Abstract
Although imatinib mesylate (IM) has transformed the treatment of gastrointestinal stromal tumors (GIST), many patients experience primary/secondary drug resistance. In a previous study, we identified a gene signature, consisting mainly of Kruppel-associated box (KRAB) domain containing zinc finger (ZNF) transcriptional repressors that predict short-term response to IM. To determine if these genes have functional significance, a siRNA library targeting these genes was constructed and applied to GIST cells in vitro. These screens identified seventeen “IM sensitizing genes” in GIST cells (sensitization index (SI) <0.85 ratio of drug/vehicle) with a false discovery rate (FDR) <15%, including twelve ZNF genes, the majority of which are located within the HSA19p12–13.1 locus. These genes were shown to be highly specific to IM and another tyrosine kinase inhibitor (TKI), sunitinib, in GIST cells. In order to determine mechanistically how these ZNFs might be modulating response to IM, RNAi approaches were used to individually silence genes within the predictive signature in GIST cells and expression profiling was performed. Knockdown of the 14 IM-sensitizing genes (10 ZNFs) universally led to downregulation of six genes, including TGFb3, periostin, and NEDD9. These studies implicate a role of KRAB-ZNFs in modulating response to TKIs in GIST.
Introduction
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumors with an estimated annual incidence of ∼6,000 new cases in the United States [1]. Mazur and Clark originally described these tumors in 1983, noting they contained smooth muscle and neural elements [2]. GISTs are recognized immunohistochemically by CD117, the 145 kDa transmembrane glycoprotein KIT. They are believed to arise from the Interstitial Cells of Cajal (ICC) [3] or from interstitial mesenchymal precursor stem cells [4]. The majority (∼70%) of GISTs possess gain-of-function KIT mutations in exons 9, 11, 13 or 17. Smaller subsets of GISTs possess either gain-of-function mutations in PDGFRA (exons 12, 14, or 18) (∼10%), BRAF (exon 15) (∼1–2%) or no mutations, commonly referred to as wild-type (WT) GISTs (∼10–15%) [5], [6] [1], [7]. Recently, germline inactivating mutations in the genes encoding succinate dehydrogenase subunits A, B, C and D (SDHA, SDHB, SDHC or SDHD) have been reported in GISTs lacking KIT/PDGFRA mutations [8], [9] correlating with overexpression of IGF1R in these tumors [10], [11], [12]. The most common primary site for these neoplasms is the stomach (∼55%) [1], [13], followed by small intestine (∼30%), colon, rectum, anus, esophagus, mesentery and omentum (∼15% total) [14], [15], [16]. GISTs occur most frequently in patients over 50, with a median age of presentation of 58 years; however, GISTs have also been observed in the pediatric population [15].
Imatinib (IM), an oral 2-phenylaminopyrimidine derivative that works as a selective inhibitor against mutant forms of type III tyrosine kinases such as KIT, PDGFRA, and BCR/ABL, has significantly improved the clinical outcome of patients with advanced GIST. IM has become the standard treatment for patients with metastatic and/or unresectable GIST, with objective responses or stable disease obtained in >80% of patients and a median time to progression of 2 years [17], [18], [19]. Response to IM has been correlated to the genotype of a given tumor [20]. GIST patients with exon 11 KIT mutations have the best response and disease-free survival, while other KIT mutation types and WT GIST have low response rates to IM. For patients whose GISTs fail IM therapy, sunitinib malate, a multi-targeted tyrosine kinase inhibitor, with activity against KIT, PDGFRA, VEGFR, and FLT-1, is utilized as second line therapy. Clinical trials of sunitinib have demonstrated an objective response rate of ∼10% with further disease stabilization in 50% of patients with IM-refractory disease, and a median progression-free survival of 6 months [21]. While it is not clear what the final result will be for agents currently in clinical trials of GIST patients, e.g., dasatinib, nilotinib, masatinib, and regorafenib, they too are likely to have limited action as monoagents.
Although IM has revolutionized the treatment of patients with GISTs; clinical resistance to IM has become a reality, despite the initial efficacy observed. Furthermore, very limited options exist for patients (∼20%) that are refractory at the start of treatment. We have focused on determining why some GISTs respond initially to IM, while others are refractory, sometimes regardless of mutational status. Using clinical pre-treatment biopsy samples from a prospective neoadjuvant phase II trial (RTOG 0132), we previously identified a 38-gene signature that includes KRAB-ZNF 91 subfamily members that can predict rapid response to IM [22]. This was the first neoadjuvant trial of IM in GIST patients and the first molecular study to examine gene expression changes associated with tumor response following drug treatment in both primary and metastatic GISTs. Here we demonstrate that 17 of these IM-sensitizing genes, including 12 ZNFs, are not only predictive of IM response but mediate the drug's activity. In order to determine mechanistically how these ZNFs might be modulating response to IM, RNAi approaches were used to silence the expression of the genes within the predictive signature (including 10 ZNFs) in GIST cells and individually assess their effect on global gene expression in order to find common regulatory pathways.
Methods
Cell Cultures
GIST-T1, a tumor cell line possessing a heterozygous mutation in KIT exon 11 kindly provided by Takahiro Taguchi [7], [23], [24], and A2780 [25], an ovarian cancer cell line were grown as previously described [26]. For drug treatment, drugs were added directly to the cell medium at the indicated final concentration for the specified period of time. Imatinib mesylate (Gleevec™), ifosfamide, doxorubicin and sunitinib malate were purchased from the Fox Chase Cancer Center pharmacy.
siRNA transfection and IM sensitivity
The custom siRNA library targeting 53 genes, 25 of which were identified in our previous study [22], was designed and synthesized with four independent siRNAs pooled for each target (Qiagen, Valencia, CA). Transfection conditions were determined for GIST T1 cells using siRNA smart pools against KIT and GL-2 (Dharmacon) controls to achieve Z′ factor of 0.5 or greater. We used a reverse transfection protocol in which the final concentration of siRNA was 50 nM. Forty-eight hours after transfection, vehicle only or vehicle+IM (45 nM) were added to two plates. After twenty-four hours cell viability was assessed using the Alamar blue assay. This assay is based on the ability of living cells to convert the redox dye, resazurin, into the fluorescent end product, resorufin. Alamar blue was added to all wells and incubated for three hours followed by data recording using an EnVision microplate reader (PerkinElmer). Hits identified by pooled siRNAs were validated by confirming that 2 or more (of 4) independent siRNAs (at 12.5 nM) targeting the same gene in each case provided sensitization to IM. Ifosfamide (1 µM), doxorubicin (100 nM) or sunitinib (40 nM) were also applied to the deconvoluted siRNAs to identify sensitizing targets.
