Recently, the role of the activated fibroblasts in cancer is gradually recognized and the evidence is increasing that the subpopulation of fibroblasts—the so-called cancer associated fibroblasts (CAFs)—are important promoters of tumor growth and progression.58, 59, 60, 61 A 3D microfluidic device incorporated with six co-culture units was designed to investigate the heterotypic interaction between CAFs and adenoid cystic carcinoma (ACC-M) cells62 (Fig. 3c). In this work, CAFs were primarily isolated from the carcinoma associated tissues, and then co-cultured with ACC-M cells in 3D matrix with close communication via medium diffusion. It was observed that CAFs could promote the invasion of ACC-M cells in a spheroid fashion and the invasion ability could be evaluated quantitatively and in real-time. Meanwhile, the process of CAF-promoted cancer invasion could be prohibited by MMP inhibitor, suggesting that the MMP inhibitor might be a powerful candidate against CAF-target anti-invasion therapy.63, 64 This microdevice provides a physiologically relevant tumor microenvironment for the modeling of cancer progression and potential drug testing in a 3D format.
Endothelial cells
Endothelial cell is one of the key determinants in tumor microenvironment, which can interact with tumor cells, ECM, and immune cells in the cellular niches. Commonly, the endothelial cells can assemble to form the linings of the walls of capillaries and blood vessels with barrier function, representing a vital component involving in the process of tumor intravasation, extravasation, and angiogenesis.65
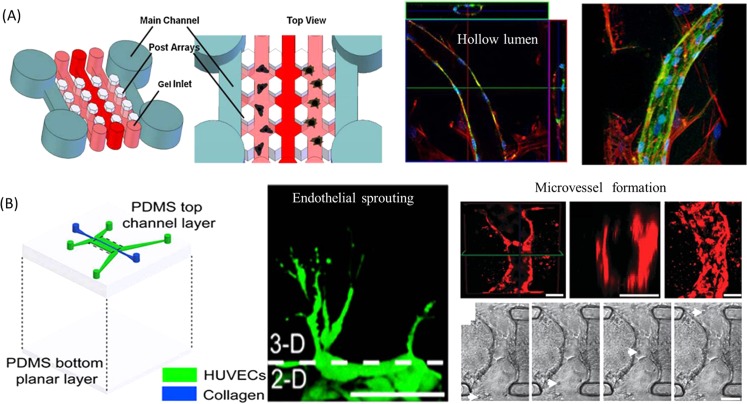
Several works have been performed on a microfluidic device by using endothelial cells as a major component and study some biological processes related to angiogenesis and tumor invasion. Putnam et al. designed a microfluidic device which was composed of paralleled channels connected by a series of postarrays and investigated the endothelial cells with the supporting ability for new blood vessel generation.66 This device can facilitate the cell loading process sequentially by using the periodic hexagonal posts designed to contain different types of cells in hydrogels (Fig. 4a). Bone marrow-derived mesenchymal stem cells (MSCs) suspended within 3D fibrin gels were patterned in the channel adjacent to endothelial cells and they executed a morphogenetic process akin to vasculogenesis, forming a primitive vascular plexus and maturing into a robust capillary network with hollow well-defined lumens. In addition, a typical endothelial sprouting process from an intact vessel could be observed on a microfluidic platform by designing the device with two parallel endothelial loaded channels and 3D collagen matrix on the abluminal side.67 As shown in Fig. 4b, the process of blood vessel dilation could be induced by the negative vascular endothelial growth factor (VEGF) gradient and filopodial extensions could be caused by the positive VEGF gradient in the sprouting of new vessel. Furthermore, the authors reproduced the dynamics of vascular anastomosis68 using the same device. The obvious advantages obtained by this device are that it can reproduce anastomosis process with functional vessel, by accurately lining the endothelial cells adjacent the collagen gel and precise control of the physiological flow within microstructure, thus providing a novel approach to study the mechanisms of angiogenesis in cancer development.
Figure 4.
The endothelial sprouting and new blood vessel formation in biomimetic tumor microenvironment. (a) Schematic representation of a microfluidic device showing two parallel main channels, which provide media and nutrients to the gel channels to support cell co-culture between MSCs and endothelial cells.66 Confocal images shown in an orthogonal display confirmed the presence of hollow lumens in the forming capillary-like structure. Reprinted with permission from B. Carrion et al., Biotechnol. Bioeng. 107, 1020 (2010). Copyright 2010 John Wiley and Sons. (b) Microfluidic device with localized 3D ECM used for endothelial sprounting67 and new functional microvessel formation by anastomosis in vitro. Particle (white arrow) advection through the lumen can be seen from pictures captured from the video.68 Reprinted with permission from J. W. Song and L. L. Munn, Proc. Natl Acad. Sci. U.S.A. 108, 15342 (2011). Copyright 2011 National Academy of Sciences; J. W. Song et al., Integr. Biol. 4, 857 (2012). Copyright 2012 The Royal Society of Chemistry.
