Summary
Enhancers play a central role in precisely regulating the expression of developmentally regulated genes. However, the machineries required for enhancer-promoter communication have remained largely unknown. We have found that Ell3, a member of the Ell (Eleven-nineteen Lysine-rich Leukemia gene) family of RNA Pol II elongation factors, occupies enhancers in embryonic stem cells. Ell3's association with enhancers is required for setting up proper Pol II occupancy at the promoter proximal regions of developmentally regulated genes and for the recruitment of the Super Elongation Complex (SEC) to these loci following differentiation signals. Furthermore, Ell3 binding to inactive or poised enhancers is essential for stem cell specification. We have also detected the presence of Pol II and Ell3 in germ cell nuclei. These findings raise the possibility that transcription factors could prime gene expression by marking enhancers in ES cells or even as early as in the germ cell state.
Introduction
A full molecular understanding of how transcriptional networks are regulated in a pluripotent stem cell resulting in coordinated differentiation into a complex organism remains as one of the greatest challenges in biology. Enhancers play pivotal roles in modulating gene expression in a tissue-specific pattern during development and are renowned for their ability to communicate with their associated genes from great distances. Physical interaction between an enhancer and a promoter has been proposed to explain how an enhancer influences gene activation (Bulger and Groudine, 2011).
Several factors, including cohesin and CTCF, have been shown to be involved in this process. CTCF can serve either as a barrier to protect a gene from position effect variegation or as a blocker to prevent long-range enhancer-promoter interactions (Engel et al., 2004; Noonan and McCallion, 2010; Wendt and Peters, 2009). Most of what we know about cohesin and gene expression involves the interplay of cohesin and CTCF(Dorsett, 2011; Hadjur et al., 2009; Hou et al., 2010; Mishiro et al., 2009; Nativio et al., 2009). Recently, a second class of cohesin sites, without CTCF, was described. The presence of Mediator together with cohesin at enhancers was proposed to help bridge interactions between enhancer-bound transcription factors and RNA Pol II at the core promoter of active genes (Kagey et al., 2010).
In order to better predict and define the signatures of cis-regulatory elements and modifications functioning as enhancers, genome-wide sequencing analyses of chromatin occupancy and histone modifications have been used. The analysis of genomic DNA has focused on the identification of clusters of transcription factor motifs (Markstein and Levine, 2002) and resulted in the identification of highly occupied target/transcribed (HOT) regions on DNA functioning as enhancers (Gerstein et al., 2010; Kvon et al., 2012; Moorman et al., 2006). Reduced nucleosome occupancy has also been used as a signature for enhancer identification (Khoueiry et al., 2010). Additionally, genome-wide chromatin modification studies have uncovered possible signatures for identifying enhancers. Over 100,000 putative enhancers can be identified in the human genome by combining the histone modifications and transcriptional coactivator, p300 (Creyghton et al., 2010; Heintzman et al., 2009). For example, the presence of p300, H3K4me1, and H3K27ac is proposed to mark active enhancers, whereas p300 with H3K4me1 alone or with H3K27me3, mark poised or inactive enhancers (Creyghton et al., 2010; Heintzman et al., 2009; Rada-Iglesias et al., 2011). Recent studies have demonstrated that Trr (in Drosophila) and MLL3/4 COMPASS-like complexes (in mammalian cells) are essential for enhancer function and monomethylation/acetylation (Herz et al., 2012)
Fundamental transcriptional studies over the past decade have pointed to the elongation stage of transcription as a major regulatory step in controlling gene expression (Levine, 2011; Sims et al., 2004; Smith et al., 2011; Smith and Shilatifard, 2010). In embryonic stem (ES) cells, many developmentally regulated genes have paused Pol II at their promoters (Core et al., 2008; Guenther et al., 2007; Rahl et al., 2010). Many of these genes carry a bivalent chromatin mark consisting of both H3K4 and H3K27 trimethylation status (Bernstein et al., 2006; Mikkelsen et al., 2007). Recent studies classifying active and poised enhancers have shown that in ES cells, genes neighboring H3K27me3-marked enhancers are enriched for this bivalent chromatin mark and have lower expression than genes associated with active enhancers (Rada-Iglesias et al., 2011). Although the available genome-wide data has been successfully used to categorize enhancers and promoters into a limited number of predictive states, how these different classes of enhancers are used to regulate developmental gene expression is largely unknown.
Recently, we identified a novel elongation complex, named the Super Elongation Complex (SEC), comprised of the ELL family of RNA Pol II elongation factors, ELL1–3, many of the most frequent MLL translocation partners, and the positive transcription elongation factor, P-TEFb (Lin et al., 2010; Luo et al., 2012). Individual ELL family members can have distinct cellular roles within different forms of SEC (Luo et al., 2012). For example, in mouse ES cells, Ell2 has a prominent role as a component of SEC in the rapid yet synchronous activation of genes in response to extracellular signals in the presence or absence of paused Pol II (Lin et al., 2011). In contrast, in addition to its association within SEC, Ell1 is also a component of the Little Elongation Complex (LEC) involved in snRNA gene expression in ES cells (Smith et al., 2011). ELL3 was originally discovered as a Pol II elongation factor enriched in testis and sequence alignment indicated that ELL3 is more divergent from ELL1 and ELL2 (Miller et al., 2000).
Here, we report a unique role for Ell3 at enhancers for the priming of gene activation during stem cell specification. Ell3 occupies enhancers that are in a poised, active, or inactive state. Ell3 is required for establishing proper Pol II occupancy at developmentally regulated genes in a cohesin-dependent manner, and is required for priming these genes for later recruitment of SEC for transcriptional activation during differentiation. Ell3 and RNA Pol II seem to be preloaded on germ cell chromatin which may indicate that Ell3 could prime gene activation by marking enhancers as early as in the germ cells.