Statistical Analysis
Alamar blue raw luminescence values collected from the high throughput siRNAs screen were normalized to negative GL2 control samples on the same plate. Each siRNA was then assigned a sensitization index (SI) as SI = (Vdrug/GL2drug)/(VDMSO/GL2DMSO), where V was viability measure in wells transfected with targeting siRNA and GL2 was the median viability of five wells with non-targeting negative control siRNA on the same plate. All calculations were automated using the cellHTS2 [27] package within the Bioconductor open source software package (http://bioconductor.org) [28]. Drug effect was measured using a linear model [29] encoded in the limma R/Bioconductor package [30]. Hits were identified on the basis of statistical significance as well as biological significance. Statistical significance was determined from the linear model with Benjamini-Hochberg correction [31]. Hits showing an FDR of less than 15% were considered statistically significant. Biological significance was defined as a decrease in SI greater than 15%. Hits identified as statistically and biologically significant were further validated.
RNA isolation & Microarray Gene Expression Arrays
Total RNA was isolated from cells using TRIzol reagent according to the protocols provided by the manufacturer (Invitrogen Corp., Carlsbad, CA). RNA quantification and quality assessment were assessed using the 2100 Bioanalyser (Agilent Technologies, Santa Clara, CA). Affymetrix Human Exon 1.0 ST arrays were used for hybridization, arrays were scanned using an Affymetrix GeneChip Scanner 3000 and the scanned images were quantified using Affymetrix GeneChip Command Console Software (AGCC). The expression data was obtained in the form of CEL files.
Microarray Data analysis
CEL files were loaded into R/Bioconductor [28] and analyzed with the oligo package [32]. Data were normalized using RMA and summarized to the gene level using the core probes. The data comprised estimates for 22011 genes across 16 samples. Given the low number of control samples, we identified genes based on mean expression changes relative to controls. We defined a log2 fold change thresholds of 1.0. Then we focused on genes found to be down- or up-regulated in all samples with this cutoff, which included six downregulated genes all with mean log2 fold change less than −1.5. One gene was upregulated in all knockdowns beyond the threshold, and it had a mean log fold change of 1.3.
To explore the impact of the global changes in gene expression, we analyzed the data using a t-test comparing controls to siRNA knockdowns with pooled variance to account for the low number of control samples. Gene set analysis using a Wilcoxon gene set test from the limma R package [30] was used with the t-statistic to identify chromosomal regions and curated pathways (KEGG, Biocarta, Reactome) showing significant changes between knockdowns and controls. All significance was defined as Bonferroni corrected p-value<0.05. Microarray data are available through the Gene Expression Omnibus (accession number GSE40080).
Quantitative RT-PCR
RNA was reverse transcribed to cDNA by SuperScript II reverse transcriptase (Invitrogen). Expression of mRNA for the 14 IM-sensitizing genes, periostin, TGFb3 and two endogenous control genes (HPRT and 18S) was measured in each sample by real-time PCR (with TaqMan Gene Expression Assay products on an ABI PRISM 7900 HT Sequence Detection System, Applied Biosystems, Foster, CA) following protocols recommended by the manufacturer and as previously described [33]. The relative mRNA expressions were adjusted with either HPRT or actin. The primer/probe (FAM) sets were obtained from Applied Biosystems.
Results
Functional Evaluation of a Predictive Gene Set Associated with Response to IM in GIST
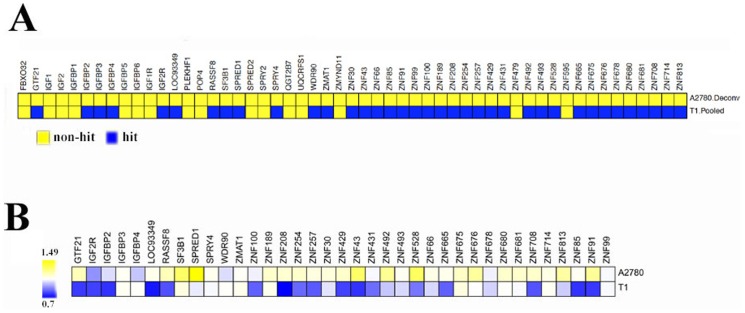
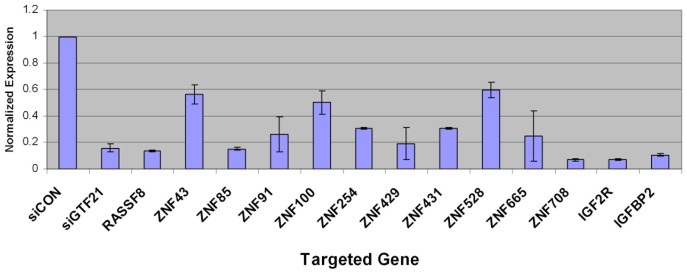
Using clinical samples from a prospective neoadjuvant/adjuvant phase II trial we had previously identified a genetic signature which includes KRAB-ZNF 91 subfamily members that may be predictive of rapid response to short-term IM treatment and which may provide a prognostic biomarker identifying patients who will benefit from it [22]. This gene signature is composed of thirty-eight genes that were expressed at significantly lower levels in the pre-treatment samples of tumors that rapidly responded to IM. Eighteen of these genes encoded KRAB domain containing zinc finger (KRAB-ZNF) transcriptional repressors, ten of which of which mapped to a single locus on chromosome 19p. In order to determine if modifying expression of genes within this predictive signature could enhance the sensitivity to IM, we designed a custom siRNA library containing 53 genes, 25 of which showed differential expression between stable disease and rapid response groups in the aforementioned study (Table S1) [22]. There were thirteen genes identified in the previous study that were not represented in the siRNA library mainly because they were not annotated or had high homology to an already included ZNF. In addition to those 25 genes, we included 13 additional genes located in that same locus, i.e., HSA19p12–19p13.1 that showed a pattern of differential expression but did not reach statistical significance in our previous study (Table S2). The remaining 15 genes included in the current screen were of interest as a result of our previous studies that elucidated the role of IGF1R and KIT signaling in the pathogenesis of GIST, i.e., SPRY2 & 4, SPRED1 & 2, FBXO32, IGF1R, IGF2R, IGF1 & 2 and IGFBP1–6 (Table S3) [10], [11], [34]. We screened GIST T1 [23], an IM sensitive GIST cell line, and an unrelated ovarian cancer cell line, A2780 [25], with the custom siRNA library (four siRNAs pooled/gene) in combination with vehicle (PBS) or small molecule inhibitor, IM at IC30 concentration. Viability was measured with Alamar blue, a metabolic indicator of cell viability. From these screens, 37 of the 53 genes were identified as “primary IM sensitizing hits” in GIST T1 cells ( Figure 1A , bottom row, blue boxes). These hits are defined as genes that when expression was silenced, decreased control-normalized viability by at least 15% in the presence of IM compared to the viability in the presence of vehicle (defined as the Sensitization Index (SI) <0.85) with a false discovery rate (FDR) <15%. No hits were identified in the A2780 cell line, indicating GIST-specificity ( Figure 1A , top row, yellow boxes). Seventeen of the primary 37 hits were validated by confirming that 2 or more (of 4) independent siRNAs targeting the same gene in each case provided sensitization to IM (SI<0.85, FDR<15%) ( Figure 1B , bottom row). Interestingly, 12 of the 17 (71%) validated hits were ZNF genes, 10 (59%) of which are located within the HSA19p12–13.1 locus. In addition, two members of the insulin-like growth factor (IGF) family, IGF2R and IGFBP2, were identified as sensitizing hits. Quantitative PCR analysis confirmed knockdown of the target in 15/17 validated genes ( Figure 2 ).