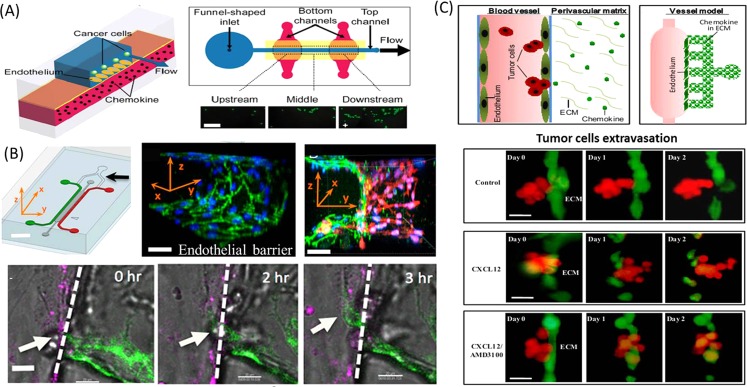
Microfluidic platform can offer a versatile and quantitative way to study the participation of endothelial cells in tumor progression by constructing controllable cues.69 Takayama et al. developed a suitable microdevice to recapitulate the adhesion behavior of breast cancer cells on endothelial layers in the intravasation process under physiological conditions.70 As shown in Fig. 5a, the device consists of two PDMS layers sandwiched by a thin porous polyester membrane cultured with endothelial cells. When the cancer cells flow into the top channel, the chemokines within the bottom channel can site-specifically stimulate the middle endothelium and induce the intravasation of cancer cells (Fig. 5a). Another more complicated tumor-vascular system was developed on a chip with the involvement of tumor cells, endothelial cells, and macrophage cells.71 This device consists of two independently addressable microchannels, in which tumor and endothelial cells are seeded (Fig. 5b). The two channels are interconnected via a 3D hydrogel incorporating into multiple functional regions, thus, enabling the observation, real-time imaging, and precise quantification of endothelial barrier function simultaneously. The intravasation of tumor cells could be enhanced with increased rates and fast dynamics when the endothelial barrier was impaired by TNF-α secreted from macrophages. Another microfluidic device with unique structures was proposed to recapitulate the bioengineered blood vessels in 3D microenvironment and investigate the trans-endothelial invasion of tumor aggregates in the process of extravasation.72 This work closely resembled the blood vessel model in tumor microenvironment by incorporating vessel cavity, endothelium, perivascular ECM, and chemokine gradient (Fig. 5c). The results demonstrated that the ACC-M tumor cell aggregates with positive expression of CXCR4 receptor could transmigrate across the endothelium under the stimulation of gradient chemokine CXCL12 and damage the endothelial integrity in an irreversible manner, indicating the significance of CXCL12/CXCR4 signal pathway in the process of extravasation. Some work also tried to probe the cell migration behavior in the mutual interaction between tumor cells and endothelial cells by mechanical assembly method73 or pneumatic microvalve control.74
Figure 5.
The process of intravasation and extravasation recapitulated on the biomimetic microfluidic devices. (a) The adhesion of cancer cells on the endothelium layer was region-specifically investigated under physiological flow conditions on the microfluidic vasculature.70 Reprinted with permission from J. W. Song et al., PLoS ONE 4, e5756 (2009). Copyright 2009 Public Library of Science. (b) Microfluidic tumor-vascular interface model for tumor cells intravasation study. The confluent endothelial monolayer can be formed on the 3D ECM. In the process of intravasaton, breast carcinoma cell (white arrow) migrated across the HUVEC mono layer (magenta) in the presence of macrophage.71 Reprinted with permission from I. K. Zervantonakis et al., Proc. Natl Acad. Sci. U.S.A. 109, 13515 (2012). Copyright 2012 National Academy of Sciences. (c) Mimicking the extravasation process of tumor aggregates from the endothelial layer based on the bioengineering blood vessel model.72 Reprinted with permission from Q. Zhang et al., Lab Chip 12, 2837 (2012). Copyright 2012 The Royal Society of Chemistry.
Mesenchymal stem cells
MSCs are progenitors of stromal cells and fibroblasts, which have been found to interact with multiple types of tumors in microenvironment, and have aroused great concerns in cancer biology. Nowadays, there are two sides regarding to the participation of MSCs in tumor progression, one is positive and the other is negative.75 For the positive side, MSCs hold a promise avenue for tumor-targeted delivery of therapeutic agents, and can be engineered to release drugs for some specific tumor therapy.76 Recently, a microfluidic platform is also explored to investigate the role of MSCs in salivary gland cancer progression by using a series of functional microdevices in an in vivo like tumor microenvironment.77 These devices are able to perform the study of heterotypic interaction between MSCs and ACC-M cells in 2D and 3D assays by precise cells patterning, stable chemokine gradient formation, and real-time evaluation of cell migration quantitatively. MSCs were observed to be recruited by ACC-M cells significantly. Particularly, MSCs exhibited the ability to enhance the invasion of cancer cells under a chemokine CXCL12 gradient, indicating the involvement of CXCL12–CXCR4 pathway and the role of MSCs in cancer progression. The results emphasized the potential risk of using MSCs as drug delivery carriers for therapeutic purposes in cancer treatment by mimicking the complex components in microenvironment.
Inflammation cells
The inflammation cells are important constituents of the local environment in tumors, which are able to support the survival of malignant cells, promote the angiogenesis and metastasis, and subvert adaptive immune responses.78, 79 Multiple discrete constructs of 3D cell-laden hydrogels have been patterned on microfluidic device for real-time imaging of the interactions between RAW 264.1 macrophage cells and metastatic breast cancer cells after exposure to both autocrine and paracrine signaling molecules.80 Using this device, a series of regularly spaced posts were functionalized as geometric capillary burst valves in order to selectively fill with different types of cells embedded in 3D matrix. The RAW cells were observed to invade into neighboring gels containing MDA-MB-231 cells. Also, the effect of macrophage cells in the participation of tumor-endothelial interaction could be studied on the microdevice cultured with multiple types of cells.71 Macrophages could accelerate the tumor migration across the endothelial barrier by secretion of TNF-α under the involvement of two activated mode (M1 and M2).
