Abstract
Many Gram-negative and Gram-positive bacteria recycle a significant proportion of the peptidoglycan components of their cell walls during their growth and septation. In many—and quite possibly all—bacteria, the peptidoglycan fragments are recovered and recycled. While cell-wall recycling is beneficial for the recovery of resources, it also serves as a mechanism to detect cell-wall–targeting antibiotics and to regulate resistance mechanisms. In several Gram-negative pathogens, anhydro-MurNAc-peptide cell-wall fragments regulate AmpC β-lactamase induction. In some Gram-positive organisms, short peptides derived from the cell wall regulate the induction of both β-lactamase and β-lactam-resistant penicillin-binding proteins. The involvement of peptidoglycan recycling with resistance regulation suggests that inhibitors of the enzymes involved in the recycling might synergize with cell-wall-targeted antibiotics. Indeed, such inhibitors improve the potency of β-lactams in vitro against inducible AmpC β-lactamase-producing bacteria. We describe the key steps of cell-wall remodeling and recycling, the regulation of resistance mechanisms by cell-wall recycling, and recent advances toward the discovery of cell-wall recycling inhibitors.
Keywords: AmpD, AmpG, AmpR, autolysin, BlaR1, Escherichia coli, lytic transglycosylase, MRSA, NagZ, PBP2a, Staphylococcus aureus
Introduction
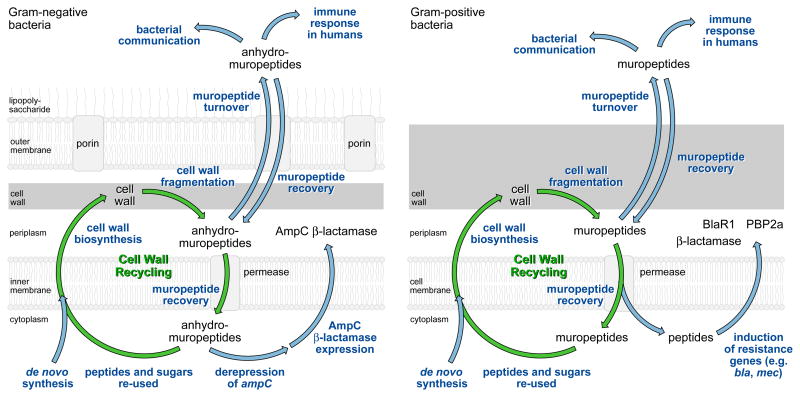
The bacterial cell wall is an elastic macromolecule that defines the shape of the bacterium and enables the bacterium to resist lysis as a result of its high intracellular osmotic pressure. Since the cell wall is structurally unique to bacteria, the steps involved in cell-wall biosynthesis are the targets of numerous antibiotics, including the β-lactams.1,2 Cell-wall synthesis is a complex process that is carefully coordinated with the cell-wall remodeling required for cell growth and division. Many bacteria remodel as much as half of their cell wall per generation and this process would represent a significant loss of resources if the liberated cell-wall fragments were not recovered and recycled (Fig. 1).
Figure 1.
Simple representations of cell-wall biosynthesis, recycling, and turnover in Gram-negative and Gram-positive bacteria. The biosynthesis of all precursors for the peptidoglycan occurs in the bacterial cytoplasm. The final intermediate, Lipid II, is translocated into the periplasm for de novo peptidoglycan polymer synthesis as catalyzed by the bifunctional penicillin-binding protein enzymes (PBPs). Remodeling of the peptidoglycan polymer occurs during both growth and septation, liberating peptidoglycan fragments called muropeptides. Peptidoglycan cell wall recycling refers to the recovery and removal of these muropeptides to the cytoplasm, for integration into the biosynthesis of the peptidoglycan precursors. Peptidoglycan cell wall turnover refers to the loss of muropeptides from the bacterium to its media. Muropeptides lost to turnover may re-enter the periplasm for recycling. Notwithstanding the clear distinction between recycling and turnover, within the literature the two terms often are used interchangeably. In this review we preserve the distinction.
While the recovery of cell-wall fragments is not essential for in vitro planktonic growth, there are other important reasons for cell-wall recycling.3 The cell-wall fragments (muropeptides) have important messenger functions in bacterial communication, and as signal molecules in spore resuscitation and germination in some Gram-positive bacteria.4,5,6,7 In eukaryotes, the detection of muropeptides (e.g. via peptidoglycan-recognizing proteins and NOD receptors) initiates an immune response, and the recovery of cell-wall muropeptides suppresses this response.8,9
Of special interest is the relationship between cell-wall recycling and antibiotic resistance. While the phenomenon of β-lactamase induction in Gram-negative bacteria has been known for many years as a resistance response to the presence of β-lactam antibiotics, the links between β-lactamase induction and cell-wall recycling have been revealed only recently. In some Gram-negative organisms, the presence ofβ-lactam antibiotics is sensed by perturbations in the cytoplasmic pool of muropeptides, resulting in derepression of the ampC gene that encodes the AmpC β-lactamase. In some Gram-positive organisms, the modification of specific β-lactam-sensing proteins (e.g. BlaR1) by β-lactam antibiotics initiates signal transduction that culminates in expression of a β-lactamase (e.g. PC1). An important implication of the link between recycling and resistance is that inhibitors of cell-wall recycling pathways might be combined with cell-wall-targeting antibiotics in order to overcome resistance. Several recent studies support this possibility. Such a strategy could prove useful (or necessary) in overcoming resistance in infections due to (for example) AmpC-β-lactamase-hyperproducing Pseudomonas aeruginosa in cystic fibrosis,10 or due to methicillin-resistant Staphylococcus aureus (MRSA), which produces the β-lactam-resistant penicillin-binding protein PBP2a.
In this review, we evaluate the current understandings with respect to bacterial cell-wall recycling and turnover, with an emphasis on their relationship to antibiotic resistance. We summarize the important early studies, describe recent work linking recycling to resistance induction, and highlight efforts toward the discovery of recycling inhibitors.
Peptidoglycan structure, biosynthesis, and remodeling
The bacterial cell wall consists of glycan strands cross-linked through peptide stems to form a “peptidoglycan” (or “murein”) polymer. The glycan strands are composed of alternating N-acetylglucosamine (GlcNAc, NAG) and N-acetylmuramic acid (MurNAc, NAM) saccharides, with peptide stems originating on the lactyl moiety of the MurNAc saccharide. Polymerization of the glycan strand and cross-linking of the peptide stem are catalyzed in separate domains of bifunctional enzymes called penicillin-binding proteins (PBPs), which were named in recognition of the ability of β-lactam antibiotics to inactivate the transpeptidation reaction used in the cross-linking step. The three-dimensional mesh-like network of the peptidoglycan is strong enough to counteract the high osmotic pressure of the bacterial cell.11,12 While numerous variations in the structure of the pentapeptide stem are known, the core structure of the un-cross-linked stem attached to the lactyl moiety is –L-Ala–γ-D-Glu–m-DAP–D-Ala–D-Ala in Gram-negative bacteria (where m-DAP is meso-1,6-diaminopimelate) and –L-Ala–γ-D-Glu–L-Lys–D-Ala–D-Ala in most Gram-positive bacteria.13,14
Cell-wall biosynthesis begins in the cytoplasm with the conversion of fructose-6-phosphate (1) to UDP-GlcNAc by the sequential activity of the GlmS, GlmM, and GlmU enzymes. UDP-GlcNAc is converted to UDP-MurNAc-pentapeptide (2) by the sequential activity of the MurA enolpyruvyl transferase, MurB reductase, and MurC–MurF ligases (Figure 2). On the cytoplasmic face of the cell (inner) membrane, MraY catalyzes the reaction of UDP-MurNAc-pentapeptide with undecaprenyl pyrophosphate to generate Lipid I, which is then coupled with GlcNAc by the MurG transferase to yield Lipid II. The cytoplasmic steps leading to these lipid-linked intermediates in cell-wall biosynthesis are reviewed elsewhere.15,16
Figure 2.
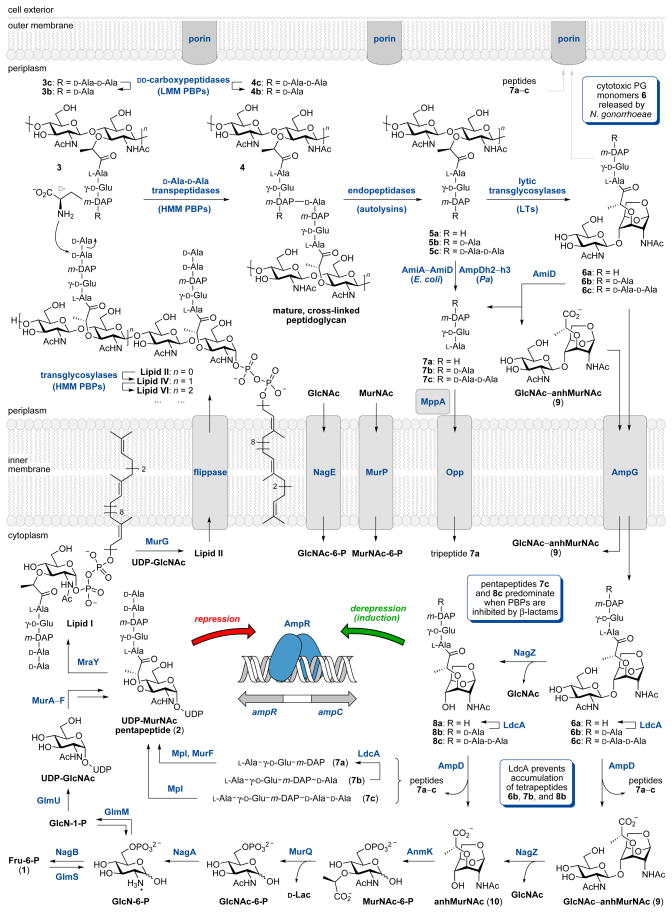
Summary of selected steps in cell-wall biosynthesis (left-hand side) and recycling (periplasm) in Gram-negative bacteria and regulation of β-lactamase (AmpC) production by cell-wall precursors (2) and anhydromuropeptide fragments (8). While the individual enzymes of the recycling pathways are most often isolated from E. coli, this organism does not possess a complete AmpC β-lactamase induction system. Studies of β-lactamase induction in E. coli use plasmids with ampC and ampR genes from an inducible bacterial strain.
The identity of the enzyme that translocates Lipid II across the cell membrane remains a matter of debate. Using bioinformatics methods, Ruiz suggested that MurJ (MviN) might function as the Lipid II flippase in Escherichia coli.17 MviN orthologs with low sequence homology, including YtgP, were later identified in Bacillus subtilis. Although YtgP complemented the growth defect of a MurJ-depleted strain of E. coli,18 Fay and Dworkin demonstrated that these genes are not essential because strains of B. subtilis that lack all four proteins grow normally.19 More recent work implicates FtsW as the Lipid II flippase (translocase).20
In the periplasm, Lipid II is the disaccharide donor for glycan strand growth, which is catalyzed in the transglycosylase domain of the HMM PBPs. Cross-linking of the peptide stems occurs in the separate transpeptidase domain of the HMM PBPs.21 Computational study of the transpeptidase reaction shows displacement of the terminal D-Ala residue of one pentapeptide strand by the terminal amine residue of the L-Lys (or m-DAP) residue of a neighboring strand (Figure 2).22β-Lactam antibiotics mimic the D-Ala-D-Ala terminus of the donor strand and inhibit transpeptidation by the functionally irreversible acylation of an active-site serine.
While the transpeptidase reaction is generally considered to be the last step in peptidoglycan biosynthesis, the structure of the cell wall undergoes constant remodeling. A peptidoglycan modification that may occur subsequently is O-acetylation, as a mechanism against peptidoglycan-cleaving lysozymes and as a mechanism to regulate glycan strand integrity against lytic transglycosylase cleavage.23 Peptidoglycan-O-acetyltransferases (PATs) acetylate the C-6 hydroxyl group of the MurNAc residue and O-acetylpeptidoglycan esterases (APEs) remove the acetyl group. Recent work indicates that both enzymes are potential antibiotic targets.24,25 Other glycan modifications include δ-lactam formation in Gram-positive spore peptidoglycan, N-deacetylation, N-glycolylation, and the attachment of surface polymers to the C6-hydroxyl group (Figure 3).26
Figure 3.
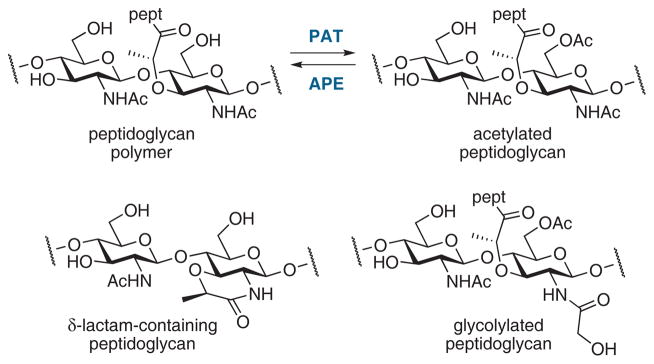
Examples of peptidoglycan structural modifications.26
Important modifications occur to the peptide stem. D-Ala-D-Ala carboxypeptidases (an activity of many LMM PBPs) hydrolytically remove the terminal D-Ala from the pentapeptide stem (e.g. 3c→3b, Figure 2).27,28 This remodeling controls the degree of final cross-linking as the resulting tetrapeptide is no longer capable as a substrate for cross-linking by D-Ala-D-Ala transpeptidases.29 DD-Carboxypeptidases are also involved in cell division30 and AmpC β-lactamase induction (discussed below), although the exact functional roles for all of the LMM PBPs are not understood.