Figure 1. siRNA screen identifies GIST-specific IM sensitizing genes.
(A) Primary IM sensitizing hits (SI<0.85, blue) identified using pooled siRNAs in A2780 (0 hits) and T1 cells (37 hits). (B) Validation of the primary IM sensitizing hits using deconvoluted siRNA screen. 17 of the primary hits were validated. Heatmap shows the average SI values from two or more confirmed individual siRNAs. SI values on log scale are plotted.
Figure 2. Validation of target gene knockdown by quantitative RT-PCR.
RNA expression of indicated target genes in GIST T1 cells after knockdown with corresponding siRNA. Expression levels were normalized to HPRT. Results are of three independent experiments.
Drug Specificity of Sensitizing Genes
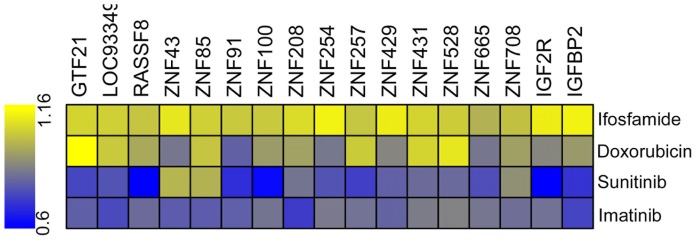
The validation set has also been tested with other agents to measure IM specificity. We selected doxorubicin (anthracycline antibiotic, a DNA intercalating agent) and ifosfamide (nitrogen mustard alkylating agent), both relatively ineffective chemotherapeutic agents used prior to IM to treat GISTs, as well as sunitinib, a small molecule kinase inhibitor (targets KIT, PDGFRA, VEGFR, and FLT-1) which has shown some success in the treatment of GISTs. For this small screen, the best two siRNAs from the previous screen were pooled in a 96-well format, and cells were transfected and treated (at IC30 concentrations) as previously described. SI values were calculated as previously described. None of these genes were sensitizing hits for ifosfamide. Knockdown of 5 out of 17 (29%) of these genes sensitized GIST T1 cells to doxorubicin. In contrast, knockdown of 14 out of 17 genes (82%) sensitized to sunitinib ( Figure 3 ). These findings are consistent with the fact that sunitinib has many of the same targets as IM and indicate that these sensitizing genes are highly specific to IM and sunitinib.
Figure 3. Heatmap showing sensitizing index (SI) ranging from 0.6 (blue) to 1.16 (yellow).
Sensitization was tested in GIST T1 cells in the presence of four drugs including separately: Ifosfamide (top), Doxorubicin (second row), Sunitinib (third row) and IM (bottom). SI values on log scale are plotted.
Elucidating Targets of these “IM Sensitizing Genes”
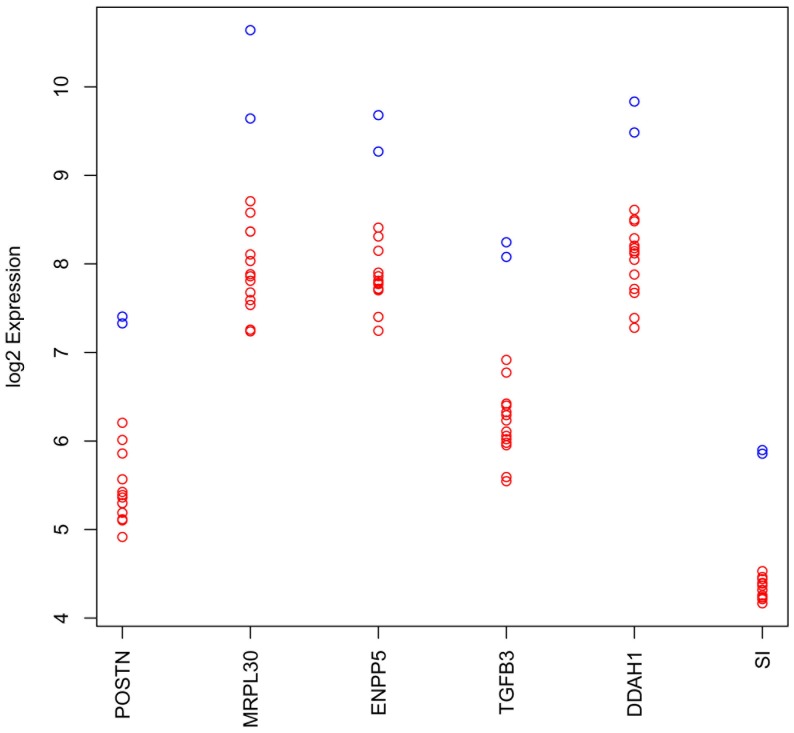
To identify downstream targets of these ZNFs in the hope of elucidating the mechanisms by which they are modulating response to IM, Affymetrix Exon 1.0 ST arrays were used to examine gene expression on GIST T1 cells that have been transfected with siRNAs against 14 of the “sensitizing genes” (10 of which were ZNFs) as well as siCON (non-targeting siRNA). All samples were compared to the GIST T1 cells transfected with siCON to identify genes that changed on knockdown, and we generated our candidate list using minimum log2 fold change of 1.0 (equivalent to a halving of expression). Six genes were found to be downregulated by this amount in all samples with median log2 fold changes >−1.5. Due to other ongoing studies, we also identified NEDD9, which was downregulated in all samples though not above our threshold with a mean log fold change of −0.91 ( Table 1 , Figure 4 ). These data have been confirmed by qRT-PCR (data not shown). In addition, only one gene, TMCO1, was upregulated (log fold change = +1.33) universally across the samples with these thresholds ( Table 1 ). These data are intriguing since two of the downregulated genes, periostin and NEDD9, have been shown to be transactivated by a third downregulated gene, TGFb3 [35], [36].