BIOPHYSICAL REGULATIONS OF TUMOR MICROENVIRONMENT
As mentioned above, the importance of the biochemical regulations in the process of cancer progression and tumor microenvironment is undisputed; however, the significance of biophysical regulations involving in this microenvironment remains poorly understood. In solid tumor tissue, the cancer cells embedded in ECM are often exposed to biochemical cues and biophysical cues as well. In recent years, the influences of the biophysical factors, such as mechanical fluidic flows and pressures on the diverse cell behaviors in tumor microenvironment have been increasingly acknowledged.81 Microfluidic technology enables the precise control of dynamic fluidic flows and pressure on the microscale and offers the diverse mechanical microenvironments of living cells, which has made it possible to create cancer microenvironment under various physical cues as described in Secs. 3A–3C.
Physical properties of ECM
In solid tumor tissue, the ECM substrate surrounding the cells can provide not only architectural support to cells but also physical cues to the cells. The physical properties of ECM, especially the mechanical elasticity/stiffness, are highly associated with the cell movement mode in the process of tumor invasion.82, 83 The motion pattern adopted by tumor cells through variable ECM microenvironment is dependent on the specific biophysical interactions between tumor cells and ECM component.84, 85, 86 For example, breast cancer cells could activate the actomyosin contractility and increased spreading area by modifying the ECM stiffness, and the changes in ECM stiffness could strongly affect the tumor growth, proliferation, and migration.87
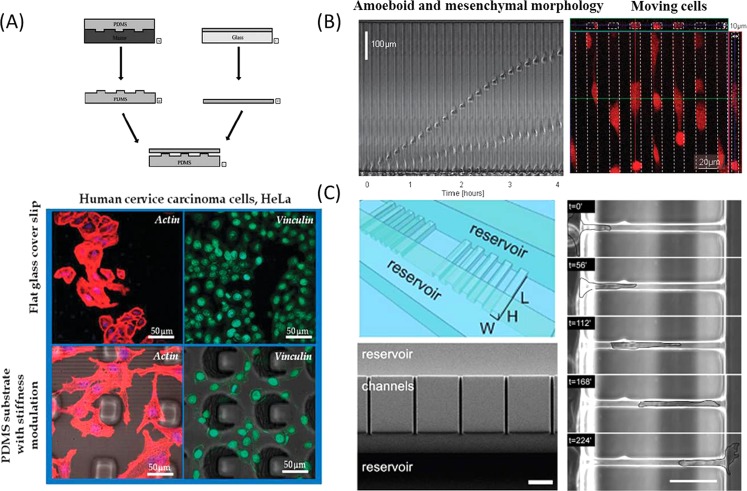
Gigli et al. proposed a “double-sheet” composite structure using soft lithography to introduce a gradient rigidity on the uniform PDMS surface.88 By modifying the shape distribution of microstructure underneath the PDMS membrane and the elasticity, the thickness of PDMS membrane and the gradient of surface rigidity can be changed. Furthermore, another study was tested on a type of cervical cancer HeLa cells. A rapid cytoskeleton remodeling was found to be associated with a more elongated shape characterized by many actin cytoplasmic protrusions on the PDMS substrate with different stiffness modulation (Fig. 7a). Besides, hydrogel presursor solution can be photopolymerized with controllable gradient in the crosslinker concentration by the integration of gradient generator on a microfluidic device. It can effectively generate substrates with well controlled micro-gradient profiles and tunable properties at microscale.89 In this work, the cells cultured on the gradient-compliance gel showed a sharp increase in the spread area, which indicated a tight correlation between the matrix properties and the cellular response. Other researchers also reported to generate variable stiffness or elasticity in the microchannel by modifying the hydrogels with chloroacetic acid, and provided the ECM with versatile properties.90
Figure 7.
The effect of ECM properties and microstructure confinement on tumor invasion. (a) “Double-sheet” composite structure was used to introduce a gradient rigidity on the uniform surface. The rapid cytoskeleton remodeling of HeLa cells can be seen with elongated shape and many actin cytoplasmic protrusions on the PDMS substrate with different stiffness modulation.88 Reprinted with permission from I. E. Palama et al., Integr. Biol. 4, 228 (2012). Copyright 2012 The Royal Society of Chemistry. (b) Sequential frames showing the displacement of two cells confined in the narrow channel, one cell with amoeboid and the other with mesenchymal morphology.105 Reprinted with permission from D. Irimia and M. Toner, Integr. Biol. 1, 506 (2009). Copyright 2009 The Royal Society of Chemistry. (c) The microchannel with subcellular dimension strongly affects the migration phenotype and the spatial cytoskeletal keratin organization related with the tumor cells invasive potential.106 Reprinted with permission from C. G. Rolli et al., PLoS ONE 5, e8726 (2010). Copyright 2010 Public Library of Science.