Results
Ell3 occupies enhancer regions in mouse embryonic stem cells
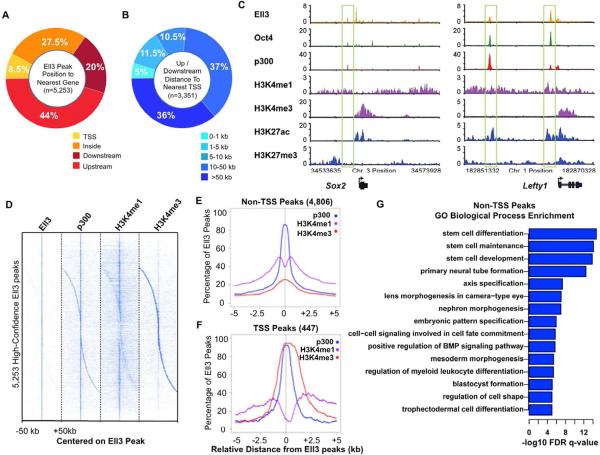
In order to further understand the functional diversity of the Ell family of proteins, we mapped the genome-wide distribution of Ell3 in mouse embryonic stem cells by ChIP-sequencing. In contrast to the enrichment of Ell1 mostly at snRNA genes and Ell2 at other highly transcribed genes in ES cells, Ell3 is preferentially found at intergenic regions in these cells (Figure S1A). We identified 5,253 high-confidence regions bound by Ell3 with an FDR < 0.05. The analyses of peak distributions show that the majority of Ell3 peaks are located long distances (more than 10 kb) from the core promoter regions (Figures 1A and 1B). Examination of well-characterized enhancers, such as the Sox2 and Lefty1 enhancers, shows co-occupancy of Ell3 with known enhancer-binding factors including p300 and Oct4 (Figure 1C).
Figure 1. Ell3 occupies enhancer regions in mouse embryonic stem cells.
(A) Pie chart showing that the percentages of the Ell3 peaks that are overlap with a Transcription Start Site (TSS), within a gene, and upstream or downstream of the nearest gene. (B) Upstream and downstream peaks were further categorized by their distance from the TSS. (C) Ell3 co-localizes with p300 at enhancer regions. Genome browser track examples for the occupancy profiles: for Ell3; transcription factor, Oct4; histone modifications, H3K4me1, H3K27ac, H3K4me3, and H3K27me3; and transcriptional coactivator, p300 (Creyghton et al., 2010; Marson et al., 2008; Mikkelsen et al., 2007). (D) Binding profiles for Ell3, p300, H3K4me1, and H3K4me3 are shown for regions 50 kb upstream and downstream of all 5,253 high-confidence Ell3 peaks. Color indicates enrichment at FDR < 0.05. The majority of the Ell3-occupied regions are also enriched for the enhancer signature of p300 with H3K4me1, but not the H3K4me3 (Creyghton et al., 2010). (E–F) Profiles of p300, H3K4me1, and H3K4me3 centered on Ell3 peaks, show 5 kb around the Ell3 peak summit. Majority of Ell3 peaks (4,806) are found upstream or downstream of a TSS, and these are co-enriched for p300 and H3K4me1. In contrast, only 447 Ell3 peaks are found at a TSS that is enriched for p300 and H3K4me3. (G) Functional annotation of Ell3-bound non-TSS peaks, as reported by GREAT (McLean et al., 2010), indicates enrichment for developmental processes. The logarithmic x-axis values correspond to binomial FDR corrected –log10 q-values. See also Figure S1.
In order to investigate whether Ell3 is generally associated with enhancers in ES cells, we analyzed the co-occurrences of p300, H3K4me1, and H3K4me3 at Ell3-bound sites (Figure 1D). The majority (~90%) of these high confidence Ell3 sites are enriched for the enhancer markers p300 and H3K4me1, with only ~10% overlapping with Transcription Start Site (TSS) regions highly occupied by H3K4me3 (Figures 1D–F), indicating a global association of Ell3 with enhancers. We find that Ell3 can co-occur with H3K27ac on active enhancers and with H3K27me3 on poised or inactive enhancers (Figure S1B). However, there is another class of Ell3-bound putative enhancers that do not have either H3K27ac or H3K27me3; and genes associated with this class of enhancers have an intermediate level of expression (Figure S1B, column 3). Gene Ontology analysis of the nearest genes to the non-TSS Ell3 peaks demonstrates that many of these genes are involved in developmental processes, including stem cell development, primary neural tube formation, embryonic pattern specification, and regulation of myeloid leukocyte differentiation (Figures 1G and S1C).
Ell3 binding to enhancers is required for the expression of a subset of neighboring genes
To identify genes that are regulated by Ell3, we performed total RNA-sequencing analyses following shRNA-mediated Ell3 knockdown in mouse ES cells. About 887 genes are significantly down-regulated with an FDR < 0.05 and fold change > 1.5 in Ell3-depleted ES cells including Sox9 and St3gal1 (Table S1). Gene ontology analysis demonstrates that many of the Ell3 affected genes are involved in developmental processes, such as anatomical structure development, epithelium development, tube development, and chordate embryonic development (Figure S2A). Many genes involved in cell adhesion, cell communication, signal transduction, and response to chemical stimulus are also affected by Ell3 knockdown.
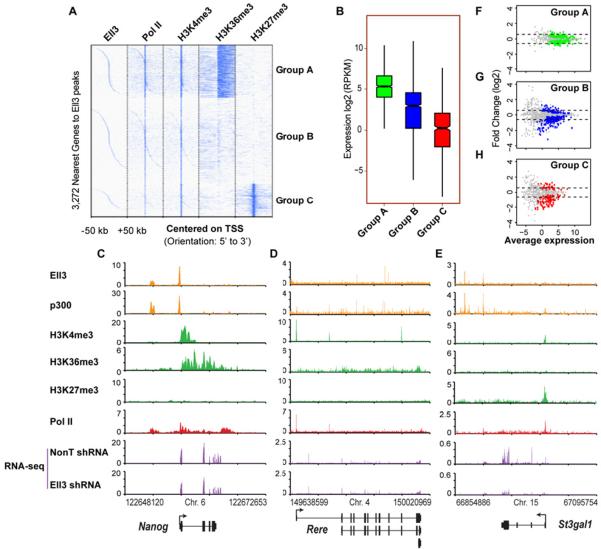
In order to further investigate the extent to which Ell3 differentially regulates the expression of specific classes of genes, we first clustered the ChIP-seq enrichment profiles of the genes nearest to the high-confidence Ell3 peaks based on the co-occurrence of Pol II and the histone modifications H3K4me3, H3K36me3, and H3K27me3. The 3,272 genes proximal to Ell3-occupied peaks were clustered into three major groups, A–C (Figure 2A). Group A (or “Active”) genes show enrichment of Pol II and the active transcription marks H3K4me3 and H3K36me3, and have the highest transcription levels (Figures 2B). Stem cell self-renewal genes, such as Sox2 and Nanog, fall into this group (Figures 1C and 2C). Group B (or “Basal”) genes, such as Rere and Bbx genes (Figures 2D and S2B), are characterized by low or no detectable levels of histone modifications H3K36me3 and H3K27me3, and by low transcription (Figures 2B). Group C genes, such as St3gal1 and Sox9 genes (Figures 2E and S2C), which are marked by both H3K27me3 and H3K4me3, have an even lower “Constrained” expression level as assayed by RNA-seq (Figure 2B). Group C contains many of the previously described “bivalent” genes that are marked by both H3K4me3 and H3K27me3 in embryonic stem cells and are proposed to be in a poised state (Bernstein et al., 2006). Gene ontology analysis shows that Group C is enriched for genes involved in various developmental processes (Figure S2D).