In the late 1990s de Pedro et al. described the incorporation of exogenous D-amino acids such as D-cysteine into the cell wall of E. coli.31 Their experiments enabled the visualization of murein segregation, wherein the new peptidoglycan was inserted over the cylindrical surface and at the cell division site while peptidoglycan at the distal poles was inert. Recently, the biosynthesis of several noncanonical D-amino acids by many bacteria was correlated to important messenger functions in the regulation of peptidoglycan biosynthesis, composition, and strength.32,33,34,35
Discovery of cell-wall recycling in gram-negative bacteria and its link with β-lactamase induction
Cell-wall turnover—the release of muropeptides to the medium—was discovered in the early 1960s using Bacillus megaterium having a 14C-labeled cell wall.36,37 Later, the occurrence of turnover was seen in both Gram-positive and Gram-negative bacteria.38 By the mid-1980s, a variety of Gram-positive and Gram-negative organisms were shown to lose up to 50% of their cell walls per generation, although turnover in E. coli was much less.39 When Goodell and Schwarz reinvestigated this phenomenon using E. coli murein labeled with 3H-DAP, they confirmed that the cell-wall peptidoglycan lost 6–8% of the label to the culture medium per generation.40 Three forms of the 3H-DAP-containing material were identified from the medium: tetrapeptide 7b (L-Ala–D-γ-Glu–m-DAP–D-Ala), the tripeptide 7a (L-Ala–D-γ-Glu–m-DAP), and a dipeptide (m-DAP–D-Ala). 3H-DAP was also found in the cytoplasm, in the form of 3H-labeled UDP-MurNAc-pentapeptide (2). This discovery led to the proposal that peptidoglycan fragments were being recycled.41 Goodell calculated that 50% of the cell wall was being degraded and recycled per generation. Hence, the majority of the cell-wall fragments created during remodeling was recovered and recycled. Since the polar caps are inert to turnover, more than 60% of the peptidoglycan of the side walls is degraded in one generation.42
DAP-containing peptide fragments 7a and 7b are generated naturally in the periplasm of E. coli by the hydrolysis of peptidoglycan peptide stems by MurNAc–L-Ala amidases AmiA, AmiB, AmiC, and AmiD (e.g. 5→7 and 6→7, Figure 2). Uptake of the tripeptide 7a via the oligopeptide permease OppA requires the muropeptide binding protein MppA and other components OppB, OppC, OppD, and OppF.43 However, the recycling of these short peptide fragments represents only a minor recycling pathway in E. coli since the extent of peptidoglycan turnover was unchanged in mutants lacking Opp permease.44 Instead, the major recycling pathway is now known to involve saccharide-containing anhydromuropeptide fragments which enter the cytoplasm through the AmpG permease.45
The determination of AmpG as the permease essential for recycling was an important discovery because it linked peptidoglycan recycling to β-lactamase induction.45 This correlation built upon previous recognition that the transmembrane AmpG permease,46,47 the transcriptional regulator AmpR,48,49 and the cytosolic protein AmpD,45 were all required for AmpC β-lactamase induction in Citrobacter freundii and Enterobacter cloacae. As mutants in the ampD gene gave semiconstitutive overproduction of AmpC β-lactamase,50 while ampG ampD double mutants were non-inducible,47 it was thought that AmpG must be a signal transducer or transporter that allows entry of a ligand that activates the AmpR regulator in the absence of AmpD activity. The discovery that a DAP-labeled muropeptide, anhMurNAc-tripeptide (8a), accumulated in the cytoplasm of ampD mutants led to the conclusion that tripeptide 8a was the signal molecule, and that AmpD acted as a negative regulator of AmpC β-lactamase expression by preventing accumulation of 8a.51
These seminal studies form the basis for our current understanding of the relationship between cell-wall recycling and β-lactamase induction. Muropeptides 6, generated by the lytic transglycosylase activity of cell-wall recycling, enter the cytoplasm through the AmpG permease and are hydrolyzed by the glucosaminidase NagZ to give anhydromuramyl peptides (8a–c). Under normal circumstances—in the absence of β-lactam antibiotics—these anhydromuropeptides are hydrolyzed by the amidase AmpD and recycled through a series of reactions into cell-wall precursors for reincorporation into the cell wall (Figure 2). As described above, anhydromuropeptides (either 8a or 8c) were proposed51,52 as the signaling molecules that bind to AmpR to induce AmpC β-lactamase production. When UDP-MurNAc-pentapeptide (2) is bound to AmpR, AmpC expression is repressed. Thus, regulation of AmpC depends on the relative concentrations of 8 and 2. In the presence of β-lactam antibiotics, anhydromuropeptides accumulate and displace UDP-MurNAc-pentapeptide (2) from the AmpR regulator. AmpC β-lactamase is expressed and exported to the periplasm to confront and neutralize the β-lactam threat.
Autolysins: peptidoglycan hydrolases and lytic transglycosylases
Cell-wall biosynthesis must be coordinated with cell-wall remodeling in a carefully controlled manner, since any loss of cell-wall integrity could lead to lysis and death. Cell-wall remodeling involves peptidoglycan (murein) hydrolases, enzymes that are also called autolysins because they are potentially autolytic if their activity is uncontrolled.53 Autolysins include peptide-cleaving carboxypeptidases, endopeptidases (4→5, Figure 2), and N-acetylmuramyl-L-alanine amidases (5→7), and glycan-cleaving lytic transglycosylases (5→6). These enzymes are ubiquitous in bacteria and have important roles in cell division and in creating space within the peptidoglycan polymer to accomodate structures such as secretion systems, flagella, and pili.54
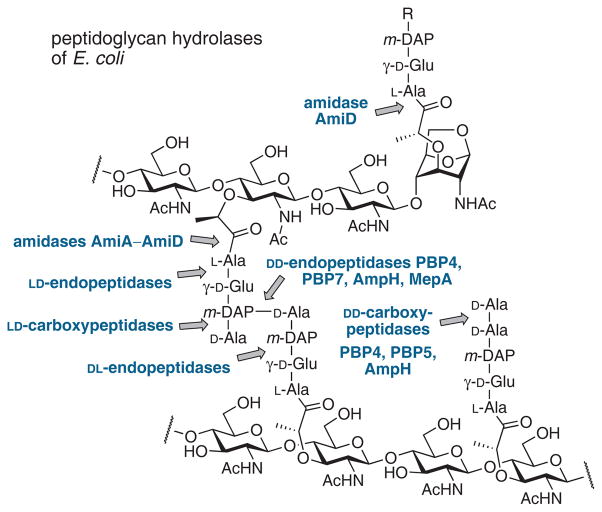
The peptide stem of peptidoglycan is hydrolyzed by a number of amidases, penicillin-sensitive carboxypeptidases (i.e. low-molecular-mass PBPs), and penicillin-insensitive carboxypeptidases and endopeptidases (Figure 4).53 Bacteria have several PBPs of each subclass. Mutants lacking one or more PBPs often grow normally, although mutants lacking multiple autolysins exhibit abnormal morphologies.55,56 Detailed reviews on the peptidoglycan hydrolases and penicillin-binding proteins have been published.29,53,57
Figure 4.
Hydrolytic cleavage sites in E. coli peptidoglycan for periplasmic carboxypeptidases and endopeptidases.14,53
The glycan strand of the peptidoglycan is cleaved at the β-(1→4) glycosidic bond between the MurNAc and GlcNAc residues by an important class of autolysins called lytic transglycosylases (LTs). LTs catalyze a non-hydrolytic cyclization wherein the MurNAc C6-hydroxyl adds to its anomeric carbon to form a 1,6-anhydrosaccharide (Figure 5), cleaving the glycosidic bond.54 Bacteria typically encode multiple lytic transglycosylases. Interactions between PBPs and LTs have been identified in E. coli58,59 and P. aeruginosa60 and it is thought these enzymes function cooperatively during cell wall biosynthesis.
Figure 5.
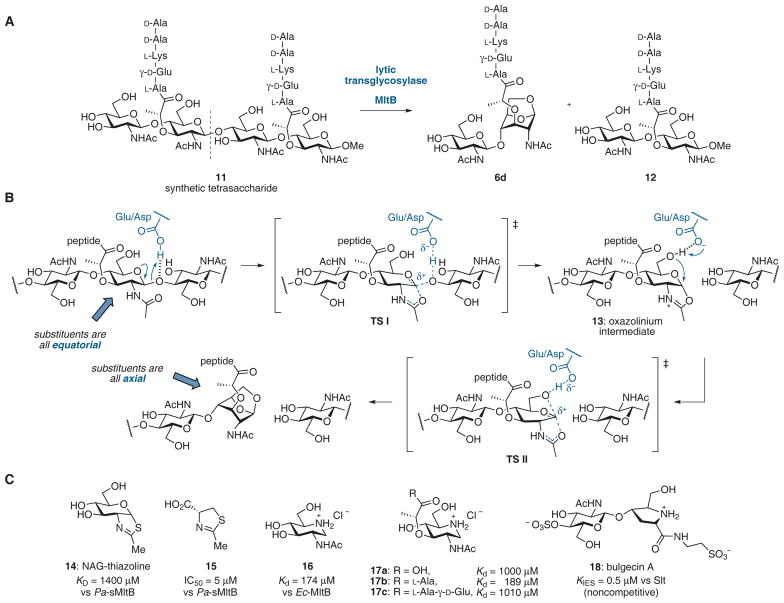
Lytic transglycosylase (LT) reactivities toward the peptidoglycan. (A) Cleavage of a synthetic peptidoglycan fragment by MltB of E. coli with formation of anhydromuropeptide 6d.70 (B) Mechanism proposed for the LT reaction.54 (C) Inhibitors of LTs.
Escherichia coli produces one soluble LT (Slt70) and six membrane-anchored LTs (MltA–MltF) that are believed to bind to the inner leaflet of the outer membrane. Based on the sequences of lytic transglycosylases from a wide variety of bacteria, LTs are divided into four families.61 Family 1 is further divided into five subfamilies (1A–1E) and includes E. coli Slt70 (1A), MltF (1B), MltC (1C), MltD (1D), and MltE (1E), with each having some sequence similarity to goose-type lysozymes. Families 2 and 3 include the E. coli enzymes MltA and MltB, respectively, and family 4 LTs are primarily endolysins of λ bacteriophage.54 Pseudomonas aeruginosa produces at least seven LTs, including four different family 3 enzymes (MltB, SltB1, SltB2, and SltB3), but unlike E. coli, P. aeruginosa does not appear to encode family 1C or 1E LTs.62 Neisseria gonorrhoeae produces five LTs, LtgA–LtgE, that have homology with E. coli enzymes, and most strains also produce LtgX and AtlA, which are involved in type IV secretion.63 Multiple LT deletion preserves normal growth and morphology, but overexpression leads to spheroplast formation or lysis. While deletion of six of the LTs of E. coli gave a normal planktonic phenotype, deletion of all seven failed to give a viable phenotype.56,64 Interestingly, certain deletion mutants were more sensitive to β-lactam antibiotics (vide infra).65,66
Among the seven LTs of E. coli, most appear to be exolytic enzymes that release GlcNAc-anhMurNAc-peptides 6 from the ends of glycan strands, but MltE shows endolytic activity and seems to cleave only internal β-(1→4) glycosidic bonds.67,68 MltA is unlike the other LTs in its ability to accept peptide-free glycan strands as substrates69 and MltB was the only E. coli LT able to cleave the short synthetic peptidoglycan fragment 11 (Figure 5A).70 While it is clear that most LTs have exolytic activity, the direction from which glycosidic cleavage occurs is not established.
Crystal structures of the E. coli LTs Slt70,71 MltA,72,73,74 Slt35 (a naturally-occurring product of MltB),75,76,77 and MltE,68 have been solved and complexes of Slt70,71 MltA,74 and Slt3578 with peptidoglycan fragments are especially informative. X-ray crystal structures of N. gonorrhoeae MltA73 and SltB1 of P. aeruginosa79 are described and an NMR structure of the potential peptidoglycan-binding domain of MltD has been determined.80
Despite differences in sequence and overall structure between families, each of the LTs displays a prominent groove or cleft, which accommodates the glycan backbone of the peptidoglycan polymer, and a single glutamic acid residue (Asp in MltA) positioned between subsites +1 and −1. The Glu/Asp is protonated at physiological pH and acts as a general acid in the first step of the reaction and as a general base in the second step of the reaction (Figure 5B). The developing positive charge in the transition state (TS I) may be stabilized by the neighboring amide, leading to a protonated oxazoline intermediate (13). The glutamate/aspartate (now in a deprotonated state) activates the C6-OH for intramolecular attack at the anomeric carbon, and this process is accompanied by a large conformational change in which the anhydrosaccharide product has all of its substituents in an axial orientation.
The notion that an oxazolinium species is encountered during the LT reaction is supported by reports that thiazoline derivatives 14 and 15 are inhibitors of MltB (Figure 5C). NAG-thiazoline (14) is a knownβ-hexosaminidase inhibitor that mimics the transition states (or intermediates) that resemble TS I or 13. Although 14 had only weak affinity (KD = 1.4 mM) for Pa-sMltB, a soluble form of P. aeruginosa MltB, it caused morphological changes in treated cells.81 The simple thiazoline 15 had more potent activity than 14 against sMltB (IC50 = 5 μM).82
Yamaguchi et al. reported a series of piperidine iminosaccharide as possible mimetics of the oxocarbenium species generated in the course of the LT reaction.83 Iminosaccharides 16 and 17b showed moderate affinity toward E. coli MltB (Kd = 170 and 190 μM, respectively) while 17a and 17c bound to MltB less tightly.