Mechanical forces
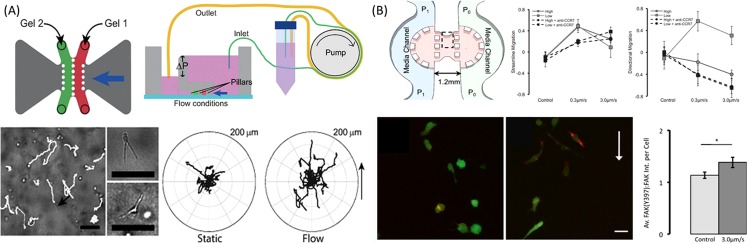
Interstitial fluid flow is one of the most important mechanical forces, which represents the movement of fluid through tissues in vivo. Compared to the normal tissues, interstitial flow can be elevated in tumor tissues because of abnormalities in tumor vascular networks, such as the unregulated vascular permeability,91 tumor-associated angiogenesis, and lymphangiogenesis, as well as changes in the tumor stroma.92, 93 Interstitial flow not only does apply physical forces to cells directly but also creates gradients of soluble signals in the tumor microenvironment, thus influencing cell behavior and modulating cell–cell interactions. As one of the most important mechanical forces, interstitial flow has an essential role in regulating tumor progression and development by modulating various cellular processes.94, 95, 96 Microfluidic technology provides the capabilities to provide the dynamic mechanical cues, such as interstitial flow, to the cells by creating the micro-scale cell microenvironment that goes beyond the current in vitro models. Swartz et al. developed a new and functional microfluidic device to examine the effects of interstitial flow on the morphology and migration in breast cancer cells.97 The device was composed of adjacent matrix regions and two parallel medium reservoirs flanked aside. The cells were initially cultured in the 3D gel with medium, and interstitial flow could be stably formed by maintaining the different pressures across the medium channels.98 Using this device, the breast cancer cells were found to become migratory at the percentage of 20% under the stimulation of interstitial flow (Fig. 6a). The subpopulation of tumor cells that responded to the fluid flow with high directness might act as the leader cells to enhance the tumor invasion and then towards the draining lymphatic vessels.99 In addition, Kamm et al.100 proposed a microfluidic culture system in which the directional and dynamics of cell migration influenced by interstitial flow could be observed and quantified in a physiologically relevant 3D matrix. The data demonstrated that CCR-7 chemokine receptor could respond to the flow-induced gradient of an autocrine chemokine signal and then stimulate the migration of MDA-MB-231 cells in the direction of flow. At the same time, a competing mechanism of tumor cells migrating to the opposite direction of the interstitial flow was found to be associated with integrin and focal adhesion kinase (FAK) activation (Fig. 6b). This microfluidic device can mimic the physiological fluid force in a micro-scale, demonstrating the two signal pathways mediated the migration of breast cancer cells to the interstitial flow. Thus, it can provide new insights for understanding and treating metastatic tumors in the tissue-engineered constructs.
Figure 6.
The biomechanical forces mediated tumor cell invasion. (a) A single unit containing two gel compartments and medium reservoirs with different pressure was headed for forming the interstitial flow to direct tumor cell invasion drive flow. Individual cells tracks can be observed in real time with extended shape.97 Reprinted with permission from U. Haessler et al., Integr. Biol. 4, 401 (2012). Copyright 2012 The Royal Society of Chemistry. (b) Interstitial flow influences direction of tumor cell migration through competing mechanisms. Besides autocrine chemokine signal in the direction of flow, a competing mechanism of tumor cells migrating to the opposite direction of the interstitial flow was found to be associated with FAK Activation.100 Reprinted with permission from W. J. Polacheck et al., Proc. Natl Acad. Sci. U.S.A. 108, 11115 (2011). Copyright 2011 National Academy of Sciences.
Microstructure confinement
Microstructure confinement can be served as one of the physical regulators to tumor cells in the microenvironment. The motility behaviors of tumor cells migrated in 3D microstructure mostly exhibit as mesenchymal, amoeboid, and other hybrid modes, which are different from the flat morphology commonly exhibited on 2D substrates.101, 102 Meanwhile, cell heterogeneity can be conveniently explored by introducing the microfabricated microstructure in the cell culture system with hundreds of cells.103 The microfluidic channels with defined wall stiffness and geometry are able to gain insight into the underlying molecular mechanism of tumor cell migration in the micrometer channels. It was found that human glioma cells confined in the narrow channels surprisingly migrated faster than in wide channels or on the 2D surfaces, which can be attributed to the increased polarization of cell-microstructure traction forces.104 The findings demonstrated that restriction of cells to narrow structure strips can significantly increase the migration speed of cells, which is in accordance with the results observed in other type of tumors including breast cancer, lung carcinoma, and so on.105 Interestingly, a small number of cancer cells were still able to migrate through the narrow channels after the stimulation with drugs targeting the microtubules, indicating that a subpopulation of cells could remain ability to migrate large distances in tissues and establish metastasis despite treatment. The microchannel with subcellular dimension could also be used to investigate the impact of 3D channel structure on the dynamics of tumor migration, invasion, and metastasis,106 suggesting the strong influences of the environmental dimensionality on the migration phenotype of the cells. Particularly, the spatial cytoskeletal keratin organization demonstrated a close relationship with the tumor cells invasive potential (Fig. 7c).