Figure 2. Ell3 binding to enhancers is required for the expression of a subset of neighboring genes.
(A) Cluster diagram of the 3,272 nearest genes to high-confidence Ell3 peaks. ChIP-seq enrichment profiles of Ell3-associated genes for the factor or modification indicated was K-means (K=3) classified into three groups, A (Active), B (Basal), and C (Constrained), which are mainly distinguished by the profiles of H3K36me3 and H3K27me3 in ES cells (Marson et al., 2008; Mikkelsen et al., 2007). (B) Gene expression (RPKM) analyses of the Ell3 nearest Group A–C genes. Only genes with statistically sufficient coverage by RNA-seq are included in this plot (see Methods). (C–E) Genome browser track examples of Groups A–C genes. Ell3 co-localizes with p300 at enhancer regions. RNA-seq analysis shows reduced expression of the Group C gene, St3gal1 upon Ell3 knockdown. (F–H) Gene expression MA plots show the differential expression of Groups A–C genes in Ell3-depleted ES cells vs. control cells. Significantly changed genes as reported by Cufflinks are shown in color. Dotted lines indicate log2 fold changes of −0.5 and 0.5. See also Figure S2 and Table S1.
We next asked how the different groups of Ell3-associated genes were affected by loss of Ell3. Gene expression MA-plots show that depletion of Ell3 reduces the expression of a subset of Group B and C genes, but has minor effects on Group A genes (Figures 2F–2H). For example, although Ell3 occupies the active enhancer regions of the critical stem cell self-renewal genes Sox2, Nanog, and Pou5f1 (Figures 1C and 2C and data not shown), Ell3 is largely dispensable for their expression (Figures 2C and S2E–S2F) and for stem cell self-renewal (Figure S2G). Furthermore, fold changes of expression among Groups A–C were directly compared, indicating that Group C genes are the most significantly affected by Ell3 (Figure S2H). For example, the expression level of the Group C gene, St3gal1, is dramatically reduced after Ell3 knockdown, when compared with the Group A gene, Nanog, and the Group B gene, Rere (Figures 2C–2E, purple box). Therefore, enhancer-associated Ell3 mainly affects the expression of a subset of the “constrained” genes which are enriched for developmental processes, with a subtle effect on the basal expression of the Group B genes involved in cellular processes (Figure S2D and S2H).
Ell3 regulates Pol II occupancy at promoter-proximal regions of neighboring genes
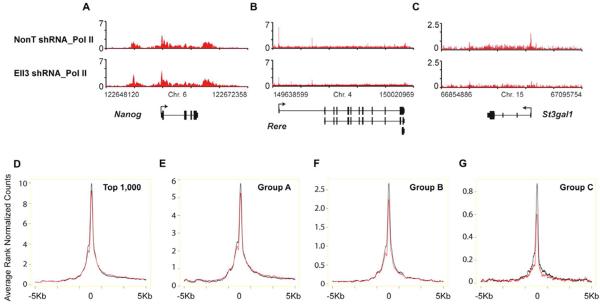
Ell3 was initially identified as a member of the ELL family of RNA Pol II elongation factors, which can increase the transcription elongation rate catalyzed by Pol II (Miller et al., 2000). ELL family members can directly interact with Pol II, and are proposed to facilitate the proper alignment of the 3' terminus of the nascent transcript with the Pol II active site (Elmendorf et al., 2001; Shilatifard et al., 2003). Since Ell3 binding to enhancers is required for the proper expression of a subset of Group B and C genes (Figure 2), we asked whether Ell3 is required for the proper occupancy of Pol II at its nearest genes by performing Pol II ChIP-seq after Ell3 knockdown. Loss of Ell3 leads to reduction of Pol II in many Group C genes, such as Sox9 and St3gal1, and Group B genes, such as Rere and Bbx, with a lesser effect on Group A genes, like Nanog (Figures 3A–3C and S2B–S2C). The effect on promoter-proximal Pol II occupancy is specific to Ell3 knockdown, as the depletion of the related protein Ell2 does not result in changes in Pol II occupancy at these genes (Figure S3A).
Figure 3. Ell3 regulates Pol II occupancy at promoter-proximal regions of neighboring genes.
(A–C) Genome browser profiles of Pol II occupancy in control and Ell3-depleted cells. Pol II levels are reduced at the Rere and St3gal1 genes, but not significantly altered on the Nanog gene. (D–G) Average Pol II occupancy plots for the top 1,000 highly expressed genes and Ell3 nearest genes from the Figure 2A group analysis. Rank normalized average Pol II levels within 5 kb of the TSS are shown in control (black line) and Ell3 knockdown (red line) ES cells. Pol II is reduced at the TSS region of Ell3-associated genes, with strong effects on Group C genes. See also Figure S3.
In order to further investigate whether Ell3 differentially regulates the Pol II occupancy in Groups A–C genes genome-wide, we directly compared the occupancy levels of Pol II at promoter-proximal regions of genes nearest to Ell3-bound peaks in control and Ell3-depleted ES cells. Compared with Group A and B, Group C genes show the largest fold reduction in Pol II occupancy (Figure 3A–3C and 3D–3G). We note that group C genes are expressed at a very low level and it therefore may be easier to observe a larger fold change in Pol II occupancy and expression after Ell3 RNAi than at highly expressed genes. However, our data suggest that during the process of gene activation genes may achieve a state where they no longer require Ell3 at their enhancers for the maintenance of expression. To rule out the possibility that Ell3 might affect the assembly of the basal transcriptional machinery at an early stage, the level of the basal factor TFIIB loading was also examined in Ell3-depleted cells (Figures S3B and S3C). We find that the basal transcription factor TFIIB is properly recruited to the promoter regions of the Group C genes after Ell3 knockdown. Thus, Ell3's presence at enhancers appears to be specific for establishing Pol II at neighboring genes.