Table 1. Differentially expressed genes in all 14 samples.
| Gene Symbol | Description | Cytoband | Mean LogFold Change |
| POSTN | periostin, osteoblast specific factor | 13q13.3 | −2.21 |
| MRPL30 | mitochondrial ribosomal protein L30 | 2q11.2 | −1.98 |
| ENPP5 | ectonucleotide pyrophosphatase/phosphodiesterase 5 | 6p12.3 | −1.95 |
| TGFb3 | transforming growth factor, beta 3 | 14q24.3 | −1.65 |
| DDAH1 | dimethylarginine dimethylaminohydrolase 1 | 1p22.3 | −1.62 |
| SI | sucrase-isomaltase | 3q26.1 | −1.53 |
| NEDD9/HEF1/Cas-L | Human enhancer of filamentation | 6p25-p24 | −0.91 |
| TMCO1 | transmembrane and coiled-coil domains 1 | 1q24.2 | +1.33 |
Figure 4. Log2 expression estimates for the six genes identified as downregulated by at least a factor of 2 in all samples relative to mean control value.
For each gene, blue circles indicate log2 expression estimates for siCON samples and red circles indicate values for knockdown samples.
Pathway Analysis
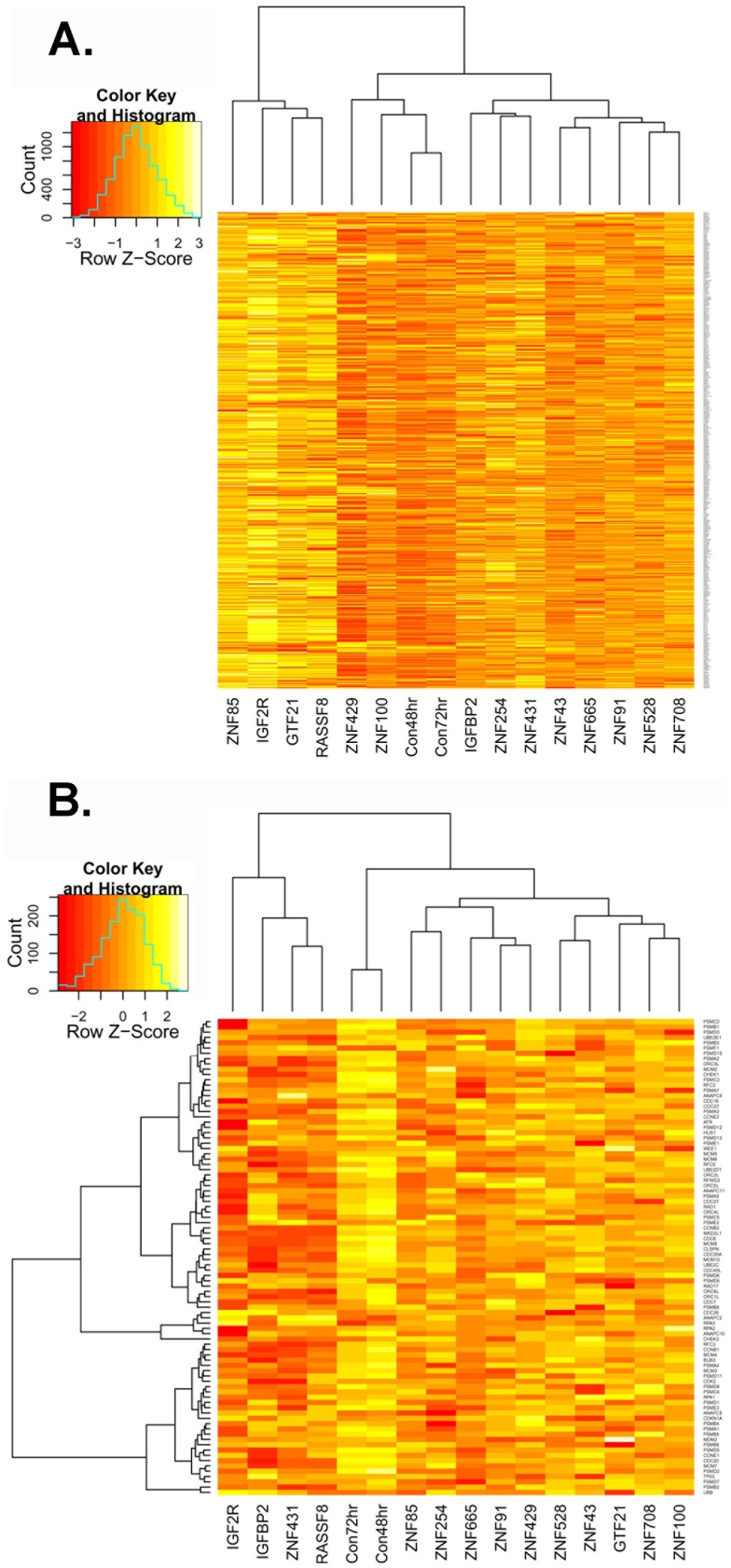
Gene set analysis of t-statistics revealed significant changes at chromosomal locations 19p13, 16q13, 19q13, 3p21, 17q21, 11p15, and 6p22. As 19q13 was by far the strongest signal (corrected p<4.4×10−21), we examined changes in expression for all genes in the locus ( Figure 5A ). As can be seen, most siRNA knockdowns show slightly increased expression across the full locus relative to controls, with ZNF85, IGF2R, GTF21, and RASSF8 knockdowns showing the strongest signals, and ZNF429 and ZNF100 being most similar to controls.