SUMMARY AND CHALLENGES
Microscale engineering and microfluidic technology have proved to be a valuable platform for the exploration of tumor microenvironment containing biochemical and biophysical cues. The technologies reviewed here have the potential to be more predictive human relevant tumor models for effective drug testing, and to provide insights into the mechanisms of action at cellular and tissue levels. Despite the considerable advances in the recreation of tumor microenvironment on a chip, the integration of multiple biochemical and biophysical factors in a physiologically relevant manner to reconstitute realistic whole tumor physiology still remains a huge challenge. There are still more questions and bottlenecks to be addressed, such as (1) developing more accurate approaches to extract the limited number of cells from the microfluidic device for sequential PCR or western-blotting and help understand the regulation of related gene or protein expressions in the microenvironment; (2) closely mimicking what is actually happening in the natural setting of tumor microenvironment, especially for the specified type of cancer; (3) paying more attention to the tumor environment changes when the cells are embedded within 3D matrix for a long time and respond in a cell-mediated way; (4) investigating the single cell behavior and cancer stem cell niche upon environmental perturbations and address the intrinsically heterogenic tumor microenvironment; and (5) coalescing with automated instruments that provide high-level microenvironmental control and real-time analysis of multiple factors to ensure adoption of the techniques in a practical way. As a field, we have to move beyond to confront these challenges that are necessary to overcome and translate the research from laboratory to practical applications. In near future, it is envisaged that the micro-scale technology may well represent the platform of choice towards next generation research in cancer biology, potential therapeutic intervention, and personalized medicine.
ACKNOWLEDGMENTS
This work was supported by the funding of Knowledge Innovation Program of the Chinese Academy of Sciences (KJCX2-YW-H18), Mainland and Hong Kong Joint Research Fund (1026/N_HKUST601/11), and the National Nature Science Foundation of China (No. 81201689).
References
- van Kempen L., Ruiter D. J., van Muijen G. N. P., and Coussens L. M., Eur. J. Cell Biol. 82, 539 (2003). 10.1078/0171-9335-00346 [DOI] [PubMed] [Google Scholar]
- Fukumura D. and Jain R. K., J. Cell. Biochem. 101, 937 (2007). 10.1002/jcb.21187 [DOI] [PubMed] [Google Scholar]
- Shieh A. C., Rozansky H. A., Hinz B., and Swartz M. A., Cancer Res. 71, 790 (2011). 10.1158/0008-5472.CAN-10-1513 [DOI] [PubMed] [Google Scholar]
- Spano D. and Zollo M., Clin. Exp. Metastasis 29, 381 (2012). 10.1007/s10585-012-9457-5 [DOI] [PubMed] [Google Scholar]
- Saha S., Lo P.-K., Duan X., Chen H., and Wang Q., Integr. Biol. 4, 897 (2012). 10.1039/c2ib20034a [DOI] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg R. A., Cell 100, 57 (2000). 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Ruan K., Song G., and Ouyang G., J. Cell. Biochem. 107, 1053 (2009). 10.1002/jcb.22214 [DOI] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg R. A., Cell 144, 646 (2011). 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Pietras K. and Ostman A., Exp. Cell Res. 316, 1324 (2010). 10.1016/j.yexcr.2010.02.045 [DOI] [PubMed] [Google Scholar]
- Tlsty T. D., Semin. Cancer Biol. 11, 97 (2001). 10.1006/scbi.2000.0361 [DOI] [PubMed] [Google Scholar]
- Shieh A. C., Ann. Biomed. Eng. 39, 1379 (2011). 10.1007/s10439-011-0252-2 [DOI] [PubMed] [Google Scholar]
- Breward C. J. W., Byrne H. M., and Lewis C. E., Eur. J. Appl. Math. 12, 529 (2001). 10.1017/S095679250100448X [DOI] [Google Scholar]
- Ciavarra R. P., Holterman D. A., Brown R. R., Mangiotti P., Yousefieh N., Wright G. L., Schellhammer P. F., Glass W. F., and Somers K. D., J. Immunother. 27, 13 (2004). 10.1097/00002371-200401000-00002 [DOI] [PubMed] [Google Scholar]
- Mitsiades C. S., Mitsiades N. S., Richardson P. G., Munshi N. C., and Anderson K. C., J. Cell. Biochem. 101, 950 (2007). 10.1002/jcb.21213 [DOI] [PubMed] [Google Scholar]
- Krasny L., Shimony N., Tzukert K., Gorodetsky R., Lecht S., Nettelbeck D. M., and Haviv Y. S., Nephrol. Dial. Transplant. 25, 373 (2010). 10.1093/ndt/gfp525 [DOI] [PubMed] [Google Scholar]