Ell3-dependent enhancer-promoter communication requires the cohesin complex
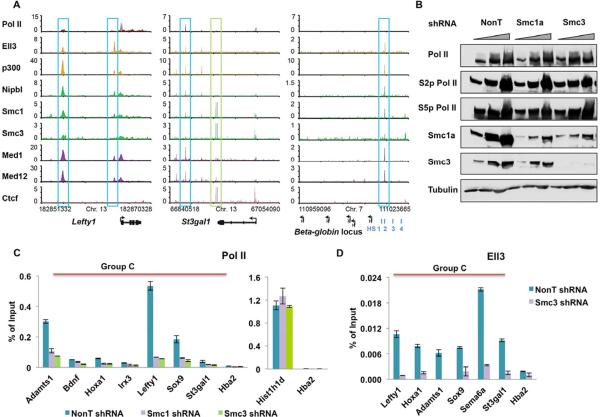
One of the well-accepted models for the regulation of neighboring gene activities by enhancers is enhancer-promoter looping (Bulger and Groudine, 2011; Li et al., 2012; Lieberman-Aiden et al., 2009; Montavon et al., 2011). It has recently been shown that the cohesin and Mediator complexes occupy both the enhancer and promoter regions, promoting loop formation between the enhancer-promoter pairs at active genes (Kagey et al., 2010). Interestingly, the cohesin and Mediator complexes are found on many of the Ell3-bound enhancers, including the Lefty1, St3gal1, as well as the DNase I hypersensitive site HS2 enhancer of the beta globin locus, which is known to be regulated by cohesin in erythroid cells (Hou et al., 2010) (Figure 4A). Beta globin genes are completely silent genes in ES cells, suggesting that cohesin and Mediator complexes might also function in enhancer-promoter communication at both poised and inactive genes.
Figure 4. Ell3-dependent enhancer-promoter communication requires the cohesin complex.
(A) Genome browser profiles for Ell3, p300, cohesin (Nipbl, Smc1(a), and Smc3), Mediator components (Med1 and Med12), and Ctcf (Kagey et al., 2010). Ell3 is found to colocalize with cohesin at sites that are enriched for Mediator and have low Ctcf occupancy (blue boxes). Ell3 is not enriched at cohesin sites that have high Ctcf and low Mediator occupancy (green box). (B) Knockdown of cohesin components Smc1a or Smc3 does not affect cellular Pol II levels. The unphosphorylated (8wg16 antibody), Ser5 phosphorylated (H14), and Ser2 phosphorylated (H5) forms of RNA Pol II levels remain unchanged upon the knockdown of the cohesin components. Triangles indicate increasing amounts of cell lysates. Tubulin serves as a loading control. (C) Knockdown of the cohesin components reduces the promoter-proximal Pol II occupancy at many Ell3-responsive genes. Histone H1 (Histh1d) and alpha globin (Hba2) serve as highly expressed and non-expressed control genes. (D) Knockdown of Smc3 reduces Ell3 occupancy at the enhancer regions of Ell3-responsive genes. Ell3-bound putative enhancer regions were chosen based on the co-occupancy with p300, Cohesin, and H3K4me1. The Hba2 gene serves as a non-transcribed control gene. The error bars stand for the standard deviation of three independent measurements. See also Figure S4.
To explore whether the cohesin complex is also required for the establishment of promoter-proximal Pol II at Ell3-responsive genes, Pol II occupancy was first examined in cohesin subunit-depleted cells. The Smc1a and Smc3 proteins are significantly reduced by the Smc1a and Smc3 shRNAs, respectively (Figure 4B). Smc3 knockdown also leads to a reduction of the protein levels of Smc1a, but not the Smc1a mRNA levels, suggesting that Smc3 affects Smc1a protein stability (Figures 4B and S4A). The depletion of the cohesin complex does not have much of an effect on global Pol II levels, whether looking at the total, phosphorylated or unphosphorylated forms (Figure 4B). However, Pol II occupancy at promoters of the genes nearest to Ell3-bound peaks was greatly reduced after cohesin knockdown as shown by ChIP-qPCR (Figure 4C), indicating an important role of the cohesin complex in establishing promoter-proximal engaged Pol II at Ell3 target genes.
To further assess whether the role of Ell3 in promoter-proximal Pol II occupancy is mediated through the cohesin complex, we tested Ell3-binding profiles in the cohesin-depleted cells. The results revealed that depletion of the cohesin complex (Smc3 knockdown) also greatly reduces Ell3 occupancy at the enhancer regions (Figure 4D). This is not due to the reduced expression of the Ell3 gene, as quantitative RT-PCR shows that Ell3 mRNA levels remain unchanged after Smc3 knockdown (Figure S4B). Therefore, the cohesin complex appears to play a role in the localization/stability of Ell3 on these enhancers and for Ell3's effect on RNA Pol II at promoter-proximal regions.
To further explore if Ell3's binding at enhancers could stabilize enhancer-promoter interactions, we performed a Chromosome Conformation Capture (3C) assay by anchoring on an Ell3 binding site at the Hoxa locus. We observe a broad domain of interactions at the Hoxa locus in the ES cell state, consistent with the published Hi-C data reporting this region as encompassing a topological domain (Figures S4C–S4E) (Dixon et al., 2012). Interestingly, after 24 hours of RA treatment, these interactions become more local and specific, and this transition in interactions requires Ell3 (Figure S4F).
Ell3 binding at enhancers is required for future gene activation by SEC
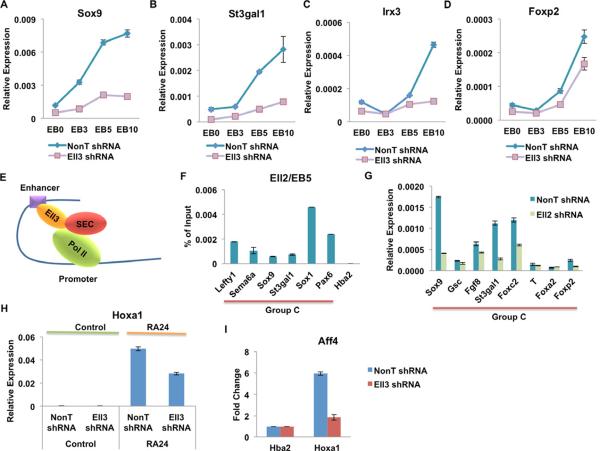
Many of the bivalent genes can be activated during differentiation (Bernstein et al., 2006). Since the Ell3-bound Group C is enriched for bivalently marked genes, we asked whether their induction requires Ell3. Embryoid bodies (EBs) were derived from shRNA-mediated control and Ell3 knockdown of ES cells for three, five, and ten day periods. As shown by quantitative RT-PCR analyses, the activation of Sox9, Irx3, St3gal1, and Foxp2 were significantly reduced in the Ell3-depleted EBs, especially in day 5 and day 10 EBs (Figures 5A–5D). Thus, apart from Ell3's role in ES cells in regulating the constrained expression of its proximal Group C genes, Ell3 is also seems to be required for their further transcriptional activation during differentiation.