Bulgecin A (18), a GlcNAc-containing natural product, is a noncompetitive LT inhibitor.65,84 X-ray structures of bulgecin A bound to Slt7071,85 and Slt3578 show that bulgecin A binds to the −1 and −2 saccharide-binding subsites, with the hydroxymethyl group of the pyrrolidine ring hydrogen-bonded to the catalytic Glu162 in the same way that the substrate is proposed to interact (i.e. TS II). Bulgecin A reduced the MICs of β-lactams against E. coli to the same level as accomplished by LT deletion mutants.65 Inhibition of LTs therefore may represent an important strategy for extending the life of β-lactam antibiotics.10,66 Alternatively, LT inhibitors may attenuate the pathogenesis of bacteria such as Bordetella pertussis and N. gonorrhoeae, which release cytotoxic anhydromuropeptides that cause inflammation.63,86 The mechanism by which LTs cleave peptidoglycan is distinct from that of human lysozymes and glucosaminidases, and this difference also makes the LTs attractive as targets.54
Muropeptide recovery and recycling in gram-negative organisms
As mentioned above, early studies of peptidoglycan recycling with E. coli mutants lacking Opp, the permease required for DAP uptake, revealed that only 0–8% of their cell wall-derived muropeptides were released to the medium per generation.44 Since the extent of turnover was the same as that of the parent strain, it was clear that Opp had an inconsequential role in recycling. Identification of the permease used for muropeptide recycling focused on the AmpG membrane protein.39 AmpG was found essential for high-level AmpC β-lactamase induction in Enterobacter cloacae46 and Citrobacter freundii.47 A central role for AmpG in cell-wall recycling was confirmed by the discovery that mutants lacking ampG released up to 40% of their peptidoglycan per generation, in contrast to the <10% turnover with AmpG functional.45 Since these results were reported in the early 1990s, AmpG was shown to be required for β-lactamase induction and for muropeptide recycling in other Gram-negative organisms and mutations in AmpG affect muropeptide turnover.63,87,88,89 P. aeruginosa carries two AmpG permease homologs, AmpG and AmpP (also called AmpGh1).90 Both are required for maximum β-lactamase induction but AmpG is the more important.91
A topological model indicates that AmpG contains ten transmembrane segments and two large cytoplasmic loops.92 A study of the substrate specificity AmpG, using freeze-thawed E. coli cells and 3H-labeled muropeptides, revealed that the GlcNAc–anhMurNAc disaccharide substructure is a requirement for transport.93 Disaccharide GlcNAc-anhMurNAc-peptides (8a–c) and the free disaccharide GlcNAc-anhMurNAc (9) were taken up efficiently while tripeptide 7a, disaccharide GlcNAc-MurNAc-peptides, and the monosaccharides anhMurNAc (10) and anhMurNAc-tripeptide (8a) were taken up poorly. AmpG is thought to be a single-component permease and dependent on the proton motive force because its activity was sensitive to carbonyl cyanide m-chlorophenyl hydrazone.93 Nearly all of the cell-wall fragments that are recycled pass through AmpG. Since inactivation of AmpG fully restores β-lactam susceptibility in β-lactam-resistant strains of P. aeruginosa,94 AmpG is an obvious target for the development of inhibitors of cell-wall recycling.
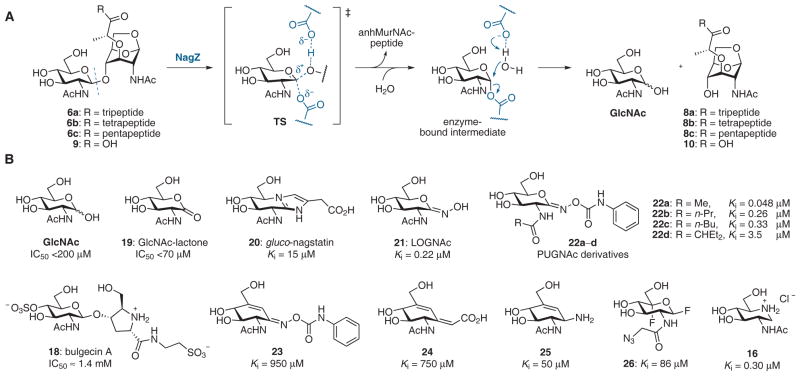
Following internalization of the GlcNAc-anhMurNAc muropeptides (6a–c and 9) into the cytoplasm through AmpG permease, the anhydrodisaccharides encounter NagZ, the first cytoplasmic enzyme of the muropeptide recycling pathway. NagZ is an N-acetylglucosaminidase that hydrolytically cleaves the β-(1→4) glycosidic bond of anhydrodisaccharides 6 and 9 to generate GlcNAc and anhydrosaccharides 8 and 10 (Figure 6A).
Figure 6.
Muropeptide cleavage by the N-acetylglucosaminidase NagZ. (A) The double-displacement mechanism for the NagZ-catalyzed hydrolysis of GlcNAc-MurNAc disaccharides.101 (B) NagZ inhibitors.
An N-acetylglucosaminidase of E. coli was isolated by Yem and Wu in 1976 as a 36 kDa cytoplasmic enzyme.95,96 The central role of this enzyme in cell-wall recycling and β-lactamase induction was realized in mid-1990s with the discovery by Jacobs et al. that anhMurNAc-tripeptide accumulates in the cytoplasm of ampD mutants.45 Since AmpG was thought to act as a permease for the GlcNAc-anhMurNAc-tripeptide (6a) and the anhMurNAc-tripeptide (8a) was an AmpD substrate, the participation of a cytosolic N-acetylglucosaminidase was implicated for the removal of GlcNAc from 6a.97 Subsequent studies confirmed that nagZ encodes a cytosolic enzyme, is active on GlcNAc-anhMurNAc-containing muropeptides and is required for cell-wall recycling but not required for normal cell growth.98,99 A nagZ deletion mutant totally lacked N-acetylglucosaminidase activity, and GlcNAc-anhMurNAc disaccharides accumulated in the cytoplasm, indicating that NagZ is the only N-acetylglucosaminidase expressed in E. coli.98 E. coli strains lacking nagZ induced only about 25% as much β-lactamase (from a plasmid carrying the ampC-ampR system of E. cloacae) compared to the control strain, and were more sensitive to β-lactam antibiotics.99 Regarding substrate specificity, NagZ cleaves GlcNAc from anhydromuropeptides (e.g. 6a and 9), and surprisingly also from GlcNAc-MurNAc-peptides (e.g. 12) that NagZ would not normally encounter in the cytoplasm.83,98
The requirement of NagZ for AmpC β-lactamase induction and, more specifically, its role as the enzyme that produces the anhMurNAc-peptide signal molecule(s), makes NagZ a key target for inhibitor development. It was noted in early characterization studies of NagZ that its activity was weakly inhibited by glucosamine, muramic acid, MurNAc, and bulgecin A, while GlcNAc (the reaction product) and N-acetylglucosaminolactone (19) exhibited more potent inhibition (Figure 6B).95,99 More recent efforts to discover NagZ inhibitors have involved the screening of known inhibitors of other glucosaminidases and Stubbs et al. found moderate inhibition of Vibrio colerae NagZ (VcNagZ) by gluco-nagstatin (20) and potent inhibition by oximes LOGNAc (21) and PUGNAc (22a).100 An X-ray crystal structure of VcNagZ with 22a showed a large open pocket in NagZ that is notably different than the analogous binding pocket of human glucosaminidases. A series of PUGNAc analogues (e.g. 22b–22d) was prepared in order to exploit this structural difference and the evaluation of these derivatives against VcNagZ (a family GH3 glucosaminidase) and human β-hexosaminidase (family GH20) and O-GlcNAcase (family GH84) showed that selectivity was achieved. While PUGNAc, the most potent compound of the series, is nonselective, inhibitors 22c and 22d showed more than 100-fold selectivity toward NagZ over human enzymes.100 Crystal structures of 22b and 22c bound to VcNagZ have also been published.101
The structural differences in the binding pockets of NagZ and human (GH20 and GH84) enzymes reflect differences in mechanism.101 While family GH3 enzymes (e.g. NagZ) employ a double-displacement mechanism that involves a covalent enzyme-bound intermediate (Figure 6A), the human glucosaminidases rely on substrate-assisted catalysis in which the neighboring 2-acetamido group acts as a nucleophile to form an oxazolinium intermediate. Accordingly, the binding pocket of the human enzymes is more restrictive than that of NagZ in order to orient the acetamido group appropriately and promote its participation.
Carbocyclic analogues of PUGNAc, 23 and 24, were explored as inhibitors of NagZ but only modest activity was observed.102 The 1-acetamido analogue of 6-epi-valienamine, 25, showed moderate inhibition of NagZ (Ki = 50 μM) but this compound was also active against GH20 and GH84 enzymes.103 The azide-containing glucosamine fluoride derivative 26, which was prepared as an activity-based proteomics probe for the profiling of exo-glycosidases, inactivated VcNagZ with a Ki value of 50 μM and kinact value of 0.74 s−1.104 Iminosaccharide 16, which showed only moderate affinity toward the lytic transglycosylase MltB, was a potent competitive inhibitor of P. aeruginosa NagZ (Ki = 300 nM).83 Iminosaccharide analogues of MurNAc and MurNAc-peptides (17a–17c) were less potent (Ki = 51, 35, and 33 μM, respectively).
Inactivation of NagZ (by 22d) not only attenuated inducible AmpC-mediated β-lactam resistance in P. aeruginosa,105 but its inhibition (by 22a) restored wild-type MICs for β-lactams against strains of P. aeruginosa with ampD and dacB (PBP4) mutations.106 As an important benefit, NagZ inhibition reduced the ability of P. aeruginosa to develop ceftazidime resistance.106 Together, these studies support NagZ as an attractive target for inhibitors of cell wall recycling.
The pool of anhydromuropeptides generated by NagZ is controlled by the amidase AmpD. As mentioned above, the relative concentrations of anhMurNAc-peptides (8a and 8c) and UDP-MurNAc-pentapeptide (2) are critical for the regulation of AmpC β-lactamase, and AmpD therefore occupies an important position at the crossroads of peptidoglycan recycling and antibiotic resistance. An important role for AmpD in resistance was recognized in the late 1980s, when semi-constitutive expression of AmpC in C. freundii and E. cloacae resulted from ampD mutation.50 Its essential role in peptidoglycan recycling was discovered when ampD mutants were found to accumulate anhMurNAc-tripeptide (8a).45 AmpD is a cytoplasmic zinc-dependent amidase that cleaves anhydromuropeptides between the D-lactate moiety and L-alanine residue (Figure 7). AmpD was highly selective for anhydromuropeptides and hydrolyzed UDP-MurNAc-pentapeptide (2) at least 10,000-fold more slowly than anhMurNAc-tripeptide (8a).97 This selectivity ensures that AmpD participates in muropeptide recycling without degrading peptidoglycan precursors. Detailed studies with synthetic muropeptides show that anhMurNAc-peptides 8a and 8c are hydrolyzed much more efficiently than GlcNAc-anhMurNAc-peptide 6c.107,108
Figure 7.