Figure 5. Heatmaps of relative expression levels for genes in two gene sets using row (gene) averaging to provide relative differences.
(A) The heatmap for the genes at 19q13 are shown clustered by sample. The four samples on the left (knockdowns of ZNF85, IGF2R, GTF21, and RASSF8) show the strongest upregulation of gene relative to the siCON samples (Con48 hr, Con72 hr). Knockdowns of ZNF429 and ZNF100 behave much more like controls, as shown by the dendrogram, than do other knockdowns. (B) The heatmap for the Reactome Cell Cycle Checkpoints gene set is provided with clustering on both columns and genes. There is overall downregulation of cell cycle checkpoint genes in knockdowns relative to siCON samples, with knockdowns of IGF2R, IGFBP2, ZNF431, and RASSF8 showing the largest overall changes. In this case, no knockdown samples cluster closely with the controls.
A similar analysis of curated KEGG, Biocarta, and Reactome pathways identified nine pathways as significantly changed after Bonferroni multiple testing correction: KEGG Neuroactive Ligand Receptor Interaction, KEGG ECM Receptor Interaction, Reactome Cell Cycle Checkpoints, Reactome Cell Cycle Mitotic, Reactome M G1 Transition, Reactome Mitotic Prometaphase, Reactome Trasmembrane Transport of Small Molecules, Reactome Transmission Across Chemical Synapses, Reactome Mitotic M M G1 Phases. Most significant pathways involve cell cycle changes. We looked at all genes in the overarching class (Reactome Cell Cycle Checkpoints), which shows significant downregulation of checkpoint genes in all knockdowns relative to controls, with the strongest signals in knockdowns of IGF2R, IGFBP2, ZNF431, and RASSF8 ( Figure 5B ).
Discussion
As a follow-up to our 2009 article [22] reporting the KRAB-ZNF-centric gene signature that predicts rapid response to IM in GIST, we set out to determine whether these genes could have a functional role in modulating drug response. We designed a customized siRNA library containing many members of the predictive gene signature as well as some additional genes that were of interest in other areas of focus. We found that seventeen of these predictive genes, when silenced, conferred sensitivity to TKIs, IM and sunitinib, in GIST cells. Gene set analysis revealed that the siRNA knockdowns affected many of the genes within the HSA19p12–19p13.1 locus previously identified as biomarkers of initial tumor response to imatinib. In order to more thoroughly determine the biological effects of the knockdowns, we performed gene set analysis on manually curated KEGG, Biocarta, and Reactome pathways. We determined that cell cycle pathways were highly affected and further analysis demonstrated that downregulation of cell cycle checkpoint genes appeared to play a role, suggesting a potential mode of action. These findings are intriguing and indicate potential therapeutic implications in GIST. Interestingly, Strauss et al. [37] recently showed significant correlations with these ZNFs and SUV and SUVmax as determined by dynamic PET studies. In addition, two recent reports have implicated ZNFs in modulation of therapeutic response in cancer cells. Duan et al. [38] showed overexpression of ZNF 93 and ZNF 43 in two human chondrosarcoma cell lines resistant to ET-743 (trabectedin; Yondelis) and PM00104 (Zalypsis), compared to their parental sensitive counterparts. They also found ZNF 93 overexpressed in two ET-743 resistant Ewing sarcoma cell lines and in a cisplatin-resistant ovarian cancer cell line. They went on to show that these cells could be sensitized by silencing ZNF 93. Although not TKIs, ET-473 and PM00104 both have shown activity in a variety of solid and hematological tumors. In addition, a 2012 report from Seyhan et al. [39], implicated a number of ZNFs in modulating response to neratinib, a TKI of the ERBB family, in SKBR-3, a breast cancer cell line overexpressing ERBB2. Also, of great interest is the fact that the majority (59%) of ZNFs identified from our siRNA screen are located within the 19p12–13.1 locus and belong to the ZNF 91 subfamily. 19p12–13.1 has recently been described as a breast and ovarian cancer susceptibility locus [40], [41], [42].
Not much is currently known regarding the function of the ZNF 91 subfamily members, which include 64 genes, 37 of which are found on chromosome 19 [43]. In an attempt to elucidate for the first time the down-stream targets of these ZNFs, RNAi approaches were used to silence these IM-sensitizing genes (including 10 ZNFs) in GIST-T1 cells followed by expression profiling with Affymetrix Human Exon 1.0 ST arrays. One interesting discovery arising from these studies is that the knockdown of the 14 IM-sensitizing genes universally led to downregulation of 3 genes, TGFb3, periostin, and NEDD9. Interestingly, TGFb3 is a known inducer of both periostin and NEDD9, both of which have been reported as frequently overexpressed in a variety of human cancers [35], [36], [44], [45], [46]. Periostin, also called osteoblast-specific factor 2, is a secreted protein originally isolated from osteoblast cells and has been shown to mediate cell adhesion as an extracellular matrix-associated protein [47] Periostin was subsequently shown to play a role in new bone formation and wound healing, normally regulated by TGFb [48]. There are a number of studies identifying periostin as overexpressed in a wide variety of tumor tissues including breast [45], pancreas [49], ovary [50], colon [51] and head and neck [52]. The PI3-K/AKT pathway has been described in several reports to be the critical pathway involved in regulation of periostin-induced tumorigenesis and prevention of stressed-induced tumor cell apoptosis in both colon and pancreatic cancer [49], [51]; specifically, periostin can induce the phosphorylation of AKT by binding a6b4 integrins [49]. Upregulation of periostin was reported to be stimulated by hypoxia-responsive growth factors, TGFa and bFGF, followed by activation of the PI3-K signaling pathway [35]. This is fascinating because hypoxia is a common condition within solid tumors, allowing cancer cells to undergo genetic and adaptive changes leading to escape from apoptosis and increased proliferation and survival [53], [54]. Also of interest, TGFb3 was recently described as a candidate gene for resistance of human glioblastoma multiforme cell lines to erlotinib, another tyrosine kinase inhibitor targeting EGFR [55]. NEDD9 (also known as HEF1, Cas-L) is a scaffolding protein whose overexpression has been shown to promote cell growth, migration and invasion in a number of cancers like melanoma and glioblastoma [46], [56], [57]. Intriguingly, like periostin, TGFb is known to induce transcription of NEDD9 mRNA [58], [59] and NEDD9 expression has been shown to be regulated by hypoxia [60], [61]. Therefore, it is plausible to imagine both periostin and NEDD9 regulated in a similar mechanism by TGFb and hypoxia, controlled by ZNFs, to modulate IM response in GIST cells. In addition, NEDD9 was shown to be overexpressed and SRC hyper-activated in an IM-resistant GIST cell line; when NEDD9 was knocked down, IM sensitivity was restored [62].