Figure 5. Ell3 binding at enhancers is required for future gene activation by SEC.
(A–D) qRT-PCR analyses of the activation time-course of four bivalently marked genes in the control and Ell3 knockdown EBs. Control and Ell3 knockdown ES cells were induced to form EB for 0 (EB0), 3 (EB3), 5 (EB5), and 10 (EB10) days, as indicated. The expression levels were normalized to Actb. (E) Schematic model for Ell3 pre-binding at enhancers primes future gene activation by SEC. (F) Ell2 is recruited to the promoters of Ell3-regulated genes in 5-day EBs, as shown by ChIP. The Hba gene serves as a non-transcribed control gene. (G) Ell2 is required for the activation of many of the genes regulated by Ell3 in EBs. The control and Ell2 knockdown ES cells were induced to form EBs in the petri dishes for 5 days before the qRT-PCR analyses. The expression levels were normalized to Actb. (H) Ell3 binding to enhancers is required for the activation of Hox genes by retinoic acid (RA). Control and Ell3 knockdown ES cells were untreated (Control) or treated with RA for 24 hours (RA24) before harvesting for the qRT-PCR analysis. (I) Ell3 is required for the recruitment of SEC (Aff4) to the Hoxa1 gene after RA treatment. ChIP signal is normalized to the non-transcribed Hba2 gene. The error bar stands for the standard deviation of three independent measurements.
Previously, we demonstrated that Ell2 within SEC plays important roles in the activation of developmentally regulated genes in ES cells (Lin et al., 2011). Many of these Ell2/SEC-responsive genes contain engaged Pol II at their promoter-proximal regions in the undifferentiated state. Therefore, we asked if Ell2/SEC is required for the activation of Ell3-regulated genes (Figure 5E). Chromatin immunoprecipitation in day-5 EB samples shows that Ell2 is indeed recruited to the promoter regions of the Sox9 and St3gal1 genes during differentiation (Figure 5F). Furthermore, qRT-PCR analysis of day 5 Ell2-depleted EBs revealed that Ell2 is also required for the activation of many Ell3-responsive genes, including Sox9, St3gal1, and Foxp2 during development (Figure 5G). Taken together, these results suggested that Ell3 might function upstream of Ell2/SEC in transcriptional programs during development.
To further investigate whether pre-binding of Ell3 to enhancers is essential for future gene activation through recruitment of SEC, we first examined the requirement of Ell3 for the activation of Hoxa1 by retinoic acid (RA), a gene activated by SEC (Lin et al., 2011). Quantitative RT-PCR analysis indicates that the activation of Hoxa1 by RA is reduced after Ell3 knockdown (Figure 5H). In addition, depletion of Ell3 impairs the recruitment of Aff4, the central factor of SEC, to the Hoxa1 promoter after 24 hours of RA treatment (Figure 5I). Our previous biochemical studies indicated that ELL3 can interact with AFF4 and P-TEFb to form a complex similar to ELL2-containing SEC in 293 cells (Lin et al., 2010). We have also found that Ell3 can interact with Aff4 and P-TEFb when overexpressed in ES cells (data not shown). Therefore, we propose that Ell3's binding to enhancers is required for the full assembly of SEC on the promoter of genes, and thus, future gene activation by SEC at the Ell3 target genes.
Ell3 is essential for stem cell specification
Interestingly, in addition to the above-mentioned Group A–C genes, some inactive or “Dormant” lineage-specific genes are also associated with Ell3/p300-bound enhancers, but have no detectable Pol II, H3K4me3, and H3K36me3 in their transcription units. For example, Ell3 and p300 are present at the above-mentioned well-characterized HS2 enhancer element of the beta globin locus, which is silent in ES cells (Figure 4A and data not shown). Therefore, we consider HS2-like enhancers to be in an “inactive/dormant state” or Group D.
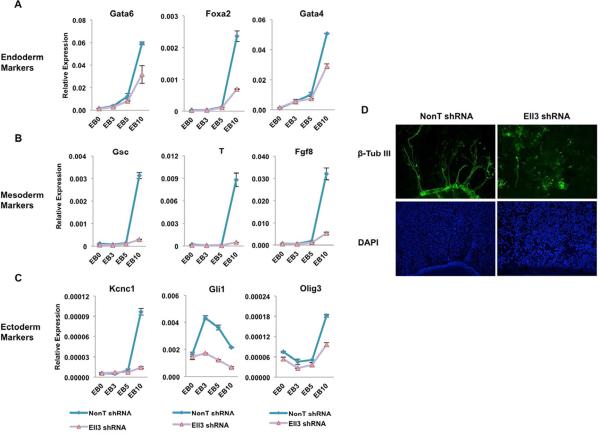
To explore if Ell3 is required for the induction of lineage-specific genes, we measured transcript levels in the embryoid bodies derived from control and Ell3-depleted ES cells. Our analyses demonstrate that many endoderm markers (Foxa2, Gata4, and Gata6), mesoderm markers (Gsc, T, and Fgf8), and ectoderm specific genes (Kcnc1, Gli1, and Olig3) are significantly down-regulated in Ell3-depleted EB samples compared with control EB samples (Figures 6A–6C). Examination of Ell3 occupancy shows that Ell3 associates with many of the inactive or poised enhancers of these lineage-specific genes (Figure S5). Moreover, many of these genes do not contain detectable Pol II at their promoter-proximal regions (Figure S5). Further RNA-seq analysis on the differentiated day-5 EBs indicates that 2,862 genes were up-regulated with an FDR < 0.05 and fold change > 1.5 (Table S2). Of these, 510 of them contain Ell3 at their enhancers in the ES state (Table S2), suggesting the involvement of Ell3 in stem cell differentiation. Many of these genes with Ell3-bound enhancers are required for neuronal specification, including Slit3, Pax6, Ephb2, Ephb3, Sema6a, Ank3, Nfasc, Ripply2, Slitrk5, and Klf7. Therefore, we next assessed the effect of Ell3 knockdown on neural induction by retinoic acid in ES-derived embryoid bodies. The β-tubulin III positive neural fibers are significantly reduced in the Ell3-depleted EBs compared with the control EBs (Figure 6D). Taken together, these results suggest that enhancer-associated Ell3 is essential for stem cell specification.