- Jeffers M., Rong S., and VandeWoude G. F., J. Mol. Med. 74, 505 (1996). 10.1007/BF00204976 [DOI] [PubMed] [Google Scholar]
- Zhang G. X., He B., and Weber G. F., Mol. Cell. Biol. 23, 6507 (2003). 10.1128/MCB.23.18.6507-6519.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivonen S.-K. and Kahari V.-M., Int. J. Cancer 121, 2119 (2007). 10.1002/ijc.23113 [DOI] [PubMed] [Google Scholar]
- Frank D. Y. and Lowery J., Cancer Metastasis Rev. 31, 479 (2012). 10.1007/s10555-012-9380-x [DOI] [PubMed] [Google Scholar]
- Mocellin S., Provenzano M., Rossi C. R., Pilati P., Nitti D., and Lise M., J. Immunol. Methods 280, 1 (2003). 10.1016/S0022-1759(03)00274-6 [DOI] [PubMed] [Google Scholar]
- Ma X.-J., Dahiya S., Richardson E., Erlander M., and Sgroi D. C., Breast Cancer Res. 11, R7 (2009). 10.1186/bcr2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm C. A., Stevens J. W., Xie H., Vanin E. F., Morcuende J. A., Abdulkawy H., Seftor E. A., Sredni S. T., Bischof J. M., Wang D., Malchenko S., d. M. Bonaldo F., Casavant T. L., Hendrix M. J. C., and Soares M. B., BMC Cancer 10, 471 (2010). 10.1186/1471-2407-10-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K.-J. and Suh K.-Y., Lab Chip 10, 36 (2010). 10.1039/b907515a [DOI] [PubMed] [Google Scholar]
- Breslauer D. N., Lee P. J., and Lee L. P., Mol. Biosyst. 2, 97 (2006). 10.1039/b515632g [DOI] [PubMed] [Google Scholar]
- El-Ali J., Sorger P. K., and Jensen K. F., Nature 442, 403 (2006). 10.1038/nature05063 [DOI] [PubMed] [Google Scholar]
- Yi C. Q., Li C. W., Ji S. L., and Yang M. S., Anal. Chim. Acta 560, 1 (2006). 10.1016/j.aca.2005.12.037 [DOI] [Google Scholar]
- Sims C. E. and Allbritton N. L., Lab Chip 7, 423 (2007). 10.1039/b615235j [DOI] [PubMed] [Google Scholar]
- Sia S. K. and Whitesides G. M., Electrophoresis 24, 3563 (2003). 10.1002/elps.200305584 [DOI] [PubMed] [Google Scholar]
- Walker G. M., Zeringue H. C., and Beebe D. J., Lab Chip 4, 91 (2004). 10.1039/b311214d [DOI] [PubMed] [Google Scholar]
- Yeon J. H. and Park J.-K., Biochip J. 1, 17 (2007). [Google Scholar]
- Chao T.-C. and Ros A., J. R. Soc., Interface 5, S139 (2008). 10.1098/rsif.2008.0233.focus [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud D., Mehta G., Mehta K., Linderman J., Takayama S., and Mycek M.-A., J. Biomed. Opt. 11, 050504 (2006). 10.1117/1.2355665 [DOI] [PubMed] [Google Scholar]
- Mehta G., Mehta K., Sud D., Song J. W., Bersano-Begey T., Futai N., Heo Y. S., Mycek M.-A., Linderman J. J., and Takayama S., Biomed. Microdevices 9, 123 (2007). 10.1007/s10544-006-9005-7 [DOI] [PubMed] [Google Scholar]
- Grist S. M., Chrostowski L., and Cheung K. C., Sensors 10, 9286 (2010). 10.3390/s101009286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan J., Leung D. Y. C., Leung M. K. H., Ni M., and Wang H., Int. J. Hydrogen Energy 36, 9231 (2011). 10.1016/j.ijhydene.2011.04.151 [DOI] [Google Scholar]
- Zervantonakis I. K., Kothapalli C. R., Chung S., Sudo R., and Kamm R. D., Biomicrofluidics 5, 013406 (2011). 10.1063/1.3553237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C. P., Hinz B., and Swartz M. A., J. Cell Sci. 118, 4731 (2005). 10.1242/jcs.02605 [DOI] [PubMed] [Google Scholar]
- Chung B. G. and Choo J., Electrophoresis 31, 3014 (2010). 10.1002/elps.201000137 [DOI] [PubMed] [Google Scholar]
- Keenan T. M. and Folch A., Lab Chip 8, 34 (2008). 10.1039/b711887b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim H. J., and Jeon N. L., Integr. Biol. 2, 584 (2010). 10.1039/c0ib00055h [DOI] [PubMed] [Google Scholar]
- Meert A. P., Martin B., Delmotte P., Berghmans T., Lafitte J. J., Mascaux C., Paesmans M., Steels E., Verdebout J. M., and Sculier J. P., Eur. Respir. J. 20, 975 (2002). 10.1183/09031936.02.00296502 [DOI] [PubMed] [Google Scholar]
- Christensen M. E., Dan. Med. Bull. 45, 121 (1998).9587699 [Google Scholar]
- Reuter C. W. M., Morgan M. A., and Eckardt A., Br. J. Cancer 96, 408 (2007). 10.1038/sj.bjc.6603566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R. H., Gao P., Chen L., Ma D. L., Wang J. M., Oppenheim J. J., and Zhang N., Cancer Res. 65, 1433 (2005). 10.1158/0008-5472.CAN-04-1163 [DOI] [PubMed] [Google Scholar]
- Spano J.-P. and Vignot S., Bull. Cancer 94, F171 (2007). [PubMed] [Google Scholar]
- Wang S. J., Saadi W., Lin F., Nguyen C. M. C., and Jeon N. L., Exp. Cell Res. 300, 180 (2004). 10.1016/j.yexcr.2004.06.030 [DOI] [PubMed] [Google Scholar]
- Saadi W., Wang S. J., Lin F., and Jeon N. L., Biomed. Microdevices 8, 109 (2006). 10.1007/s10544-006-7706-6 [DOI] [PubMed] [Google Scholar]