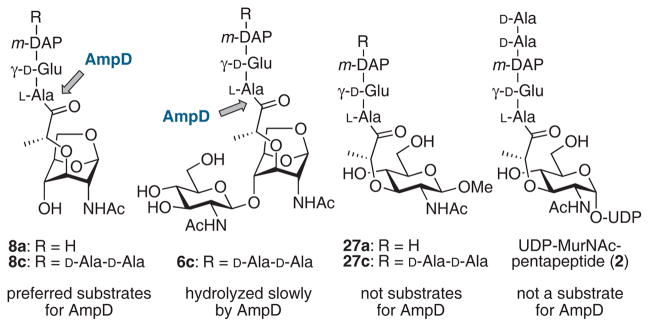
anhMurNAc-peptide substrates for AmpD and MurNAc-peptide non-substrates.107,108
An NMR structure of AmpD from C. freundii, reported in 2003, revealed similarities between AmpD and bacteriophage T7 lysozyme and domains of eukaryotic peptidoglycan-recognizing proteins (PGRPs).109 Recent X-ray crystal structures of the C. freundii AmpD, however, show that the solid-state conformation of AmpD is very different from that of the NMR structure, with domain motions as high as 17 Å.110 In addition to structures of the apoenzyme (without zinc) and the holoenzyme (with its zinc cofactor), crystals of AmpD that were soaked with a substrate, anhMurNAc-tripeptide (8a), gave a structure with the bound reaction products (6a and 7a: PDB code 2y2b). Since the crystals of AmpD showed catalytic activity, the solid-state conformation was proposed to be the active conformation while the NMR solution structure, which has a ‘closed’ conformation, is an inactive form.110 This large conformational change may serve to prevent adventitious proteolytic degradation within the cytoplasm.110 The interconversion between the closed (NMR) and open (X-ray) conformations was proposed as an activation mechanism for this cytosolic protein.110
In addition to AmpD, five other zinc-dependent enzymes with N-acetylmuramyl-L-alanine amidase activity were identified in E. coli: AmiA, AmiB, AmiC, AmiD, and a partially-characterized 39-kDa amidase.53 AmiA–C are periplasmic amidases involved in cell division and separation.111,112,113 Mutants lacking one of these amidases grew normally but multiple deletions caused cell separation defects. AmiA, AmiB, and AmiC amidases have narrow substrate specificities and are only active on isolated peptidoglycan. AmiD, on the other hand, has a broader substrate scope and acts on the purified sacculus, low-molecular-weight muropeptides, and both MurNAc-peptides and anhydromuropeptides.114 Unlike AmiA–AmiC, AmiD is anchored to the periplasmic face of the outer membrane and does not participate in cell separation.114 Crystal structures of AmiD complexed with the anhMurNAc-L-Ala-γ-D-Glu-L-Lys substrate and with the tripeptide reaction product were solved.115
In recent studies of AmpC hyperproduction in clinical strains of P. aeruginosa, Juan et al. discovered that the level of AmpC derepression could not be fully attributed to mutations in ampD only.116 Mutations were detected in the region of ampE, which encodes a cytoplasmic membrane protein, but the role of ampE remains unknown. It was later found that P. aeruginosa expresses three AmpD homologs—AmpD (also called AmpD1), AmpDh2, and AmpDh3—and that these three homologs are responsible for stepwise upregulation of AmpC expression.117 AmpDh2 and AmpDh3 show 50% and 47% similarity to E. coli AmiD, respectively, and they are also annotated to be periplasmic.118,119 Single mutations (e.g. ΔD) and double mutants (e.g. ΔDDh3) gave progressively higher-level (derepressed) inducible expression and the triple mutant (ΔDDh2Dh3) showed fully derepressed, non-inducible expression (>1000-fold vs wild-type). This stepwise upregulation mechanism is advantageous because there is a fitness cost for constitutive AmpC production and high-level β-lactam resistance can be achieved with only partial derepression of ampC.120 The clinical relevance of this model is still unclear, however. It is possible that additional unknown factors contribute to the complex AmpC regulation mechanisms in P. aeruginosa.121
Following the processing of anhydromuropeptides (8) by AmpD, the anhMurNAc product (10) is recycled and converted to glucosamine-6-phosphate (GlcN-6-P) by the action of AnmK kinase, MurQ etherase, and NagA deacetylase (Figure 2). Anhydro-N-acetylmuramic acid kinase (AnmK) was identified in 2005 by Uehara et al. and X-ray crystal structures of anhMurNAc (10) and ADP bound to AnmK were published recently.122,123 Superimposition of these structures indicates that the terminal phosphorus atom of ATP and the C6-oxygen atom of anhMurNAc (10) lie in close proximity to each other within the active site. This model suggests that the Mg(II)- and ATP-dependent AnmK-catalyzed hydrolysis and phosphorylation occurs in a single step.123 murQ was identified as a gene required for the growth of E. coli on N-acetylmuramic acid as the only carbon source,124 and MurQ was confirmed to be the hypothetical MurNAc-6-phosphate etherase that had been predicted previously.122,125,126 D-Lactate is released during the MurQ reaction,124 and studies with 18O-labeled water and 2H-labeled MurNAc-6-P indicate that the elimination of D-Lac is followed by hydration to give the GlcNAc-6-P product.127 The next step in the recycling process involves N-acetylglucosamine-6-phosphate deacetylase (NagA), an essential enzyme for the recycling of GlcNAc and anhMurNAc.39 Following deacetylation by NagA, glucosamine-6-phosphate can be converted to fructose-6-phosphate by NagB deaminase or isomerized to GlcN-1-P by GlmM (Figure 2).15
The hydrolysis of anhydromuropeptides 8a–c by AmpD releases the free tripeptide 7a, the free pentapeptide 7c, and to a lesser extent tetrapeptide 7b. Tetrapeptides 6b, 7b, and 8b do not accumulate in the cytoplasm because of the activity of the L,D-carboxypeptidase LdcA, a D-Ala-releasing enzyme that is essential for normal cell growth.128 Deletion mutants of ldcA in E. coli lyse in the stationary phase of growth. Since the UDP-MurNAc-tetrapeptide accumulated in the mutant, it is thought that tetrapeptides were incorporated into the cell wall and lysis resulted from an inability to cross-link the peptide stems.
The major pathway for the recycling of peptides generated by AmpD (7a and 7c) involves their direct use by the murein peptide ligase (Mpl) to generate peptidoglycan precursors.39 For example, Mpl catalyzes the ligation of UDP-MurNAc with tripeptide 7c to produce the UDP-MurNAc-tripeptide, which is coupled with D-Ala-D-Ala by MurF to generate the UDP-MurNAc-pentapeptide (2).129 However, Mpl is known to accept the tri-, tetra-, and pentapeptides 7a, 7b, and 7c equally well and the ligation with 7c produces 2 directly.130 A crystal structure of Mpl was solved recently.131 A minor pathway for recycling of the tripeptide 7c involves its breakdown into the individual amino acids by the sequential action of MpaA, YcjG, and PepD. MpaA amidase cleaves the γ-D-Glu–m-DAP bond of 7c to generate L-Ala-γ-D-Glu and m-DAP. The L-alanyl-γ-D/L-glutamate isomerase YcjG produces L-Ala-γ-L-Glu, which can be cleaved by the dipeptidase PepD.
AmpR and the induction of AmpC β-lactamase in Gram-negative organisms
In many Gram-negative organisms, such as P. aeruginosa, N. gonorrhoeae, E. cloacae, and C. freundii, but not E. coli (which lacks ampR and expresses AmpC β-lactamase constitutively), exposure to β-lactam antibiotics induces high-level AmpC β-lactamase production. Several genes (including ampG, nagZ, ampD, and ampR) are required for inducibility, and the link between peptidoglycan recycling and β-lactamase induction is now well established. In studies of β-lactamase induction with ampD mutants, Jacobs et al. found that the anhMurNAc-tripeptide (8a) accumulated in the cytoplasm and concluded that the anhydromuropeptide 8a was the signal molecule (effector) that binds to the transcriptional regulator AmpR and permits AmpC production.45,51 However, Dietz et al. have argued that the anhMurNAc-pentapeptide (8c) is more likely to be the signal molecule, since 8c accumulated in β-lactam-treated ampD-lacking mutants of E. coli.52 The enhancement of β-lactam resistance in P. aeruginosa caused by the inactivation of PBP4, a noncritical carboxypeptidase encoded by dacB, seems to implicate the pentapeptide 8c as a more likely candidate, but the identity of the true signal molecule(s) has not yet been determined unequivocally.132,133 The tetrapeptide 8b is unlikely to be the signal molecule because it is known that tetrapeptides do not accumulate in the cytoplasm. Park and Uehara also suggest that the free pentapeptide 7c may be an effector of AmpR.39 The anhydromuropeptide effector molecules (8a, 8c, or 7c) are thought to compete for binding to AmpR with UDP-MurNAc-pentapeptide (2), a cell-wall precursor that acts as a corepressor with AmpR to repress β-lactamase expression in the absence of β-lactam antibiotics.51,134 Lipid II has also been proposed as a possible repressor of AmpR.39
The ampR and ampC genes form a divergent operon with overlapping promoters, and AmpR regulates the transcription of both genes.49 AmpR in P. aeruginosa also regulates genes for virulence factors.135 Mutations in ampR may lead to constitutive hyperexpression of AmpC, but ampR mutations are relatively rare compared to mutations in ampD.116
AmpR is a 32-kDa protein that belongs to a large family of LysR-type transcriptional regulators (LTTR) and has a C-terminal effector-binding domain (EBD) and an N-terminal DNA-binding domain. By analogy with other LTTR proteins, it has long been thought that the binding of the effector ligand to the EBD of AmpD leads to conformational changes in the protein that alter its affinity for the DNA, and in turn convert AmpD from a repressor into a transcriptional activator.47 A model for the binding of the anhMurNAc-pentapeptide (8c) to AmpR, based on crystal structures of the effector-binding domain, has been proposed recently.136 The EBD of AmpR, which is dimeric in the crystal structure, has two subdomains. The effector molecule is thought to bind in a pocket between the two subdomains. Aminoacid substitutions (Thr103Val, Ser221Ala, and Tyr264Phe) at the base of the interdomain pocket eliminate the ability of AmpR to induce ampC while the Gly102Glu mutant is unable to repress AmpC production. Previous proposals that a conformational change is necessary for AmpR derepression are supported by circular dichroism experiments. These indicate that wild-type AmpR and the Thr103Val, Ser221Ala, and Tyr264Phe mutants favor one conformation while the Gly102Glu mutant favors another conformation.136 The structural insights provided by these AmpR crystal structures may enable the design of small molecules that could block the binding of the anhMurNAc-peptides (8a or 8c) to AmpR or at least prevent the conformational change necessary for the activation of ampC transcription.10
Peptidoglycan recycling and turnover in Gram-positive bacteria
In all key aspects the pathways and purposes of peptidoglycan recycling are less well understood in Gram-positive compared to Gram-negative bacteria. Nonetheless, in those Gram-positive species where the possibility of cell-wall turnover was examined (including Bacillus, Lactobacillus, and Listeria species; and Staphylococcus aureus), robust turnover during vegetative growth was observed.137 Nonetheless, the possibility that robust turnover would correlate to robust recycling by these Gram-positive species is recent.138 The basis for the initial uncertainty concerning this correlation followed from the contrasting character of the Gram-positive and Gram-negative cell walls. The Gram-positive cell wall comprises an exoskeleton, and is significantly more massive than that of the Gram-negative bacterium. Moreover, the inside-to-outside model for peptidoglycan synthesis gives a presumptive requirement for autolysin-dependent “relaxation” of the outermost cell wall, so as to enable circumferential expansion.139,140 These characteristics, coupled with the observation of significant quantities of cell-wall fragments in the medium of Gram-positive cultures,141 led to the assumption that cell-wall recycling by the Gram-positive bacterium was not consequential. This presumption is now called into question, both by direct experimental studies on cell-wall recycling by Gram-positives undertaken recently by Mayer and colleagues, and by an emerging understanding of a common purpose—in both Gram-negatives and Gram-positives—for cell-wall recycling as a means of controlling resistance and virulence responses.
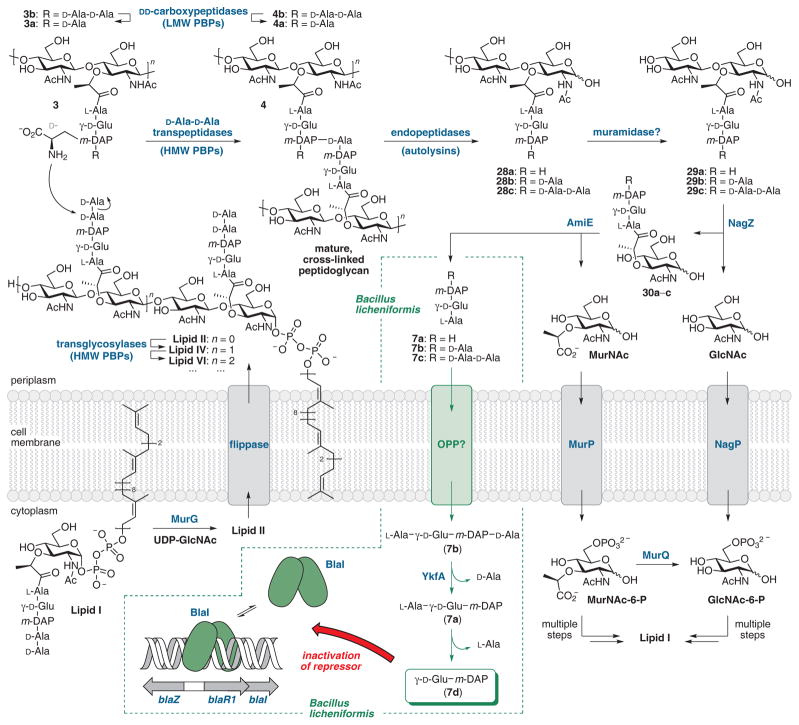
A key experiment supporting the presence of pathways for Gram-positive peptidoglycan recycling was the identification by Litzinger et al. in B. subtilis of a six gene-cluster (original annotation ybbIHFEDC) wherein the first five gene products were orthologs of the E. coli recycling proteins murQ (etherase), murR (transcriptional repressor), murP (MurNAc phosphotransferase), amiE (an N-acetylmuramyl-L-alanine-specific amidase) and nagZ (glucosaminidase).142 The function of the protein encoded by the sixth gene ybbC is unknown. Mutation of ybbI (Bs murQ) and ybbF (Bs murP) impaired growth of B. subtilis on MurNAc, while mutation of ybbD (Bs nagZ) resulted in the accumulation in the spent medium of muropeptides having a structure consistent with the proven enzymatic activity of E. coli NagZ. Functional and structural confirmation (by high-resolution crystallography) of the B. subtilis ybbD gene product as a NagZ glucosaminidase was obtained.143 B. subtilis NagZ enzyme is secreted under signal peptide control during both exponential and stationary growth resulting in non-covalent attachment (as evidenced by its liberation by either salt or lysozyme) to the peptidoglycan.142 Its catalytic motif is an unusual His-Asp dyad. Its possible role within a functional B. subtilis peptidoglycan recycling system is shown in Figure 8.138,143
Figure 8.
Summary of selected steps in cell-wall biosynthesis and recycling in the Gram-positive organism Bacillus subtilis incorporating mechanistic features proposed by Amoroso et al.181 for BlaI inactivation in Bacillus licheniformis (green arrows) by the dipeptide γ-D-Glu-m-DAP (m-DAP = meso-1,6-diaminopimelate).
This formulation is not, however, general. Whereas orthologs of the B. subtilis nagZ gene are found in other representative Gram-positive bacteria (including those of cytoplasmic enzymes of Clostridium acetobutylicum and Streptomyces avermitilis, and as a secreted enzyme of Lactococcus lactis), orthologs are not found in other bacteria (Bacillus cereus, Enterococcus faecalis, and Staphylococcus aureus).138 An initial evaluation of the contrasting features of the B. subtilis and C. acetobutylicum recycling systems (NagZ secreted in the former, but cytoplasmic in the latter) was undertaken by Reith et al.144,145 In addition to encoding a NagZ ortholog, the C. acetobutylicum genome also encodes orthologs of AmiE and MurQ as cytoplasmic enzymes, and of the MurP and MurR proteins. These genes strongly suggest the presence of a muropeptide recovery system that is similar, other than the cytoplasmic location for each of these identified component proteins, to the system present in B. subtilis. A key difference is the mechanism for muropeptide entry to the cytoplasm. Additional protein components that are inferred as part of the recycling system are cytoplasmic MurK kinase, catalyzing ATP-dependent 6′-phosphorylation of GlcNAc and MurNAc, and a GlmA glucosamine and glucosamine-containing muropeptide N-acetyl transferase.144,145 B. subtilis and E. coli encode dipeptide epimerases, catalyzing the formation of D-Ala and D-Glu dipeptides, and presumptively involved in peptidoglycan recycling.146 An orthologous gene corresponding to the AmpG permease is not found in Gram-positives, suggesting that lytic transglycosylase liberation of anhydromuropeptides is unimportant to recycling, and by inference is also unimportant as a Gram-positive virulence mechanism.138 In contrast, lytic transglycosylases have essential roles during the extensive peptidoglycan remodeling that occurs during Bacillus spore germination147,148,149,150 and in S. aureus septation.151,152 These studies do not suggest, however, the pathway used for muropeptide recycling by S. aureus.