In addition to the ZNFs, IGF2R and IGFBP2, were identified as IM sensitizing genes in our study. Several reports, from our lab and others, have demonstrated roles for IGF family members in GIST pathogenesis and response to IM, prompting many groups to begin evaluating the use of IGF inhibitors as a potential therapy for GIST [10], [11], [63], [64]. The findings reported in this study lend further support for the testing of IGF inhibitors in GIST. Also of interest, IGF2R has been shown to directly complex with the latent form of TGFb contributing to its activation [65], [66]. Therefore, it is not surprising that knockdown of IGF2R leads to downregulation of TGFb3 and indicates that TGFb3, independent of the ZNFs, may be of importance in response to IM in GIST.
The studies reported here, for the first time, define the functional role of a family of KRAB-ZNF transcriptional repressors in IM sensitivity. These results also uncover that several members of the IGF signaling pathway, IGF2R and IGFBP2 are similarly associated with IM sensitivity in KIT mutant GIST cells. This is interesting given aberrant IGF signaling is a key oncogenic driver in 10 to 15% of GISTs that lack tyrosine kinase mutations and are less sensitive to IM and Sunitinib [10], [11]. This report describes potential downstream targets of a family of KRAB-ZNF, potent transcriptional repressors that for the most part remain completely uncharacterized. Overall, these studies implicate a role of KRAB-ZNFs in modulating response to TKIs and that TGFb3 may be a key downstream mediator of these proteins and thus IM sensitivity in GIST.
Supporting Information
Additional target genes from chromosome 19 showing differential expression but not reaching statistical significance in previous study [22].
(DOC)
Acknowledgments
We would like to acknowledge Drs. Joseph R. Testa and Yuesheng-Li of the Fox Chase Cancer Center Genomics Services for processing of the expression arrays and Dr. Margret Einarson of the Fox Chase Cancer Center High Throughput Screening Facility for help with siRNA screens.
Funding Statement
This work was supported in part by grants from the NCI R01 CA106588 (MvM & AKG), and K99 CA158061-01A1 (LR), the NIH UL1 TR000001-02S1 (AKG) and the National Institutes of Health core grant CA-06927. The authors would especially like to thank the GIST Cancer Research Fund for their continued support. The authors would also like to acknowledge support from the Kansas Bioscience Authority Eminent Scholar Program and the Chancellors Distinguished Chair in Biomedical Sciences endowment at KUMC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Corless CL, Heinrich MC (2008) Molecular Pathobiology of Gastrointestinal Stromal Sarcomas. Annu Rev Pathol 3: 557–586. [DOI] [PubMed] [Google Scholar]
- 2. Mazur MT, Clark HB (1983) Gastric stromal tumors. Reappraisal of histogenesis. Am J Surg Pathol 7: 507–519. [DOI] [PubMed] [Google Scholar]
- 3. Sircar K, Hewlett BR, Huizinga JD, Chorneyko K, Berezin I, et al. (1999) Interstitial cells of Cajal as precursors of gastrointestinal stromal tumors. Am J Surg Pathol 23: 377–389. [DOI] [PubMed] [Google Scholar]
- 4. Kindblom LG, Remotti HE, Aldenborg F, Meis-Kindblom JM (1998) Gastrointestinal pacemaker cell tumor (GIPACT): gastrointestinal stromal tumors show phenotypic characteristics of the interstitial cells of Cajal. Am J Pathol 152: 1259–1269. [PMC free article] [PubMed] [Google Scholar]
- 5. Heinrich MC, Corless CL (2005) Gastric GI stromal tumors (GISTs): the role of surgery in the era of targeted therapy. J Surg Oncol 90: 195–207; discussion 207. [DOI] [PubMed] [Google Scholar]
- 6. Subramanian S, West RB, Corless CL, Ou W, Rubin BP, et al. (2004) Gastrointestinal stromal tumors (GISTs) with KIT and PDGFRA mutations have distinct gene expression profiles. Oncogene 23: 7780–7790. [DOI] [PubMed] [Google Scholar]
- 7. Tarn C, Merkel E, Canutescu AA, Shen W, Skorobogatko Y, et al. (2005) Analysis of KIT mutations in sporadic and familial gastrointestinal stromal tumors: therapeutic implications through protein modeling. Clin Cancer Res 11: 3668–3677. [DOI] [PubMed] [Google Scholar]
- 8. Janeway KA, Kim SY, Lodish M, Nose V, Rustin P, et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc Natl Acad Sci U S A 108: 314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pantaleo MA, Astolfi A, Indio V, Moore R, Thiessen N, et al. (2011) SDHA loss-of-function mutations in KIT-PDGFRA wild-type gastrointestinal stromal tumors identified by massively parallel sequencing. J Natl Cancer Inst 103: 983–987. [DOI] [PubMed] [Google Scholar]
- 10. Tarn C, Rink L, Merkel E, Flieder D, Pathak H, et al. (2008) Insulin-like growth factor 1 receptor is a potential therapeutic target for gastrointestinal stromal tumors. Proc Natl Acad Sci U S A 105: 8387–8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Belinsky MG, Rink L, Cai KQ, Ochs MF, Eisenberg B, et al. (2008) The insulin-like growth factor system as a potential therapeutic target in gastrointestinal stromal tumors. Cell Cycle 7: 2949–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chou A, Chen J, Clarkson A, Samra JS, Clifton-Bligh RJ, et al. (2012) Succinate dehydrogenase-deficient GISTs are characterized by IGF1R overexpression. Mod Pathol 25: 1307–1313. [DOI] [PubMed] [Google Scholar]
- 13. El-Rifai W, Sarlomo-Rikala M, Andersson LC, Knuutila S, Miettinen M (2000) DNA sequence copy number changes in gastrointestinal stromal tumors: tumor progression and prognostic significance. Cancer Res 60: 3899–3903. [PubMed] [Google Scholar]