- Walker G. M., Sai J. Q., Richmond A., Stremler M., Chung C. Y., and Wikswo J. P., Lab Chip 5, 611 (2005). 10.1039/b417245k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abhyankar V. V., Toepke M. W., Cortesio C. L., Lokuta M. A., Huttenlocher A., and Beebe D. J., Lab Chip 8, 1507 (2008). 10.1039/b803533d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosadegh B., Huang C., Park J. W., Shin H. S., Chung B. G., Hwang S.-K., Lee K.-H., Kim H. J., Brody J., and Jeon N. L., Langmuir 23, 10910 (2007). 10.1021/la7026835 [DOI] [PubMed] [Google Scholar]
- Liu T., Li C., Li H., Zeng S., Qin J., and Lin B., Electrophoresis 30, 4285 (2009). 10.1002/elps.200900289 [DOI] [PubMed] [Google Scholar]
- Bhowmick N. A., Neilson E. G., and Moses H. L., Nature 432, 332 (2004). 10.1038/nature03096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R. and Zeisberg M., Nat. Rev. Cancer 6, 392 (2006). 10.1038/nrc1877 [DOI] [PubMed] [Google Scholar]
- Ma H., Liu T., Qin J., and Lin B., Electrophoresis 31, 1599 (2010). 10.1002/elps.200900776 [DOI] [PubMed] [Google Scholar]
- Hsu T.-H., Xiao J.-L., Tsao Y.-W., Kao Y.-L., Huang S.-H., Liao W.-Y., and Lee C.-H., Lab Chip 11, 1808 (2011). 10.1039/c1lc20090a [DOI] [PubMed] [Google Scholar]
- Sung K. E., Yang N., Pehlke C., Keely P. J., Eliceiri K. W., Friedl A., and Beebe D. J., Integr. Biol. 3, 439 (2011). 10.1039/c0ib00063a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Li L., Wang X., Ren L., Wang X., Wang J., Tu Q., Huang X., and Wang J., Lab Chip 10, 1717 (2010). 10.1039/c001049a [DOI] [PubMed] [Google Scholar]
- Mueller M. M. and Fusenig N. E., Nat. Rev. Cancer 4, 839 (2004). 10.1038/nrc1477 [DOI] [PubMed] [Google Scholar]
- Orimo A. and Weinberg R. A., Cell Cycle 5, 1597 (2006). 10.4161/cc.5.15.3112 [DOI] [PubMed] [Google Scholar]
- Olumi A. F., Grossfeld G. D., Hayward S. W., Carroll P. R., Tisty T. D., and Cunha G. R., Cancer Res. 59, 5002 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing F., Saidou J., and Watabe K., Front. Biosci. 15, 166 (2010). 10.2741/3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Lin B., and Qin J., Lab Chip 10, 1671 (2010). 10.1039/c000022a [DOI] [PubMed] [Google Scholar]
- Micke P. and Ostman A., Lung Cancer 45, S163 (2004). 10.1016/j.lungcan.2004.07.977 [DOI] [PubMed] [Google Scholar]
- Micke P. and Ostman A., Exp. Opin. Ther. Targets 9, 1217 (2005). 10.1517/14728222.9.6.1217 [DOI] [PubMed] [Google Scholar]
- Srigunapalan S., Lam C., Wheeler A. R., and Simmons C. A., Biomicrofluidics 5, 13409 (2011). 10.1063/1.3530598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion B., Huang C. P., Ghajar C. M., Kachgal S., Kniazeva E., Jeon N. L., and Putnam A. J., Biotechnol. Bioeng. 107, 1020 (2010). 10.1002/bit.22891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. W. and Munn L. L., Proc. Natl Acad. Sci. U.S.A. 108, 15342 (2011). 10.1073/pnas.1105316108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. W., Bazou D., and Munn L. L., Integr. Biol. 4, 857 (2012). 10.1039/c2ib20061a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M. K., Kim S. K., and Jung H., Lab Chip 11, 3880 (2011). 10.1039/c1lc20671k [DOI] [PubMed] [Google Scholar]
- Song J. W., Cavnar S. P., Walker A. C., Luker K. E., Gupta M., Tung Y.-C., Luker G. D., and Takayama S., PLoS ONE 4, e5756 (2009). 10.1371/journal.pone.0005756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervantonakis I. K., Hughes-Alford S. K., Charest J. L., Condeelis J. S., Gertler F. B., and Kamm R. D., Proc. Natl Acad. Sci. U.S.A. 109, 13515 (2012). 10.1073/pnas.1210182109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Liu T., and Qin J., Lab Chip 12, 2837 (2012). 10.1039/c2lc00030j [DOI] [PubMed] [Google Scholar]
- Kaji H., Yokoi T., Kawashima T., and Nishizawa M., Lab Chip 9, 427 (2009). 10.1039/b812510d [DOI] [PubMed] [Google Scholar]
- Zheng C., Zhao L., Chen G. E., Zhou Y., Pang Y., and Huang Y., Anal. Chem. 84, 2088 (2012). 10.1021/ac2032029 [DOI] [PubMed] [Google Scholar]
- Yen B. L. and Yen M.-L., J. Cancer Mol. 4, 5 (2008). [Google Scholar]
- Nakamizo A., Marini F., Amano T., Khan A., Studeny M., Gumin J., Chen J., Hentschel S., Vecil G., Dembinski J., Andreeff M., and Lang F. F., Cancer Res. 65, 3307 (2005). 10.1158/0008-5472.CAN-04-1874 [DOI] [PubMed] [Google Scholar]
- Ma H., Zhang M., and Qin J., Integr. Biol. 4, 522 (2012). 10.1039/c2ib20026k [DOI] [PubMed] [Google Scholar]