S. aureus coordinates autolysin expression (for the purpose of muropeptide turnover) with expression of its other virulence mechanisms, as a probable event in its commensal to pathogen transition.153,154 While this observation alone does not implicate a requirement for peptidoglycan recycling, S. aureus uses other mechanisms to suppress a premature peptidoglycan-dependent immune response during infection.155,156,157 Peptidoglycan recycling, by minimizing extracellular peptidoglycan, would abet these mechanisms. Two further observations more directly implicate peptidoglycan recovery (if not recycling) by S. aureus. A particular nosocomial methicillin-resistant S. aureus (MRSA) strain found in Japan requires the presence of β-lactam antibiotics in order to secure vancomycin resistance.158 While the origin of the strain is easily rationalized as a consequence of the extensive use combined β-lactam-vancomycin chemotherapy for MRSA infection, the requirement that one cell-wall targeting antibiotic be present—this MRSA strain suppresses β-lactamase expression to this purpose159—to withstand a second cell-wall targeting antibiotic is seemingly paradoxical. Moreover, the role of the β-lactam is indirect: addition of a β-lactam antibiotic to a culture of this strain results in release of muropeptides into the medium. A particular structural class of these muropeptides replicates the ability of the β-lactam to induce vancomycin resistance.160 Vancomycin resistance is presumed to arise as a consequence of biosynthesis of a cell wall exhibiting enhanced, non-productive vancomycin affinity similar to what is observed in other vancomycin-resistant S. aureus strains.161 The signaling mechanism is conjectured to initially involve β-lactam-dependent inactivation of a PBP, resulting in the appearance in the medium of particular muropeptide structures reflecting the loss of PBP activity. The resulting muropeptides would then be transported into the S. aureus cell to function as a signal for enhanced cell-wall synthesis to result in vancomycin resistance.160
A second observation—one that also involves manifestation of resistance—likewise implicates the presence of specific mechanism(s) for peptidoglycan recovery by S. aureus. Two mechanisms are used by S. aureus to secure resistance against the β-lactam antibiotics: inducible expression of β-lactamase activity, and inducible expression of a β-lactam-resistant, PBP2a transpeptidase giving the MRSA phenotype. The two mechanisms share remarkable similarities, coincidence, and complexity.162 Of the two induction mechanisms, the β-lactamase pathway is the experimentally more tractable and is relevant to both, as its key components suffice for successful PBP2a induction.163 A central component of these induction pathways is the constitutive (but very low copy) expression of the sensor protein, BlaR1, having in addition to the exposed domain a transmembrane segment and a cytoplasmic domain. The key events of the mechanism by which the cell surface domain of BlaR1—acting as a sensor and signal transducer protein for β-lactam resistance) initiates β-lactamase expression are understood. Moreover, this emerging understanding now places BlaR1 (and the homologous sensor/signal transducer that may substitute for BlaR1 in PBP2a induction in MRSA, MecR1) as pivotal not simply to β-lactam resistance expression, but to cell-wall recycling.
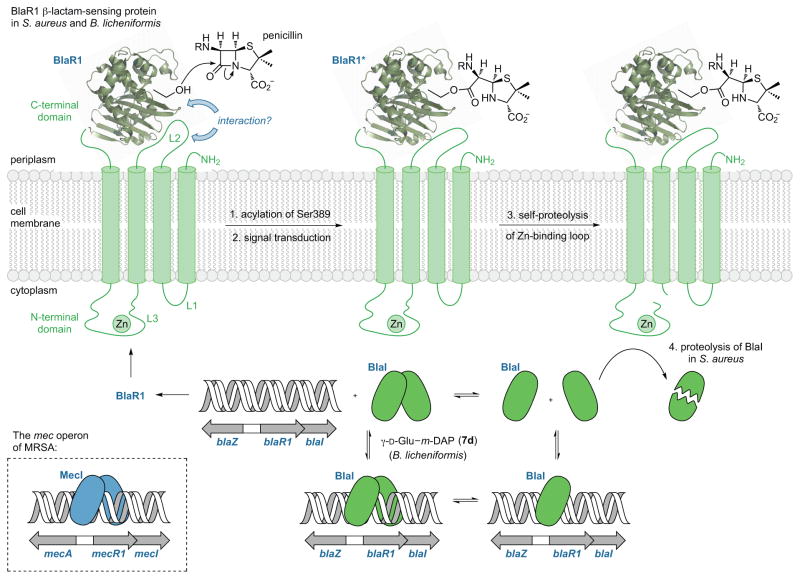
The initiating event in BlaR1 signal transduction is irreversible acylation of an active-site serine of its cell surfaced-exposed sensor domain.164,165 Activation of the serine uses an active-site lysine-derived carbamate functional group,164,166 in a mechanism that parallels directly the acylation mechanism used by the Class D “OXA” β-lactamases.167,168,169,170,171,172,173 The structural consequence of serine acylation by β-lactam antibiotics on the sensor domain has been studied extensively, with particular focus on the temporal relationship of acylation, lysine decarboxylation (which renders the acylation irreversible),174,175,176 signal transduction through the transmembrane domains to the L3 cytoplasmic loop,177 activation of the L3 loop as a metalloprotease, and recovery from the mobilization.178 In both Bacillus licheniformis179 and S. aureus,180 an immediate consequence of BlaR1 sensor domain acylation and transmembrane signal transduction is the autoproteolytic activation of the L3 loop of the BlaR1 protein as a zinc metalloprotease. The ultimate consequence of this proteolytic activation is release from the DNA of the BlaI (MecI) repressor protein so as to derepress β-lactamase (PBP2a) expression. Figure 9 summarizes these events, using for illustration a generic penicillin structure for the β-lactam involved in the initial serine acylation reaction.
Figure 9.
The bla system of S. aureus and B. licheniformis and the mec system of methicillin-resistant S. aureus.
Identification of a point of union between BlaI dissociation (Figure 9) and cell-wall recycling (Figure 8) in B. licheniformis was made recently by Amoroso et al.181 These authors show that a cell wall-derived dipeptide fragment, γ-D-Glu-m-DAP (7d), controls the release of the BlaI (MecI) repressor from DNA. Use of a peptidoglycan fragment to control resistance expression in this Gram-positive bacterium through repressor protein binding conceptually parallels what is seen for AmpC β-lactamase expression system of Gram-negative bacteria. While a pathway leading to γ-D-Glu-m-DAP formation is conjectured easily (Figure 8), the relationship within the pathway to the activation of the BlaR1 L3 loop as a protease, is not. The outstanding question is the identity of the substrate for this new proteolytic activity. Amoroso et al.181 suggest as this substrate the tripeptide L-Ala-γ-D-Glu-m-DAP, with the L3 loop acting as an L-Ala aminopeptidase to generate the γ-D-Glu-m-DAP dipeptide used to derepress β-lactamase expression through its binding to BlaI (Figure 8).
The relevance of such a mechanism to induction of the β-lactamase in S. aureus through its BlaR1/BlaI system, and PBP2a in MRSA through its MecR/MecI system, is uncertain. In contrast to B. licheniformis, proteolytic degradation of the released BlaI and MecI proteins sustains the resistance responses.182 The cleavage site is such that prior BlaI (MecI) dissociation from the DNA is necessary.183 Direct evidence supporting BlaI as a substrate for the activated L3 loop protease of S. aureus BlaR1 is in hand.184 As the BlaI proteins of S. aureus and B. licheniformis are highly homologous,183 a reasonable assumption for experimental design is that release of S. aureus BlaI from its DNA correlates to the composition of the cytoplasmic pool of muropeptides in S. aureus. The structure of the relevant muropeptide, and its relationship to cell-wall recycling, are not known.
This summary greatly oversimplifies what is known concerning the regulation of these Gram-positive resistance responses. While the possible pathways for cell-wall recycling in B. licheniformis and S. aureus clearly likely have key points of contrast with each other, and moreover with that of B. subtilis, the identification181 of γ-D-Glu-m-DAP as a pivotal entity controlling β-lactamase expression in B. licheniformis is a fundamental advance in our evolving appreciation of the possible relationships in Gram-positive bacteria between resistance and cell-wall recycling.
Conclusions
The structural uniqueness of the bacterial cell wall is complemented by the extraordinary small molecule structures perfected by Nature to recognize both this polymer and the enzymes involved in its maintenance. As mankind continues to exploit these small molecules as antibiotics, and as there is an increasing prevalence of life-threatening infections caused by bacteria with resistance mechanisms to these same antibiotics, new strategies for overcoming antibiotic resistance are required. For cell-wall-targeting antibiotics, the emerging connection between cell-wall recycling and the expression of antibacterial resistance enzymes represents a promising opportunity. Bacteria succumb to an antibiotic only after the myriad pathways that bacteria have to detect and respond to the presence of the antibiotic are compromised. Especially for the β-lactam antibiotics—now and for the foreseeable a mainstay of antibacterial chemotherapy—understanding the relationship between cell-wall recycling and resistance has great potential for the preservation of their value as antibiotics.