- 14. Miettinen M, Lasota J (2003) Gastrointestinal stromal tumors (GISTs): definition, occurrence, pathology, differential diagnosis and molecular genetics. Pol J Pathol 54: 3–24. [PubMed] [Google Scholar]
- 15. Corless CL, Fletcher JA, Heinrich MC (2004) Biology of gastrointestinal stromal tumors. J Clin Oncol 22: 3813–3825. [DOI] [PubMed] [Google Scholar]
- 16. Miettinen M, Lasota J (2001) Gastrointestinal stromal tumors–definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Arch 438: 1–12. [DOI] [PubMed] [Google Scholar]
- 17. Demetri GD (2002) GIST 1, chemotherapy 0, with a brand new hitter up next. Cancer Invest 20: 853–854. [DOI] [PubMed] [Google Scholar]
- 18. Verweij J, van Oosterom A, Blay JY, Judson I, Rodenhuis S, et al. (2003) Imatinib mesylate (STI-571 Glivec, Gleevec) is an active agent for gastrointestinal stromal tumours, but does not yield responses in other soft-tissue sarcomas that are unselected for a molecular target. Results from an EORTC Soft Tissue and Bone Sarcoma Group phase II study. Eur J Cancer 39: 2006–2011. [PubMed] [Google Scholar]
- 19. Blanke CD, Demetri GD, von Mehren M, Heinrich MC, Eisenberg B, et al. (2008) Long-term results from a randomized phase II trial of standard- versus higher-dose imatinib mesylate for patients with unresectable or metastatic gastrointestinal stromal tumors expressing KIT. J Clin Oncol 26: 620–625. [DOI] [PubMed] [Google Scholar]
- 20. Debiec-Rychter M, Sciot R, Le Cesne A, Schlemmer M, Hohenberger P, et al. (2006) KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer 42: 1093–1103. [DOI] [PubMed] [Google Scholar]
- 21. Demetri GD, van Oosterom AT, Garrett CR, Blackstein ME, Shah MH, et al. (2006) Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet 368: 1329–1338. [DOI] [PubMed] [Google Scholar]
- 22. Rink L, Skorobogatko Y, Kossenkov AV, Belinsky MG, Pajak T, et al. (2009) Gene expression signatures and response to imatinib mesylate in gastrointestinal stromal tumor. Mol Cancer Ther 8: 2172–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taguchi T, Sonobe H, Toyonaga S, Yamasaki I, Shuin T, et al. (2002) Conventional and molecular cytogenetic characterization of a new human cell line, GIST-T1, established from gastrointestinal stromal tumor. Lab Invest 82: 663–665. [DOI] [PubMed] [Google Scholar]
- 24. Tarn C, Skorobogatko YV, Taguchi T, Eisenberg B, von Mehren M, et al. (2006) Therapeutic effect of imatinib in gastrointestinal stromal tumors: AKT signaling dependent and independent mechanisms. Cancer Res 66: 5477–5486. [DOI] [PubMed] [Google Scholar]
- 25. Eva A, Robbins KC, Andersen PR, Srinivasan A, Tronick SR, et al. (1982) Cellular genes analogous to retroviral onc genes are transcribed in human tumour cells. Nature 295: 116–119. [DOI] [PubMed] [Google Scholar]
- 26. Godwin AK, Meister A, O'Dwyer PJ, Huang CS, Hamilton TC, et al. (1992) High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc Natl Acad Sci U S A 89: 3070–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boutros M, Bras LP, Huber W (2006) Analysis of cell-based RNAi screens. Genome Biol 7: R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- 30.Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman R, Carey RV, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and BioConductor. New York: Springer. pp. 397–420.
- 31. Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125: 279–284. [DOI] [PubMed] [Google Scholar]
- 32. Carvalho BS, Irizarry RA (2010) A framework for oligonucleotide microarray preprocessing. Bioinformatics 26: 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen X, Arciero CA, Wang C, Broccoli D, Godwin AK (2006) BRCC36 is essential for ionizing radiation-induced BRCA1 phosphorylation and nuclear foci formation. Cancer Res 66: 5039–5046. [DOI] [PubMed] [Google Scholar]
- 34. Frolov A, Chahwan S, Ochs M, Arnoletti JP, Pan ZZ, et al. (2003) Response markers and the molecular mechanisms of action of Gleevec in gastrointestinal stromal tumors. Mol Cancer Ther 2: 699–709. [PubMed] [Google Scholar]
- 35. Ouyang G, Liu M, Ruan K, Song G, Mao Y, et al. (2009) Upregulated expression of periostin by hypoxia in non-small-cell lung cancer cells promotes cell survival via the Akt/PKB pathway. Cancer Lett 281: 213–219. [DOI] [PubMed] [Google Scholar]
- 36. Ruan K, Bao S, Ouyang G (2009) The multifaceted role of periostin in tumorigenesis. Cell Mol Life Sci 66: 2219–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Strauss LG, Dimitrakopoulou-Strauss A, Koczan D, Pan L, Hohenberger P (2012) Correlation of Dynamic PET and Gene Array Data in Patients with Gastrointestinal Stromal Tumors. ScientificWorldJournal 2012: 721313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duan Z, Choy E, Harmon D, Yang C, Ryu K, et al. (2009) ZNF93 increases resistance to ET-743 (Trabectedin; Yondelis) and PM00104 (Zalypsis) in human cancer cell lines. PLoS One 4: e6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seyhan AA, Varadarajan U, Choe S, Liu W, Ryan TE (2012) A genome-wide RNAi screen identifies novel targets of neratinib resistance leading to identification of potential drug resistant genetic markers. Mol Biosyst 8: 1553–1570. [DOI] [PubMed] [Google Scholar]
- 40. Micci F, Skotheim RI, Haugom L, Weimer J, Eibak AM, et al. (2010) Array-CGH analysis of microdissected chromosome 19 markers in ovarian carcinoma identifies candidate target genes. Genes Chromosomes Cancer 49: 1046–1053. [DOI] [PubMed] [Google Scholar]