- Mantovani A., Allavena P., Sica A., and Balkwill F., Nature 454, 436 (2008). 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- Coussens L. M. and Werb Z., Nature 420, 860 (2002). 10.1038/nature01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. P., Lu J., Seon H., Lee A. P., Flanagan L. A., Kim H.-Y., Putnam A. J., and Jeon N. L., Lab Chip 9, 1740 (2009). 10.1039/b818401a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental K. R., Yu H., Kass L., Lakins J. N., Egeblad M., Erler J. T., Fong S. F. T., Csiszar K., Giaccia A., Weninger W., Yamauchi M., Gasser D. L., and Weaver V. M., Cell 139, 891 (2009). 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich T. A., d. E. M. Pardo J., and Kumar S., Cancer Res. 69, 4167 (2009). 10.1158/0008-5472.CAN-08-4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harunaga J. S. and Yamada K. M., Matrix Biol. 30, 363 (2011). 10.1016/j.matbio.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus B., Marga F., Jakab K., Sharpe-Timms K. L., and Forgacs G., Biophys. J. 91, 2708 (2006). 10.1529/biophysj.105.077834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra I. and Beningo K. A., J. Cell. Biochem. 112, 3151 (2011). 10.1002/jcb.23241 [DOI] [PubMed] [Google Scholar]
- Carey S. P., Kraning-Rush C. M., Williams R. M., and Reinhart-King C. A., Biomaterials 33, 4157 (2012). 10.1016/j.biomaterials.2012.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman R. W., Cowan C. R., Mih J. D., Koryakina Y., Gioeli D., Slack-Davis J. K., Blackman B. R., Tschumperlin D. J., and Parsons J. T., PLoS ONE 5, e12905 (2010). 10.1371/journal.pone.0012905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palama I. E., D'Amone S., Coluccia A. M. L., Biasiuccia M., and Gigli G., Integr. Biol. 4, 228 (2012). 10.1039/c2ib00116k [DOI] [PubMed] [Google Scholar]
- Zaari N., Rajagopalan P., Kim S. K., Engler A. J., and Wong J. Y., Adv. Mater. 16, 2133 (2004). 10.1002/adma.200400883 [DOI] [Google Scholar]
- Dong R., Jensen T. W., Engberg K., Nuzzo R. G., and Leckband D. E., Langmuir 23, 1483 (2007). 10.1021/la062738l [DOI] [PubMed] [Google Scholar]
- Swartz M. A. and Lund A. W., Nat. Rev. Cancer 12, 210 (2012). 10.1038/nrc3186 [DOI] [PubMed] [Google Scholar]
- Carl-Henrik Heldin K. R., Pietras K., and Östman A., Nat. Rev. Cancer 4, 806 (2004). 10.1038/nrc1456 [DOI] [PubMed] [Google Scholar]
- Simonsen T. G., Gaustad J.-V., Leinaas M. N., and Rofstad E. K., PLoS ONE 7, e40006 (2012). 10.1371/journal.pone.0040006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z.-D., Wang H., and Tarbell J. M., PLoS ONE 6, e15956 (2011). 10.1371/journal.pone.0015956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Buckley M., Cohen I., Bonassar L., and Awad H. A., Biomech. Model. Mechanobiol. 11, 689 (2012). 10.1007/s10237-011-0343-x [DOI] [PubMed] [Google Scholar]
- Shieh A. C. and Swartz M. A., Phys. Biol. 8, 015012 (2011). 10.1088/1478-3975/8/1/015012 [DOI] [PubMed] [Google Scholar]
- Haessler U., Teo J. C. M., Foretay D., Renaud P., and Swartz M. A., Integr. Biol. 4, 401 (2012). 10.1039/c1ib00128k [DOI] [PubMed] [Google Scholar]
- Bonvin C., Overney J., Shieh A. C., Dixon J. B., and Swartz M. A., Biotechnol. Bioeng. 105, 982 (2010). [DOI] [PubMed] [Google Scholar]
- Khalil A. A. and Friedl P., Integr. Biol. 2, 568 (2010). 10.1039/c0ib00052c [DOI] [PubMed] [Google Scholar]
- Polacheck W. J., Charest J. L., and Kamm R. D., Proc. Natl Acad. Sci. U.S.A. 108, 11115 (2011). 10.1073/pnas.1103581108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayanan B., Kim Y., and Kumar S., Biomaterials 32, 7913 (2011). 10.1016/j.biomaterials.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest J. M., Califano J. P., Carey S. P., and Reinhart-King C. A., Macromol. Biosci. 12, 12 (2012). 10.1002/mabi.201100264 [DOI] [PubMed] [Google Scholar]
- Domenech M., Yu H., Warrick J., Badders N. M., Meyvantsson I., Alexander C. M., and Beebe D. J., Integr. Biol. 1, 267 (2009). 10.1039/b823059e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak A. and Kumar S., Proc. Natl. Acad. Sci. U.S.A. 109, 10334 (2012). 10.1073/pnas.1118073109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia D. and Toner M., Integr. Biol. 1, 506 (2009). 10.1039/b908595e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolli C. G., Seufferlein T., Kemkemer R., and Spatz J. P., PLoS ONE 5, e8726 (2010). 10.1371/journal.pone.0008726 [DOI] [PMC free article] [PubMed] [Google Scholar]