Figure 6. Ell3 is essential for stem cell specification.
(A–C) qRT-PCR analyses of the activation kinetics of lineage-specific genes in control and Ell3-knockdown EBs. Control and Ell3-knockdown ES cells were induced to form EB for the indicated time points. Expression levels were normalized to Actb. The error bar stands for the standard deviation of three independent measurements. (D) Ell3 is essential for the proper neural differentiation of mouse ES cells. The 5-day differentiated EBs from control and Ell3-knockdown ES cells were further differentiated into neural cells by exposure to retinoic acid for 14 days. Neural-differentiation competence was visualized by immunostaining for the neuronal marker class III, β-tubulin (β-tub III, green), and the DNA marker, DAPI (blue). See also Figure S5 and Table S2.
Ell3 is present on the chromatin of germ cells
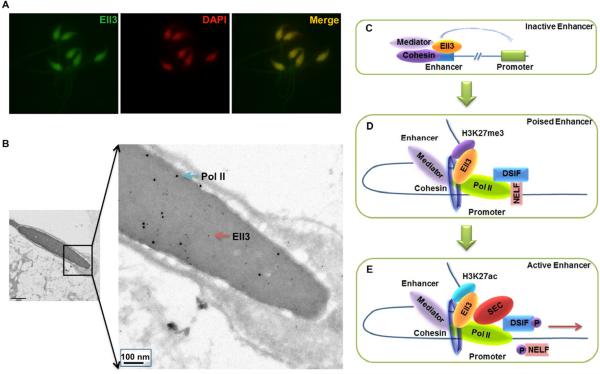
The presence of Ell3 at enhancers in ES cells of many dormant lineage-specific genes (both Group C and Group D genes) raises the question of at what stage is Ell3 recruited to mark these enhancers. Interestingly, our previous northern blot analyses indicated that Ell3 is highly enriched in testes (Miller et al., 2000). We, therefore, performed immunofluorescence staining of mouse sperm and observe that Ell3 localizes to sperm nuclei (Figure 7A). Immunogold labeling of Ell3 in mouse sperm by Ell3 antibodies raised against either the N-terminus or the C-terminus of Ell3 further validates the nuclear localization of Ell3 in sperm (Figures 7B and S6). Interestingly, we also detect Pol II in sperm nuclei by immunogold labeling (Figure 7B). While Ell3 N-and C-terminal antibodies co-localize well (within 5–10nm) in sperm nuclei, antibodies to Ell3 and Pol II appear to occupy different regions on the sperm cell chromatin (Figures 7B).
Figure 7. Ell3 and Pol II are found in germ cell nuclei.
(A) Immunofluorescence staining of Ell3 in mouse sperm. Mouse sperm were fixed and stained with antibodies raised against the C-terminus of mouse Ell3, and were counterstained with DAPI. (B) Immunogold labeling of Ell3 and Pol II in mouse sperm. Mouse sperm were fixed, cryo-sectioned, and double stained with Ell3 N-terminal and Pol II antibodies. Both Ell3 (red arrow, 6 nm gold particles) and Pol II (blue arrow, 12 nm gold particles) localize to the nucleus of the sperm. Co-localization of Ell3 and Pol II was largely not observed compared to the co-localization of the N- and C-terminally raised Ell3 antibodies, which are frequently found within 5–10 nm of each other (See also Figure S6). (C–E) A model for the enhancer-associated Ell3 in coordinated transcriptional induction by SEC. H3K4me1, p300, Mediator, and cohesin can be found with Ell3 at inactive, poised, and active enhancers. At inactive/dormant enhancers (Group D), Ell3 is prebound with Mediator and cohesin, but Pol II is not found at the promoter. In the poised state (Group B and C), a subset of developmental regulators is in a constrained state of expression, with both H3K4me3 and H3K27me3 at the promoter. Pol II's presence at these promoters depends on the interactions between cohesin, Mediator, and Ell3. Bottom panel, upon receiving the proper activating signals, SEC is recruited and stabilized at the promoter region through interaction with Mediator and Ell3 (Group A). SEC phosphorylates RNA Pol II CTD, Spt5, and Nelf, thus resulting in the release of Pol II and gene activation.
Discussion
Regulatory elements play a central role in establishing promoter-proximal engaged Pol II, as previously demonstrated for the Drosophila Hsp70 gene and the murine Ig kappa gene (Lee et al., 1992; Levine, 2011; Raschke et al., 1999). The deletion of the GAGA element upstream of the Hsp70 core promoter or the deletion of the intron enhancer and C regions of the Ig kappa gene abolished the occupancy of Pol II at their respective promoters. However, whether there are enhancer-associated factors that are more generally required for the establishment of Pol II at developmental genes was not known. Here, we report that the elongation factor Ell3 preferentially binds to enhancers, mediates the promoter-proximal occupancy of RNA Pol II at many of the developmentally regulated genes in mouse embryonic stem cells, and is required for their future activation during stem cell specification (Figures 7C–E). The Ell3-mediated enhancer function in promoter-proximal occupancy by Pol II requires the cohesin complex, revealing a novel step in the establishment of the “paused Pol II” state that is pervasive in ES cells and the regulation of gene activation during early embryo development.
A Model for the enhancer-associated Ell3 in the coordinated induction of transcription by SEC
Cohesin and mediator complexes were initially shown to be involved in the enhancer-promoter communication at active genes (Newman and Young, 2010). Interestingly, cohesin and mediator are also present at Ell3-bound “inactive” enhancers in ES cells, such as the beta globin locus (Figure 7C). It is possible that during differentiation, lineage-specific transcription factors such as GATA1 and NFE2 (Bulger and Groudine, 2011; Deng and Blobel, 2010) can interact with enhancer and promoter elements to help bridge cohesin and mediator communication with Pol II at the promoter, a process that can be further stabilized by Ell3 (Figure 7D). Setting up looped chromosomal domains could form a constrained transcriptional state associated with bivalent mark of H3K4 and H3K27 methylation (Bernstein et al., 2006) in a progenitor cell before full transcriptional activation. In addition, we find that many of the genes showing reduced paused Pol II after Ell3 knockdown were also in our Group B, which lacked the bivalent mark. Group B genes may constitute a transition state between constrained expression of bivalently marked genes and activated transcription (Cui et al., 2009; Zhang et al., 2012).