Acknowledgments
This work was supported by the NIH grant GM61629.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Bugg TDH, Braddick D, Dowson CG, Roper DI. Bacterial cell wall assembly: Still an attractive antibacterial target. Trends Biotechnol. 2011;29:167–173. doi: 10.1016/j.tibtech.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Koch AL. Bacterial wall as target for attack: Past, present, and future research. Clin Microbiol Rev. 2003;16:673–687. doi: 10.1128/CMR.16.4.673-687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park JT. Why does Escherichia coli recycle its cell wall peptides? Mol Microbiol. 1995;17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 4.Dworkin J, Shah IM. Exit from dormancy in microbial organisms. Nat Rev Microbiol. 2010;8:890–896. doi: 10.1038/nrmicro2453. [DOI] [PubMed] [Google Scholar]
- 5.Keep NH, Ward JM, Cohen-Gonsaud M, Henderson B. Wake Up! Peptidoglycan lysis and bacterial non-growth states. Trends Microbiol. 2006;14:271–276. doi: 10.1016/j.tim.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee M, Hesek D, Shah IM, Oliver AG, Dworkin J, Mobashery S. Synthetic peptidoglycan motifs for germination of bacterial spores. ChemBioChem. 2010;11:2525–2529. doi: 10.1002/cbic.201000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boudreau MA, Fisher JF, Mobashery S. Messenger functions of the bacterial cell wall-derived muropeptides. Biochemistry. 2012;51:2974–2990. doi: 10.1021/bi300174x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho S, Wang Q, Swaminathan CP, Hesek D, Lee M, Boons GJ, Mobashery S, Mariuzza RA. Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins. Proc Natl Acad Sci, USA. 2007;104:8761–8766. doi: 10.1073/pnas.0701453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mark BL, Vocadlo DJ, Oliver A. Providing β-lactams a helping hand: Targeting the AmpC β-lactamase induction pathway. Future Microbiol. 2011;6:1415–1427. doi: 10.2217/fmb.11.128. [DOI] [PubMed] [Google Scholar]
- 11.Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci USA. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovering AL, Safadi SS, Strynadka NCJ. Structural perspective of peptidoglycan biosynthesis and assembly. Ann Rev Biochem. 2012;81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 13.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 14.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 15.Barreteau H, Kovač A, Boniface A, Sova M, Gobec S, Blanot D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:168–207. doi: 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- 16.Bouhss A, Trunkfield AE, Bugg TDH, Mengin-Lecreulx D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008;32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz N. Streptococcus pyogenes YtgP (Spy_0390) Complements Escherichia coli strains depleted of the putative peptidoglycan flippase. MurJ Antimicrob Agents Chemother. 2009;53:3604–3605. doi: 10.1128/AAC.00578-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fay A, Dworkin J. Bacillus subtilis homologs of MviN (MurJ), the putative Escherichia coli lipid II flippase, are not essential for growth. J Bacteriol. 2009;191:6020–6028. doi: 10.1128/JB.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammadi T, van Dam V, Sijbrandi R, Vernet T, Zapun A, Bouhss A, Bruin M Diepeveen-de, Nguyen-Distèche M, de Kruijff B, Breukink E. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to β-lactam antibiotics: Compelling opportunism, compelling opportunity. Chem Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 22.Shi Q, Meroueh SO, Fisher JF, Mobashery S. A computational evaluation of the mechanism of penicillin-binding protein-catalyzed cross-linking of the bacterial cell wall. J Am Chem Soc. 2011;133:5274–5283. doi: 10.1021/ja1074739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moynihan PJ, Clarke AJ. O-Acetylated peptidoglycan: Controlling the activity of bacterial autolysins and lytic enzymes of innate immune systems. Int J Biochem Cell Biol. 2011;43:1655–1659. doi: 10.1016/j.biocel.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Hadi T, Pfeffer JM, Clarke AJ, Tanner ME. Water-soluble substrates of the peptidoglycan-modifying enzyme O-acetylpeptidoglycan esterase (Ape1) from Neisseria gonorrhoeae. J Org Chem. 2011;76:1118–1125. doi: 10.1021/jo102329c. [DOI] [PubMed] [Google Scholar]
- 25.Pfeffer JM, Clarke AJ. Identification of the first known inhibitors of O-acetylpeptidoglycan esterase: A potential new antibacterial target. ChemBioChem. 2012;13:722–731. doi: 10.1002/cbic.201100744. [DOI] [PubMed] [Google Scholar]
- 26.Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 2008;32:287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Shi Q, Meroueh SO, Vakulenko SB, Mobashery S. Catalytic mechanism of penicillin-binding protein 5 of E. coli. Biochemistry. 2007;46:10113–10121. doi: 10.1021/bi700777x. [DOI] [PubMed] [Google Scholar]
- 28.Shi Q, Meroueh SO, Fisher JF, Mobashery S. Investigation of the mechanism of the cell wall DD-carboxypeptidase reaction of penicillin-binding protein 5 of Escherichia coli by QM/MM calculations. J Am Chem Soc. 2008;130:9293–9303. doi: 10.1021/ja801727k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 30.Potluri LP, de Pedro MA, Young KD. Escherichia coli Low-molecular-weight penicillin-binding proteins help orient septal FtsZ, and their absence leads to asymmetric cell division and branching. Mol Microbiol. 2012;84:203–224. doi: 10.1111/j.1365-2958.2012.08023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Pedro MA, Quintela JC, Höltje JV, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. D-Amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1556. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 2011;30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cava F, Lam H, de Pedro MA, Waldor MK. Emerging knowledge of regulatory roles of D-amino acids in bacteria. Cell Mol Life Sci. 2011;68:817–831. doi: 10.1007/s00018-010-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horcajo P, de Pedro MA, Cava F. Peptidoglycan plasticity in bacteria: Stress-induced peptidoglycan editing by noncanonical D-amino acids. Microb Drug Resist. 2012;18:306–313. doi: 10.1089/mdr.2012.0009. [DOI] [PubMed] [Google Scholar]
- 36.Chaloupka J, Křečková P, Říhová L. Changes in the character of the cell wall in growth of Bacillus megaterium cultures. Folia Microbiol. 1962;7:269–274. doi: 10.1007/BF02928656. [DOI] [PubMed] [Google Scholar]
- 37.Chaloupka J, Křečková P, Říhová L. The mucopeptide turnover in the cell walls of growing cultures of Bacillus megaterium KM. Experientia. 1962;18:362–363. doi: 10.1007/BF02172250. [DOI] [PubMed] [Google Scholar]
- 38.Doyle RJ, Chaloupka J, Vinter V. Turnover of cell walls in microorganisms. Microbiol Rev. 1988;52:554–567. doi: 10.1128/mr.52.4.554-567.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park JT, Uehara T. How bacteria consume their own exoskeletons (Turnover and recycling of cell wall peptidoglycan) Microbiol Mol Biol Rev. 2008;72:211–227. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodell EW, Schwarz U. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J Bacteriol. 1985;162:391–397. doi: 10.1128/jb.162.1.391-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodell EW. Recycling of murein by Escherichia coli. J Bacteriol. 1985;163:305–310. doi: 10.1128/jb.163.1.305-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uehara T, Park JT. Growth of Escherichia coli: Significance of peptidoglycan degradation during elongation and septation. J Bacteriol. 2008;190:3914–3922. doi: 10.1128/JB.00207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park JT, Raychaudhuri D, Li H, Normark S, Mengin-Lecreulx D. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide L-alanyl-γ-D-glutamyl-meso-diaminopimelate. J Bacteriol. 1998;180:1215–1223. doi: 10.1128/jb.180.5.1215-1223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park JT. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: Indirect evidence for an alternative permease system and for a monolayered sacculus. J Bacteriol. 1993;175:7–11. doi: 10.1128/jb.175.1.7-11.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobs C, Huang L-j, Bartowsky E, Normark S, Park JT. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 1994;13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korfmann G, Sanders CC. ampG is essential for high-level expression of AmpC β-lactamase in Enterobacter cloacae. Antimicrob Agents Chemother. 1989;33:1946–1951. doi: 10.1128/aac.33.11.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindquist S, Weston-Hafer K, Schmidt H, Pul C, Korfmann G, Erickson J, Sanders C, Martin HH, Normark S. AmpG, a signal transducer in chromosomal β-lactamase induction. Mol Microbiol. 1993;9:703–715. doi: 10.1111/j.1365-2958.1993.tb01731.x. [DOI] [PubMed] [Google Scholar]
- 48.Honoré N, Nicolas MH, Cole ST. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 1986;5:3709–3714. doi: 10.1002/j.1460-2075.1986.tb04704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindquist S, Lindberg F, Normark S. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC β-lactamase gene. J Bacteriol. 1989;171:3746–3753. doi: 10.1128/jb.171.7.3746-3753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindberg F, Lindquist S, Normark S. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii β-lactamase. J Bacteriol. 1987;169:1923–1928. doi: 10.1128/jb.169.5.1923-1928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacobs C, Frère JM, Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in Gram-negative bacteria. Cell. 1997;88:823–832. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 52.Dietz H, Pfeifle D, Wiedmann B. The signal molecule for β-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob Agents Chemother. 1997;41:2113–2120. doi: 10.1128/aac.41.10.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Heijenoort J. Peptidoglycan hydrolases of. Escherichia coli. 2011;75:636–663. doi: 10.1128/MMBR.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheurwater E, Reid CW, Clarke AJ. Lytic transglycosylases: Bacterial space-making autolysins. Int J Biochem Cell Biol. 2008;40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 55.Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: Viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol. 1999;181:3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heidrich C, Ursinus A, Berger J, Schwarz H, Höltje JV. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol. 2002;184:6093–6099. doi: 10.1128/JB.184.22.6093-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Höltje JV. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch Microbiol. 1995;164:243–254. doi: 10.1007/BF02529958. [DOI] [PubMed] [Google Scholar]
- 58.Vollmer W, von Rechenberg M, Höltje JV. Demonstration of molecular interactions Between the murein polymerase PBP1b, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J Biol Chem. 1999;274:6726–6734. doi: 10.1074/jbc.274.10.6726. [DOI] [PubMed] [Google Scholar]
- 59.Romeis T, Höltje J-V. Specific interaction of penicillin-binding proteins 3 and 7/8 with soluble lytic transglycosylase in Escherichia coli. J Biol Chem. 1994;269:21603–21607. [PubMed] [Google Scholar]
- 60.Legaree BA, Clarke AJ. Interaction of penicillin-binding protein 2 with soluble lytic transglycosylase B1 in Pseudomonas aeruginosa. J Bacteriol. 2008;190:6922–6927. doi: 10.1128/JB.00934-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blackburn NT, Clarke AJ. Identification of four families of peptidoglycan lytic transglycosylases. J Mol Evol. 2001;52:78–84. doi: 10.1007/s002390010136. [DOI] [PubMed] [Google Scholar]
- 62.Blackburn NT, Clarke AJ. Characterization of soluble and membrane-bound family 3 lytic transglycosylases from Pseudomonas aeruginosa. Biochemistry. 2002;41:1001–1013. doi: 10.1021/bi011833k. [DOI] [PubMed] [Google Scholar]
- 63.Chan YA, Hackett KT, Dillard JP. The lytic transglycosylases of Neisseria gonorrhoeae. Microb Drug Resist. 2012;18:271–279. doi: 10.1089/mdr.2012.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scheurwater EM, Clarke AJ. The C-terminal domain of Escherichia coli YfhD functions as a lytic transglycosylase. J Biol Chem. 2008;283:8363–8373. doi: 10.1074/jbc.M710135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Templin MF, Edwards DH, Höltje JV. A murein hydrolase is the specific target of bulgecin in Escherichia coli. J Biol Chem. 1992;267:20039–20043. [PubMed] [Google Scholar]
- 66.Kraft AR, Prabhu J, Ursinus A, Höltje JV. Interference with murein turnover has no effect on growth but reduces β-lactamase induction in Escherichia coli. J Bacteriol. 1999;181:7192–7198. doi: 10.1128/jb.181.23.7192-7198.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kraft AR, Templin MF, Höltje JV. Membrane-bound lytic endotransglycosylase in Escherichia coli. J Bacteriol. 1998;180:3441–3447. doi: 10.1128/jb.180.13.3441-3447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Artola-Recolons C, Carrasco-López C, Llarrull LI, Kumarasiri M, Lastochkin E, Martínez de Ilarduya I, Meindl K, Usón I, Mobashery S, Hermoso JA. High-resolution crystal structure of MltE, an outer membrane-anchored endolytic peptidoglycan lytic transglycosylase from Escherichia coli. Biochemistry. 2011;50:2384–2386. doi: 10.1021/bi200085y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romeis T, Vollmer W, Höltje JV. Characterization of three different lytic transglycosylases in Escherichia coli. FEMS Microbiol Lett. 1993;111:141–146. doi: 10.1111/j.1574-6968.1993.tb06376.x. [DOI] [PubMed] [Google Scholar]
- 70.Suvorov M, Lee M, Hesek D, Boggess B, Mobashery S. Lytic transglycosylase MltB of Escherichia coli and its role in recycling of peptidoglycan strands of bacterial cell wall. J Am Chem Soc. 2008;130:11878–11879. doi: 10.1021/ja805482b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Asselt EJ, Thunnissen AMWH, Dijkstra BW. High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment. J Mol Biol. 1999;291:877–898. doi: 10.1006/jmbi.1999.3013. [DOI] [PubMed] [Google Scholar]
- 72.van Straaten KE, Dijkstra BW, Vollmer W, Thunnissen AMWH. Crystal structure of MltA from Escherichia coli reveals a unique lytic transglycosylase fold. J Mol Biol. 2005;352:1068–1080. doi: 10.1016/j.jmb.2005.07.067. [DOI] [PubMed] [Google Scholar]
- 73.Powell AJ, Liu ZJ, Nicholas RA, Davies C. Crystal structures of the lytic transglycosylase MltA from N. gonorrhoeae and E. coli: Insights into interdomain movements and substrate binding. J Mol Biol. 2006;359:122–136. doi: 10.1016/j.jmb.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 74.van Straaten KE, Barends TRM, Dijkstra BW, Thunnissen AMWH. Structure of Escherichia coli lytic transglycosylase MltA with bound chitohexaose. J Mol Biol. 2007;282:21197–21205. doi: 10.1074/jbc.M701818200. [DOI] [PubMed] [Google Scholar]