- 41. Couch FJ, Gaudet MM, Antoniou AC, Ramus SJ, Kuchenbaecker KB, et al. (2012) Common variants at the 19p13.1 and ZNF365 loci are associated with ER subtypes of breast cancer and ovarian cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Epidemiol Biomarkers Prev 21: 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stevens KN, Fredericksen Z, Vachon CM, Wang X, Margolin S, et al. (2012) 19p13.1 is a triple-negative-specific breast cancer susceptibility locus. Cancer Res 72: 1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hamilton AT, Huntley S, Tran-Gyamfi M, Baggott DM, Gordon L, et al. (2006) Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res 16: 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Streit S, Michalski CW, Erkan M, Friess H, Kleeff J (2009) Confirmation of DNA microarray-derived differentially expressed genes in pancreatic cancer using quantitative RT-PCR. Pancreatology 9: 577–582. [DOI] [PubMed] [Google Scholar]
- 45. Shao R, Bao S, Bai X, Blanchette C, Anderson RM, et al. (2004) Acquired expression of periostin by human breast cancers promotes tumor angiogenesis through up-regulation of vascular endothelial growth factor receptor 2 expression. Mol Cell Biol 24: 3992–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tikhmyanova N, Little JL, Golemis EA CAS proteins in normal and pathological cell growth control. Cell Mol Life Sci 67: 1025–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takeshita S, Kikuno R, Tezuka K, Amann E (1993) Osteoblast-specific factor 2: cloning of a putative bone adhesion protein with homology with the insect protein fasciclin I. Biochem J 294 Pt 1: 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Horiuchi K, Amizuka N, Takeshita S, Takamatsu H, Katsuura M, et al. (1999) Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res 14: 1239–1249. [DOI] [PubMed] [Google Scholar]
- 49. Baril P, Gangeswaran R, Mahon PC, Caulee K, Kocher HM, et al. (2007) Periostin promotes invasiveness and resistance of pancreatic cancer cells to hypoxia-induced cell death: role of the beta4 integrin and the PI3k pathway. Oncogene 26: 2082–2094. [DOI] [PubMed] [Google Scholar]
- 50. Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, et al. (2002) Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res 62: 5358–5364. [PubMed] [Google Scholar]
- 51. Bao S, Ouyang G, Bai X, Huang Z, Ma C, et al. (2004) Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell 5: 329–339. [DOI] [PubMed] [Google Scholar]
- 52. Kudo Y, Ogawa I, Kitajima S, Kitagawa M, Kawai H, et al. (2006) Periostin promotes invasion and anchorage-independent growth in the metastatic process of head and neck cancer. Cancer Res 66: 6928–6935. [DOI] [PubMed] [Google Scholar]
- 53. Pouyssegur J, Dayan F, Mazure NM (2006) Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 441: 437–443. [DOI] [PubMed] [Google Scholar]
- 54. Bristow RG, Hill RP (2008) Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer 8: 180–192. [DOI] [PubMed] [Google Scholar]
- 55. Halatsch ME, Low S, Mursch K, Hielscher T, Schmidt U, et al. (2009) Candidate genes for sensitivity and resistance of human glioblastoma multiforme cell lines to erlotinib. Laboratory investigation. J Neurosurg 111: 211–218. [DOI] [PubMed] [Google Scholar]
- 56. Natarajan M, Stewart JE, Golemis EA, Pugacheva EN, Alexandropoulos K, et al. (2006) HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene 25: 1721–1732. [DOI] [PubMed] [Google Scholar]
- 57. Kim M, Gans JD, Nogueira C, Wang A, Paik JH, et al. (2006) Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell 125: 1269–1281. [DOI] [PubMed] [Google Scholar]
- 58. Zheng M, McKeown-Longo PJ (2002) Regulation of HEF1 expression and phosphorylation by TGF-beta 1 and cell adhesion. J Biol Chem 277: 39599–39608. [DOI] [PubMed] [Google Scholar]
- 59. Liu X, Elia AE, Law SF, Golemis EA, Farley J, et al. (2000) A novel ability of Smad3 to regulate proteasomal degradation of a Cas family member HEF1. Embo J 19: 6759–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sasaki T, Iwata S, Okano HJ, Urasaki Y, Hamada J, et al. (2005) Nedd9 protein, a Cas-L homologue, is upregulated after transient global ischemia in rats: possible involvement of Nedd9 in the differentiation of neurons after ischemia. Stroke 36: 2457–2462. [DOI] [PubMed] [Google Scholar]
- 61. Martin-Rendon E, Hale SJ, Ryan D, Baban D, Forde SP, et al. (2007) Transcriptional profiling of human cord blood CD133+ and cultured bone marrow mesenchymal stem cells in response to hypoxia. Stem Cells 25: 1003–1012. [DOI] [PubMed] [Google Scholar]
- 62. Thao le B, Vu HA, Yasuda K, Taniguchi S, Yagasaki F, et al. (2009) Cas-L was overexpressed in imatinib-resistant gastrointestinal stromal tumor cells. Cancer Biol Ther 8: 683–688. [DOI] [PubMed] [Google Scholar]
- 63. Dupart JJ, Trent JC, Lee HY, Hess KR, Godwin AK, et al. (2009) Insulin-like growth factor binding protein-3 has dual effects on gastrointestinal stromal tumor cell viability and sensitivity to the anti-tumor effects of imatinib mesylate in vitro. Mol Cancer 8: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Italiano A, Chen J, Zhang L, Hajdu M, Singer S, et al. (2012) Patterns of deregulation of insulin growth factor signalling pathway in paediatric and adult gastrointestinal stromal tumours. Eur J Cancer [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ghosh P, Dahms NM, Kornfeld S (2003) Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol 4: 202–212. [DOI] [PubMed] [Google Scholar]
- 66. Chang M, Kuo W, Chen R, Lu M, Tsai F, et al. (2008) IGFII/mannose 6-phosphate receptor activation induces metalloproteinase-9 matrix activity and increases plasminogen activator expression in H9c2 cardiomyoblast cells. J Mol Endocrinology 41: 65–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional target genes from chromosome 19 showing differential expression but not reaching statistical significance in previous study [22].
(DOC)