The presence of Ell3 could be particularly important at genes requiring an Ell2 version of SEC to release poised Pol II through phosphorylation of DSIF/NELF and the Pol II CTD (Figure 7E) (Lin et al., 2011; Luo et al., 2012). Upon differentiation, Ell2 within SEC is recruited to the genes with Ell3-occupied enhancers and is required for their activation. This can be attributed to the function of Ell2 within SEC in releasing paused Pol II (Lin et al., 2011). Therefore, in certain circumstances, the ability of Ell2 and Ell3 to form similar and dynamic complexes could underlie a mechanism for the transition from Ell3's presence at poised enhancers in ES cells to Ell2's role in the release of paused Pol II during gene activation. Enhancer-promoter interactions could facilitate local assembly and/or recruitment of functional SEC complexes for rapid, but regulated, gene activation. This model is supported by observations that: 1) Ell3 binding to enhancers is required for the recruitment of SEC during differentiation; 2) Mediator occupies Ell3-bound enhancers, and that it has been shown that the MED26 subunit is required for the recruitment of SEC to the HSP70 and MYC genes (Kagey et al., 2010; Takahashi et al., 2011).
Ell3 as a candidate for priming future gene activation
The activation of the zygotic genome, leading to the control of development by both the paternal and the maternal genomes, is a key event during the maternal to zygotic transition (MZT) following fertilization. Recent studies have identified the zinc finger protein Zelda as a factor that marks the promoter and enhancer regions of both active and inactive genes in the Drosophila early zygotic genome (Harrison et al., 2011; Liang et al., 2008; Nien et al., 2011). The association of Zelda with inactive genes is required for their future activation, indicating an essential role of Zelda as a “pioneer transcription factor” (Harrison et al., 2011). However, how Zelda binding at the promoter and enhancer regions of inactive genes regulates their future expression and how the cofactors working together with Zelda to activate the zygotic genome remain largely unknown.
In this study, we found that mammalian Ell3 not only binds to the enhancer regions of active genes, but also marks the enhancers of inactive genes in mouse embryonic stem cells, many of which are lineage-specific genes, such as beta-globins, Gsc and T. The marking of Ell3 at the inactive enhancers of mammalian cells is required for the future activation of their associated genes, analogous to what has been reported for Zelda in Drosophila embryogenesis. While there is no Zelda homolog known outside of arthropods (Liang et al., 2008), the mechanism by which Ell3 is so generally recruited to enhancers of genes with varying chromatin states and transcriptional activities is currently unknown. Instead of a single Zelda in mammals, there could conceivably be a large family of zinc finger proteins that can recognize various enhancer sequences and help recruit Ell3.
Intriguingly, both Ell3 and Pol II seem to be associated in the nuclei of mouse sperm, but do not co-localize, which might be explained if Ell3 was occupying inactive enhancers and Pol II was present at TSS regions, similar to what we observe by ChIP-seq analyses in the ES cell state. These data suggest that Ell3 might serve as an epigenetic marker in germ cells by bookmarking the inactive enhancers of genes for future activation in the embryo. Although sperm chromatin is highly compacted due to replacement of histones with protamines during spermatogenesis, it was recently discovered that histones can mark promoters of developmentally regulated genes and that histone modifications at these promoters are predictive of early developmental expression (Hammoud et al., 2009). Our work suggests that it might also be possible that regulatory regions of key developmental genes are bound by transcription factors such as Ell3 or by RNA Pol II. ChIP-sequencing analyses of Ell3 and Pol II in sperm and oocytes will be required to test this model and could reveal interesting information on mechanisms of epigenetic inheritance.
A potential role of Ell3 in cancer pathogenesis
Many of the SEC components are among the most frequent MLL (myeloid/lymphoid or mixed-lineage leukemia) translocation partners found in human AML and ALL leukemia patients (Mohan et al., 2010; Smith et al., 2011). SEC is broadly recruited to MLL chimera target genes in both MLL-SEC and MLL-non-SEC-translocated leukemic cells (Lin et al., 2010; Yokoyama et al., 2010). Many of the commonly and highly mis-regulated genes by different MLL chimeras including the Runx1, Ebf1, Cdk6, Meis1/2, and Hoxa cluster genes (Dawson et al., 2011; Krivtsov and Armstrong, 2007; Lin et al., 2010) are occupied by Ell3 on their enhancers in the ES state. It is likely that the MLL chimeras bypass the tight regulation of these ELL3-associated genes in hematopoietic cells, contributing to leukemogenesis. Therefore, investigating the extent to which Ell3 functions in other stem/progenitor cells could have implications in SEC function in other developmental pathways and the mis-regulation of SEC in disease.
EXPERIMENTAL PROCEDURES
ES Cell Culture and Differentiation
Mouse embryonic stem cells (KH2 and V6.5) were cultured on irradiated mouse embryonic fibroblast (MEF) feeder layers in 0.1% gelatin-coated tissue culture flask. Cells were grown in DMEM (D6546, Sigma) supplemented with 15% ES-certified fetal bovine serum (Hyclone), 2 mM L-glutamine, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol, and recombinant LIF (Millipore). For ChIP and RNA analysis, cells were grown for one passage off feeders on tissue culture plates for 30 minutes. Embryoid bodies (EBs) were formed by culturing 150,000/ml ES cells in ES medium without LIF (ES differentiation medium) on non-adhesive bacteriological petri dishes for the indicated days (Kurosawa, 2007). Media were changed daily after two days. For neural differentiation, day-5 EBs were plated on laminin-coated 6-well tissue culture plate in ES differentiation medium with 1 mM all-trans retinoic acid (Sigma) for an additional 14 days. Media were changed every other day. On day 14, the RA-treated EBs were immunostained with anti-β-Tubulin III (Covance).
Cryo-immuno electron microscopy
Sperms were fixed in 2% paraformaldehyde/0.01% glutaraldehyde (PBS) and embedded in 3% gelatin. Samples were infiltrated with 2.3 M sucrose at 4°C overnight, and then mounted on aluminium stubs, frozen and sectioned. Thin sections (50 – 70 nm) were picked up in drops of 2.3M sucrose and collected on formvar and carbon coated mesh grids. After blocking in 1% BSA in PBS the sections were incubated with primary antibodies and subsequently incubated with secondary antibody conjugated to 6 nm and/or 12 nm gold particles (Jachson ImmunoResearch Laboratories, Inc.). The sections were fixed in 1% glutaraldehyde and stained with ice cold 0.4% uranyl acetate/1 % methyl cellulose (pH 4) and dried. The samples were viewed in a FEI Tecnai transmission electron microscope operated at 80kv.