- 75.van Asselt EJ, Perrakis A, Kalk KH, Lamzin VS, Dijkstra BW. Accelerated X-ray structure elucidation of a 36 kDa muramidase/transglycosylase using wARP. Acta Cryst. 1998;D54:58–73. doi: 10.1107/s0907444997010330. [DOI] [PubMed] [Google Scholar]
- 76.van Asselt EJ, Dijkstra AJ, Kalk KH, Takacs B, Keck W, Dijkstra BW. Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-Hand. Structure. 1999;7:1167–1180. doi: 10.1016/s0969-2126(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 77.van Asselt EJ, Dijkstra BW. Binding of calcium in the EF-Hand of Escherichia coli lytic transglycosylase Slt35 is important for stability. FEBS Lett. 1999;458:429–435. doi: 10.1016/s0014-5793(99)01198-9. [DOI] [PubMed] [Google Scholar]
- 78.van Asselt EJ, Kalk KH, Dijkstra BW. Crystallographic studies of the interactions of Escherichia coli lytic transglycosylase Slt35 with peptidoglycan. Biochemistry. 2000;39:1924–1934. doi: 10.1021/bi992161p. [DOI] [PubMed] [Google Scholar]
- 79.Nikolaidis I, Izore T, Job V, Thielens N, Breukink E, Dessen A. Calcium-dependent complex formation between PBP2 and lytic transglycosylase SltB1 of Pseudomonas aeruginosa. Microb Drug Resist. 2012;18:298–305. doi: 10.1089/mdr.2012.0006. [DOI] [PubMed] [Google Scholar]
- 80.Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- 81.Reid CW, Blackburn NT, Legaree BA, Auzanneau FI, Clarke AJ. Inhibition of membrane-bound lytic transglycosylase B by NAG-thiazoline. FEBS Lett. 2004;574:73–79. doi: 10.1016/j.febslet.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 82.Reid CW. PhD Thesis. University of Guelph; Guelph, Ontario, Canada: 2005. Bacterial lytic transglycosylases: Biochemical characterization and rational design of inhibitors. [Google Scholar]
- 83.Yamaguchi T, Blázquez B, Hesek D, Lee M, Llarrull LI, Boggess B, Oliver AG, Fisher JF, Mobashery S. Inhibitors for bacterial cell-wall recycling. ACS Med Chem Lett. 2012;3:238–242. doi: 10.1021/ml2002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bonis M, Williams A, Guadagnini S, Werts C, Boneca IG. The effect of bulgecin A on peptidoglycan metabolism and physiology of Helicobacter pylori. Microb Drug Resist. 2012;18:230–239. doi: 10.1089/mdr.2011.0231. [DOI] [PubMed] [Google Scholar]
- 85.Thunnissen AMWH, Rozeboom HJ, Kalk KH, Dijkstra BW. Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A. Implications for the enzymatic mechanism. Biochemistry. 1995;34:12729–12737. doi: 10.1021/bi00039a032. [DOI] [PubMed] [Google Scholar]
- 86.Cloud-Hansen KA, Hackett KT, Garcia DL, Dillard JP. Neisseria gonorrhoeae uses two lytic transglycosylases to produce cytotoxic peptidoglycan monomers. J Bacteriol. 2008;190:5989–5994. doi: 10.1128/JB.00506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Garcia DL, Dillard JP. Mutations in ampG or ampD affect peptidoglycan fragment release from Niesseria gonorrhoeae. J Bacteriol. 2008;190:3799–3807. doi: 10.1128/JB.01194-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adin DM, Engle JT, Goldman WE, McFall-Ngai MJ, Stabb EV. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol. 2008;191:2012–2022. doi: 10.1128/JB.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nigro G, Fazio LL, Martino MC, Rossi G, Tattoli I, Liparoti V, De Castro C, Molinaro A, Philpott DJ, Bernardini ML. Muramylpeptide shedding modulates cell sensing of Shigella flexneri. Cell Microbiol. 2008;10:682–695. doi: 10.1111/j.1462-5822.2007.01075.x. [DOI] [PubMed] [Google Scholar]
- 90.Zhang Y, Bao Q, Gagnon LA, Huletsky A, Oliver A, Jin S, Langaee T. ampG gene of Pseudomonas aeruginosa and its role in β-lactamase expression. Antimicrob Agents Chemother. 2010;54:4772–4779. doi: 10.1128/AAC.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kong KF, Aguila A, Schneper L, Mathee K. Pseudomonas aeruginosa β-lactamase induction requires two permeases, AmpG and AmpP. BMC Microbiol. 2010;10:328. doi: 10.1186/1471-2180-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chahboune A, Decaffmeyer M, Brasseur R, Joris B. Membrane topology of the Escherichia coli AmpG permease required for recycling of cell wall anhydromuropeptides and AmpC β-lactamase induction. Antimicrob Agents Chemother. 2005;49:1145–1149. doi: 10.1128/AAC.49.3.1145-1149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng Q, Park JT. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J Bacteriol. 2002;184:6434–6436. doi: 10.1128/JB.184.23.6434-6436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zamorano L, Reeve TM, Juan C, Moyá B, Cabot G, Vocadlo DJ, Mark BL, Oliver A. AmpG inactivation restores susceptibility of pan-β-lactam-resistant Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother. 2011;55:1990–1996. doi: 10.1128/AAC.01688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yem DW, Wu HC. Purification and properties of β-N-acetylglucosaminidase from Escherichia coli. J Bacteriol. 1976;125:324–331. doi: 10.1128/jb.125.1.324-331.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yem DW, Wu HC. Isolation of Escherichia coli K-12 mutants with altered levels of β-N-acetylglucosaminidase. J Bacteriol. 1976;125:372–373. doi: 10.1128/jb.125.1.372-373.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jacobs C, Joris B, Jamin M, Klarsov K, Beeumen JV, Mengin-Lecreulx D, van Heijenoort J, Park JT, Normark S, Frère JM. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-L-alanine amidase. Mol Microbiol. 1995;15:553–559. doi: 10.1111/j.1365-2958.1995.tb02268.x. [DOI] [PubMed] [Google Scholar]
- 98.Cheng Q, Li H, Merdek K, Park JT. Molecular characterization of the β-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J Bacteriol. 2000;182:4836–4840. doi: 10.1128/jb.182.17.4836-4840.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vötsch W, Templin MF. Characterization of a β-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and β-lactamase induction. J Biol Chem. 2000;275:39032–39038. doi: 10.1074/jbc.M004797200. [DOI] [PubMed] [Google Scholar]
- 100.Stubbs KA, Balcewich M, Mark BL, Vocadlo DJ. Small molecule inhibitors of a glycoside hydrolase attenuate inducible AmpC-mediated β-lactam resistance. J Biol Chem. 2007;282:21382–21391. doi: 10.1074/jbc.M700084200. [DOI] [PubMed] [Google Scholar]
- 101.Balcewich MD, Stubbs KA, He Y, James TW, Davies GJ, Vocadlo DJ, Mark BL. Insight into a strategy for attenuating AmpC-mediated β-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ. Protein Sci. 2009;18:1541–1551. doi: 10.1002/pro.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scaffidi A, Stubbs KA, Vocadlo DJ, Stick RV. The synthesis and biological evaluation of some carbocyclic analogues of PUGNAc. Carbohydr Res. 2008;343:2744–2753. doi: 10.1016/j.carres.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 103.Scaffidi A, Stubbs KA, Dennis RJ, Taylor EJ, Davies GJ, Vocadlo DJ, Stick RV. A 1-acetamido derivative of 6-epi-valienamine: An inhibitor of a diverse group of β-N-acetylglucosaminidases. Org Biomol Chem. 2008;5:3013–3019. doi: 10.1039/b709681j. [DOI] [PubMed] [Google Scholar]
- 104.Stubbs KA, Scaffidi A, Debowski AW, Mark BL, Stick RV, Vocadlo DJ. Synthesis and use of mechanism-based protein-profiling probes for retaining β-D-glucosaminidases facilitate identification of Pseudomonas aeruginosa NagZ. J Am Chem Soc. 2008;130:327–335. doi: 10.1021/ja0763605. [DOI] [PubMed] [Google Scholar]
- 105.Asgarali A, Stubbs KA, Oliver A, Vocadlo DJ, Mark BL. Inactivation of the glycoside hydrolase NagZ attenuates antipseudomonal β-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53:2274–2282. doi: 10.1128/AAC.01617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zamorano L, Reeve TM, Deng L, Juan C, Moyá B, Cabot G, Vocadlo DJ, Mark BL, Oliver A. NagZ inactivation prevents and reverts β-lactam resistance, driven by AmpD and PBP4 mutations, in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010;54:3557–3563. doi: 10.1128/AAC.00385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hesek D, Lee M, Zhang W, Noll BC, Mobashery S. Total synthesis of N-acetylglucosamine-1,6-anhydro-N-acetylmuramylpentapeptide and evaluation of its turnover by AmpD from Escherichia coli. J Am Chem Soc. 2009;131:5187–5193. doi: 10.1021/ja808498m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee M, Zhang W, Hesek D, Noll BC, Boggess B, Mobashery S. Bacterial AmpD at the crossroads of peptidoglycan recycling and manifestation of antibiotic resistance. J Am Chem Soc. 131:8742–8743. doi: 10.1021/ja9025566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liepinsh E, Généreaux C, Dehareng D, Joris B, Otting G. NMR structure of Citrobacter freundii AmpD: comparison with bacteriophage T7 lysozyme and homology with PGRP domains. J Mol Biol. 2003;327:833, 842. doi: 10.1016/s0022-2836(03)00185-2. [DOI] [PubMed] [Google Scholar]
- 110.Carrasco-López C, Rojas-Altuve A, Zhang W, Hesek D, Lee M, Barbe S, André I, Ferrer P, Silva-Martin N, Castro GR, Martínez-Ripoll M, Mobashery S, Hermoso JA. Crystal structures of bacterial peptidoglycan amidase AmpD and an unprecedented activation mechanism. J Biol Chem. 2011;286:31714–31722. doi: 10.1074/jbc.M111.264366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Heiderich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, de Pedro MA, Höltje J-V. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-Induced autolysis of Escherichia coli. Mol Microbiol. 2001;41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- 112.Priyadarshini R, de Pedro MA, Young KD. Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J Bacteriol. 2007;189:5334–5347. doi: 10.1128/JB.00415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garcia DL, Dillard JP. AmiC functions as an N-acetylmuramyl-L-alanine amidase necessary for cell separation and can promote autolysis in Neisseria gonorrhoeae. J Bacteriol. 2007;188:7211–7221. doi: 10.1128/JB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Uehara T, Park JT. An anhydro-N-acetylmuramyl-L-alanine amidase with broad specificity tethered to the outer membrane of Escherichia coli. J Bacteriol. 2007;189:5634–5641. doi: 10.1128/JB.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kerff F, Petrella S, Mercier F, Sauvage E, Herman R, Pennartz A, Zervosen A, Luxen A, Frère JM, Joris B, Charlier P. Specific structural features of the N-acetylmuramoyl-L-alanine amidase AmiD from Escherichia coli and mechanistic implications for enzymes of this family. J Mol Biol. 2010;397:249–259. doi: 10.1016/j.jmb.2009.12.038. [DOI] [PubMed] [Google Scholar]
- 116.Juan C, Maciá MD, Gutiérrez O, Vidal C, Pérez JL, Oliver A. Molecular mechanisms of β-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother. 2005;49:4733–4738. doi: 10.1128/AAC.49.11.4733-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Juan C, Moyá B, Pérez JL, Oliver A. Stepwise Upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level β-lactam resistance involves three AmpD homologues. Antimicrob Agents Chemother. 2006;50:1780–1787. doi: 10.1128/AAC.50.5.1780-1787.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.ampDh2 gene product [Pseudomonas aeruginosa PAO1] [accessed August 23, 2012]; http://www.ncbi.nlm.nih.gov/protein/NP_254172.1.
- 119.ampDh3 gene product [Pseudomonas aeruginosa PAO1] [accessed August 23, 2012]; http://www.ncbi.nlm.nih.gov/protein/NP_249498.1.
- 120.Moyá B, Juan C, Albertí S, Pérez JL, Oliver A. Benefit of having Multiple ampD genes for acquiring β-lactam resistance without losing fitness and virulence in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52:3694–3700. doi: 10.1128/AAC.00172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schmidtke AJ, Hanson ND. Role of ampD homologs in overproduction of AmpC in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52:3922–3927. doi: 10.1128/AAC.00341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Uehara T, Suefuji K, Valbuena N, Meehan B, Donegan M, Park JT. Recycling of the anhydro-N-acetylmuramic acid derived from cell wall murein involves a two-Step conversion to N-acetylglucosamine-phosphate. J Bacteriol. 2005;187:3643–3649. doi: 10.1128/JB.187.11.3643-3649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bacik JP, Whitworth GE, Stubbs KA, Yadav AK, Martin DR, Bailey-Elkin BA, Vocadlo DJ, Mark BL. Molecular basis of 1,6-anhydro bond cleavage and phosphoryl transfer by Pseudomonas aeruginosa 1,6-anhydro-N-acetylmuramic acid kinase. J Biol Chem. 2011;286:12283–12291. doi: 10.1074/jbc.M110.198317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jaeger T, Arsic M, Mayer C. Scission of the lactyl ether bond of N-acetylmuramic acid by Escherichia coli “etherase”. J Biol Chem. 2005;280:30100–30106. doi: 10.1074/jbc.M502208200. [DOI] [PubMed] [Google Scholar]
- 125.Dahl U, Jaeger T, Nguyen BT, Sattler JM, Mayer C. Identification of a phosphotransferase system of Escherichia coli required for growth on N-acetylmuramic acid. J Bacteriol. 2004;186:2385–2392. doi: 10.1128/JB.186.8.2385-2392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Uehara T, Suefuji K, Jaeger T, Mayer C, Park JT. MurQ etherase is required by Escherichia coli in order to metabolize anhydro-N-acetylmuramic acid obtained either from the environment or from its own cell wall. J Bacteriol. 2006;188:1660–1662. doi: 10.1128/JB.188.4.1660-1662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hadi T, Dahl U, Mayer C, Tanner ME. Mechanistic studies on N-acetylmuramic acid 6-phosphate hydrolase (MurQ): An etherase involved in peptidoglycan recycling. Biochemistry. 2008;47:11547–11558. doi: 10.1021/bi8014532. [DOI] [PubMed] [Google Scholar]
- 128.Templin MF, Ursinus A, Höltje JV. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 1999;18:4108–4117. doi: 10.1093/emboj/18.15.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mengin-Lecreulx D, van Heijenoort J, Park JT. Identification of the mpl gene encoding UDP-N-acetylmuramate:L-alanyl-γ-D-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J Bacteriol. 1996;178:5347–5352. doi: 10.1128/jb.178.18.5347-5352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hervé M, Boniface A, Gobec S, Blanot D, Mengin-Lecreulx D. Biochemical characterization and physiological properties of Escherichia coli UDP-N-acetylmuramate:L-alanyl-γ-D-glutamyl-meso-diaminopimelate ligase. J Bacteriol. 2007;189:3987–3995. doi: 10.1128/JB.00087-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Das D, Hervé M, Feuerhelm J, Farr CL, Chiu HJ, Elsliger MA, Knuth MW, Klock HE, Miller MD, Godzik A, Lesley SA, Deacon AM, Mengin-Lecreulx D, Wilson IA. Structure and function of the first full-length murein peptide ligase (Mpl) cell wall recycling protein. PLoS One. 2011;6:e17624. doi: 10.1371/journal.pone.0017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moyá B, Dötsch A, Juan C, Blázquez J, Zamorano L, Haussler S, Oliver A. β-Lactam resistance response triggered by inactivation of a nonessential Penicillin-Binding Protein. PLoS Pathog. 2009;5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zamorano L, Moyá B, Juan C, Oliver A. Differential β-lactam resistance response driven by ampD or dacB (PBP4) inactivation in genetically diverse Pseudomonas aeruginosa strains. J Antimicrob Chemother. 2010;65:1540–1542. doi: 10.1093/jac/dkq142. [DOI] [PubMed] [Google Scholar]