Quantitative RT-PCR and Total RNA-Seq analysis
ES cells (V6.5) were infected with lentivirus carrying either Non-targeting shRNA or Ell3 shRNA in the presence of 8 ug/ml of polybrene (Sigma). 24 hours later, ES cells were selected with 2 ug/ml of puromycin for an additional 48 hours and then were grown one passage off feeders for 30 min before harvest. Total RNA was isolated with the RNeasy (Qiagen) kit, treated with DNase I (NEB), and re-purified with RNeasy. cDNAs were synthesized with High Capacity RNA-to-cDNA Kit from Applied Biosystems. The expression levels were measured with iQ SYBR Green Supermix from Bio-Rad on MyIQ (Bio-Rad). Relative expression to housekeeping genes was calculated assuming 2-fold primer efficiencies. For RNA-seq, total RNA was depleted of rRNA using Ribozero (Illumina) before library preparation using Tru-seq (Illumina). Details of RNA-seq data analysis are provided in the Supplemental Information.
Chromosome Conformation Capture (3C) assay
The 3C assay was performed as previously described with minor modifications (Hagege et al., 2007). Briefly, 1 × 107 cells were crosslinked with 2% of paraformaldehyde at room temperature for 10 min, followed by glycine quenching and cell lysis. The nuclei were digested with HindIII overnight at 37 °C and then ligated with T4 DNA ligase at 16 °C for 4h. DNA was purified by phenol-chloroform extraction. Primer efficiencies were monitored by serial dilution. Digestion efficiencies were examined by primer pairs amplifying genomic regions spanning or devoid of HindIII sites. A bacterial artificial chromosome (BAC) containing the entire Hoxa locus (RP23-20F21) was digested with HindIII and religated to prepare the control template. To compare results between samples, the 3C signals were normalized to a control locus Ercc3.
ChIP-Seq
5×107 cells were used per ChIP assay according to the previously described protocol (Lee et al., 2006). Briefly, cells were cross-linked with 1% paraformaldehyde for 10 min at room temperature; cross-linking was quenched by glycine. Fixed chromatin was sonicated and immunoprecipitated with a specific antibody. Libraries were prepared with Illumina's ChIP-Seq sample prep kit for the further next-generation sequencing. ChIP sequencing and other data analyses can be found in the Supplementary Materials and Methods. ChIP-seq and expression data have been deposited at GEO under the accession number GSE38148.
Supplementary Material
Highlights
Ell3 is preferentially found at active, poised, and inactive enhancers in ES cells.
Ell3 is required for promoter-proximal Pol II occupancy at neighboring genes.
Ell3-associated enhancer promoter communication requires cohesin.
Ell3 is required for the recruitment of P-TEFb within SEC during differentiation.
Acknowledgments
We are grateful to Dr. Edwin Smith for conversations and for critical reading of the manuscript. We also thank Boris Rubenstein for mathematical support, and Stacy Marshall and Ka Chun Lai for technical assistance. We are also thankful to Laura Shilatifard for editorial assistance. This work was performed to fulfill, in part, requirements for C.L.'s Ph.D. thesis research as a student registered with the Open University. Studies in this manuscript were supported in part by funds provided by the Alex's Lemonade Stand Foundation and from the National Institute of Health, R01CA89455 and R01CA150265 to A.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION Supplemental Information includes six figures, two tables, and the Supplemental Experimental Procedures.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Blobel GA. Do chromatin loops provide epigenetic gene expression states? Curr Opin Genet Dev. 2010;20:548–554. doi: 10.1016/j.gde.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev. 2011;21:199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmendorf BJ, Shilatifard A, Yan Q, Conaway JW, Conaway RC. Transcription factors TFIIF, ELL, and Elongin negatively regulate SII-induced nascent transcript cleavage by non-arrested RNA polymerase II elongation intermediates. J Biol Chem. 2001;276:23109–23114. doi: 10.1074/jbc.M101445200. [DOI] [PubMed] [Google Scholar]
- Engel N, West AG, Felsenfeld G, Bartolomei MS. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is uncovered by CpG mutations. Nat Genet. 2004;36:883–888. doi: 10.1038/ng1399. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz H-M, Mohan M, Garruss AS, Takahashi Y-H, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 mono-methylation by Drosophila Trithorax-related/COMPASS, the Drosophila homolog of Mll3/Mll4 complexes. Genes Dev. 2012 doi: 10.1101/gad.201327.112. in pres. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoueiry P, Rothbacher U, Ohtsuka Y, Daian F, Frangulian E, Roure A, Dubchak I, Lemaire P. A cis-regulatory signature in ascidians and flies, independent of transcription factor binding sites. Curr Biol. 2010;20:792–802. doi: 10.1016/j.cub.2010.03.063. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Kurosawa H. Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. J Biosci Bioeng. 2007;103:389–398. doi: 10.1263/jbb.103.389. [DOI] [PubMed] [Google Scholar]
- Kvon EZ, Stampfel G, Yanez-Cuna JO, Dickson BJ, Stark A. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev. 2012;26:908–913. doi: 10.1101/gad.188052.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kraus KW, Wolfner MF, Lis JT. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 2012;13:543–547. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- Markstein M, Levine M. Decoding cis-regulatory DNAs in the Drosophila genome. Curr Opin Genet Dev. 2002;12:601–606. doi: 10.1016/s0959-437x(02)00345-3. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Williams K, Johnstone RW, Shilatifard A. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J Biol Chem. 2000;275:32052–32056. doi: 10.1074/jbc.M005175200. [DOI] [PubMed] [Google Scholar]
- Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Moorman C, Sun LV, Wang J, de Wit E, Talhout W, Ward LD, Greil F, Lu XJ, White KP, Bussemaker HJ, et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:12027–12032. doi: 10.1073/pnas.0605003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JJ, Young RA. Connecting transcriptional control to chromosome structure and human disease. Cold Spring Harb Symp Quant Biol. 2010;75:227–235. doi: 10.1101/sqb.2010.75.016. [DOI] [PubMed] [Google Scholar]
- Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7:e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP, McCallion AS. Genomics of long-range regulatory elements. Annu Rev Genomics Hum Genet. 2010;11:1–23. doi: 10.1146/annurev-genom-082509-141651. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke EE, Albert T, Eick D. Transcriptional regulation of the Ig kappa gene by promoter-proximal pausing of RNA polymerase II. J Immunol. 1999;163:4375–4382. [PubMed] [Google Scholar]
- Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu Rev Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Peters JM. How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res. 2009;17:201–214. doi: 10.1007/s10577-008-9017-7. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.