- 134.Uehara T, Park JT. Role of the murein precursor UDP-N-acetylmuramyl-L-Ala-γ-D-Glu-meso-diaminopimelic acid-D-Ala-D-Ala in repression of β-lactamase induction in cell division mutants. J Bacteriol. 2002;184:4233–4239. doi: 10.1128/JB.184.15.4233-4239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Balasubramanian D, Schneper L, Merighi M, Smith R, Narasimhan G, Lory S, Mathee K. The regulatory repertoire of Pseudomonas aeruginosa AmpC β-lactamase regulator AmpR includes virulence genes. PLoS One. 2012;7:e34067. doi: 10.1371/journal.pone.0034067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Balcewich MD, Reeve TM, Orlikow EA, Donald LJ, Vocadlo DJ, Mark BL. Crystal structure of the AmpR effector binding domain provides insight into the molecular regulation of inducible AmpC β-lactamase. J Mol Biol. 2010;400:998–1010. doi: 10.1016/j.jmb.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 137.Litzinger S, Mayer C. The Murein Sacculus. In: König H, Claus H, Varma A, editors. Prokaryotic Cell Wall Compounds—Structure and Biochemistry. Springer; Berlin: 2010. pp. 3–54. [Google Scholar]
- 138.Reith J, Mayer C. Peptidoglycan turnover and recycling in Gram-positive bacteria. Appl Microbiol Biotechnol. 2011;92:1–11. doi: 10.1007/s00253-011-3486-x. [DOI] [PubMed] [Google Scholar]
- 139.Höltje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Delaune A, Poupel O, Mallet A, Coic YM, Msadek T, Dubrac S. Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLoS One. 2011;6:e17054. doi: 10.1371/journal.pone.0017054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mauck J, Chan L, Glaser L. Turnover of the cell wall of Gram-positive bacteria. J Biol Chem. 1971;246:1820–1827. [PubMed] [Google Scholar]
- 142.Litzinger S, Duckworth A, Nitzsche K, Risinger C, Wittmann V, Mayer C. Muropeptide rescue in Bacillus subtilis involves sequential hydrolysis by β-N-acetylglucosaminidase and N-acetylmuramyl-L-alanine amidase. J Bacteriol. 2010;192:3132–3143. doi: 10.1128/JB.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Litzinger S, Fischer S, Polzer P, Diederichs K, Welte W, Mayer C. Structural and kinetic analysis of Bacillus subtilis N-acetylglucosaminidase reveals a unique Asp–His dyad mechanism. J Biol Chem. 2010;285:35675–35684. doi: 10.1074/jbc.M110.131037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Reith J, Berking A, Mayer C. Characterization of an N-acetylmuramic acid/N-acetylglucosamine kinase of Clostridium acetobutylicum. J Bacteriol. 2011;193:5386–5392. doi: 10.1128/JB.05514-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Reith J, Mayer C. Characterization of a glucosamine/glucosaminide N-acetyltransferase of Clostridium acetobutylicum. J Bacteriol. 2011;193:5393–5399. doi: 10.1128/JB.05519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lukk T, Sakai A, Kalyanaraman C, Brown SD, Imker HJ, Song L, Fedorov AA, Fedorov EV, Toro R, Hillerich B, Seidel R, Patskovsky Y, Vetting MW, Nair SK, Babbitt PC, Almo SC, Gerlt JA, Jacobson MP. Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily. Proc Natl Acad Sci USA. 2012;109:4122–4127. doi: 10.1073/pnas.1112081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Heffron JD, Orsburn B, Popham DL. Roles of germination-specific lytic enzymes CwlJ and SleB in Bacillus anthracis. J Bacteriol. 2009;191:2237–2247. doi: 10.1128/JB.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Heffron JD, Sherry N, Popham DL. In vitro studies of peptidoglycan binding and hydrolysis by the Bacillus anthracis germination-specific lytic enzyme SleB. J Bacteriol. 2011;193:125–131. doi: 10.1128/JB.00869-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li Y, Jin K, Setlow B, Setlow P, Hao B. Crystal structure of the catalytic domain of the Bacillus cereus SleB protein, important in cortex peptidoglycan degradation during spore germination. J Bacteriol. 2012;194:4537–4545. doi: 10.1128/JB.00877-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jing X, Robinson HR, Heffron J, Popham DL, Schubot FD. The catalytic domain of the germination-specific lytic transglycosylase SleB from Bacillus anthracis displays a unique active site topology. Proteins. 2012 doi: 10.1002/prot.24140. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sakata N, Terakubo S, Mukai T. Subcellular location of the soluble lytic transglycosylase homologue in Staphylococcus aureus. Curr Microbiol. 2005;50:47–51. doi: 10.1007/s00284-004-4381-9. [DOI] [PubMed] [Google Scholar]
- 152.Stapleton MR, Horsburgh MJ, Hayhurst EJ, Wright L, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J Bacteriol. 2007;189:7316–7325. doi: 10.1128/JB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dubrac S, I, Boneca G, Poupel O, Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol. 2007;189:8257–8269. doi: 10.1128/JB.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Delauné A, Dubrac S, Blanchet C, Poupel O, Mäder U, Hiron A, Leduc A, Fitting C, Nicolas P, Cavaillon JM, Adib-Conquy M, Msadek T. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect Immun. 2012 doi: 10.1128/IAI.00195-12. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bera A, Biswas R, Herbert S, Kulauzovic E, Weidenmaier C, Peschel A, Gotz F. Influence of wall teichoic acid on lysozyme resistance in Staphylococcus aureus. J Bacteriol. 2007;189:280–283. doi: 10.1128/JB.01221-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shimada T, Park BG, Wolf AJ, Brikos C, Goodridge HS, Becker CA, Reyes CN, Miao EA, Aderem A, Götz F, Liu GY, Underhill DM. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1β secretion. Cell Host Microbe. 2010;7:38–49. doi: 10.1016/j.chom.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Figueiredo TA, Sobral RG, Ludovice AM, Almeida JM, Bui NK, Vollmer W, Lencastre H, Tomasz A. Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathog. 2012;8:e1002508. doi: 10.1371/journal.ppat.1002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yanagisawa C, Hanaki H, Matsui H, Ikeda S, Nakae T, Sunakawa K. Rapid depletion of free vancomycin in medium in the presence of β-lactam antibiotics and growth restoration in Staphylococcus aureus strains with β-lactam-induced vancomycin resistance. Antimicrob Agents Chemother. 2009;53:63–68. doi: 10.1128/AAC.00762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hirao Y, Ikeda-Dantsuji Y, Matsui H, Yoshida M, Hori S, Sunakawa K, Nakae T, Hanaki H. Low level β-lactamase production in methicillin-resistant Staphylococcus aureus strains with β-lactam antibiotics-induced vancomycin resistance. BMC Microbiol. 2012;12:69. doi: 10.1186/1471-2180-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ikeda S, Hanaki H, Yanagisawa C, Ikeda-Dantsuji Y, Matsui H, Iwatsuki M, Shiomi K, Nakae T, Sunakawa K, Omura S. Identification of the active component that induces vancomycin resistance in MRSA. J Antibiot. 2010;63:533–538. doi: 10.1038/ja.2010.75. [DOI] [PubMed] [Google Scholar]
- 161.Pereira PM, Filipe SR, Tomasz A, Pinho MG. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:3627–3633. doi: 10.1128/AAC.00431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Llarrull LI, Fisher JF, Mobashery S. Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new β-lactams that meet the challenge. Antimicrob Agents Chemother. 2009;53:4051–4063. doi: 10.1128/AAC.00084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Oliveira DC, de Lencastre H. Methicillin-resistance in Staphylococcus aureus is not affected by the overexpression in trans of the mecA gene repressor: a surprising observation. PLoS One. 2011;6:e23287. doi: 10.1371/journal.pone.0023287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Birck C, Cha JY, Cross J, Schulze-Briese C, Meroueh SO, Schlegel HB, Mobashery S, Samama JP. X-ray crystal structure of the acylated β-lactam sensor domain of BlaR1 from Staphylococcus aureus and the mechanism of receptor activation for signal transduction. J Am Chem Soc. 2004;126:13945–13947. doi: 10.1021/ja044742u. [DOI] [PubMed] [Google Scholar]
- 165.Wilke MS, Hills TL, Zhang HZ, Chambers HF, Strynadka NC. Crystal structures of the apo and penicillin-acylated forms of the BlaR1 β-lactam sensor of Staphylococcus aureus. J Biol Chem. 2004;279:47278–47287. doi: 10.1074/jbc.M407054200. [DOI] [PubMed] [Google Scholar]
- 166.Cha J, Vakulenko SB, Mobashery S. Characterization of the β-lactam antibiotic sensor domain of the MecR1 signal sensor/transducer protein from methicillin-resistant Staphylococcus aureus. Biochemistry. 2007;46:7822–7831. doi: 10.1021/bi7005459. [DOI] [PubMed] [Google Scholar]
- 167.Golemi D, Maveyraud L, Vakulenko S, Samama JP, Mobashery S. Critical involvement of a carbamylated lysine in catalytic function of class D β-lactamases. Proc Natl Acad Sci USA. 2001;98:14280–14285. doi: 10.1073/pnas.241442898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Schneider KD, Bethel CR, Distler AM, Hujer AM, Bonomo RA, Leonard DA. Mutation of the active site carboxy-lysine (K70) of OXA-1 β-lactamase results in a deacylation-deficient enzyme. Biochemistry. 2009;48:6136–6145. doi: 10.1021/bi900448u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vercheval L, Bauvois C, di Paolo A, Borel F, Ferrer JL, Sauvage E, Matagne A, Frere JM, Charlier P, Galleni M, Kerff F. Three factors that modulate the activity of class D β-lactamases and interfere with the post-translational carboxylation of Lys70. Biochem J. 2010;432:495–504. doi: 10.1042/BJ20101122. [DOI] [PubMed] [Google Scholar]
- 170.Docquier JD, Benvenuti M, Calderone V, Giuliani F, Kapetis D, De Luca F, Rossolini GM, Mangani S. Crystal structure of the narrow-spectrum OXA-46 class D β-lactamase: Relationship between active-site lysine carbamylation and inhibition by polycarboxylates. Antimicrob Agents Chemother. 2010;54:2167–2174. doi: 10.1128/AAC.01517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Che T, Bonomo RA, Shanmugam S, Bethel CR, Pusztai-Carey M, Buynak JD, Carey PR. Carboxylation and decarboxylation of active site Lys 84 controls the activity of OXA-24 β-lactamase of Acinetobacter baumannii: Raman crystallographic and solution evidence. J Am Chem Soc. 2012;134:11206–11215. doi: 10.1021/ja303168n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Verma V, Testero SA, Amini K, Wei W, Liu J, Balachandran N, Monoharan T, Stynes S, Kotra LP, Golemi-Kotra D. Hydrolytic mechanism of OXA-58 enzyme, a carbapenem-hydrolyzing class D β-lactamase from Acinetobacter baumannii. J Biol Chem. 2011;286:37292–37303. doi: 10.1074/jbc.M111.280115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kumarasiri M, Llarrull LI, Borbulevych O, Fishovitz J, Lastochkin E, Baker BM, Mobashery S. An amino-acid position at the crossroads of evolution of protein function: antibiotic-sensor domain of the BlaR1 protein from Staphylococcus aureus vs. class D β-lactamases. J Biol Chem. 2012;287:8232–8241. doi: 10.1074/jbc.M111.333179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Thumanu K, Cha J, Fisher JF, Perrins R, Mobashery S, Wharton C. Discrete steps in sensing of β-lactam antibiotics by the BlaR1 protein of the methicillin-resistant Staphylococcus aureus bacterium. Proc Natl Acad Sci USA. 2006;103:10630–10635. doi: 10.1073/pnas.0601971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cha JY, Mobashery S. Lysine Nζ-decarboxylation in the BlaR1 protein from Staphylococcus aureus at the root of its function as an antibiotic sensor. J Am Chem Soc. 2007;129:3834–3835. doi: 10.1021/ja070472e. [DOI] [PubMed] [Google Scholar]
- 176.Borbulevych O, Kumarasiri M, Wilson B, Llarrull LI, Lee M, Hesek D, Shi Q, Peng J, Baker BM, Mobashery S. Lysine Nζ-decarboxylation switch and activation of the β-lactam sensor domain of BlaR1 protein of methicillin-resistant Staphylococcus aureus. J Biol Chem. 2011;286:31466–31472. doi: 10.1074/jbc.M111.252189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang HZ, Hackbarth CJ, Chansky KM, Chambers HF. A proteolytic transmembrane signaling pathway and resistance to β-lactams in staphylococci. Science. 2001;291:1962–1965. doi: 10.1126/science.1055144. [DOI] [PubMed] [Google Scholar]
- 178.Llarrull LI, Toth M, Champion MM, Mobashery S. Activation of BlaR1 protein of methicillin-resistant Staphylococcus aureus, its proteolytic processing, and recovery from induction of resistance. J Biol Chem. 2011;286:38148–38158. doi: 10.1074/jbc.M111.288985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Berzigotti S, Benlafya K, Sepulchre J, Amoroso A, Joris B. Bacillus licheniformis BlaR1 L3 loop is a zinc metalloprotease activated by self-proteolysis. PLoS One. 2012;7:e36400. doi: 10.1371/journal.pone.0036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Llarrull LI, Mobashery S. Dissection of events in the resistance to β-lactam antibiotics mediated by the protein BlaR1 from Staphylococcus aureus. Biochemistry. 2012;51:4642–4649. doi: 10.1021/bi300429p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Amoroso A, Boudet J, Berzigotti S, Duval V, Teller N, Mengin-Lecreulx D, Luxen A, Simorre JP, Joris B. A peptidoglycan fragment triggers β-lactam resistance in Bacillus licheniformis. PLoS Pathog. 2012;8:e1002571. doi: 10.1371/journal.ppat.1002571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gregory PD, Lewis RA, Curnock SP, Dyke KG. Studies of the repressor (BlaI) of β-lactamase synthesis in Staphylococcus aureus. Mol Microbiol. 1997;24:1025–1037. doi: 10.1046/j.1365-2958.1997.4051770.x. [DOI] [PubMed] [Google Scholar]
- 183.Berzigotti S, Benlafya K, Sepulchre J, Amoroso A, Joris B. Bacillus licheniformis BlaR1 L3 loop is a zinc metalloprotease activated by self-proteolysis. PLoS One. 2012;7:e36400. doi: 10.1371/journal.pone.0036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Llarrull LI, Mobashery S. Dissection of events in the resistance to β-lactam antibiotics mediated by the protein BlaR1 from Staphylococcus aureus. Biochemistry. 2012;51:4642–4649. doi: 10.1021/bi300429p. [DOI] [PMC free article] [PubMed] [Google Scholar]